SUMMARY
Macroautophagy is a lysosomal degradative pathway essential for neuron survival. Here we show that macroautophagy requires the Alzheimer's disease (AD) - related protein, presenilin-1 (PS1). In PS1-null blastocysts, neurons from mice hypomorphic for PS1 or conditionally depleted of PS1, substrate proteolysis and autophagosome clearance during macroautophagy are prevented due to a selective impairment of autolysosome acidification and cathepsin activation. These deficits are caused by failed PS1-dependent targeting of the v-ATPase subunit to lysosomes. N-glycosylation of the V0a1 subunit, essential for its efficient ER-to-lysosome delivery, requires the selective binding of PS1 holoprotein to the unglycosylated subunit and the Sec61alpha/oligosaccharyltransferase complex. PS1 mutations causing early-onset AD produce a similar lysosomal/autophagy phenotype in fibroblasts from AD patients. PS1 is therefore essential for v-ATPase targeting to lysosomes, lysosome acidification, and proteolysis during autophagy. Defective lysosomal proteolysis represents a basis for pathogenic protein accumulations and neuronal cell death in AD and suggests novel therapeutic targets.
INTRODUCTION
Macroautophagy, the major lysosomal degradative pathway in cells, is responsible for degrading long-lived cytoplasmic constituents and is the principal mechanism for turning over cellular organelles and protein aggregates too large to be degraded by the proteasome (Klionsky, 2007; Mizushima, 2007; Rubinsztein, 2006).
Macroautophagy, hereafter referred to as autophagy, involves the sequestration of a region of cytoplasm within an enveloping double-membrane structure to form an autophagosome. Autophagosome formation is induced by inhibition of mTOR (mammalian target of Rapamycin)(Schmelzle and Hall, 2000) or AMP-activated protein kinase (AMPK) (Samari and Seglen, 1998). Autophagosomes and their contents are cleared upon fusing with late endosomes or lysosomes containing cathepsins, other acid hydrolases, and vacuolar [H+] ATPase (v-ATPase) (Yamamoto et al., 1998), a proton pump that acidifies the newly created autolysosome. Acidification of autolysosomes is crucial for activating cathepsins and effecting proteolysis of substrates; however, these late digestive steps of autophagy remain relatively uncharacterized.
Autophagic vacuoles (AVs), the general term for intermediate vesicular compartments in the process of autophagy, accumulate in several neurodegenerative diseases (Cuervo et al., 2004; Nixon et al., 2005; Ravikumar et al., 2004) but autophagy pathology in Alzheimer’s disease (AD) is exceptionally robust. AVs, many containing amyloid-β peptide, collect in massive numbers within grossly distended portions of axons and dendrites of affected neurons (Yu et al., 2005), likely reflecting defective AV clearance (Boland et al., 2008). This lysosome-related pathology, along with neuronal loss and amyloid deposition, are greatly accentuated in early-onset familial AD (FAD) due to mutations of PS1, the most common cause of FAD (Cataldo et al., 2004).
Presenilin 1 (PS1), a ubiquitous transmembrane protein, has diverse putative biological roles in cell adhesion, apoptosis, neurite outgrowth, calcium homeostasis, and synaptic plasticity (Kim and Tanzi, 1997; Shen and Kelleher, 2007). A portion of the PS1 holoprotein, a ~45 kDa protein, is cleaved in the endoplasmic reticulum (ER) to create a two-chain form (Zhang et al., 1998). Many known PS1 functions, but not all, involve the cleaved form of PS1 as the catalytic subunit of the gamma (γ)-secretase enzyme complex, which mediates the intramembranous cleavage of many type 1 membrane proteins, including APP and Notch (Citron et al., 1997; De Strooper et al., 1998). Although the pathogenic effects of PS1 mutations in AD are commonly ascribed to increased generation of the neurotoxic Aβ peptide from APP, not all of the disease-causing PS1 mutations have this effect (Junichi et al., 2007). Additional contributions to AD pathogenesis may involve loss of one or more of the other suspected biological functions of PS1 (Naruse et al., 1998).
In this report, we show that PS1 is required for lysosomal turnover of autophagic and endocytic protein substrates. PS1 deletion causes virtually complete loss of macroautophagy while having minimal influence on non-lysosomal types of proteolysis. We have identified the molecular basis for this requirement to be a novel action of PS1 holoprotein in the ER as an ER chaperone to facilitate maturation and targeting of the v-ATPase V0a1 subunit to lysosomes, which is essential for acidification, protease activation, and degradation of autophagic/lysosomal substrates. We demonstrate defects in these processes in cells lacking PS1, which are completely reversed by introducing wild type (WT) human PS1 into the cells. Similar autophagy pathology and deficits in lysosomal acidification are demonstrated in neurons of mice hypomorphic for PS1 or conditional PS knockout mice. Of particular clinical significance, we further show that mutations of PS1 that cause early-onset FAD disrupt the same lysosomal/autophagic functions that are more severely affected in PS1 KO cells. Our findings underscore the pathogenic importance of lysosomal proteolytic dysfunction seen in all forms of AD (Nixon and Cataldo, 2006; Nixon 2008) and provide a basis for the accelerated autophagy dysfunction and defective neuronal protein clearance seen in PS-FAD.
RESULTS
PS1 gene deletion selectively inhibits macroautophagic turnover of proteins
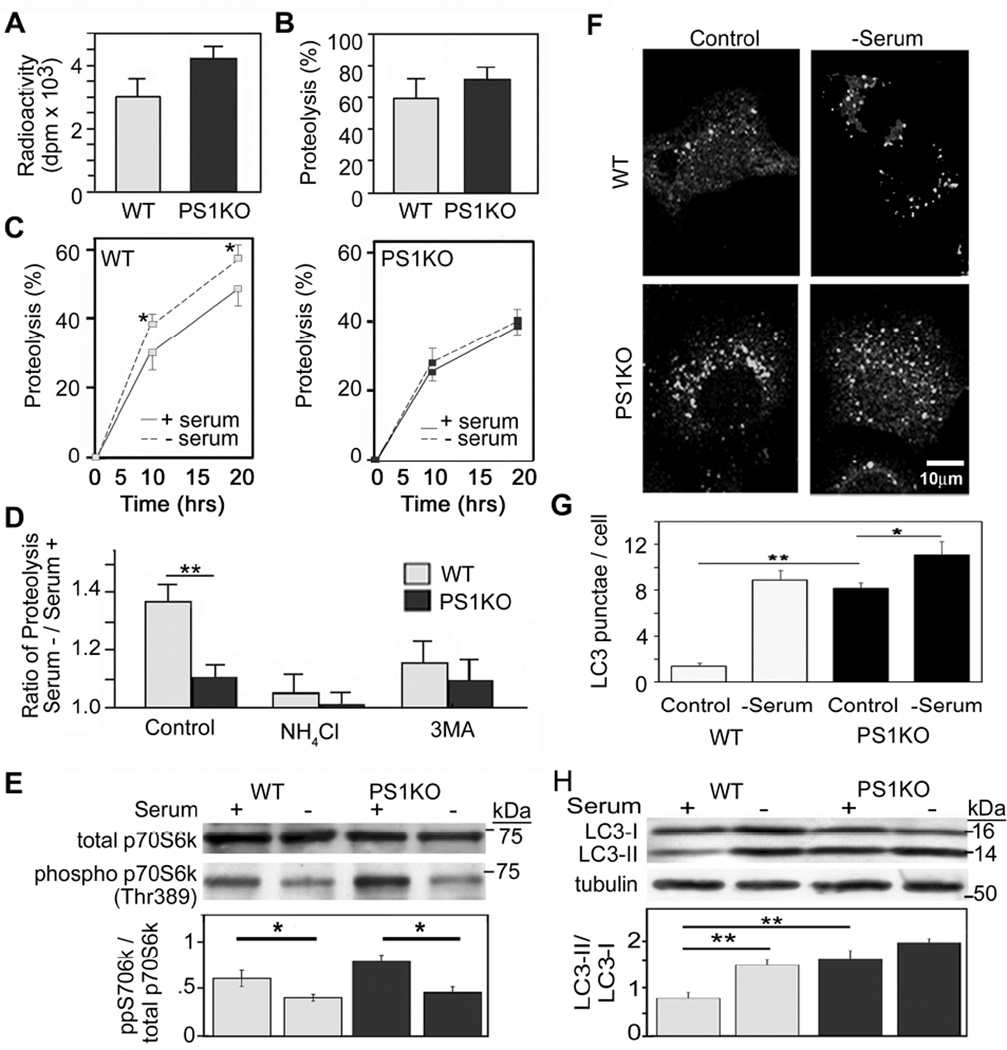
We investigated the competence of proteolytic systems in blastocysts from WT mice and constitutive PS1 KO mice using a well-established metabolic labeling procedure (Auteri et al., 1983). Neither incorporation of [3H]-leucine into proteins, used as a measurement of protein synthesis (Figure 1A), nor proteolysis of short-lived proteins, reflective mainly of ubiquitin-proteasome-dependent degradation (Figure 1B), were significantly altered in PS1 KO cells. However, the turnover of long-lived proteins was decreased in PS1 KO cells compared to WT under serum-supplemented media (Figure 1C). When autophagic/lysosomal degradation was induced through serum withdrawal, proteolysis increased 15–20% in WT cells (p < 0.05) but was not significantly changed in PS1 KO cells (Figure 1C). Under these induced conditions, NH4Cl (20 mM) entirely blocked the increase of proteolysis in both cell types as expected (Figure 1D). When macroautophagy was selectively inhibited with 3-methyladenine (3MA, 10 mM) (Seglen and Gordon, 1982), only the increased proteolysis in WT cells was blocked (Figure 1D), indicating that the residual increase in lysosome-related degradation in response to serum removal in PS1 KO cells is not due to macroautophagy. Together, these findings demonstrate that PS1 deletion selectively affects macroautophagic turnover of proteins.
Figure 1.
