Abstract
Prokaryotic ubiquitin-like protein (Pup) is the first identified prokaryotic protein that is functionally analogous to ubiquitin. Despite using the proteasome as the end point for proteolysis, Pup differs from ubiquitin both biochemically and structurally. We will discuss these differences that have been highlighted by several recent studies. Finally, we will speculate on the possible interactions between the two analogous pathways in pathogen and host.
Introduction
The addition and removal of various functional groups on proteins expands the biochemical and structural diversity of proteins beyond the sequences encoded by genomic DNA. Examples of protein modifications include glycosylation, lipid attachment, phosphorylation, disulfide bond formation, cofactor attachment, degradation tags, and proteolytic processing. These and other modifications alter protein regulation, stability, localization, and function. Due to the large number of diverse modifications currently identified, a protein’s chemical and structural properties, as well as its half-life, are often not predictable based on amino acid sequence alone.
Modifications that target proteins for proteolysis, and thereby affect protein stability, have been extensively studied. These modifications can serve as quality control features that regulate protein synthesis, remove defective proteins, recycle amino acids, and inactivate proteins. Bacteria co-translationally tag stalled or interrupted translational products with the SsrA peptide. This 11-amino acid signal, AANDENYALAA, targets the defective translation product to cellular proteases, including ClpA/XP (Lies et al., 2008). The SsrA signal is encoded by a small, stable RNA containing an alanyl-tRNA domain and mRNA domain encoding the open reading frame of the tag (tmRNA) (Karzai et al., 2000, Keiler et al., 1996). Upon encountering stalled or incomplete translation, the tmRNA and accessory factors are recruited to the ribosome to rescue the idle translation complex by replacing it with the tmRNA encoding the SsrA tag. The ssrA mRNA serves as the template for the addition of the SsrA peptide. The SsrA tag is added co-translationally to the carboxy (C-) terminus of incomplete proteins to target them for degradation and thereby rescues stalled ribosomes by releasing mistranslated mRNA.
The best-studied protein modification targeting proteins for degradation in eukaryotes is ubiquitin. Ubiquitin is a highly conserved, 76 amino acid protein that covalently attaches to substrate lysines though its C-terminal di-glycine (GG) motif. Ubiquitin conjugates to target proteins in a sophisticated multi-step activation and ligation pathway that ultimately delivers doomed proteins to the proteasome protease, where they are degraded (Hochstrasser, 1996). Unlike the SsrA tag in bacteria, ubiquitin is added to target proteins post-translationally and it is removed prior to their degradation.
Another protein modifier called Pup was recently characterized in Mycobacterium tuberculosis (Mtb) (Pearce et al., 2008) and its non-pathogenic relative Mycobacterium smegmatis (Msm) (Burns et al., 2009). Similar to ubiquitin, Pup post-translationally attaches to proteins on lysine residues through its C-terminus, ultimately targeting them to the proteasome protease. Although the ubiquitin and Pup modification systems appear to be functionally analogous, new studies on Pup shed light on key differences between the two pathways. This review will highlight these recent studies and will discuss the similarities and differences between the ubiquitin and Pup proteasome systems. In addition, we will discuss the significance of the pathways in disease and the possible consequences of having proteasome-targeting machineries in both the host and the pathogen.
Proteasomes
For detailed information on the eukaryotic ubiquitin-proteasome pathway, the reader is referred to a number of excellent reviews on the topic (Finley, 2009, Pickart et al., 2004b, Schmidt et al., 2005, Pickart et al., 2004c, Hochstrasser, 1996, Hochstrasser, 2009). The proteasome is a mini-compartment that contains proteolytic active sites enclosed within a chamber and are essential for eukaryotic life. The eukaryotic proteasome core (20S) is composed of four rings: two hetero-heptameric rings of beta (β) subunits sandwiched between two hetero-heptameric rings of alpha (α) subunits that restrict access to the catalytic core. Eukaryotic proteasome cores are highly complex and the entry of substrates into the proteasome core requires a hetero-hexameric ring of regulatory particle ATPases (Rpts) that cap the ends of 20S core particles (CP). In addition to the ATPases, regulatory particle non-ATPases (Rpns) and de-ubiquitinases (DUBs), participate in the recognition, unfolding and degradation of substrates. Together these proteasome-associated factors form the 19S regulatory particle (RP). The barrel- shaped architecture of the proteasome and RP help regulate proteolysis, which is irreversible (Schrader et al., 2009, Finley, 2009).
