SUMMARY
ISWI-family enzymes remodel chromatin by sliding nucleosomes along DNA, but the nucleosome translocation mechanism remains unclear. Here we use single-molecule FRET to probe nucleosome translocation by ISWI-family remodelers. Distinct ISWI-family members translocate nucleosomes with a similar stepping pattern maintained by the catalytic subunit of the enzyme. Nucleosome remodeling begins with a 7-bp step of DNA translocation followed by 3-bp subsequent steps towards the exit side of nucleosomes. These multi-bp, compound steps are comprised of 1-bp substeps. DNA movement on the entry side of the nucleosome occurs only after 7 bp of exit-side translocation and each entry-side step draws in a 3-bp equivalent of DNA that allows three additional base pairs to be moved to the exit side. Our results suggest a remodeling mechanism with precise coordination at different nucleosomal sites featuring DNA translocation towards the exit side in 1-bp steps preceding multi-bp steps of DNA movement on the entry side.
INTRODUCTION
The packaging of genomic DNA into nucleosomes and higher-order chromatin structures represses many essential DNA transactions including transcription, DNA repair, replication and recombination. DNA accessibility during these processes is regulated in part by ATP-dependent chromatin remodeling enzymes, which utilize the energy from ATP hydrolysis to assemble, disassemble, mobilize, or restructure nucleosomes. These remodelers typically possess a catalytic subunit and one or more accessory subunits. The catalytic subunits contain a conserved ATPase domain that shares sequence homology with superfamily 2 (SF2) helicases, as well as unique flanking domains that give rise to four distinct remodeler families: SWI/SNF, ISWI, CHD/Mi2, and INO80 (Clapier and Cairns, 2009; Gangaraju and Bartholomew, 2007). The ATPase domain binds to and translocates DNA at a site internal to the nucleosome, which is two helical turns (or 20 bp) from the dyad and referred to as the SHL2 site (Dang and Bartholomew, 2007; Kagalwala et al., 2004; Lorch et al., 2005; Saha et al., 2002, 2005; Schwanbeck et al., 2004; Whitehouse et al., 2003; Zofall et al., 2006). Depending on the subunit composition, remodelers can display divergent remodeling activities. For example, ISWI-family enzymes reposition nucleosomes while maintaining their canonical structure, whereas SWI/SNF family enzymes can not only translocate nucleosomes but can also change the nucleosome structure, alter histone compositions, or eject histone octamers altogether (Clapier and Cairns, 2009). Within the ISWI family, remodelers such as human ACF and yeast ISW2 help generate regularly-spaced nucleosomal arrays (Ito et al., 1997; Langst et al., 1999; Tsukiyama et al., 1999; Varga-Weisz et al., 1997), whereas yeast ISW1b largely lacks nucleosome spacing activity (Stockdale et al., 2006; Vary et al., 2003).
The mechanism by which remodeling enzymes couple ATP hydrolysis to nucleosome translocation remains incompletely understood. Various models have been proposed for how remodelers reposition nucleosomes along DNA. The “twist diffusion” model hypothesizes that remodelers generate a twist defect in the DNA, which propagates around the histone octamer, shifting the position of the nucleosome 1 base pair (bp) at a time (Flaus and Owen-Hughes, 2003; Kulic and Schiessel, 2003a; Richmond and Davey, 2003; Suto et al., 2003; van Holde and Yager, 2003). The “loop propagation” model involves DNA being pushed into the nucleosome at the entry side, forming a loop that propagates around the nucleosome and resolves at the exit side (Flaus and Owen-Hughes, 2003; Kulic and Schiessel, 2003b; Langst and Becker, 2004; Lorch et al., 2005; Narlikar et al., 2002; Schwanbeck et al., 2004; Strohner et al., 2005; Widom, 2001). As a third alternative, the “octamer swiveling” model proposes that remodelers disrupt major contacts between the DNA and histone octamer, and allow a concerted swiveling of the DNA relative to the histone core (Bowman, 2010; Lorch et al., 2010). It remains unclear whether the true remodeling mechanism involves one of the above models, a combination of aspects from multiple models, or a model distinct from any of the above. Given their distinct remodeling outcomes, different remodeler families or different members within the same family may also utilize distinct mechanisms to mobilize nucleosomes.
Single-molecule experiments can provide valuable insights into chromatin remodeling. These experiments can resolve transient intermediate states of the nucleosome during remodeling and reveal how DNA movement at different nucleosomal sites is coordinated, allowing various models to be tested directly. Single-molecule techniques have been applied to study DNA or nucleosome translocation by remodeling enzymes in real time (Amitani et al., 2006; Blosser et al., 2009; Lia et al., 2006; Prasad et al., 2007; Shundrovsky et al., 2006; Sirinakis et al., 2011; Zhang et al., 2006). These studies have shown that SWI/SNF enzymes can both induce DNA loop formation on DNA and nucleosome substrates (Lia et al., 2006; Zhang et al., 2006), and generate canonically repositioned nucleosomes (Shundrovsky et al., 2006). Two recent studies have revealed that ACF, an ISWI remodeler, moves nucleosomes in ~7 or ~3 bp steps (Blosser et al., 2009), whereas RSC, a SWI/SNF remodeler, translocates DNA substrates with a step size of ~2 bp (Sirinakis et al., 2011). While these results place constraints on the remodeling mechanisms, it remains unclear how DNA is moved into the nucleosome at the entry side, propagated around the octamer, and released at the exit side, and how these events are coordinated.
Moreover, the observed multi-bp translocation steps also raised a question about their underlying mechanism. The ATPase domains of remodeling enzymes are homologous to those of SF2 family helicases, even though remodelers typically do not exhibit helicase activity. An SF2 helicase has been shown to unwind DNA or RNA duplexes in bursts of multiple base pairs (Dumont et al., 2006; Myong et al., 2007), while translocating along the oligonucleotide backbone with 1-bp steps (Cheng et al., 2011; Myong et al., 2007). The translocation step size of SF1 helicases has also been reported to be 1 bp (Dillingham et al., 2000; Lee and Yang, 2006; Park et al., 2010). Based on these results, one may hypothesize that DNA translocation during nucleosome remodeling also occurs in 1-bp steps. However, such 1-bp steps have not been observed for any chromatin remodeler. Does this discrepancy indicate that remodelers and helicases have evolved divergent DNA translocation mechanisms or was the resolution of previous measurements insufficient to detect 1-bp steps? The elementary step size of chromatin remodelers remains an unresolved question for this family of molecular motors.