Protein turnover in PS1 KO cells: (A) Incorporation of [3H]-leucine in blastocysts from WT or PS1 KO mice. (B) Following labeling, the proteolysis of short-lived proteins was measured after a chase period. (C) Degradation of long-lived proteins was measured in WT (left panel) and PS1 KO cells (right panel). After incorporation of [3H]-leucine, cells were incubated in serum-supplemented or -deprived medium during the chase period (up to 20 hrs) (* for p <0.05, n=9). (D) The increase in proteolysis at 12 hrs after removal of serum relative to serum-replete conditions was determined for WT and PS1 KO cells that were untreated (control) or treated with NH4Cl or 3MA (** for p <0.001, n=9). (E) Total p70S6K and phospho (Thr389)-p70S6K levels quantified by densitometry, following growth in the presence or absence of serum for 6 hrs (* for p <0.05, n=3). (F) LC3 immunostaining after incubation in the presence and absence of serum. (G) Percentages of cell area occupied by LC3 puncta analyzed using ImageJ software (see Methods) (* for p <0.05 and ** for p <0.001, n=50). (H) LC3-II and LC3-I immunoreactivity and LC3-II/LC3-I ratios by Western blot analysis using tubulin as a loading control (** for p <0.001, n=3). See also Figure S1. All values are the mean ±S.E.M.
We next assessed the competence of the nutrient-related signaling pathway leading to mTOR-mediated induction of autophagy. For this, we measured the phosphorylation state of p70S6 kinase (p70S6K) before and after serum deprivation. Levels of both total p70S6K and its phospho–epitope (Thr389), measured by quantitative immunoblotting analyses, were comparable in WT and PS1 KO cells grown in serum-containing medium (Figure 1E). Moreover, phospho-p70S6K levels declined comparably in both cell types after inducing autophagy by serum withdrawal for 6 hrs, indicating that mTOR is inhibited normally in response to nutrient deprivation in PS1 KO cells (Figure 1E).
To evaluate autophagosome formation, we used immunofluorescence labeling with antibodies to LC3. LC3-positive vesicular profiles of sizes 0.5 – 2.0 µm, were significantly more numerous in PS1 KO cells than in WT cells grown in serum and were slightly increased after serum withdrawal (Figure 1F,G; S1). Consistent with immunocytochemical findings, LC3 western blot analyses showed that ratios of LC3-II to LC3-I, or LC3-II levels alone, were more than 2-fold higher (p < 0.001) in PS1 KO cells than WT cells grown in the presence of serum. Serum withdrawal resulted in higher LC3-II levels in both WT cells and PS1 KO cells although, in the latter cells, the proportional increase over the already elevated level of LC3-II was less than in WT cells (Figure 1H). Inducing autophagy by inhibiting mTOR directly with rapamycin yielded similar results (data not shown).
AVs were identified ultrastructurally (Figure S1A) on the basis of their size and morphology (Figure S1B). Autophagosomes and early autolysosomes were more numerous in PS1 KO cells whereas most AVs in WT cells were late autolysosomes. Despite abnormally high baseline AV numbers, PS1 KO cells exhibited modestly elevated AV numbers after serum withdrawal (Figure S1C). AVs and lysosomes isolated from cells on metrizamide gradients confirmed that engulfed materials were less degraded in PS1 KO cells compared to WT cells, where most materials were extensively digested, and lysosomes were mainly small and electron-dense, representing a terminal stage of degradation (Figure S1D). These data suggested that, in PS1 KO cells, autophagic protein degradation was impaired after fusion with lysosomes.
Defective clearance of autophagic vacuoles in PS1 KO blastocysts
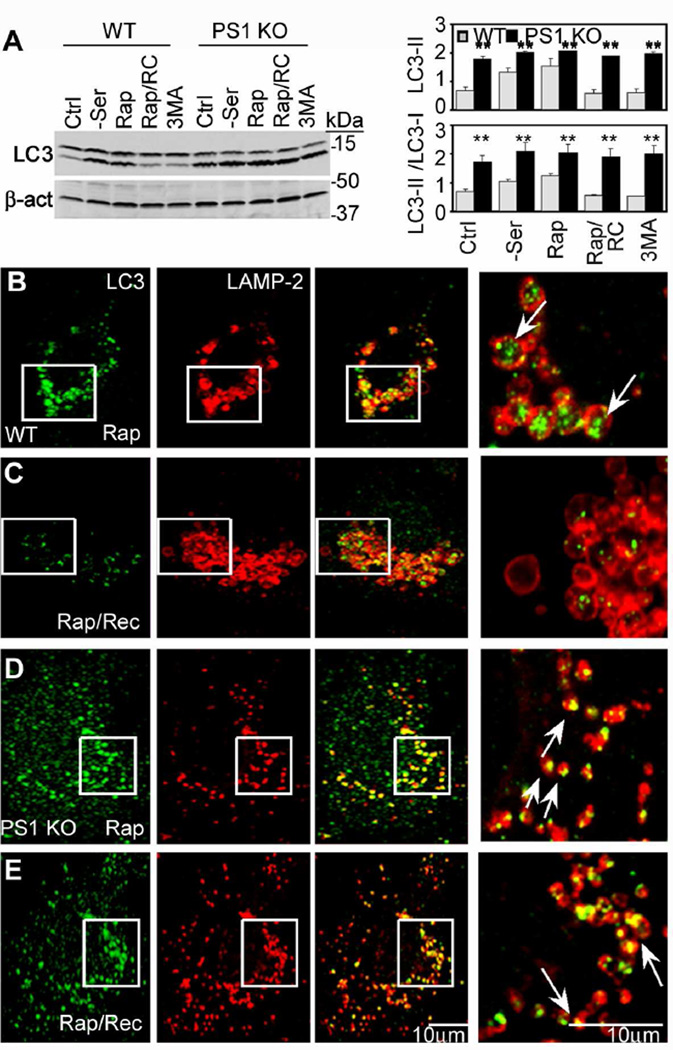
Further analyses of autolysosome maturation showed that clearance of LC3 after fusion, a measure of autophagy degradative competence, was greatly impaired in PS1 KO cells. In WT cells, acute autophagy induction with rapamycin elevated LC3-II levels by immunoblot analysis. These levels returned to pretreatment baseline levels within 6 hrs after removing rapamycin from the medium; however, in PS1 KO cells, LC3-II levels remained significantly elevated (Figure 2A). Double-immunofluorescence labeling with LC3 and LAMP-2 antibodies confirmed that LC3 accumulated in LAMP-2-positive vesicles after rapamycin exposure of WT (Figure 2B) and PS1 KO cells (Figure 2D), but 6 hrs after rapamycin was removed, LC3 remained only in LAMP vesicles of PS1 KO cells (Figure 2C,E). Despite this evidence that autophagosomes can fuse with lysosomes, inspection at higher magnification revealed that LC3 distributed more peripherally along the membrane of the fused vesicles in PS1 KO cells compared with those in WT cells, suggesting that the handling of LC3 after autophagosome-lysosome fusion is impaired (Figure 2, compare B and D).
Figure 2.
Impaired clearance of LC3-II from autolysosomes in PS1 KO cells: (A) Immunoblot analysis of LC3-I and –II levels in cells under conditions of no treatment (Ctrl), serum starvation (-Ser), rapamycin (Rap), rapamycin treatment followed by rapamycin removal (Rap/RC), and 3MA. WT (B,C) and PS1 KO cells (D,E) analyzed by double-immunofluorescence using LC3 and LAMP-2 antibodies after rapamycin (B,D) or later removal of rapamycin (C,E). Right panels depict enlarged images of the boxed areas seen in the left panels. Scale bar represents 10 µm. ** for p <0.001. See also Figure S2. All values are the mean ±S.E.M.
Using an alternative approach to investigate autophagosome clearance, we observed that LC3 levels remained abnormally high in PS1 KO cells even when autophagosome formation was blocked for 6 hrs with 3MA (Figure 2A; Figure S1E). Moreover, treatment with leupeptin (0.3 mM, 6 hrs) to inhibit cysteine proteases in autolysosomes, significantly elevated LC3-II levels (Figure S2A) and LC3-positive puncta in WT cells (Figure S2C), whereas in PS1 KO cells, this treatment did not increase LC3 levels beyond the elevated baseline evident in these cells before leupeptin addition (Figure S2D,E). As an alternative assessment of LC3-II turnover, p62/SQSTM1 degradation may also be used to evaluate impairments of autophagic protein degradation (Bjorkoy et al., 2005). In addition to LC3-II accumulation, p62 levels also increased in PS1 KO cells (Figure S2F). Each of these three lines of evidence consistently supports a defect in autophagic vacuole clearance.