The bacterial proteasome, which shares sequence and structural homology with its eukaryotic counterpart, has been identified in all sequenced Archaea but is limited to bacteria of the order Actinomycetales, including Streptomyces, Rhodococcus, Frankia, and Mycobacterium (Lupas et al., 1997, Darwin, 2009). Bacterial CPs are barrel-shaped proteases with two rings of homo-heptameric alpha (PrcA, α) subunits and two rings of homo-heptameric beta (PrcB, (β) subunits (Lin et al., 2006, Nagy et al., 1998, Hu et al., 2006). Similar to their eukaryotic counterparts, bacterial proteasomes contain amino-terminal (N-terminal) threonine nucleophiles in the β-subunits (Baumeister et al., 1998) that are responsible for its chymotrypsin-like catalytic activity. Bacterial CPs have been shown to degrade model peptide substrates, however, protease activity on full-length, folded, native substrates has been elusive (Lin et al., 2006, Pouch et al., 2000, Tamura et al., 1995, Nagy et al., 1998).
A gene (mpa) encoding an AAA ATPase similar to those in the RP in eukaryotes co-localizes with the proteasome CP genes in proteasome-bearing Actinobacteria (Wolf et al., 1998). Mutations in mpa sensitize Mtb to nitrosative stress, a similar phenotype to that observed with the addition of proteasome inhibitor (Darwin et al., 2003). Additionally, mpa mutants displayed altered levels of certain proteins compared to the wild-type strain, also implicating this gene product in a protein degradation pathway (Pearce et al., 2006). Biochemical studies with Mpa and ARC (AAA ATPase forming ring-shaped complexes), an orthologous ATPase from Rhodococcus erythropolis, showed that the ATPases formed hexameric or dodecameric rings with ATPase activity (Wolf et al., 1998, Zhang et al., 2004, Darwin et al., 2005). It is hypothesized that Mpa and ARC perform an analogous function to the AAA ATPases of the eukaryotic RP, however, robust interactions between the bacterial ATPases and CPs have not been observed (Wolf et al., 1998, Wang et al., 2009). This suggests that the interactions are either transient or require additional factors.
In addition to the AAA ATPase mpa, genes for other factors believed to be involved in proteasome function co-localize with the proteasome CP genes in proteasome-bearing Actinobacteria and were initially annotated as “proteasome-associated genes”. A few of these genes have been characterized and will be discussed below; the others could potentially provide the additional factors required for optimal ATP-dependent proteasome activity.
Ubiquitin
In the eukaryotic proteasome pathway, ubiquitin is the primary modification that provides the recognition and specificity required to deliver target proteins to the proteasome for proteolysis (Pickart et al., 2004b, Elsasser et al., 2005, Finley, 2009). Ubiquitin is a small protein that is first activated in a multi-step process prior to conjugation to lysine residues on protein substrates. All ubiquitin and ubiquitin-like modifiers have an absolutely conserved GG motif at the C-terminus. Ubiquitin is usually translated as a precursor and is cleaved to expose the GG motif (Reyes-Turcu et al., 2009, Ozkaynak et al., 1987, Komander et al., 2009). The terminal glycine carboxyate is first activated through adenylation by an E1 enzyme, followed by thioesterification with a cysteine residue resulting in the formation of a covalent ubiquitin-E1 intermediate (Figure 1a). This is followed by another thioesterification reaction with conjugating enzymes (E2s), whereby ubiquitin can either be directly attached to protein substrates, or transferred to ubiquitin ligases (E3s), which can also facilitate the conjugation (Pickart et al., 2004c). Ubiquitin is usually conjugated to lysine side chains of substrates via an isopeptide bond, however, variations in this linkage have been identified (Ciechanover et al., 2004, Cadwell et al., 2005).
Fig. 1. Comparison of the ubiquitin and Pup proteasome pathways.