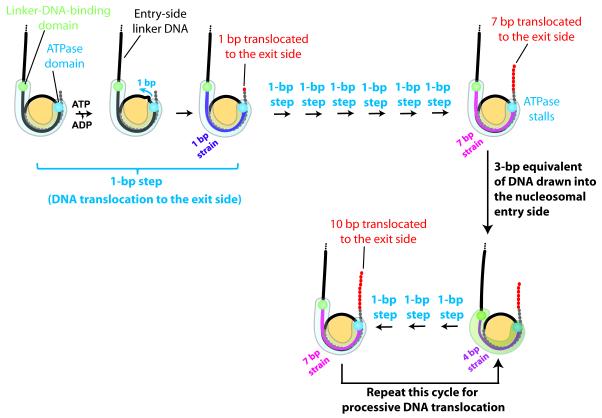
In this study, we used single-molecule fluorescence resonance energy transfer (FRET) (Ha et al., 1996; Stryer and Haugland, 1967; Zhuang et al., 2000) to probe nucleosome translocation by several ISWI-family remodelers. Despite their distinct accessory subunits and remodeling outcomes, we observed a common stepping pattern for these enzymes. DNA translocation at the exit side of the nucleosome occurred with an initial 7-bp step followed by 3-bp subsequent steps. This stepping pattern was preserved even when all accessory subunits of the enzyme were removed. The multi-bp steps were further comprised of 1-bp elementary steps. Surprisingly, DNA movement at the nucleosomal entry side appeared to occur only after 7 bp of DNA were translocated toward the exit side and to proceed in 3-bp increments, in accordance with the 3-bp steps observed at the exit side after the initial 7-bp step. Our results suggest a remodeling mechanism as follows: DNA is first translocated towards the nucleosomal exit side by the ATPase domain, 1 bp at a time, generating strain on the entry-side DNA; after 7 bp of translocation, the strain becomes sufficiently strong to trigger an enzyme action at the nucleosomal entry side that draws DNA into the nucleosome; this action partially releases the strain and allows three additional base pairs of DNA to be translocated to the exit side; this 3-bp step then repeats to generate processive DNA translocation across the nucleosome.
RESULTS
Monitoring ISWI-induced nucleosome remodeling dynamics at the exit side
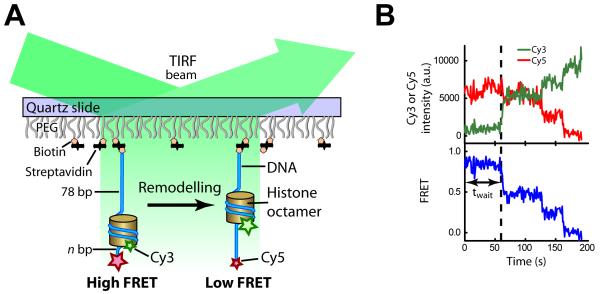
To monitor the remodeling dynamics of individual nucleosomes using single-molecule FRET, we reconstituted mononucleosomes using histone octamers labeled with the FRET donor dye, Cy3, on histone H2A and double-stranded DNA labeled with the acceptor dye, Cy5 (Figure 1A). The 601 nucleosome positioning sequence (Lowary and Widom, 1998) was used to place the octamer at a well-defined position, such that the DNA wrapped around the histone octamer in ~ 1.7 turns (Chua et al., 2012; Luger et al., 1997; Makde et al., 2010; Vasudevan et al., 2010), leaving n bp of linker DNA on the exit side and 78 bp of linker DNA on the entry side (Table S1). The nucleosomes were anchored onto a PEG-coated quartz surface and fluorescence signals from individual nucleosomes were monitored using a total-internal-reflection fluorescence (TIRF) geometry (Figure 1A). Although the presence of two H2A subunits on each histone octamer led to three different donor labeling configurations (donor on the H2A subunit proximal to the acceptor on the DNA, donor on the distal H2A, and donors on both H2A subunits), their distinct FRET values allowed us to clearly distinguish these populations at the single-nucleosome level and to specifically select the first population for further analyses (Figure S1A) (Blosser et al., 2009). Addition of remodelers, such as yeast ISW2, and ATP to nucleosomes caused a decrease in FRET that was not observed when the enzyme was added without ATP (Figures 1B and S1A-C), consistent with the ability of ISW2 and similar enzymes to mobilize the histone octamer toward the center of the DNA (He et al., 2006; Kagalwala et al., 2004; Kassabov et al., 2002; Stockdale et al., 2006; Yang et al., 2006). Spontaneous fraying of DNA ends previously observed on the 0.01 – 0.05 s time scale (Li et al., 2005) was not visible in our experiments, which have a resolution of 0.3 – 1 s.
Figure 1. Probing DNA translocation on the exit side of the nucleosome by single-molecule FRET.
(A) Schematic of FRET detection for DNA translocation on the exit side of the nucleosome. The nucleosomes are labeled with the FRET donor, Cy3 (green star), and acceptor, Cy5 (red star). The histone octamer and DNA are depicted as a yellow cylinder and a blue line, respectively.
(B) Representative Cy3 (green) and Cy5 (red) fluorescence and FRET (blue) time traces showing translocation of a single nucleosome after addition of the enzyme and ATP at time zero.
To quantitatively interpret FRET changes in terms of how many base pairs of DNA were translocated to the exit side, we generated a calibration curve of FRET versus the length of the exit linker DNA (Figure S1D) by measuring the FRET values for a series of nucleosome constructs with different exit linker lengths (varying n, Table S1). To further validate this calibration for determining the exit linker length of an ISWI-induced remodeling product, we prepared another series of nucleosomes with the same initial linker lengths (78 bp on the entry side and 3 bp on the exit side), but each possessing a 2-nucleotide (nt) ssDNA gap at a specified distance (m bp) from SHL2 to stall translocation after m bp of movement (Lorch et al., 2005; Saha et al., 2005; Schwanbeck et al., 2004; Zofall et al., 2006). As expected, the m = 0 construct exhibited no remodeling even upon addition of ISW2 and ATP. The FRET value of the m > 0 constructs after remodeling by ISW2 decreased as m increased (Figure S1E). Such FRET change was not observed when the enzyme was added alone without ATP (Figure S1C). Quantitatively, the dependence of the post-remodeling FRET values on m, i.e. the amount of DNA translocation allowed, was identical to the dependence of FRET on the preset exit linker length n (Figure S1D, E), indicating that the observed FRET changes were due to DNA translocation to the exit side and that the amount of DNA translocation can be quantified based on the calibrations.