Proteolysis deficits in autolysosomes of PS1 KO blastocysts
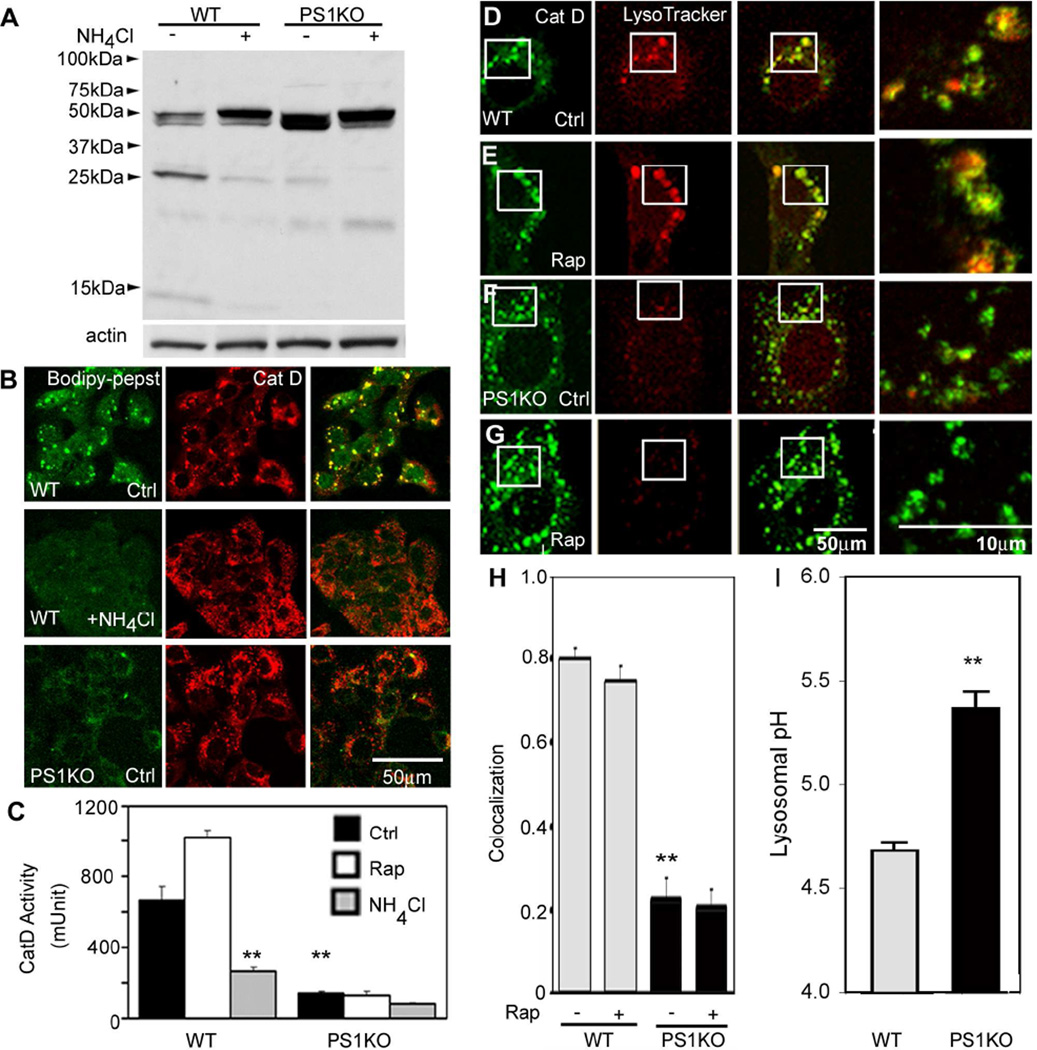
We investigated further the basis for delayed clearance of LC3 and other autophagy substrate proteins by examining the activation of cathepsins in PS1 KO cells. Western blot analyses of Cathepsin D (CatD), the major aspartyl protease of lysosomes, showed slightly elevated total Cat D immunoreactivity levels but a more rapid migration of the mature single chain enzyme on gels than the WT enzyme (46 kDa) and decreased proteolytic generation of 31- and 14-kDa forms of the mature enzyme in PS1 KO cells (Figure 3A). This deficit was similar to that seen in WT cells when lysosomal pH was neutralized by treatment with NH4Cl (Isidoro et al., 1991) or Bafilomycin A1 (Figure S3A). Metabolic labeling data confirmed that Cat D maturation was impaired in PS1 KO cells (Figure S3B). To assess Cat D activation within lysosomes, we incubated cells with Bodipy-FL-pepstatin A, which binds selectively to active Cat D (Chen et al., 2000). Dual fluorescence analyses of WT cells using Cat D antibodies showed strong double-labeling of compartments with Cat D antibodies and Bodipy-FL-pepstatin A, which was nearly completely abolished when lysosomal pH was neutralized by treatment with NH4Cl. By contrast, in PS1 KO cells, Bodipy-FL-pepstatin A labeling within Cat D-positive vesicles was markedly reduced despite normal numbers of these compartments (Figure 3B). To analyze Cat B activity in vivo, we used MR-Cat B, a cresyl-violet conjugated (Arg-Arg)2 peptide, which fluoresces only after it is cleaved by Cat B in an acidic environment. We confirmed that, in PS1 KO cells, MR-CatB signal was dramatically reduced compared to WT and was similar to levels seen in WT cells in which lysosomal acidification was inhibited using NH4Cl (Figure S3D). In vitro assays of Cat D activity in PS1 KO cells in the absence or presence of rapamycin confirmed a markedly reduced proteolytic activity relative to WT cells (Figure 3C). The activities of the cysteine proteases Cat B and Cat L were also similarly lowered in PS1 KO cells under these conditions (Figure S3E,F). Similar or more severe reductions in cathepsins B, D, and L activities were achieved in WT cells by incubating them in the presence of NH4Cl (Figure 3C and S3E,F).
Figure 3.
Cathepsin processing and activity impairment in PS1 KO cells: (A) Cat D immunoblots show reduced generation of the mature two-chain (31 kDa, 14kDa) form in PS1 KO cells, similar to NH4Cl treated (20 mM; 6 hrs) WT cells. (B) In vivo Cat D activity assays using Bodipy-FL-pepstatin A. Following Bodipy-FL-pepstatin A treatment, cells were immunolabeled with Cat D antibody. Bodipy-FL-pepstatin A binds to active Cat D of WT blastocysts and co-localize, but minimal colocalization is shown in PS1 KO cells or WT cells treated with NH4Cl. Scale bar 50 µm. (C) In vitro assays of Cat D enzyme activities in WT and PS1 KO cells with or without rapamycin or NH4Cl. ** for p <0.001. (D–H) Cells with or without rapamycin (10 nM; 6 hrs) were preincubated with LysoTracker and immunolabeled with Cat D antibody (D–G). Cat D-positive compartments were LysoTracker-positive in WT cells (D,E) but LysoTracker-negative in PS1 KO cells (F,G). Scale bars 50 or 10µm. (H) Quantitative analysis of LysoTracker and Cat D-positive compartments. ** p <0.001. (I) Lysosomal pH values were measured ratiometrically using LysoSensor yellow/blue DND-160–Dextran. ** p<0.001. See also Figure S3. Values are means ±S.E.M.
Defective lysosome acidification in PS1 KO blastocysts
The possibility that lysosome acidification may be impaired, raised by the foregoing observations, was further investigated by evaluating another process requiring lysosome acidification, namely, the dissociation of the cation-independent mannose-6-phosphate receptor (CI-MPR) from cathepsins after their delivery to late endosomes. Using double-immunofluorescence labeling with antibodies to Cat D and CI-MPR, we observed that most Cat D-positive vesicles were CI-MPR-negative in WT cells, but were nearly all CI-MPR-positive in PS1 KO cells (Figure S3C), indicating that dissociation of CI-MPR from Cat D was impaired. CI-MPR co-immunoprecipitation with Cat D revealed more Cat D bound to CI-MPR in PS1 KO cells, a phenomena reversed by human PS1 reintroduction (Figure S3C bottom). To assess lysosome acidification directly, we used LysoTracker. In WT cells, LysoTracker demonstrated strong fluorescence in virtually all Cat D-positive vesicles (Figure 3D), while in PS1 KO cells, fewer than 20% of Cat D-positive vesicles exhibited detectable fluorescence (Figure 3F). Compared to that in WT cells, LysoTracker signal in PS1 KO cells remained low after inducing autophagy with rapamycin (Figure 3E,G,H). Measurement of average lysosomal pH using LysoSensor yellow/blue DND-160–Dextran (Diwu et al. 1999) (Diwu et al., 1999) confirmed that WT cells display an average lysosomal pH of 4.7±0.08, consistent with previously published studies (Ohkuma and Poole, 1978; Ramachandran et al., 2009). By contrast, PS1KO cells displayed a substantially elevated lysosomal pH of 5.4±0.04 (p<0.001) (Figure 3I). We show that lysosomal acidification is normal in mouse fibroblasts lacking Nicastrin (Nct), a γ-secretase complex component required for secretase activity (Figure S3H). In addition, γ-secretase inhibitor (L685,458) has no effect on lysosomal acidification, Cat B activity and Cat D processing (Figure S3I,J). This suggests that the effect of PS1 on autophagy is not dependent on γ-secretase.