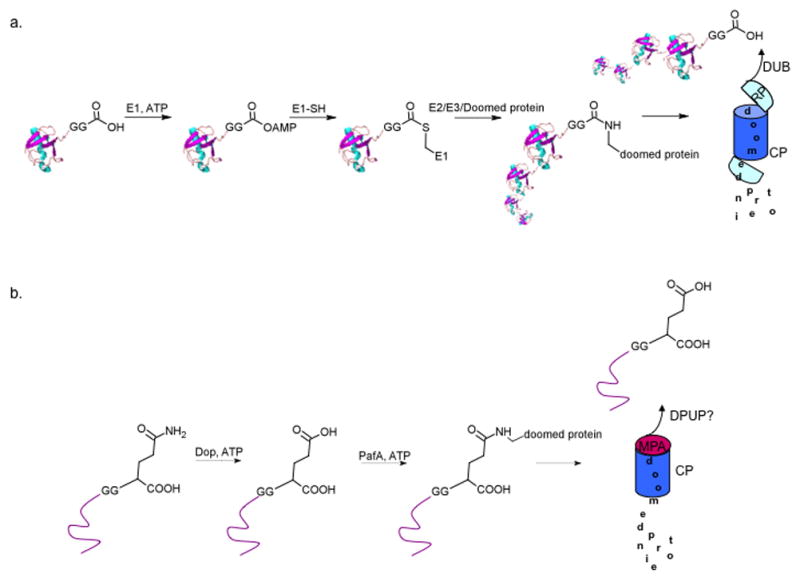
(a) Ubiquitin is adenylated at the C-terminal di-glycine, followed by a series of thioesterification reactions and finally conjugated to a doomed protein. In general, multiple ubiquitin molecules are conjugated to the protein and/or ubiquitin itself. Ubiquitin is removed by DUBs and the doomed protein is unfolded and delivered into the 20S CP by the RP. (b) Pup is first deamidated at the C-terminal glutamine and then conjugated to doomed proteins (conjugation shown at the γ-COOH for simplicity). Pup targets proteins to Mpa and the proteasome, however, it is unknown whether Pup is recycled and whether additional proteins are needed to complete the process.
More than 600 distinct mammalian proteins are thought to be involved in the ligation of ubiquitin to substrates (reviewed in (Deshaies et al., 2009)). It is the multitude, diversity and combination of these ubiquitin ligases that allows a variety of substrates to be ubiquitylated in a specific and regulated manner. In addition, multiple ubiquitin residues can conjugate to one another on any one of the seven conserved lysine residues on ubiquitin, often resulting in a different fate for the target protein (Pickart et al., 2004a). Lysine 48-linked chains primarily direct substrates for degradation. Polyubiquitin chains, or branches, are recognized by receptor proteins that associate with the 19S proteasome RP to deliver the tagged protein to the proteasome complex for degradation (reviewed in (Finley, 2009)). At the proteasome the polyubiquitin chains are usually removed by deubiquitinating enzymes (DUBs) (Figure 1a). DUBs recycle ubiquitin prior to degradation and serve as regulatory elements, ensuring that the correct proteins are targeted for destruction (Reyes-Turcu et al., 2009, Komander et al., 2009). Following deubiquitylation, the AAA ATPase unfolds the target protein that is then translocated into the proteasome.
Pup
Pup is encoded by a gene located directly upstream of the proteasome CP genes in proteasome-bearing bacteria and, similar to ubiquitin, it is a small protein. Although the end-point for both the ubiquitin degradation system in eukaryotes and the Pup degradation system in prokaryotes is the proteasome, the two functionally analogous tagging systems do not share similar methods of activation and conjugation to target proteins. Pup has a di-glycine motif at the penultimate position of the C-terminus, followed by either glutamate (Glu) or glutamine (Gln), depending on the organism. The small size of Pup and the di-glycine motif near the C-terminus are the only common features shared between Pup and ubiquitin. It was initially hypothesized that the C-terminal amino acid (Glu or Gln) of Pup was removed to expose the di-glycine motif for conjugation, which would be analogous to ubiquitin activation by C-terminal hydrolases (Pickart, 2001). Mass spectrometry on pupylated substrates in both Mtb and Msm, where the C-terminal residue is Gln, revealed that Gln was not removed; instead it was deamidated to Glu prior to conjugation to substrate lysines (Pearce et al., 2008, Burns et al., 2009). The deamidation reaction is catalyzed by Dop (deamidase of Pup), which is encoded by a gene upstream of pup and the proteasome core particle genes (Striebel et al., 2009) (Figure 1b). Dop shares no homology to ubiquitin-activating enzymes and bioinformatic analysis suggests structural homology to the carboxylate- amine/ammonia ligase super family of glutamine synthetases (Iyer et al., 2008). This family catalyzes the ligation of amine groups with carboxylates, resulting in an amide linkage. It is intriguing bacteria that encode Glu at the C-terminus of Pup and therefore do not require deamidation have retained the dop gene. This may suggest that dop plays additional roles in the pupylation pathway.