The translocation step sizes are conserved among different ISWI family members
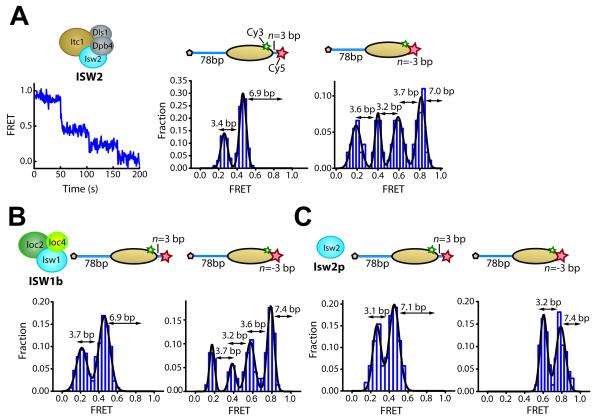
We studied nucleosome remodeling by three representative yeast ISWI enzymes with different accessory proteins: i) ISW2, which is comprised of a catalytic subunit, Isw2 (130 kDa), and three accessory subunits, Itc1 (146 kDa), Dpb4 (22 kDa) and Dls1 (18 kDa) (McConnell et al., 2004), ii) ISW1b, which is comprised of the catalytic subunit Isw1 (131 kDa), homologous to Isw2, and two different accessory subunits Ioc2 (93 kDa) and Ioc4 (55 kDa) (Vary et al., 2003), and iii) the catalytic subunit of ISW2 alone, which will be referred to as Isw2p for clarity. Isw1 and Isw2 each contain a single ATP-binding site.
We first added ISW2 and ATP to nucleosomes with 3 bp of linker DNA on the exit side (n = 3 bp; Table S1). We observed a step-wise decrease in FRET with pauses at FRET values of 0.46 ± 0.03 and 0.26 ± 0.04 (Figures 2A and S2A). These pauses correspond to a translocation of 6.9 ± 0.6 bp of DNA prior to the first pause and 3.4 ± 0.6 bp between the first and second pauses. A similar stepping behavior was also observed for nucleosomes with a different initial exit linker length (n = −3 bp, i.e. 3 bp omitted from the 601 sequence (Table S1)), except that the higher initial FRET value allowed us to observe two additional pauses that occurred after further translocation by 3.2 ± 0.5 bp and 3.6 ± 0.8 bp, respectively (Figures 2A and S2A). These data indicate that nucleosome translocation by ISW2 involves a unique first translocation step of approximately 7 bp in size and subsequent steps that are approximately 3 bp each. These step sizes are quantitatively similar to the ones previously observed for the human ISWI remodeler ACF (Blosser et al., 2009), the catalytic subunit of which shares sequence homology with that of ISW2 (Hota and Bartholomew, 2011).
Figure 2. Identical step sizes of DNA translocation on the exit side of the nucleosome induced by different ISWI-family enzymes.
(A) Remodeling of nucleosomes with initial exit linker DNA length of n = 3 bp or n = −3 bp by ISW2. Left: FRET time trace showing ISW2-induced translocation of a single n = 3 bp nucleosome. 6.2 nM ISW2 and 2 μM ATP were added at time zero. Middle: Histogram of the FRET values at translocation pauses constructed from n = 3 bp nucleosomes. Right: FRET histograms of the translocation pauses constructed from n = −3 bp nucleosomes.
(B) Remodeling of the n = 3 bp and n = −3 bp nucleosomes by ISW1b. FRET histograms of the pauses for n = 3 bp (left) and n = −3 bp (right) nucleosomes in the presence of 8.8 nM ISW1b and 10-150 μM ATP.
(C) Remodeling of the n = 3 bp and n = −3 bp nucleosomes by Isw2p. FRET distribution of the pauses for the n = 3 bp (left) and n = −3 bp (right) nucleosomes in the presence of 69 nM Isw2p and 1 mM ATP.
The nucleosome schemes display the footprint of the histone octamer (yellow oval) on the DNA (blue line). Numbers above double-headed arrows shown in the histograms represent mean step sizes.
See also Figure S2.
To test whether this stepping pattern was also shared by other ISWI enzymes with similar catalytic subunits, we probed nucleosome remodeling dynamics by ISW1b, which possesses a catalytic subunit homologous to that of ISW2. Notably, nucleosome remodeling by ISW1b exhibited translocation step sizes virtually identical to the ones observed for ISW2 (Figures 2B and S2B), despite their different compositions of accessory subunits and nucleosome spacing activities (McConnell et al., 2004; Stockdale et al., 2006; Tsukiyama et al., 1999; Vary et al., 2003).
Next, we purified the catalytic subunit of ISW2, Isw2p, without any accessory subunits. Due to its low nucleosome binding affinity and processivity (Hota and Bartholomew, 2011), nucleosomes exhibited only a limited amount of DNA translocation before enzyme dissociation. Therefore, typically only two translocation pauses were detected, independent of the initial exit linker length (n = 3 or −3 bp). Nonetheless, the first pause again occurred after ~7 bp of DNA translocation and the second pause after an additional ~3 bp (Figures 2C and S2C).
For ISW1b and Isw2p, a fraction of the remodeling traces (~36% and ~31%, respectively) displayed reversals of the translocation direction, as reflected by back-and-forth FRET changes, within the dynamic range of our measurement. Such direction reversal was rarely observed for intact ISW2 complexes. Analysis of the FRET values at direction reversal points suggests that direction reversal by Isw2p preferably occurred at the 7-bp pause (Figure S2D). In contrast, the direction reversal positions did not correlate with the translocation step sizes for the ISW1b enzyme determined from traces that did not exhibit direction reversal, but rather seemed to coincide with the 5-bp and 10-bp periodicities of the nucleosome (Figure S2E).
The multi-bp DNA translocation steps at the nucleosomal exit side are comprised of 1-bp elementary steps
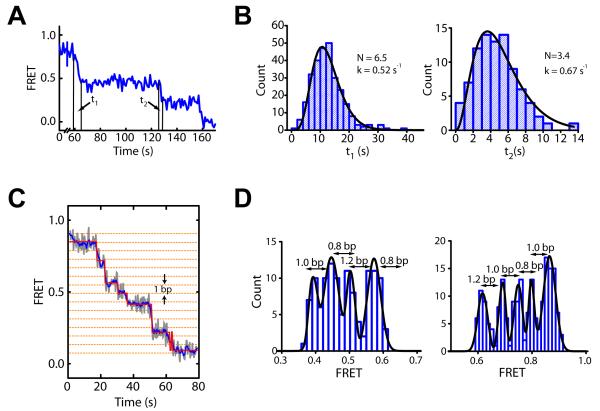
In order to explore whether the observed multi-bp steps are further comprised of hidden translocation events with a smaller step size, we measured the dwell times (t1 and t2) of the first two translocation phases during ISW2-induced remodeling for the n = 3 bp nucleosomes (Figure 3A). In a simple model where each transition consists of a series of irreversible elementary steps with an identical rate constant k, the corresponding transition time t should follow a Γ-distribution, AtN−1exp(−kt) (Dumont et al., 2006; Myong et al., 2007). Depending on whether the stepping transient itself or the wait time between steps is rate-limiting, the total number of elementary steps within the transition would be either N or N+1, respectively. Notably, the dwell time (t1 and t2) distributions were both well described by a Γ-distribution, with N = 6.5 ± 0.4 and 3.4 ± 0.4 for the first and second translocation phases, respectively (Figure 3B). Given the 6.9 ± 0.6 bp and 3.4 ± 0.6 bp of DNA translocation observed during the two phases (Figure 2A), these N values correspond to a mean elementary step size close to 1 bp (1.1 ± 0.2 bp or 0.9 ± 0.1 bp for the t1 phase, and 1.0 ± 0.3 bp or 0.8 ± 0.2 bp for the t2 phase depending on whether the number of steps equals N or N+1, respectively). These results suggest that the multi-bp steps are likely compound steps consisting of 1-bp elementary steps.