Impaired glycosylation and targeting of the v-ATPase V0a1 subunit in PS1 KO cells
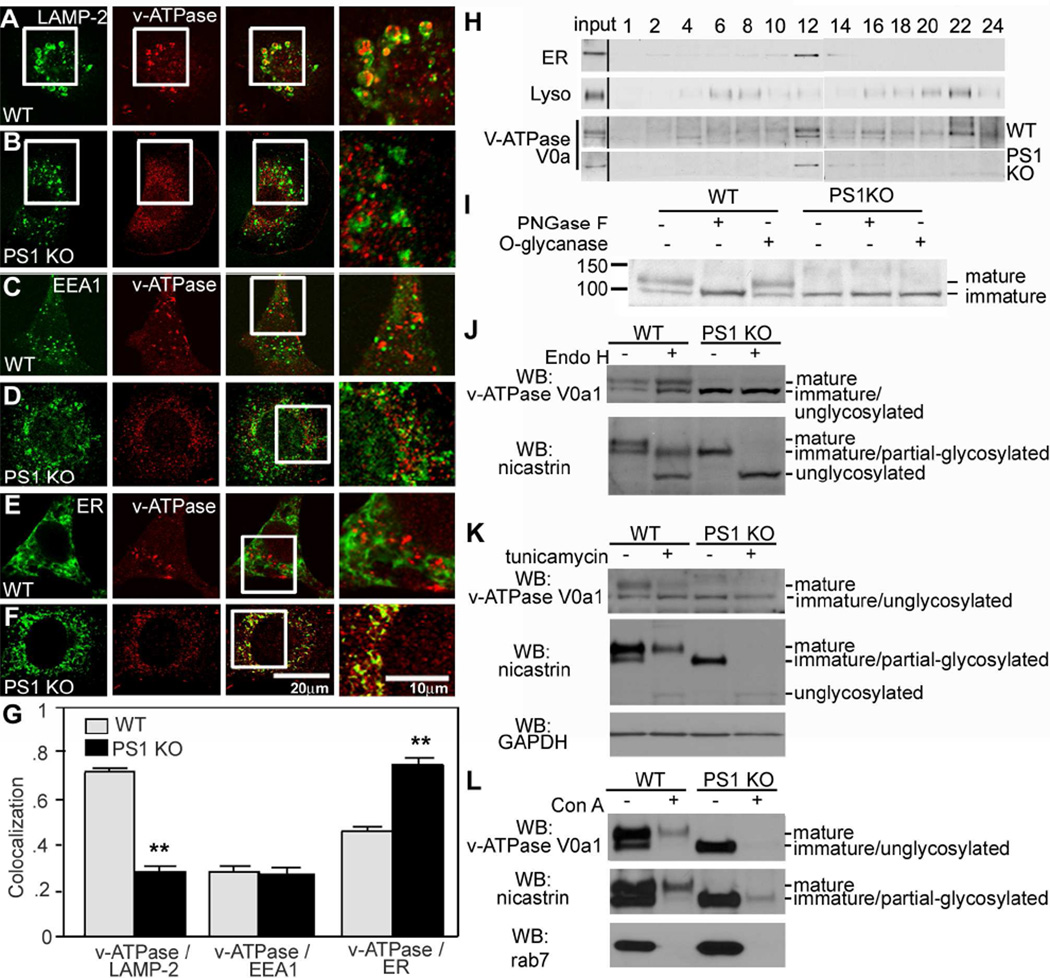
To further understand the basis for the acidification defect in PS1 KO cells, we investigated the v-ATPase V0a1 subunit as a marker of proton pump function in lysosomes. Double-immunofluorescence labeling analysis revealed strong co-localization of v-ATPase with LAMP-2-positive compartments in WT cells (Figure 4A). In PS1 KO cells, however, v-ATPase immunolabeling was concentrated in a perinuclear region remote from most of the peripherally distributed LAMP-2-positive compartments (Figure 4B). v-ATPase immunoreactivity strongly colocalized with calnexin, an ER-integral protein, in PS1KO cells (Figure 4F) but minimally in WT cells (Figure 4E). As expected, colocalization of v-ATPase with EEA1 (early endosome antigen-1) was nearly absent in early endosomes of both WT and PS1KO cells (Figure 4C,D). Quantitative analyses of marker colocalization showed a significantly greater extent of association of v-ATPase with ER than with lysosomes in PS1KO cells, converse to the pattern seen in WT cells (Figure 4G). Subcellular fractionation studies confirmed the immunocytochemical results showing that v-ATPase was retained in the ER-rich (calnexin-positive) fraction but was markedly depleted from the lysosomal (LAMP2) fraction in PS1 KO cells, whereas in WT cells, v-ATPase was enriched in both ER and lysosomal fractions (Figure 4H).
Figure 4.
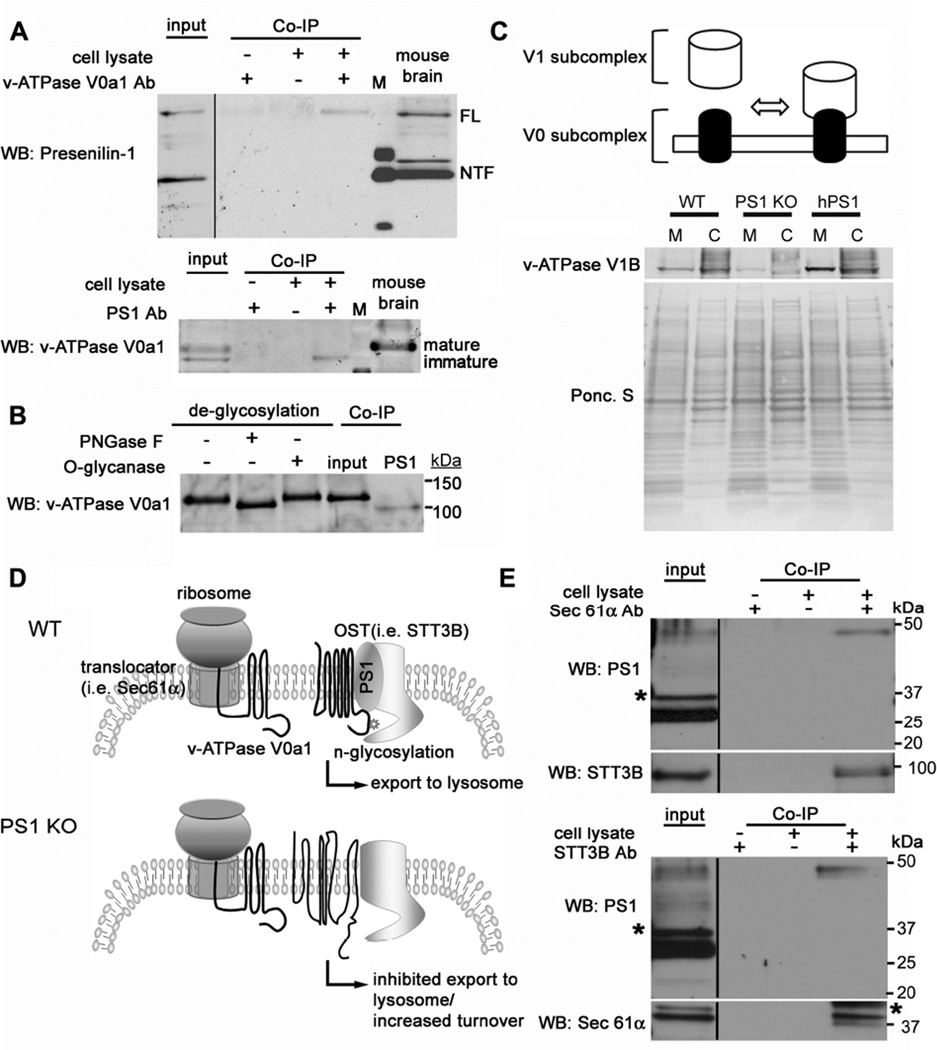
Lysosomal targeting of v-ATPase is impaired in PS1 KO blastocysts: Double-immunofluorescence labeling shows strong colocalization of v-ATPase (V0a1 subunit) and LAMP-2 in WT cells (A) but minimal colocalization in PS1 KO cells (B). v-ATPase V0a1 and early endosomal marker, EEA1, show little colocalization in WT (C) and PS1 KO cells (D). v-ATPase V0a1 and the ER marker, calnexin, strongly colocalized in PS1KO cells (F) but minimally colocalized in WT cells (E). Scale bars 20 or 10µm. (G) Quantitation analysis of v-ATPase V0a1 association with organelle markers. (**for p <0.001). (H) Immunoblots of v-ATPase V0a1 subunit distribution in subcellular fractions of WT and PS1KO cells. Calnexin, primarily localizes in fraction 12 and LAMP2 mainly in fraction 22. In WT cells, the v-ATPase V0a1 subunit, detected as 100 and 120 kDa bands was present in fraction 12 (120 kDa and 100 kDa) and fraction 22 (only in its 120 kDa form), but was primarily detected in the ER-rich fraction (only as a 100 kDa protein) of PS1KO cells. (I) WT and PS1KO cell lysates treated with PNGase F or O-glycanase. The N-glycosylated form of v-ATPase V0a1 subunit (120 kDa) was deglycosylated (100 kDa) after treatment with PNGase F but not with O-glycanase in WT cells. The v-ATPase V0a1 subunit in PS1 KO cells was not N-glycosylated, with the 100 kDa form unchanged by treatment. (J) Insensitivity to Endo H of 100- and 120-kDa bands in WT lysates. Both mature and immature glycosylated forms of nicastrin, serving as a positive control, are EndoH sensitive. (K) Cells incubated 24 hrs with tunicamycin to block glycoprotein synthesis in the ER display reduced levels of 120-kDa v-ATPase V0 subunit in WT cells but no effect on 100-kDa subunit. Nicastrin immunoblot analysis under the same conditions is shown. (L) After cell lysates were incubated with Con A beads, glycoproteins were eluted. Con A binds 120-kDa but not 100 kDa v-ATPase V0a1, and both mature and partially glycosylated nicastrin species. Rab7, a negative control, was not bound. Values are means ±S.E.M.
In subcellular fractions from PS1 KO cells, we also observed that the v-ATPase V0a1 subunit located in ER-enriched fractions exists as a single 100 kDa band, whereas, in WT cells, it was present as a double band (120/100 kDa) in the ER-enriched fraction and as a single 120 kDa band in the lysosome-enriched fraction. In cell lysates treated with either PNGase F or O-glycanase (Figure 4I), PNGase F converted the mature 120-kDa v-ATPase form in WT cells to a 100-kDa form suggesting that the 100-kD protein is a non-glycosylated core protein. Both 100- and 120-kDa bands were insensitive to Endo H (Figure 4J). Because nicastrin glycosylation patterns are well established in WT and PS1 KO cells (Herreman et al., 2003), we compared nicastrin as a positive control, confirming these previously published data and the competence of Endo H used in our experiments. The results showed that v-ATPase maturation follows a different pattern than nicastrin. Nicastrin maturation involves a glycosylated Endo H sensitive intermediate, which is the precursor to a fully mature glycosylated Endo H insensitive form. By comparison, the Endo H and PNGase F digestions indicated that the 100-kDa v-ATPase V0a1 subunit is non-glycosylated and that the 120-kD form is a complex glycosylated mature protein.
To further confirm the glycosylation state of v-ATPase variants, cells were incubated with tunicamycin and the mobility of the v-ATPase V0a1 subunit expressed in the absence or presence of tunicamycin was compared by Western blot. Tunicamycin treatment of WT cells caused a substantial loss of the 120 kDa protein and increased levels of the 100 kDa form. By contrast, tunicamycin had no effect on the mobility of the 100 kDa form in either WT cells or in the PS1 KO cells where this form was the only detectable variant (Figure 4K). Both mature and partially glycosylated immature nicastrin were markedly reduced, yielding small amounts of unglycosylated nicastrin present in both WT and PS1 KO cells. As further confirmation of the Endo H results, we also performed lectin affinity binding studies using concanavalin A (Con A). As expected, Con A bound the 120-kDa v-ATPase V0a1 species in WT cells, and the mature and partially glycosylated forms of nicastrin, but none of the 100 kDa v-ATPase species in PS1 KO cells, even though this form was present in amounts comparable to the mature forms of v-ATPase V0a1 subunit and nicastrin in WT cells. Rab7, a negative control, did not bind Con A (Figure 4L). These lines of evidence confirm that the 100 kDa protein is not glycosylated and that the 120 kDa protein is the complex glycosylated mature v-ATPase V0a1 subunit.