Similar to Dop, bioinformatic analysis suggested that PafA (proteasome-associated factor A) also has structural homology to the carboxylate-amine/ammonia ligase super family (Iyer et al., 2008). pafA co-localizes with proteasome-associated genes and its gene product catalyzes the conjugation of Pup to the known proteasome substrates FabD and PanB in vitro in the presence of ATP and Dop (Striebel et al., 2009) (Figure 1b). ATP is hydrolyzed during the course of the reaction, suggesting that the PafA-catalyzed ligation reaction proceeds through a phosphorylated intermediate, as do other members of the carboxylate-amine/ammonia ligase super family of enzymes (Iyer et al, 2008). It is unknown which C-terminal carboxylate (the backbone carboxylate or the γ-carboxylate on the Glu) is conjugated to substrates. Pup terminating in Glu (Pup-Glu) instead of Gln (Pup-Gln) is a substrate for PafA-catalyzed conjugation in the absence of Dop, suggesting deamidation precedes conjugation, and that Dop and PafA mediated reactions are not necessarily coupled in vitro.
In contrast to ubiquitin activation and conjugation, which proceeds through a series of at least four enzymatic steps prior to substrate conjugation, Pup can be activated and conjugated in two steps (Figure 1a, b). In organisms where pup encodes a C-terminal Glu, it is likely that only one enzyme is required for activation and conjugation. In Mtb, a pafA mutation abrogates pupylation (Pearce et al., 2008), suggesting that it is responsible for most, if not all pupylation in vivo. This contrasts sharply with the multitude of E3 ligases used for ubiquitin conjugation. Many of the E3 ligases in eukaryotes target ubiquitylated substrates to destinations other than the proteasome. It is currently not known whether the Pup tag has additional functions besides proteasome targeting. It is possible that additional Pup ligases exist in Mtb besides PafA; these ligases may not always be present, may require additional factors for activity, or may serve functions other than proteasome targeting. Another hypothesis is that other proteins modulate the specificity of PafA.
Both in vitro as well as in vivo experiments suggest that unlike ubiquitin, Pup does not form polymeric chains (Pearce et al., 2008, Burns et al., 2009, Striebel et al., 2009). Only a single Pup moiety has been observed conjugated to a target lysine residue. It is currently unknown whether a single substrate can have multiple pupylated lysine residues and if so, what the functional consequences would be.
Recent studies have shed light on Pup activation and conjugation to target proteins, however, nothing is known about how pupylated substrates are delivered to the proteasome. Affinity protein pull-down assays with Mycobacterium bovis BCG lysates using Pup as bait resulted in the co-purification of Mpa, Dop and PafA (Striebel et al., 2009). If Dop and PafA activities are limited to Pup activation, it is possible that Mpa binds pupylated substrates and delivers them to the proteasome. In both Mtb and Msm, mutations in mpa result in the accumulation of proteasome substrates, both pupylated and unpupylated, compared to the wild-type strain (Pearce et al., 2006, Pearce et al., 2008), confirming its role in the degradation of pupylated proteins. Additionally, NMR studies suggest that Mpa interacts with the central region of Pup, from residues 21–61 (Chen et al., 2009, Sutter et al., 2009, Liao et al., 2009), and it appears to be the N-terminal coiled-coil domain of Mpa that is bound by Pup (Sutter et al., 2009). Interestingly, Mpa forms hexameric rings which structurally resemble the eukarytotic chaperone complex p97/valosin-containing protein (mammals)/Cdc48 (yeast) (Darwin et al., 2005). p97 is an ATPase that is distantly related to the ATPases of the 19S RP (Elsasser et al., 2005) and can bind directly to polyubiquitin chains and other proteasome components (Wang et al., 2004). It functions as a binding scaffold for the ubiquitin-proteasome machinery, serving to target ubiquitylated proteins to the proteasome, and has been shown to have other functions as well (Dai et al., 2001, Thrower et al., 2000).
It is not yet know if Pup has to be removed from target proteins prior to their degradation by the proteasome. One might predict that the steric bulk of the non-linear Pup~target fusion would make it difficult for the substrate to fit in the 3 nm pore of Mpa (Darwin et al., 2005), which presumably unwinds the substrate prior to delivery into the gated CP. Additionally, a method to recycle Pup for subsequent ligations would be energetically more favorable and efficient. For these reasons, one would expect to find “depupylase” activity associated with the bacterial proteasome, however, it is currently unknown whether such an activity exists or what enzyme(s) could perform this function.