Figure 3. The multi-bp translocation steps on the exit side of the nucleosome are comprised of 1-bp elementary steps.
(A) FRET time trace of a nucleosome indicating the dwell times of the first two translocation phases (t1 and t2).
(B) Histogram of t1 and t2 values constructed from many nucleosomes. Fits to the Γ-distribution AtN−1exp(−kt) (black lines) yield N = 6.5 ± 0.4 and k = 0.52 ± 0.04 s−1 for the t1 phase (left), and N = 3.4 ± 0.4 and k = 0.67 ± 0.11 s−1 for the t2 phase (right).
(C) FRET time trace, before (grey) and after (blue) 5-point averaging, of a single n = 3 bp nucleosome in the presence of 6.2 nM ISW2, 2 μM ATP and 2 mM ATP-γ-S. A HMM fit is shown by the red line. The horizontal orange dotted lines indicate 1-bp intervals (derived from the calibration in Figure S1D).
(D) Histograms of FRET plateaus from many nucleosomes determined using the HMM analysis. Numbers above double-headed arrows represent mean step sizes.
See also Figure S3.
Considering that the assumptions underlying the Γ-distribution may not be fully satisfied for the translocation phases, we set out to detect the elementary 1-bp steps directly. To this end, we reduced the stepping rate in two alternative ways. First, we used low ATP concentrations in combination with high concentrations of the nucleotide analog ATP-γ-S, which hydrolyzes at a dramatically reduced rate. Using this approach, we directly observed 1-bp translocation steps in single-molecule FRET traces (Figure 3C). In addition to the 1-bp steps, the traces also showed larger step sizes of 2 and 3 bp. Both 1-bp and larger steps occurred randomly at varying positions in different traces, consistent with a uniform step size of 1 bp where some pauses were too short to be resolved. Indeed, a simulation based on experimentally determined stepping rate constant and FRET signal-to-noise ratio suggests that we would miss ~ 50% of the 1-bp steps. Although we cannot rule out a translocation mechanism that involves heterogeneous steps of varying sizes, we consider such a mechanism less likely given that the durations of the multi-bp steps follow a Γ-distribution with the number of elementary steps matching the number of base pairs translocated (Figure 3A, B).
For automated step identification, we utilized a hidden Markov modeling (HMM) algorithm (McKinney et al., 2006) to determine the distinct FRET states (plateaus) present in the FRET traces (Figure 3C). We separately analyzed two FRET regions, 0.32 ≤ FRET ≤ 0.62 and 0.59 ≤ FRET ≤ 1, and allowed 10 initial states in each region. The HMM analysis of the FRET traces converged to ~4-5 states in each region, suggesting that the state identification was unlikely influenced by the initial parameter setting. Moreover, we obtained nearly identical fits using an alternative step finding algorithm (Kerssemakers et al., 2006) (Figure S3A). Remarkably, the histograms of the FRET plateau values exhibit well-defined peaks each separated by 1.0 ± 0.2 bp (Figure 3D and Figure S3A).
Alternatively, to reduce the stepping rate without using ATP-γ-S, we monitored remodeling at a lower temperature of 15°C, instead of the 30°C used for the above experiments. Again, 1-bp steps were observed (Figure S3B). A histogram of the FRET plateau values shows peaks separated by 1.0 ± 0.1 bp (Figure S3B). However, because of the reduced enzyme binding affinity and slower translocation kinetics at this lower temperature, photobleaching restricted analysis to only the first four 1-bp translocation steps (Figure S3B). Taken together, our data suggest that the multi-bp translocation steps observed at the exit side of the nucleosome are compound steps comprised of 1-bp elementary steps.
Roles of ATP binding and hydrolysis during nucleosome translocation
To dissect the roles of ATP binding and hydrolysis during nucleosome translocation, we examined the dwell times associated with individual 1-bp steps at various concentrations of ATP and ATP-γ-S at 30°C. Photobleaching limited our analysis to the first nine steps. At low ATP-γ-S concentrations, the pause duration after the seventh 1-bp step, tp,7, was noticeably longer than the dwell times associated with all other 1-bp steps (Figure S4A). As the concentration of ATP-γ-S increased, the pause duration after the seventh step (tp,7) decreased (Figures 4A and S4A) whereas the dwell times of the other steps (tp) increased (Figures 4B and S4A). As a result, all steps became equal in duration at saturating ATP-γ-S concentrations (Figure S4A). At saturating ATP-γ-S concentrations, the duration of these 1-bp steps (tp) decreased with increasing ATP concentrations (Figures 4C and S4B). Given that ATP-γ-S hydrolyzes much more slowly than ATP and competes with ATP for binding to the enzyme, the above results indicate that the individual 1-bp translocation steps require ATP hydrolysis, whereas the pausing observed after 7 bp of translocation involves an additional ATP binding event of the enzyme. Binding of ATP-γ-S can thus facilitate this event.
Figure 4. Dependence of the stepping kinetics on the concentrations of ATP and ATP-γ-S.
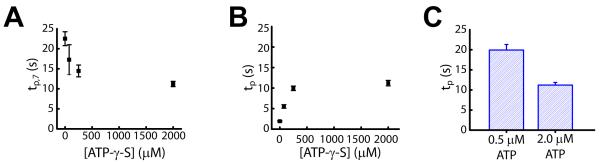
(A) The pause duration after the first, 7-bp compound step (tp,7) at various ATP-γ-S concentrations and 2 μM ATP.
(B) The pause duration between each 1-bp elementary step (tp) at various ATP-γ-S concentrations and 2 μM ATP. All pauses except for the ones after 6, 7, and 8 bp of translocation were pooled to determine tp. As shown in Figure S4, in addition to the 7th pause, the 6th and 8th pauses also appear longer than the remaining ones, likely due to errors in pause identification. The value of tp at 0 μM ATP-γ-S was derived from 1 / k value obtained from the Γ-distribution in Figure 3B.