To investigate a possible role of PS1 in v-ATPase V0a1 subunit maturation, we performed co-immunoprecipitation assays with the endogenous proteins from WT cells and mouse brain. Precipitation of endogenous v-ATPase V0a1 subunit led to co-precipitation of full length PS1 but not its more abundant N- or C- terminal cleaved forms, indicating that only full length PS1 can bind to the v-ATPase V0a1 subunit (Figure 5A, top). Importantly, PS1 preferentially co-precipitated with the immature 100 kDa form of v-ATPase V0a1 subunit (Figure 5A, bottom). In WT mouse brain, only unglycosylated immature v-ATPase bound to PS1 even though most of the v-ATPase V0a1 subunit pool in this tissue was N-glycosylated (Figure 5B). These data indicated that uncleaved PS1 binds to immature v-ATPase V0a1 to modulate its maturation in the ER and affect its delivery to lysosomes.
Figure 5.
PS1 directly binds to the v-ATPase V0a subunit affecting its maturation and assembly of the v-ATPase complex: (A) Co-immunoprecipitation of endogenous PS1 with anti-v-ATPase V0a1 antibody and v-ATPase V0a1 with anti-PS1-NTF antibody. Precipitated proteins were detected by immunoblot with either anti-PS1 (Ab14) or anti-v-ATPase V0a1. M = marker lane. (B) Lysate from WT mouse brain was treated with PNGase F or O-glycanase. The v-ATPase V0a1 subunit is highly glycosylated in mouse brain. The N-glycosylated form of v-ATPase V0a1 subunit (120 kDa) is deglycosylated (i.e., MW shift to 100 kDa after treatment with PNGase F but not with O-glycanase). The v-ATPase V0a1 subunit was immunoprecipitated with anti-PS1-NTF antibody and detected by anti-v-ATPase V0a1 antibody. Only unglycosylated v-ATPase V0a1 subunit co-precipitates with PS1. (C) Top panel is a model of v-ATPase assembly. Immunoblot shows v-ATPase V1B1 subunit distributes between the membrane and cytosolic fractions. (D) The diagram shows a hypothetical model of N-glycosylation of the v-ATPase V0a1 subunit via PS1. PS1 binding to translocon and OST complex facilitates the presentation of the v-ATPase V0a1 subunit to the OST complex. (E) Co-precipitation of endogenous PS1 with anti-Sec61α antibody and STT3B antibody. Only full length PS1 was co-precipitated with Sec61α and STT3B. Sec61α and STT3B were also co-immunoprecipitated each other. * represents a non-specific band. See also Figure S4.
The v-ATPase is a multicomplex molecule composed of a membrane bound V0 subcomplex and a cytosolic V1 subcomplex and is only active when both are assembled in the lysosomal membrane. We next examined the v-ATPase multicomplex assembly status using membrane fractionation. Relatively small amounts of the v-ATPase V1B subunit were present in the membrane fraction and negligible amounts were detected in the cytosolic fraction in PS1 KO cells relative to WT cells (Figure 5C). This observation shows that PS1 deletion impairs v-ATPase assembly and that the unassembled V1 subunit is depleted and presumably degraded at greater rates in PS1 KO cells.
We suspected that PS1 may be involved in N-glycan transfer from the oligosaccharyltransferase (OST) to the v-ATPase V0a1 subunit after its translation and translocation via the translocon (Figure 5D). Supporting this mechanism, co-immunoprecipitation data showed that endogenous full length PS1 preferentially co-precipitates with Sec61α (translocon subunit) and STT3B (OST subunit) (Figure 5E) and vice versa; however, other ER proteins, such as GRP94 and PDI, did not interact (data not shown). Collectively, these data support a mechanism in which full length PS1 holds the v-ATPase V0a1 subunit close to the OST complex, thereby facilitating posttranslational N-glycosylation of the subunit (Fig 5D), as described previously for several other membrane proteins (Bolt et al., 2005; Ruiz-Canada et al., 2009).
If PS1 is solely responsible for these deficits in autolysosome function, exogenously reintroduced PS1 should be able to rescue the PS1 KO phenotype. Stable transfection of human PS1 into PS1/2 KO cells completely restored vesicular compartment acidification, Cat D maturation, v-ATPase V0a1 glycosylation, and macroautophagy responses (Figure S4). These data strongly support the conclusion that, under conditions of PS1 ablation, the v-ATPase V0a1 subunit is not N-glycosylated and, therefore, is retained in the ER, thereby preventing acidification of lysosome-related compartments and activation of proteases in autolysosomes during autophagy.
Defective vesicle acidification and autophagic pathology in neurons of PS1 hypomorphic and PS cKO mice
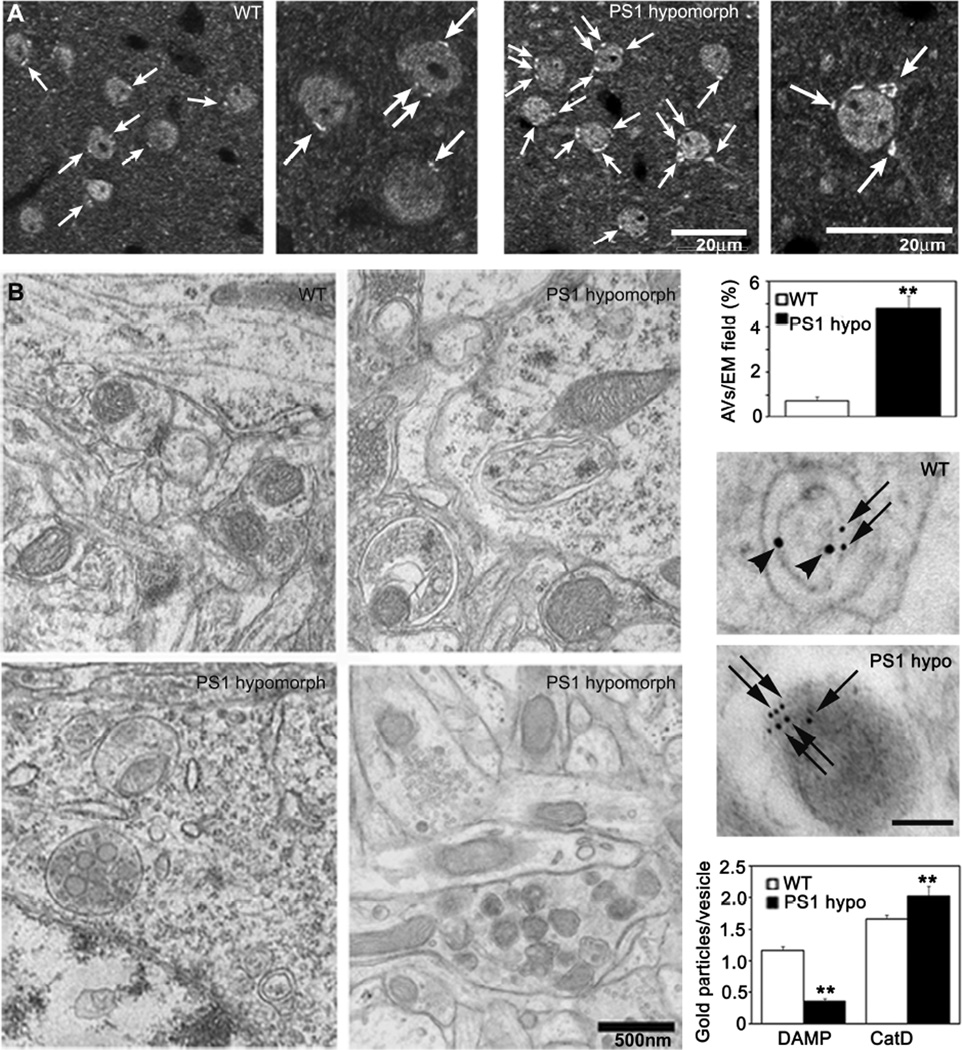
To extend these observations to neurons in vivo, we examined whether AVs accumulate in association with defective lysosome acidification in mouse models of PS1 hypofunction. PS1 hypomorphic mice, which express very low levels of PS1 protein (1%) but sufficient for brain development and normal lifespan, displayed significantly increased numbers of LC3-positive compartments in cortical and hippocampal neurons (Figure 6A) and 6-fold more immature AVs (p<0.001) quantified by EM morphometry compared to WT mice (Figure 6B,C). To assess lysosome acidification in vivo in these mice, we performed intraventricular injections of DAMP, a probe sensitive to changes in vesicular pH and whose abundance reflects the degree of acidification (Anderson et al., 1984) followed by double-immunogold EM using antibodies to CatD to identify lysosome-related vesicles and dinitrophenol to detect the presence of DAMP. These data showed that vesicle acidification was much less in Cat D-positive compartments in neurons of PS1 hypomorph mice than in WT controls (Figure 6D). Similarly, PS cKO mice, under conditions of PS1 conditional knockdown, exhibited abnormally increased numbers of LC3-positive compartments and AV in brain (Figure S5A,B) and significantly decreased acidification of lysosomal compartments compared to WT controls (p<0.001)(Figure S5C,D). These results confirm that the PS1-dependent lysosomal acidification defect observed in the cell culture system also occur in vivo in the brains of PS1-deficient mice.
Figure 6.
Defective autophagosome accumulation and acidification in PS1 hypomorphic mice. (A) LC3 immunohistochemistry of PS1 hypomorph brain shows greater LC3 staining (arrow) in the PS1 deficient mouse compared to WT. Scale bar - 20 µm. (B) EM of AVs and dystrophic neurite-like structures in brains of PS1 hypomorph mice compared to littermate controls. Scale bar - 500nm. (C) Quantitation of AVs per EM field. ** p <0.001. (D) DAMP, a marker which localizes to acidic compartments, was infused intraventricularly into the brains of mice and analyzed by immuno-EM using DNP (10 nm-gold, arrowheads) and CatD (6 nm-gold, arrows) antibodies. Graphs show quantitation of immunogold labeling for DAMP and CatD. ** p < 0.001. See also Figure S5. All values are means ±S.E.M.