Structural differences
Ubiquitin and ubiquitin-like proteins (Ubls) share a common ordered β-grasp fold (Vijay-Kumar et al., 1987, Vijay-Kumar et al., 1985). This fold is defined by a prominent β-sheet with short α-helical regions, where the β-sheet provides a scaffold for protein-protein interactions (Vijay-Kumar et al., 1985, Vijay-Kumar et al., 1987, Burroughs et al., 2007). Although Ubls share little sequence homology with ubiquitin, they are easily identified based on this common fold. Interestingly, prokaryotic sulfur carrier proteins, most likely the prokaryotic ancestors of the ubiquitin tagging system, also have this distinct fold (Iyer et al., 2006). In eukaryotes, a two component “degron” has been identified for optimal ubiquitin-dependent proteolysis. The two keys to the degron are ubiquitin, which serves as the recognition tag, and a disordered region in the target protein or associated proteins (Prakash et al., 2004). Due to the stability of ubiquitin and its compact β-grasp fold, the unstructured region in the target proteins provide an initiation site for its unfolding and subsequent degradation by the proteasome. Unstructured regions are not uncommon in eukaryotic proteins (Ward et al., 2004), suggesting that many cellular proteins are already primed for degradation after ubiquitylation.
Unstructured regions in proteins are less common in prokaryotes compared to eukaryotes (Ward et al., 2004). It was therefore surprising when recent structural studies revealed that Pup is an intrinsically disordered protein (IDP) (Chen et al., 2009, Sutter et al., 2009, Liao et al., 2009). IDPs contain labile secondary structures, specifically under physiological conditions. Due to the rare nature of disordered proteins in prokaryotes, this feature of Pup may have functional significance. IDPs are known to bind partners with specificity yet low affinity and can adapt to many protein partners (Dyson et al., 2005). As discussed previously, Mpa was shown to bind the C-terminal two-thirds of Pup. When bound to Mpa under these conditions, the N-terminal section of Pup is still disordered, with properties similar to that of a degradation initiation site (Chen et al., 2009, Liao et al., 2009, Sutter et al., 2009). It is possible that in prokaryotes, where disorder is generally not observed in proteins, Pup alone can serve as the two component degron: it is both a tag for recognition and a marker for a disordered region on the modified protein.
Perspectives
Pup is the first identified prokaryotic protein that is functionally analogous to ubiquitin. Despite the functional similarities, Pup differs from ubiquitin both biochemically and structurally. In Mtb, Pup is first deamidated by Dop and conjugated to lysine residues of target proteins via the C-terminal Glu, a reaction catalyzed by PafA. Pupylated proteins are then delivered to the proteasome, presumably unfolded by the ATPase Mpa, and processed by the proteasomal protease. Whereas ubiquitin has a highly conserved and ordered 3-dimensional structure, Pup is an intrinsically disordered protein, which may help initiate proteasome-dependent degradation.
In addition to conservation of function, both ubiquitin and Pup (in Mtb) are critical for cell maintenance and survival. The ubiquitin-proteasome system is important for general “housekeeping” functions as well as for a wide array of cellular processes including antigen presentation, DNA repair, transcription, cell cycle control, and stress response (Loureiro et al., 2006, Bergink et al., 2009, Wickliffe et al., 2009, Daulny et al., 2009). Due to the number of cellular processes that depend on the ubiquitin-proteasome system, it follows that dysregulation or malfunction of the system may contribute to disease. The ubiquitin-proteasome system tightly regulates protein levels. A mutation in any of the proteins involved in the system could result in altered protein abundance, potentially leading to deleterious effects. In humans, the ubiquitin-proteasome system has been implicated in cancer, genetic diseases such as cystic fibrosis, neurodegenerative diseases including Alzheimers, among many other diseases (Schwartz et al., 1999).
The Pup-proteasome system also appears to play a key role in the survival and persistence of Mtb. Proteasome core mutants have yet to be isolated in Mtb, whereas they have been isolated in Msm (Knipfer et al., 1997, Burns et al., 2009) a non-pathogenic relative of Mtb, and in Streptomyces lividans (Hong et al., 2005), another saprophytic Actinomycete. This suggests a critical role for the proteasome in Mtb fitness. Additionally, mutations in the proteasome-associated genes mpa and pafA render the bacteria more sensitive to nitric oxide (NO) and are attenuated for infection in mice (Darwin et al., 2003). Chemical inhibition of the proteasome also resulted in NO sensitization. NO is a host defense used to contain and kill invading pathogenic bacteria, however, Mtb can persist even in the presence of the immune response and NO. By inference, the inability to degrade pupylated proteins results in increased NO sensitivity and attenuation of virulence of Mtb, however, the mechanism of proteasome-associated NO tolerance remains unclear. It is possible that NO stress results in the accumulation of damaged proteins, which are deleterious in proteasome-defective cells. Alternatively, the Pup-proteasome system could be responsible for regulating the expression or abundance of certain virulence genes or proteins, such as transcription factors, which would otherwise counteract NO toxicity.