(C) Dependence of tp on the ATP concentration at 2 mM ATP-γ-S. At this saturating concentration of ATP-γ-S, all pauses, including the 7th one, had approximately equal durations and were pooled to determine tp.
All data are shown as the mean ± SEM (N = 15 - 100 events).
See also Figure S4.
Monitoring ISWI-induced nucleosome remodeling dynamics at the entry side
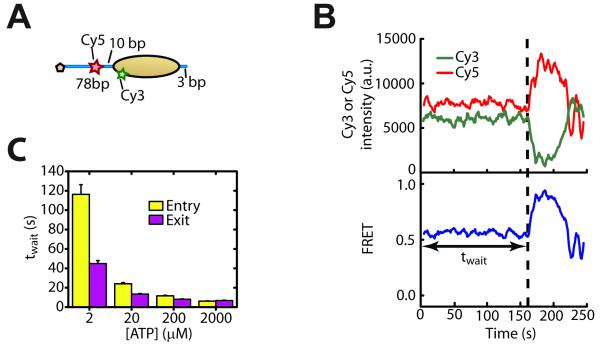
In order to monitor DNA movement on the entry side of the nucleosome, we moved the FRET acceptor dye Cy5 from the exit DNA linker to the entry DNA linker, 10 bp away from the nucleosomal edge, but kept the entry and exit linker lengths at 78 bp and 3 bp, respectively (Figure 5A). As is the case for the exit-side labeling scheme, we were able to distinguish the different donor-labeling configurations at the single-nucleosome level and select only those with a single Cy3 dye on the proximal H2A for further analysis (Figure S5A).
Figure 5. DNA movement on the entry side of the nucleosome precedes that on the exit side.
(A) Schematic of the nucleosome construct used for measuring DNA movement at the entry side.
(B) Donor signal (green), acceptor signal (red), and FRET (blue) time traces showing ISW2-induced remodeling after adding 12 nM ISW2 and 2 μM ATP at time zero. The dashed line indicates the onset of FRET change after an initial wait time, twait.
(C) Comparison of twait on the entry (yellow bars) and exit sides (purple bars) of the nucleosome under identical enzyme and ATP concentrations. Data are shown as the mean ± SEM (N = 80 - 220 events).
See also Figure S5.
Upon addition of ISW2 and ATP and after a waiting period twait, the FRET time traces displayed an increase in FRET as the Cy5 dye on the entry DNA linker was moved closer to the octamer (Figure 5B). As DNA continues to move into the nucleosome, the Cy5 dye is expected to eventually pass the Cy3 dye, causing a FRET decrease. Indeed, such a non-monotonic FRET change was observed (Figure 5B). No FRET change was observed when ISW2 was added without ATP (Figure S5B).
Coordination of the DNA movement on the entry and exit sides of the nucleosome
To investigate the coordination of entry-side and exit-side remodeling activity, we first compared the waiting times before the onset of any FRET change, twait, at both entry and exit sides (Figures 1B and 5B). Surprisingly, the average twait value measured at the entry side was substantially longer than that at the exit side (Figure 5C), suggesting that exit-side DNA translocation likely occurred prior to any DNA movement at the entry side. The delay between entry-side and exit-side movements decreased as the ATP concentration increased (Figure 5C). Since DNA translocation at the exit side occurred in 1-bp steps, we reason that this movement was caused by the SF2-homologous ATPase domain bound at the SHL2 site of the nucleosome. Our observations may thus be interpreted as DNA translocation at SHL2 by the ATPase occurring prior to any DNA movement into the nucleosomal entry side.
Since twait was measured at the entry and exit sides with differently labeled constructs, the observed time difference provides only an indirect measure of the order of these events. To further test whether DNA translocation at SHL2 by the ATPase domain is indeed required for DNA movement at the entry side, we generated entry-side labeled nucleosomes with a 2-nt ssDNA gap positioned at the SHL2 site (m = 0 bp, Figure 6A), which prevented any DNA translocation to the exit side (Figure S1E). Interestingly, addition of ISW2 and ATP to these nucleosomes did not cause any FRET change at the entry side (Figures 6B and 6C), indicating that DNA movement at the entry side was also inhibited.
Figure 6. Entry-side DNA movement occurs after 7 bp of DNA translocation towards the exit side and proceeds in 3-bp steps.
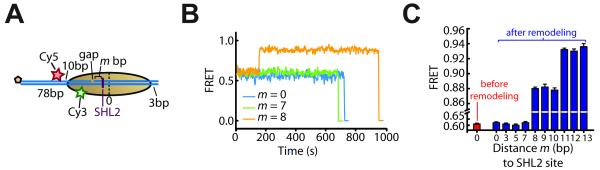
(A) Schematic of the nucleosome constructs used to monitor DNA movement at the entry side when exit-side translocation is restricted by a 2-nt ssDNA gap. The gap is located m bp away from the SHL2 site (shown as a purple line) such that m bp of DNA can be translocated to the exit side.
(B) FRET time traces of single m = 0, 7, and 8 bp nucleosomes (blue, green, and orange line, respectively) after addition of 12 nM ISW2 and 2 μM ATP at time zero.
(C) FRET values before (red bar) and after (blue bars) remodeling by ISW2 as a function of the distance m to the SHL2 site. Because the DNA path on the entry side may involve bending and/or twisting due to the direct interaction with the remodeling enzyme, we do not expect a similar linear dependence of FRET on the linker DNA length as on the exit-side where the linker DNA is largely free of enzyme-induced distortion. Data are shown as the mean ± SEM (N = 80 - 150 nucleosomes).
See also Figure S6.