PS1 mutations impair macroautophagy and v-ATPase targeting in fibroblasts from patients with familial AD
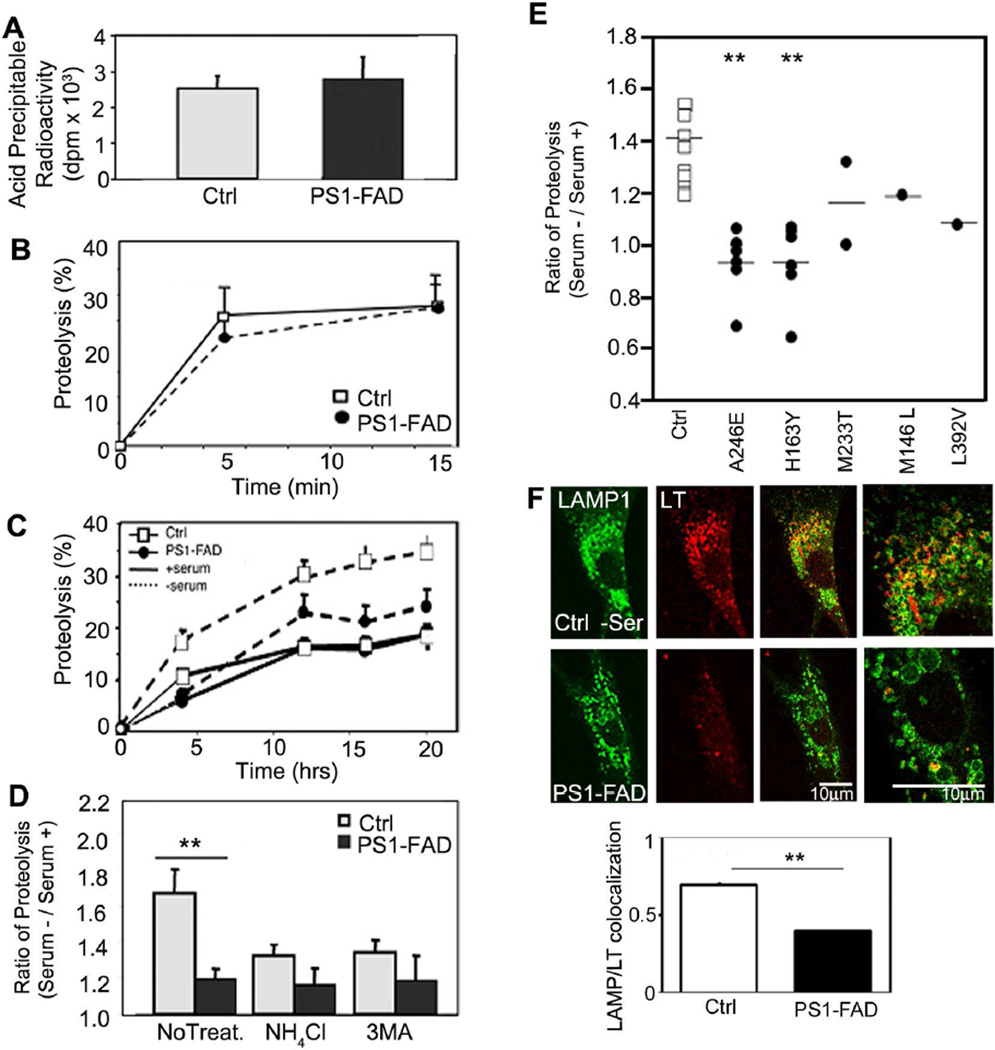
3 PS1-FAD and control human fibroblasts exhibited comparable rates of [3H]-leucine incorporation into proteins (Figure 7A), degradation of short-lived proteins (Figure 7B), and proteolysis of long-lived proteins under autophagy-suppressed conditions (Figure 7C). By contrast, when macroautophagy was induced by serum withdrawal, PS1-FAD fibroblasts exhibited a minimal rise in proteolysis compared to control fibroblasts (Figure 7C). NH4Cl treatment eliminated the difference in proteolytic rates between the two cell groups (Figure 7D), confirming that proteolysis impairment selectively involved the lysosomal system in PS1-FAD fibroblasts. In an expanded analysis including a total of 16 different PS1-FAD lines sub-classified by the specific PS1 mutation (A246E, M233T, H163Y, M146L, L392V), autophagic protein degradation was lowered compared to that in control cells (Figure 7E). The LC3-II/LC3-I ratio and also total LC3-II levels in PS1-FAD fibroblasts were also comparatively high even in serum supplemented conditions and stayed elevated compared to control cells after serum removal (Figure S6A). To assess autophagosome formation, control and PS1-FAD fibroblasts were immunostained with LC3 antibody, which revealed increased LC3 vesicular puncta in PS1-FAD compared to control fibroblasts after autophagy was induced by serum withdrawal (Figure S6B). Morphometric ultrastructural analyses revealed that immature AVs were more numerous in PS1-FAD cells than in control fibroblasts where the AVs were mainly smaller and electron-dense, representing a terminal stage of autophagic degradation (Figure S6C). We also confirmed using LysoTracker that the level of acidification of the lysosomal compartments (highlighted with LAMP-1) in PS1-FAD was markedly lower (Figure 7F).
Fig. 7.
Defective autophagy in PS1-FAD human fibroblasts. (A) [3H]-leucine incorporation into fibroblasts from 5 different PS1-FAD patients and age matched controls. (B) After [3H]-leucine labeling, proteolysis of short-lived proteins was measured following the chase period. (C) Degradation of long-lived proteins was measured after incorporation of [3H]-leucine followed by incubation in serum-supplemented or -deprived medium during the chase period (up to 20 hrs) (* for p <0.05, n=15). (D) The increase in proteolysis at 12 hrs after serum removal relative to serum-replete conditions was determined for control and PS1-FAD fibroblasts cells treated with NH4Cl (20 mM) or 3MA (10 mM) (** for p <0.001, n=15), or left untreated. (E) Increases in degradation of long-lived proteins after serum removal were compared in fibroblasts from control (n=11) and PS1-FAD patients carrying different PS1 mutations (as labeled). (F) Control or PS1-FAD fibroblasts treated in the absence of serum were preincubated with LysoTracker and immunolabeled for LAMP-1. LAMP-1-positive compartments colocalized with LysoTracker control cells, but not in PS1-FAD, as verified by quantitative analysis). ** for p <0.001. See also Figure S6, S7. Values are means ±S.E.M.
To investigate the mechanism underlying impaired autophagic protein turnover in PS1-FAD fibroblasts, we used a double-tagged mRFP-GFP-LC3 construct, enabling us to assess lysosome acidification in vitro. Almost all of the RFP and GFP signal colocalized in PS1 FAD fibroblasts under serum starved conditions (Figure S7A) indicating that acidification of lysosomes is insufficient to quench GFP fluorescence or, less likely, that transport of mRFP-GFP-LC3 to acidic compartments is delayed. By contrast, in control fibroblasts, only 10% GFP colocalized with RFP, indicating that formation of acidified autolysosomes is efficient. Accumulation of p62 immunoreactivity before or after serum starvation was disproportionately high in PS1-FAD fibroblasts by western blot analysis (Figure S7B left) or by ICC analysis of p62 localization within LC3-positive compartments (Figure S7B, right), consistent with delayed proteolytic autolysosomal clearance of this autophagy-selective substrate. Double-immunofluorescence labeling with V0a1 antibodies established that the v-ATPase was localized with CatD-positive compartments in control fibroblasts, but only a small proportion of v-ATPase colocalized with Cat D in PS1 FAD fibroblasts (Figure S7C,F); however, most v-ATPase immunoreactivity colocalized with PDI in PS1-FAD fibroblasts but not in control fibroblasts (Figure S7D,F). Additional double-label analyses showed that the v-ATPase V0a1subunit also colocalized less with V1B1/2 subunit in PS1 FAD fibroblasts compared to control cells (Figure S7E,F). Levels of v-ATPase were also significantly decreased in PS1-FAD fibroblast (Figure S7G).
DISCUSSION
Our results identify a novel, essential role of PS1 in lysosomal-dependent proteolysis directly relevant to the mechanism by which PS1 mutations accelerate the pathogenesis of AD. PS1 deletion prevented macroautophagic protein turnover while minimally affecting non-lysosomal turnover of short and long-lived proteins. We have traced this defect in autophagy-dependent proteolysis to inadequate autolysosome/lysosome acidification resulting from a failure of the V0a1 subunit of v-ATPase to become N-glycosylated in the ER and subsequently delivered to autolysosomes/lysosomes. This acidification defect explains the many other abnormalities of AV dynamics and autolysosome maturation/digestion that we and others have observed in PS1 KO cells (Esselens et al., 2004; Wilson et al., 2004). Furthermore, we also demonstrate that neurons in the brains of mouse models of PS1 hypofunction and cells from patients with AD caused by PS1 mutations show similar autolysosome maturation defects as in PS1 KO cells. Lysosomal acidification is necessary to dissociate CI-MPR from cathepsins, complete the proteolytic maturation of cathepsin D, and activate cathepsins (Kokkonen et al., 2004). All of these functions were impaired in PS1 KO cells, resulting in delayed proteolytic clearance of autophagic substrates and their accumulation in autophagic vacuoles. Transfection of WT hPS1 into PS1/2 KO cells rescued all of these deficits. We observed similar autophagic/lysosomal deficits in neurons of PS1 deficient mice, indicating a similar role for PS1 in brain.