A major obstacle in the treatment of tuberculosis is the persistent nature of the Mtb bacillus. As mentioned above, Mtb can persist even in the presence of NO and other antimicrobial stresses imposed by host cells. During starvation-induced stationary phase, prokaryotes are known to increase the degradation of nonessential proteins to provide cells with amino acids for the synthesis of essential proteins (Reeve et al., 1984). Therefore, one of the roles of the Pup-proteasome system during stationary phase and latency in Mtb could be to provide critical amino acid pools for the de novo synthesis of essential proteins which are required for survival under the respective conditions.
With the discovery of the proteasome and the ubiquitin-like protein targeting system in prokaryotes, one must ask whether Mtb can use this tagging and degradation system to interfere with host cell machinery. Several Gram-negative pathogens have been shown to secrete proteins that interfere with the host ubiquitin tagging system (Rytkonen et al., 2007). For example, both Shigella spp. and Salmonella spp. secrete effector proteins (IpaH9.8 and SspH, respectively) that act as E3 ubiquitin ligases (Rohde et al., 2007). These proteins promote the degradation of important eukaryotic gene products, thereby altering the hosts cellular machinery (Rohde et al., 2007). It would be interesting to examine whether Mtb (or other bacteria) use Pup or other small proteins to modify eukaryotic targets. Alternatively, it is possible that ubiquitylated proteins could be targets of the Mtb proteasome.
The Pup-proteasome system is essential for the pathogenesis of Mtb, one of the most deadly bacterial pathogens in the world (WHO; http://www.who.int/en). Understanding the pathway, its role in virulence, and its possible connection with the host proteasome machinery will help identify new drug targets for the development of chemotherapies for Mtb, as well as aid in understanding the physiology of other organisms.
Acknowledgments
We are grateful to K. Tahlan and R. Festa for critical review of this manuscript. Due to the abbreviated nature of this review, we regret that we were unable to cite all of the groundbreaking work that was critical for elucidating the ubiquitin and proteasome systems. This work was supported by NIH grant HL092774. K.H.D. is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases.
References
- Baumeister W, Walz J, Zuhl F, Seemuller E. The Proteasome: Paradigm of a Self-Compartmentalizing Protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- Burns KE, Liu WT, Boshoff HI, Dorrestein PC, Barry CEr. Proteasomal protein degradation in Mycobacteria is dependent upon a prokaryotic ubiquitin-like protein. Journal of Biological Chemistry. 2009;284:3069– 3075. doi: 10.1074/jbc.M808032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Balaji S, Iyer LM, Aravind L. Small but versatile: the extraordinary functional and structural diversity of the b-grasp fold. Biology Direct. 2007;2:18–45. doi: 10.1186/1745-6150-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309:127–130. doi: 10.1126/science.1110340. [DOI] [PubMed] [Google Scholar]
- Chen X, Solomon WC, Kang Y, Cerda-Maira F, Darwin KH, Walters KJ. Prokaryotic ubiquitin-like protein Pup is intrinsically disordered. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.07.018. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Ben-Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 2004;14:103–106. doi: 10.1016/j.tcb.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Dai RM, Li CC. Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat Cell Biol. 2001;3:740–744. doi: 10.1038/35087056. [DOI] [PubMed] [Google Scholar]
- Darwin KH. Prokaryotic Ubiquitin-Like Protein, Proteasomes, and Pathogenesis. Nat Rev Microbiol. 2009 doi: 10.1038/nrmicro2148. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin KH, Ehrt S, Gutierrez-Ramos J-C, Weich N, Nathan CF. The Proteasome of Mycobacterium tuberculosis Is Required for Resistance to Nitric Oxide. Science. 2003;302:1963–1966. doi: 10.1126/science.1091176. [DOI] [PubMed] [Google Scholar]
- Darwin KH, Lin G, Chen Z, Li H, Nathan CF. Characterization of a Mycobacterium tuberculosis proteasomal ATPase homologue. Molecular Microbiology. 2005;55:561–571. doi: 10.1111/j.1365-2958.2004.04403.x. [DOI] [PubMed] [Google Scholar]
- Daulny A, Tansey WP. Damage control: DNA repair, transcription, the ubiquitin-proteasome system. DNA Repair. 2009;8:444–448. doi: 10.1016/j.dnarep.2009.01.017. [DOI] [PubMed] [Google Scholar]