Next, we generated a series of entry-side labeled nucleosome constructs with ssDNA gaps at varying distances (m bp) from the SHL2 site (Figure 6A), which allow only m bp of DNA to be translocated to the exit side as shown in Figure S1E. Remarkably, after addition of ISW2 enzyme and 2 μM ATP, no entry-side FRET change was observed for nucleosomes with m ≤ 7 bp, whereas long-lasting FRET increases were observed for nucleosomes with m > 7 bp (Figures 6B and 6C). At higher ATP concentrations (≥ 200 μM), the m < 7 bp nucleosomes still showed no change in FRET. A minor fraction (~40%) of the m = 7 bp nucleosomes showed increase in FRET but mostly with only transient excursions to higher FRET, while only <20% of the nucleosomes showed a stable increase in FRET. In contrast, the vast majority (~75-90%) of m > 7 bp nucleosomes exhibited an increase in FRET, among which most (80-90%) showed stable FRET changes. These observations are consistent with DNA movement at the entry side occurring only after the ATPase has translocated 7 bp of DNA toward the exit side, likely because a certain amount of strain on the entry-side DNA is required to trigger any movement into the nucleosome. ssDNA gaps not only limit the amount of DNA translocation by the ATPase domain but can also inhibit the generation or propagation of DNA torsion. Among these two factors, entry-side DNA movement was more likely inhibited by the limited amount of ATPase translocation because the inhibition was only observed for the m ≤ 7 nucleosomes, whereas ssDNA gaps at m > 7 should also relax torsional strain on the entry-side DNA. Further supporting this interpretation, the twait values observed for the m > 7 nucleosomes were quantitatively similar to those observed for intact nucleosomes without any gap (Figure S6A), suggesting that the gaps did not perturb the strain required to trigger entry-side movement. These observations are also consistent with the long pause observed after the first 7 bp of DNA translocation to the exit side for intact nucleosomes without gaps (Figure 2A). We reason that the accumulated strain after 7 bp of DNA translocation stalls exit-side translocation momentarily, and that an entry-side movement needs to be triggered to partially relax the strain and allow additional DNA to be pumped to the exit side, giving rise to the observed pause.
Notably, the post-remodeling FRET values at the entry side were identical within error for the nucleosomes with gaps at m = 8, 9, or 10 bp, but distinct from those observed for the m = 11, 12, and 13 bp nucleosomes, which were also identical to each other (Figures 6C). These observations are consistent with a 3-bp equivalent of DNA being moved into the nucleosome per step on the entry side, which in turn allows an additional 3 bp of DNA to be translocated to the exit side. The rationale is as follows. Since 7 bp of DNA can be translocated to the exit side without any entry-side DNA movement, if each step of the entry-side movement allows an additional 3 bp of DNA to be pumped to the exit side, the cases of translocating 8, 9 and 10 bp to the exit side would all require only one entry step, leading to the same post-remodeling entry-side FRET values for the m = 8, 9, and 10 bp nucleosomes. The cases of translocating 11, 12, and 13 bp to the exit side would all require two entry steps instead, and thus the post-remodeling entry-side FRET values for the m = 11, 12, and 13 bp nucleosomes would be identical to each other but different from those of the m = 8, 9, and 10 bp nucleosomes. Our observations agree with these predictions (Figure 6C), demonstrating that the first entry-side step moved a 3-bp equivalent of DNA into the nucleosome, allowing 3 bp to be translocated to the exit side. Our data are also consistent with a second, 3-bp entry-side step though it is formally possible that the second step size is greater than 3 bp because the FRET values have not been measured for the m > 13 bp nucleosomes. However, given that after the initial 7-bp translocation step, the subsequent exit-side translocation steps are all 3 bp in size, DNA movement on the entry side likely also occurs in 3-bp increments, giving rise to the 3-bp compound steps observed at the exit side.
Interestingly, even though up to 7 bp of DNA can be translocated to the exit side without any observable action at the entry side, the exit-side translocation of the m < 7 nucleosomes appeared less stable: the majority of the m = 5 nucleosomes exhibited direction reversals (Figure S6B). In contrast, such direction reversal was rarely observed for m > 7 nucleosomes (Figure S6B), suggesting that action on the entry side helps prevent direction reversal in DNA translocation.
DISCUSSION
Chromatin remodelers utilize the energy from ATP hydrolysis to disrupt DNA-histone contacts and mobilize nucleosomes along DNA. In this work, we used single-molecule FRET to study nucleosome translocation by ISWI-family remodelers. We observed DNA movement at different sites of the nucleosome and determined how movements at these sites were coordinated. Our results suggest a novel model for nucleosome translocation by ISWI remodelers.
We showed that several representative ISWI remodelers from yeast, despite their highly distinct accessory subunits and remodeling outcomes, all translocated nucleosomes with a common stepping pattern (Figure 2). Exit-side translocation occurred with an initial 7-bp step followed by 3-bp subsequent steps. This stepping behavior was preserved even upon removal of all accessory subunits, leaving only the catalytic subunit for remodeling. These step sizes were also identical to the ones previously observed for the human ISWI remodeler ACF and previous evidence on ACF suggests that the step sizes are likely independent of the DNA sequence used (Blosser et al., 2009). Taken together, these results suggest a common remodeling mechanism for ISWI remodelers that is enabled by the catalytic subunit and conserved from yeast to humans. Notably, the step sizes observed here are not identical to the 5-bp or 10-bp periodicity of nucleosomal DNA-histone contacts (Hall et al., 2009; Luger et al., 1997), and thus are likely influenced by the remodeling enzymes, though we do not preclude the possibility that the step sizes are determined by a combination of enzyme and nucleosome properties. Moreover, the energy landscape of the nucleosomal substrates could affect the translocation kinetics quantitatively by modulating the dwell times between steps, which may explain why the remodeling products observed in ensemble biochemical analyses tend to exhibit ~10-bp intervals for DNA translocation (Schwanbeck et al., 2004; Zofall et al., 2006).
We further showed that the multi-bp steps observed on the nucleosomal exit side were compound steps comprised of 1-bp elementary steps (Figure 3). Given that the ATPase domains of the ISWI remodelers share sequence homology with SF2 helicases, which translocate DNA with 1-bp elementary steps (Cheng et al., 2011; Myong et al., 2007), the 1-bp steps of the remodelers most likely reflect an intrinsic translocation property of their ATPase domain. Since the ATPase domain binds to the SHL2 site of the nucleosome 20 bp from the dyad (Dang and Bartholomew, 2007) our results suggest that the ATPase domain translocates DNA at SHL2 1 bp at a time, which then propagates to the exit side, resulting in the observation of 1-bp steps at the exit side. Although such translocation likely tracks a DNA strand, the ATPase domain may partially disengage from the SHL2 site from time to time and hence there may not be substantial accumulation of DNA rotation during remodeling (Bowman, 2010; Cairns, 2007). It has been shown previously that ssDNA gaps placed between SHL2 and the exit site do not interfere with nucleosome sliding by ISWI enzymes (Schwanbeck et al., 2004; Zofall et al., 2006). It is thus possible that the propagation of DNA from the SHL2 site to the exit side also does not require torsional strain.
What is the mechanism underlying the multi-bp, compound steps for ISWI remodelers? Since DNA translocation at SHL2 towards the exit side occurs in 1-bp steps, it is reasonable to hypothesize that the multi-bp steps are a result of actions at the entry side. Surprisingly, we found that DNA movement at the entry side appeared to only occur after 7 bp of DNA were translocated toward the exit side (Figures 5 and 6). A net translocation to the exit side without any DNA being moved into the nucleosome will cause strain on the entry-side DNA. We thus hypothesize that a certain amount of strain needs to accumulate on the DNA before entry-side movement is triggered. Such strain may take the form of DNA stretching or transient conformational changes of the octamer or both. DNA stretching under force (Smith et al., 1996) or in nucleosome structures (Ong et al., 2007), as well as conformational changes of the octamer (Bohm et al., 2011) have been previously observed. According to this picture, as the ATP concentration increases, the strain on the entry-side DNA created by translocation at SHL2 should accumulate faster, and thus the time lag between DNA movements at the SHL2 and entry sites should decrease. Indeed, the time difference observed between entry- and exit-side movements decreased as the ATP concentration was increased (Figure 5).