Multiple lines of evidence in our study established that defective autophagy in PS1 KO cells principally reflects a failure to degrade autophagy substrates and clear AVs due to a specific defect in lysosome/autolysosome acidification. This evidence in PS1 KO cells includes abnormally elevated levels of autophagy substrates (p62, LC3), a failure to clear AVs formed after rapamycin-mediated autophagy induction, the minimal effect of a cathepsin inhibitor (leupeptin) on LC3-II accumulation in PS1 KO cells, impaired maturation of cathepsin D, reduced specific activities of multiple cathepsins in vitro and in situ within lysosomes, and impaired dissociation of MPR from cathepsins. Impaired degradation of autophagy substrates in PS1 KO cells is also indicated by the high proportions of early autolysosomes compared to electron-dense late autolysosomes in morphometric analyses and the low recovery of dense lysosomes in subcellular fractionations. All of these effects are expected outcomes of a defect in the acidification of autolysosomes/lysosomes, which was demonstrated directly by the marked decrease of LysoTracker fluorescence, ratiometric assays of lysosomal pH, DAMP labeling, and by the very low abundance of the V0a1 subunit of v-ATPase in lysosome-related compartments. Similar types of deficits and lysosomal pH increases of a similar magnitude have been observed when v-ATPase is inhibited with bafilomycin (Yoshimori et al., 1991).
In contrast to these striking effects on autophagic proteolysis, PS1 deletion did not detectably alter major upstream aspects of macroautophagy, including nutrient-dependent regulation of mTOR, a protein kinase-signaling pathway that utilizes AKT-P13K (Sarbassov et al., 2005) and is modulated in part by PS2 (Kang et al., 2005). In response to autophagy induction, p70S6K was dephosphorylated normally and LC3-positive puncta and AVs increased modestly but significantly above an already elevated level in PS1 KO cells, indicating preservation of an ability to form autophagosomes. The accumulation of p62, a known autophagy substrate (Bjorkoy et al., 2005), indicated that PS1 KO cells can sequester substrates despite the subsequent impairment of degradation. The presence of LC3/LAMP2-positive profiles in PS1 KO cells is evidence of autophagosome-late endosome fusion as seen by Wilson et al. (2004), but not by Esselens et al. (2004), and is consistent with other evidence that autophagosome-lysosome fusion is not dependent on lysosomal acidification (Jahreiss et al., 2008). Other key aspects of lysosomal biogenesis and function were also not detectably altered in PS1 KO cells, such as the delivery of cathepsins and LAMPs to late endosome/lysosomes and the distribution and levels of rab7 required for lysosome maturation (Bucci et al., 2000). Thus, the failure of v-ATPase targeting to lysosomes is a relatively selective effect of PS1 deletion on proteolytic steps in the autophagy pathway, and, as expected, the endocytic pathway.
The v-ATPases are multisubunit complexes which are composed of a membrane-bound subcomplex V0 and a cytosolic V1 subcomplex (Forgac, 2007). We established by Endo H digestion, tunicamycin treatment, and lectin affinity binding that the 100kDa immature form of v-ATPase V0a1 subunit is unglycosylated. Its physical interaction with PS1 enables the N-glycosylation required for this subunit to be efficiently delivered to lysosomes (Gillespie et al., 1991; Nishi and Forgac, 2002). This is not a generalized effect on the N-glycosylation mechanism itself since LAMP-2 was normally glycosylated in PS1KO cells. Most commonly, N-glycosylation occurs cotranslationally (Kelleher and Gilmore, 2006); however, for some proteins, glycosylation occurs after the entire polypeptide has been translocated into the ER lumen (Bolt et al., 2005) and may preferentially involve the STT3B catalytic subunit of the OST (Ruiz-Canada et al., 2009). Consistent with a posttranslational glycosylation of the v-ATPase V0a1 subunit, our co-immunoprecipitation studies showed that PS1 binds to STT3B. Given this context, our evidence that the 100 kDa v-ATPase is unglycosylated supports a model (Figure 5D) of posttranslational N-glycosylation of v-ATPase V0a1 subunit, a function lost when PS1 is deleted. Our findings implicating PS1 holoprotein provide an explanation for the observation (Esselens et al., 2004) confirmed here that PS1 effects on autophagy are not dependent on γ-secretase activity.
In conclusion, our studies define a novel essential role for PS1 in the maturation and trafficking of the v-ATPase responsible for lysosome acidification. We have demonstrated the adverse consequences of ablating this function for the normal turnover of proteins and organelles by autophagy. It is likely that a failure of PS1-dependent v-ATPase trafficking would have a range of additional effects on functions of other compartments that rely on this proton pump to acidify the intralumenal environment. Finally, we have demonstrated the clinical relevance of this PS1 function by showing that a range of mutations of PS1 causing early onset FAD also display a similar loss of lysosomal function as in PS1 KO, which can account for the marked acceleration of autophagy-related dysfunction and neuronal cell death associated with PS1-FAD. Because Aβ is both generated and degraded during autophagy (Yu et al., 2005), impaired lysosomal clearance could account for reported PS1-mediated increases in Aβ. Similarities between the severe autophagy pathology in PS1-FAD and that developing with a later onset in sporadic AD suggests that lysosomal dysfunction is also a pathogenic mechanism in the common sporadic form of AD (Nixon et al., 2008).
METHODS
Methods are described in detail in Supplemental experimental procedures.
Cell lines, mouse and reagents
Murine blastocysts with different PS1 genotypes (WT, BD6; PS1 KO BD15; PS1/2 KO, BD8) previously characterized by Lai et. al. (2003), were used in this study. In addition, human PS1 wt was stably transfected into the BD8 line (Laudon et al., 2004). Human fibroblasts lines, acquired from the Coriell Institute (Camden NJ), Karolinska Institute (Upsala, Sweden), University di Firenze (Italy) and University of Western Australia (Perth). PS1 hypomorph (Rozmahel et al., 2002) and PS cKO mice (Saura et al., 2004) were studied at 13 month and 2–3 month, respectively, together with age-matched controls.
Intracellular protein degradation measurements
Confluent cells were labeled with [3H]-leucine (2µCi/ml) for 48 hrs at 37°C in order to preferentially label long-lived proteins. Following labeling, cells were extensively washed and maintained in complete medium (DMEM + 10 % fetal bovine serum), the medium was supplemented with unlabeled 2.8 mM leucine to prevent [3H]-leucine reincorporation into newly synthesized proteins. Total protein degradation measured by pulse-chase were performed as described previously (Auteri et al., 1983).
Ultrastructural and morphometric analyses and autophagic vacuole isolation and subcellular fractionation
Following treatments, cells were prepared for EM as previously described and AVs, identified using previous morphological criteria (Yu et al., 2005), were classified and counted on electron micrographs (7900x print magnification, of 20 EM images from each experimental group). AVs were isolated by centrifugation in a discontinuous metrizamide density gradient (Marzella et al., 1982) for each cell. For subcellular fractionation, homogenate (0.5 ml) was layered on top of an Optiprep (Sigma) step gradient (10, 15, 20, 25, and 30%, 2.3 ml each) into polyallomer tubes (Beckman) and centrifuged in a SW-40Ti rotor with a model L8–80M Beckman ultracentrifuge (100,000 g, 16 hours, 4°C).
Gel electrophoresis, immunoblotting and deglycosylation
Immunoblotting was performed as previously described (Herreman et al., 2003). Briefly, cells used for Western blot analyses were lysed in buffer containing 50 mM Tris (pH=7.4), 150 mM NaCl, 1mM EDTA, 1 mM EGTA, 1% Triton X-100 and 0.5% Tween-20 with protease and phosphatase inhibitors. Following electrophoresis on 4 −20% gradient gel (Invitrogen), proteins were transferred onto 0.45 µm PVDF membranes (Millipore) then incubated overnight in primary antibody followed by HRP-conjugated secondary antibody. Blots were developed by ECL-kit (GE Healthcare).
Enzymatic assays
Cathepsin B and L activities were assayed as described previously(Nakanishi et al., 1994) and Cathepsin D activity was assayed using [14C]methemoglobin as previously described by Dottavio-Martin and Ravel (1987).
Analytical Procedures
The quantitative colocalization analyses were performed using ImageJ software (NIH Image) with colocalization analysis plugins (Wright Cell Imaging facility). The value shown represent Pearson’s coefficient. Statistical analyses were calculated by two-tailed paired student t-test using GraphPad InStat (GraphPad Software Inc.). Error bars represent standard error of the mean (±S.E.M).