- Deshaies R, Joazeiro C. RING domain E3 ubiquitin ligases. Annual Review of Biochemistry. 2009;78 doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nature Reviews Molecular Cell Biology. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nature Cell Biology. 2005;7:742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- Finley D. Recognition and Processing of Ubiquitin-Protein Conjugates by the Proteasome. Annual Review of Biochemistry. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong B, Wang L, Lammertyn E, Geukens N, Van Mellaert L, Li Y, Anne J. Inactivation of the 20S proteasome in Streptomyces lividens and its influence on the production of heterologous proteins. Microbiology. 2005;151:3137–3145. doi: 10.1099/mic.0.28034-0. [DOI] [PubMed] [Google Scholar]
- Hu G, Lin G, Wang M, Dick L, Xu RM, Nathan CF, Li H. Structure of the Mycobacterium tuberculosis proteasome and mechanism of inhibition by a peptidyl boronate. Molecular Microbiology. 2006;59:1417–1428. doi: 10.1111/j.1365-2958.2005.05036.x. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Burroughs AM, Aravind L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like b- grasp domains. Genome Biology. 2006;7:R60.61–R60.23. doi: 10.1186/gb-2006-7-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Burroughs AM, Aravind L. Unraveling the biochemistry and provenance of pupylation: a prokaryotic analog of ubiquitination. Biol Direct. 2008;3:45. doi: 10.1186/1745-6150-3-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai AW, Roche ED, Sauer RT. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nature Structural Biology. 2000;7:449–55. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Knipfer N, Shrader TE. Inactivation of the 20S proteasome in Mycobacterium smegmatis. Molecular Microbiology. 1997;25:375–383. doi: 10.1046/j.1365-2958.1997.4721837.x. [DOI] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of deubiquitinases. Nature Reviews Molecular Cell Biology. 2009;10:550– 563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Liao S, Shang Q, Zhang X, Zhang J, Xu C, Tu X. Pup, a prokaryotic ubiquitin-like protein, is an intrinsically disordered protein. Biochem J. 2009;422:207–215. doi: 10.1042/BJ20090738. [DOI] [PubMed] [Google Scholar]
- Lies M, Maurizi MR. Turnover of Endogenous SsrA-tagged Proteins Mediated by ATP-dependent Proteases in Escherichia coli. Journal of Biological Chemistry. 2008;283:22918–22929. doi: 10.1074/jbc.M801692200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Hu G, Tsu C, Kunes YZ, Li H, Dick L, et al. Mycobacterium tuberculosis prcBA genes encode a gated proteasome with broad oligopeptide specificity. Molecular Microbiology. 2006;59:1405–1416. doi: 10.1111/j.1365-2958.2005.05035.x. [DOI] [PubMed] [Google Scholar]
- Loureiro J, Ploegh HL. Antigen presentation and the ubiquitin- proteasome system in host-pathogen interactions. Adv Immunol. 2006;92:225–305. doi: 10.1016/S0065-2776(06)92006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Zühl F, Tamura T, Wolf S, Nagy I, De Mot R, Baumeister W. Eubacterial proteasomes. Mol Biol Reports. 1997;24:125–131. doi: 10.1023/a:1006803512761. [DOI] [PubMed] [Google Scholar]
- Nagy I, Tamura T, Vanderleyden J, Baumeister W, De Mot R. The 20S Proteasome of Streptomyces coelicolor. Journal of Bacteriology. 1998;180:5448– 5453. doi: 10.1128/jb.180.20.5448-5453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkaynak E, Finley D, Solomon MJ, Varshavsky A. The yeast ubiquitin genes: a family of natural gene fusions. Embo J. 1987;6:1429–1439. doi: 10.1002/j.1460-2075.1987.tb02384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce MJ, Arora P, Festa RA, Butler-Wu SM, Gokhale RS, Darwin KH. Identification of substrates of the Mycobacterium tuberculosis proteasome. The EMBO Journal. 2006;25:5423–5432. doi: 10.1038/sj.emboj.7601405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce MJ, Mintseris J, Ferreyra J, Gygi SP, Darwin KH. Ubiquitin- like protein involved in the proteasome pathway of Mycobacterium tuberculosis. Science. 2008;322:1104–1107. doi: 10.1126/science.1163885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C, Fushman D. Polyubiquitin chains: polymeric protein signals. Current Opinion in Chemical Biology. 2004a;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms Underlying Ubiquitination. Annual Review of Biochemistry. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004b;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochemica et Biophysica Acta. 2004c;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Pouch M-N, Cournoyer B, Baumeister W. Characterization of the 20S proteasome from the actinomycete. Frankia Mol Microbiol. 2000;35:368–377. doi: 10.1046/j.1365-2958.2000.01703.x. [DOI] [PubMed] [Google Scholar]
- Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nature Structural and Molecular Biology. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- Reeve C, Bockman A, Matin A. Role of protein degradation in the survival of carbon-starved Escherichia coli and Salmonella typhimurium. Journal of Bacteriology. 1984;157:758–763. doi: 10.1128/jb.157.3.758-763.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and Cellular Roles of Ubiquitin-Specific Deubiquitinating Enzymes. Annual Review of Biochemistry. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde JR, Breitkreutz A, Chenal A, Sansonetti PJ, Parsot C. Type III Secretion Effectors of the IpaH Family Are E3 Ubiquitin Ligases. Cell Host and Microbe. 2007;1:77–83. doi: 10.1016/j.chom.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Rytkonen A, Holden DW. Bacterial interference of ubiquitination and deubiquitination. Cell Host Microbe. 2007;1:13–22. doi: 10.1016/j.chom.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Hanna J, Elsasser S, Finley D. Proteasome-associated proteins: regulation of a proteolytic machine. Biol Chem. 2005;386:725–737. doi: 10.1515/BC.2005.085. [DOI] [PubMed] [Google Scholar]
- Schrader EK, Harstad KG, Matouschek A. Targeting proteins for degradation. Nature Chemical Biology. 2009;5:815–822. doi: 10.1038/nchembio.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz AL, Ciechanover A. THE UBIQUITIN-PROTEASOME PATHWAY AND PATHOGENESIS OF HUMAN DISEASES. Annual Review of Medicine. 1999;50:57–74. doi: 10.1146/annurev.med.50.1.57. [DOI] [PubMed] [Google Scholar]
- Striebel F, Imkamp F, Sutter M, Steiner M, Mamedov A, Weber-Ban E. Bacterial ubiquitin-like modifer Pup is deamidated and conjugated to substrates by distinct but homologous enzymes. Nature Structural and Molecular Biology. 2009;16:647–651. doi: 10.1038/nsmb.1597. [DOI] [PubMed] [Google Scholar]
- Sutter M, Striebel F, Damberger FF, Allain FHT, Weber-Ban E. A distinct structural region of the prokaryotic ubiquitin-like protein (Pup) is recognized by the N-terminal domain of the proteasomal ATPase Mpa. FEBS Letters. 2009;583:3151–3157. doi: 10.1016/j.febslet.2009.09.020. [DOI] [PubMed] [Google Scholar]
- Tamura T, Nagy I, Lupas A, Lottspeich F, Cejka Z, Schoofs G, et al. The first characterization of a eubacterial proteasome: the 20S complex of Rhodococcus. Current Biology. 1995;5:766–774. doi: 10.1016/s0960-9822(95)00153-9. [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar S, Bugg CE, Cook WJ. Three-dimensional structure of ubiquitin refined at 1.8A resolution. Journal of Molecular Biology. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- Vijay-Kumar S, Bugg CE, Wilkinson KD, Cook WJ. Three-dimensional struture of ubiquitin at 2.8 A resolution. PNAS. 1985;82:3582–3585. doi: 10.1073/pnas.82.11.3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Song C, Li CCH. Molecular perspectives on p97-VCP: progress in understanding its structure and diverse biological functions. Journal of Structural Biology. 2004;146:44–57. doi: 10.1016/j.jsb.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Wang T, Li H, Lin G, Tang C, Li D, Nathan C, et al. Structural insights on the Mycobacterium tuberculosis proteasomal ATPase Mpa. Structure. 2009;17:1377–1385. doi: 10.1016/j.str.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and Functional Analysis of Native Disorder in Proteins from the Three Kingdoms of Life. Journal of Molecular Biology. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Wickliffe K, Williamson A, Jin L, Rape M. The Multiple Layers of Ubiquitin-Dependent Cell Cycle Control. Chemical Reviews. 2009;109:1537–1548. doi: 10.1021/cr800414e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf S, Nagy I, Lupas A, Pfeifer G, Cejka Z, Muller SA, et al. Characterization of ARC, a Divergent Member of the AAA ATPase Family from Rhodococcus erythropolis. Journal of Molecular Biology. 1998;277:13–25. doi: 10.1006/jmbi.1997.1589. [DOI] [PubMed] [Google Scholar]
- Zhang X, Stoffels K, Wurzbacher S, Schoofs G, Pfeifer G, Banerjee T, et al. The N-terminal coiled coil of the Rhodococcus erythropolis ARC AAA ATPase is neither necessary for oligomerization nor nucleotide hydrolysis. J Struct Biol. 2004;146:155–165. doi: 10.1016/j.jsb.2003.10.020. [DOI] [PubMed] [Google Scholar]