Interestingly, once entry-side movement was triggered, it appeared to proceed in 3-bp increments that allow 3 additional base pairs to be moved to the exit side (Figure 6), which provides a simple explanation for why exit-side DNA translocation occurred with an initial 7-bp step followed by 3-bp subsequent steps. Given that the pauses preceding the 3-bp steps on the exit side can be shortened by addition of ATP-γ-S (Figure 4), indicating that an ATP-binding event is needed for exiting the pause, one may hypothesize that this event is related to the entry-side action. The entry-side step potentially involves an enzyme action, for instance, a conformational change of a linker-DNA-binding domain that draws in a 3-bp equivalent of DNA. The HAND-SANT-SLIDE module, which binds to the linker DNA on the entry side (Dang and Bartholomew, 2007; Yamada et al., 2011), may play a role in this process. Supporting this notion, our unpublished data showed that mutations in the SLIDE domain of ISW2 inhibited DNA movement on the entry side yet still allowed a substantial amount of DNA translocation to the exit side. It has been reported recently that the DNA-translocation activity of the ISWI ATPase is inhibited by a neighboring NegC region, and that binding of the HAND-SANT-SLIDE module to linker DNA relieves this inhibition (Clapier and Cairns, 2012). Thus, the HAND-SANT-SLIDE module potentially plays two roles in nucleosome remodeling, helping both DNA translocation to the exit side and DNA movement on the entry side.
Based on the above results, we propose the following model for nucleosome remodeling by ISWI-family enzymes (Figure 7). ISWI-induced nucleosome remodeling starts with the ATPase domain translocating DNA at the SHL2 site. This translocase activity pumps DNA towards the exit side, 1 bp at a time, utilizing energy from ATP hydrolysis. Translocation at SHL2 induces strain on the entry-side DNA, which initially remains immobile. After 7 bp of DNA are translocated, the strain becomes sufficiently strong to trigger action on the entry side. This entry-side action potentially involves a conformational change between the linker-DNA-binding domain (possibly the SLIDE domain) and the ATPase domain, which pushes a 3-bp equivalent of DNA into the nucleosome, allowing an additional 3 bp of DNA to be pumped to the exit side. After these 3 bp are translocated to the exit side, the strain on the entry-side DNA becomes sufficiently strong again to trigger another action on the entry side, allowing another three base pairs to be translocated to the exit side. This cycle repeats to allow processive nucleosome translocation.
Figure 7. A model for nucleosome translocation by ISWI-family remodelers.
DNA, histone octamer and remodeler are shown in black/grey/red, yellow and blue/green, respectively. The upper and lower DNA gyres are depicted as a solid black and dashed grey line, respectively. Each base pair of DNA translocated to the exit side is shown by a red dot. A cartoon representation of the remodeler is shown as a semi-transparent light blue or light green shape, and the locations of the ATPase and linker-DNA-binding domains as blue and green spheres, respectively. ISWI-induced remodeling starts with the ATPase domain translocating DNA from the SHL2 site towards the exit side, 1 bp at a time. The translocation by the ATPase domain generates strain on the entry-side DNA (depicted by magenta/purple coloring the DNA), which initially remains immobile. After 7 bp of DNA translocation, the accumulated strain is sufficiently strong to trigger an entry-side action, possibly a conformational change of the enzyme, which pushes a 3-bp equivalent of DNA into the nucleosome. This action partially relaxes the strain and allows three additional base pairs of DNA to be translocated to the exit side. This cycle then repeats to allow processive nucleosome translocation.
EXPERIMENTAL PROCEDURES
Preparation of dye-labeled mononucleosomes
Double-stranded dye-labeled DNA constructs with varying flanking linker lengths on the two sides of the 601 nucleosome positioning sequence and/or a 2-nt ssDNA gap at specific locations were generated using a standard PCR or annealing approach. Mononucleosomes were reconstituted from Cy5-labeled DNA and histone octamers, labeled with Cy3 on histone H2A, by salt dialysis and purified by gradient ultracentrifugation (Luger et al., 1997) (see Supplemental Experimental Procedures).
Preparation of ISW2, Isw2p, and ISW1b
ISW2 and ISW1b were affinity-purified from Saccharomyces cerevisiae strains BY4742 and YTT449, respectively, as previously described (Gangaraju and Bartholomew, 2007; Tsukiyama et al., 1999) (see Supplemental Experimental Procedures). For the isolation of the catalytic subunit, Isw2p, we deleted a portion of the ITC1 gene to disrupt the ITC1-Isw2p interaction and immuno-purified the isolated catalytic subunit, Isw2p.
Single-molecule FRET
Biotinylated and dye-labeled mononucleosomes were surface-anchored on poly(ethylene glycol)-coated quartz microscope slides through biotin-streptavidin linkage, which did not inhibit the remodeling activity (Blosser et al., 2009). Immobilized nucleosomes were excited with a 532 nm Nd:YAG laser (CrystaLaser) and fluorescence emissions from Cy3 and Cy5 were detected using a prism-type TIRF microscope, filtered with a 550 nm long-pass filter (Chroma Technology), spectrally separated by a 630 nm dichroic mirror (Chroma Technology), and imaged onto the two halves of a CCD camera (Andor iXonEM+888 1024 × 1024).
In order to obtain nucleosomes labeled with a single donor (Cy3) dye and a single acceptor dye (Cy5), we reconstituted nucleosomes with a mixture of Cy3-labeled and unlabeled H2A. The presence of two H2A subunits in each histone octamer gives rise to a heterogeneous population of nucleosomes with three different labeling configurations, which can be separated in FRET measurements at the single molecule level (Figures S1 and S5) (Blosser et al., 2009). In this work, we selected nucleosomes containing a single donor on the H2A subunit proximal to the acceptor dye on DNA for further analysis. The imaging buffer contained 12 mM HEPES, 40 mM Tris pH 7.5, 60 mM KCl, 0.32 mM EDTA, 3 mM MgCl2, 10 % glycerol, 0.02% Igepal (Sigma Aldrich), an oxygen scavenging system (10 % glucose, 800 μg ml−1 glucose oxidase, 40 μg ml−1 catalase) to reduce photobleaching, 2 mM Trolox (Sigma) to reduce photoblinking of the dyes (Rasnik et al., 2006), and 0.1 mg ml−1 BSA (Promega). Imaging was performed at 30°C, unless otherwise mentioned. Remodeling was induced by infusing the sample chamber with the imaging buffer supplemented with remodeling enzyme, ATP or ATP + ATP-γ-S, and additional MgCl2 equimolar to the total amount of added nucleotide using a syringe pump (KD Scientific).