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful to Dr. Alan Bernstein (Global HIV Vaccine Enterprise, Seattle) for blastocysts, Dr. Tamotsu Yoshimori (Osaka University) for the tandem-tagged LC3 construct, Dr. Samuel E. Gandy (Mount Sinai Medical Center) for PS1 antibody, Dr. Satoshi Sato (Kyoto University) for the rabbit pAb against 116kDa subunit of the v-ATPase, Dr. Jean Gruenberg (Universite de Geneve) for anti-LBPA antibody and Drs. Richard Cowburn (Karolinska Institute), Ralph Martins (Cornell University), Sandro Sorbi (Universita degli studi di Firenze) for PS1-FAD human fibroblasts. We also thank Dr. Gert Kreibich (NYU Langone Medical Center) and members of the Nixon laboratory for valuable discussions and Nicole Piorkowski for assistance with manuscript preparation. This work supported by NIH grant number P01AG017617 (R.A.N) and the Alzheimer’s Association (R.A.N).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson RGW, Falck JR, Goldstein JL, Brown MS. Visualization of Acidic Organelles in Intact Cells by Electron Microscopy. Proc Natl Acad Sci. 1984;81:4838–4842. doi: 10.1073/pnas.81.15.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auteri JS, Okada A, Bochaki V, Dice JF. Regulation of intracellular protein degradation in IMR-90 human diploid fibroblasts. J Cell Physiol. 1983;115:167–174. doi: 10.1002/jcp.1041150210. [DOI] [PubMed] [Google Scholar]
- Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA. Autophagy Induction and Autophagosome Clearance in Neurons: Relationship to Autophagic Pathology in Alzheimer's Disease. J Neurosci. 2008;28:6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolt G, Kristensen C, Steenstrup TD. Posttranslational N-glycosylation takes place during the normal processing of human coagulation factor VII. Glycobiology. 2005;15:541–547. doi: 10.1093/glycob/cwi032. [DOI] [PubMed] [Google Scholar]
- Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B. Rab7: a key to lysosome biogenesis. Mol Biol Cell. 2000;11:467–480. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Peterhoff CM, Schmidt SD, Terio NB, Duff K, Beard M, Mathews PM, Nixon RA. Presenilin mutations in familial Alzheimer disease and transgenic mouse models accelerate neuronal lysosomal pathology. J Neuropathol Exp Neurol. 2004;63:821–830. doi: 10.1093/jnen/63.8.821. [DOI] [PubMed] [Google Scholar]
- Chen C-S, Chen W-NU, Zhou M, Arttamangkul S, Haugland RP. Probing the cathepsin D using a BODIPY FL-pepstatin A: applications in fluorescence polarization and microscopy. Journal of Biochemical and Biophysical Methods. 2000;42:137–151. doi: 10.1016/s0165-022x(00)00048-8. [DOI] [PubMed] [Google Scholar]
- Citron M, Westaway D, Xia W, Carlson G, Diehl T, Levesque G, Johnson-Wood K, Lee M, Seubert P, Davis A, et al. Mutant presenilins of Alzheimer's disease increase production of 42- residue amyloid beta-protein in both transfected cells and transgenic mice. Nat Med. 1997;3:67–72. doi: 10.1038/nm0197-67. [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant α-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature. 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- Diwu Z, Chen CS, Zhang C, Klaubert DH, Haugland RP. A novel acidotropic pH indicator and its potential application in labeling acidic organelles of live cells. Chem Biol. 1999;6:411–418. doi: 10.1016/s1074-5521(99)80059-3. [DOI] [PubMed] [Google Scholar]
- Esselens C, Oorschot V, Baert V, Raemaekers T, Spittaels K, Serneels L, Zheng H, Saftig P, De Strooper B, Klumperman J, et al. Presenilin 1 mediates the turnover of telencephalin in hippocampal neurons via an autophagic degradative pathway. J Cell Biol. 2004;166:1041–1054. doi: 10.1083/jcb.200406060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- Gillespie J, Ozanne S, Tugal B, Percy J, Warren M, Haywood J, Apps D. The vacuolar H(+)-translocating ATPase of renal tubules contains a 115-kDa glycosylated subunit. FEBS Lett. 1991;282:69–72. doi: 10.1016/0014-5793(91)80446-a. [DOI] [PubMed] [Google Scholar]
- Herreman A, Van Gassen G, Bentahir M, Nyabi O, Craessaerts K, Mueller U, Annaert W, De Strooper B. γ-Secretase activity requires the presenilin-dependent trafficking of nicastrin through the Golgi apparatus but not its complex glycosylation. J Cell Sci. 2003;116:1127–1136. doi: 10.1242/jcs.00292. [DOI] [PubMed] [Google Scholar]
- Isidoro C, Horst M, Baccino FM, Hasilik A. Differential segregation of human and hamster cathepsin D in transfected baby-hamster kidney cells. Biochem J. 1991;273(Pt 2):363–367. doi: 10.1042/bj2730363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahreiss L, Menzies FM, Rubinsztein DC. The itinerary of autophagosomes: From peripheral formation to kiss-and-run fusion with lysosomes. Traffic. 2008;9:574–587. doi: 10.1111/j.1600-0854.2008.00701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junichi S, Anastasios G, Pankaj M, Zen K, Claudia ML, Lia B, Nikolaos KR. FAD mutants unable to increase neurotoxic Aβ42 suggest that mutation effects on neurodegeneration may be independent of effects on Aβ. Journal of Neurochemistry. 2007;101:674–681. doi: 10.1111/j.1471-4159.2006.04391.x. [DOI] [PubMed] [Google Scholar]
- Kang DE, Yoon IS, Repetto E, Busse T, Yermian N, Ie L, Koo EH. Presenilins Mediate Phosphatidylinositol 3-Kinase/AKT and ERK Activation via Select Signaling Receptors: selectivity of PS2 in platelet-derived growth factor signaling. J Biol Chem. 2005;280:31537–31547. doi: 10.1074/jbc.M500833200. [DOI] [PubMed] [Google Scholar]
- Kelleher DJ, Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology. 2006;16:47R–62. doi: 10.1093/glycob/cwj066. [DOI] [PubMed] [Google Scholar]
- Kim TW, Tanzi RE. Presenilins and Alzheimer's disease. Curr Opin Neurobiol. 1997;7:683–688. doi: 10.1016/s0959-4388(97)80089-x. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- Kokkonen N, Rivinoja A, Kauppila A, Suokas M, Kellokumpu I, Kellokumpu S. Defective Acidification of Intracellular Organelles Results in Aberrant Secretion of Cathepsin D in Cancer Cells. J Biol Chem. 2004;279:39982–39988. doi: 10.1074/jbc.M406698200. [DOI] [PubMed] [Google Scholar]
- Laudon H, Mathews PM, Karlstrom H, Bergman A, Farmery MR, Nixon RA, Winblad B, Gandy SE, Lendahl U, Lundkvist J, et al. Co-expressed ]presenilin 1 NTF and CTF form functional gamma-secretase complexes in cells devoid of full-length protein. J Neurochem. 2004;89:44–53. doi: 10.1046/j.1471-4159.2003.02298.x. [DOI] [PubMed] [Google Scholar]
- Marzella L, Ahlberg J, Glaumann H. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J Cell Biol. 1982;93:144–154. doi: 10.1083/jcb.93.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Nakanishi H, Tominaga K, Amano T, Hirotsu I, Inoue T, Yamamoto K. Age-Related Changes in Activities and Localizations of Cathepsins D, E, B, and L in the Rat Brain Tissues. Experimental Neurology. 1994;126:119–128. doi: 10.1006/exnr.1994.1048. [DOI] [PubMed] [Google Scholar]
- Naruse S, Thinakaran G, Luo JJ, Kusiak JW, Tomita T, Iwatsubo T, Qian X, Ginty DD, Price DL, Borchelt DR, et al. Effects of PS1 deficiency on membrane protein trafficking in neurons. Neuron. 1998;21:1213–1221. doi: 10.1016/s0896-6273(00)80637-6. [DOI] [PubMed] [Google Scholar]
- Nishi T, Forgac M. The vacuolar (H+)-ATPases--nature's most versatile proton pumps. Nat Rev Mol Cell Biol. 2002;3:94–103. doi: 10.1038/nrm729. [DOI] [PubMed] [Google Scholar]
- Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM. Extensive involvement of autophagy in Alzheimer disease: An Immuno-Electron Microscopy Study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- Nixon RA, Yang DS, Lee JH. Neurodegenerative lysosomal disorders- a continuum from development to late age. Autophagy. 2008;4:590–599. doi: 10.4161/auto.6259. [DOI] [PubMed] [Google Scholar]
- Ohkuma S, Poole B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc Natl Acad Sci USA. 1978;75:3327–3331. doi: 10.1073/pnas.75.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran N, Munteanu I, Wang P, Aubourg P, Rilstone JJ, Israelian N, Naranian T, Paroutis P, Guo R, Ren Z-P, et al. VMA21 Deficiency Causes an Autophagic Myopathy by Compromising V-ATPase Activity and Lysosomal Acidification. Cell. 2009;137:235–246. doi: 10.1016/j.cell.2009.01.054. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, Scaravilli F, Easton DF, Duden R, O'Kane CJ, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Rozmahel R, Huang J, Chen F, Liang Y, Nguyen V, Ikeda M, Levesque G, Yu G, Nishimura M, Mathews P, et al. Normal brain development in PS1 hypomorphic mice with markedly reduced γ-secretase cleavage of betaAPP. Neurobiol Aging. 2002;23:187–194. doi: 10.1016/s0197-4580(01)00267-6. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Ruiz-Canada C, Kelleher DJ, Gilmore R. Cotranslational and Posttranslational N-Glycosylation of Polypeptides by Distinct Mammalian OST Isoforms. Cell. 2009;136:272–283. doi: 10.1016/j.cell.2008.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samari HR, Seglen PO. Inhibition of Hepatocytic Autophagy by Adenosine, Aminoimidazole-4-carboxamide Riboside, and N6-Mercaptopurine Riboside. J Biol Chem. 1998;273:23758–23763. doi: 10.1074/jbc.273.37.23758. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Saura CA, Choi SY, Beglopoulos V, Malkani S, Zhang D, Shankaranarayana Rao BS, Chattarji S, Kelleher RJ, 3rd, Kandel ER, Duff K, et al. Loss of presenilin function causes impairments of memory and synaptic plasticity followed by age-dependent neurodegeneration. Neuron. 2004;42:23–36. doi: 10.1016/s0896-6273(04)00182-5. [DOI] [PubMed] [Google Scholar]
- Schmelzle T, Hall MN. TOR, a Central Controller of Cell Growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci USA. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Kelleher RJ., III The presenilin hypothesis of Alzheimer's disease: Evidence for a loss-of-function pathogenic mechanism. Proceedings of the National Academy of Sciences. 2007;104:403–409. doi: 10.1073/pnas.0608332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CA, Murphy DD, Giasson BI, Zhang B, Trojanowski JQ, Lee VM. Degradative organelles containing mislocalized α- and β-synuclein proliferate in presenilin-1 null neurons. J Cell Biol. 2004;165:335–346. doi: 10.1083/jcb.200403061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23:33–42. doi: 10.1247/csf.23.33. [DOI] [PubMed] [Google Scholar]
- Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem. 1991;266:17707–17712. [PubMed] [Google Scholar]
- Yu WH, Cuervo AM, Kumar A, Peterhoff CM, Schmidt SD, Lee J-H, Mohan PS, Mercken M, Farmery MR, Tjernberg LO, et al. Macroautophagy -- a novel β-amyloid peptide-generating pathway activated in Alzheimer's disease. J Cell Biol. 2005;171:87–98. doi: 10.1083/jcb.200505082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kang DE, Xia W, Okochi M, Mori H, Selkoe DJ, Koo EH. Subcellular distribution and turnover of presenilins in transfected cells. J Biol Chem. 1998;273:12436–12442. doi: 10.1074/jbc.273.20.12436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.