Automated step identification analyses of FRET time traces
Nucleosome translocation steps were identified by fitting the FRET time traces before photobleaching with a staircase function using a HMM algorithm (McKinney et al., 2006) (http://bio.physics.illinois.edu/HaMMy.html) or an alternative step finding algorithm (Kerssemakers et al., 2006) (see Supplemental Experimental Procedures).
To assign a number k that identifies the position of each pause for analyses shown in Figure S4, the FRET value of the corresponding plateau was converted into the exit DNA linker length (in bp) and then rounded to the nearest integer.
Supplementary Material
RESEARCH HIGHLIGHTS.
ISWI-family remodelers translocate nucleosomes with a conserved stepping pattern
Translocation step sizes are maintained by the catalytic subunit of the remodelers
Translocation proceeds in well-defined multi-bp entry steps and 1-bp exit steps
DNA translocation to the exit side precedes any DNA movement on the entry side
ACKNOWLEDGEMENTS
We thank J. Widom for providing the plasmid containing the 601 positioning sequence and G. Narlikar for providing histone protein expression plasmids. This work is supported in part by the Howard Hughes Medical Institute (to X.Z.) and National Institutes of Health GM 70864 (to B. B.). X.Z. is a Howard Hughes Medical Institute investigator. S.D. is a Merck fellow of the Jane Coffin Childs Memorial Fund for Medical Research. W.L.H. is supported by the NIH Molecular Biophysics Training Grant and Medical Scientist Training Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Amitani I, Baskin RJ, Kowalczykowski SC. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol Cell. 2006;23:143–148. doi: 10.1016/j.molcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Blosser TR, Yang JG, Stone MD, Narlikar GJ, Zhuang X. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009;462:1022–1027. doi: 10.1038/nature08627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm V, Hieb AR, Andrews AJ, Gansen A, Rocker A, Toth K, Luger K, Langowski J. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 2011;39:3093–3102. doi: 10.1093/nar/gkq1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GD. Mechanisms of ATP-dependent nucleosome sliding. Curr Opin Struct Biol. 2010;20:73–81. doi: 10.1016/j.sbi.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat Struct Mol Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Arunajadai SG, Moffitt JR, Tinoco I, Jr., Bustamante C. Single-base pair unwinding and asynchronous RNA release by the hepatitis C virus NS3 helicase. Science. 2011;333:1746–1749. doi: 10.1126/science.1206023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua EY, Vasudevan D, Davey GE, Wu B, Davey CA. The mechanics behind DNA sequence-dependent properties of the nucleosome. Nucleic Acids Res. 2012;40:6338–6352. doi: 10.1093/nar/gks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. Regulation of ISWI involves inhibitory modules antagonized by nucleosomal epitopes. Nature. 2012 doi: 10.1038/nature11625. doi:10.1038/nature11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Bartholomew B. Domain architecture of the catalytic subunit in the ISW2-nucleosome complex. Mol Cell Biol. 2007;27:8306–8317. doi: 10.1128/MCB.01351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham MS, Wigley DB, Webb MR. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry. 2000;39:205–212. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I, Jr., Pyle AM, Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Owen-Hughes T. Mechanisms for nucleosome mobilization. Biopolymers. 2003;68:563–578. doi: 10.1002/bip.10323. [DOI] [PubMed] [Google Scholar]
- Gangaraju VK, Bartholomew B. Mechanisms of ATP dependent chromatin remodeling. Mutat Res. 2007;618:3–17. doi: 10.1016/j.mrfmmm.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proc Natl Acad Sci USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MA, Shundrovsky A, Bai L, Fulbright RM, Lis JT, Wang MD. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat Struct Mol Biol. 2009;16:124–129. doi: 10.1038/nsmb.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Fan HY, Narlikar GJ, Kingston RE. Human ACF1 alters the remodeling strategy of SNF2h. J Biol Chem. 2006;281:28636–28647. doi: 10.1074/jbc.M603008200. [DOI] [PubMed] [Google Scholar]
- Hota SK, Bartholomew B. Diversity of operation in ATP-dependent chromatin remodelers. Biochim Biophys Acta. 2011;1809:476–487. doi: 10.1016/j.bbagrm.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- Kagalwala MN, Glaus BJ, Dang W, Zofall M, Bartholomew B. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J. 2004;23:2092–2104. doi: 10.1038/sj.emboj.7600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassabov SR, Henry NM, Zofall M, Tsukiyama T, Bartholomew B. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol Cell Biol. 2002;22:7524–7534. doi: 10.1128/MCB.22.21.7524-7534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerssemakers JW, Munteanu EL, Laan L, Noetzel TL, Janson ME, Dogterom M. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- Kulic IM, Schiessel H. Chromatin dynamics: nucleosomes go mobile through twist defects. Phys Rev Lett. 2003a;91:148103. doi: 10.1103/PhysRevLett.91.148103. [DOI] [PubMed] [Google Scholar]
- Kulic IM, Schiessel H. Nucleosome repositioning via loop formation. Biophys J. 2003b;84:3197–3211. doi: 10.1016/S0006-3495(03)70044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langst G, Becker PB. Nucleosome remodeling: one mechanism, many phenomena? Biochim Biophys Acta. 2004;1677:58–63. doi: 10.1016/j.bbaexp.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Langst G, Bonte EJ, Corona DFV, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- Lee JY, Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- Lia G, Praly E, Ferreira H, Stockdale C, Tse-Dinh YC, Dunlap D, Croquette V, Bensimon D, Owen-Hughes T. Direct observation of DNA distortion by the RSC complex. Mol Cell. 2006;21:417–425. doi: 10.1016/j.molcel.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Davis B, Kornberg RD. Chromatin remodeling by DNA bending, not twisting. Proc Natl Acad Sci U S A. 2005;102:1329–1332. doi: 10.1073/pnas.0409413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Maier-Davis B, Kornberg RD. Mechanism of chromatin remodeling. Proc Natl Acad Sci U S A. 2010;107:3458–3462. doi: 10.1073/pnas.1000398107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467:562–566. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell AD, Gelbart ME, Tsukiyama T. Histone fold protein Dls1p is required for Isw2-dependent chromatin remodeling in vivo. Mol Cell Biol. 2004;24:2605–2613. doi: 10.1128/MCB.24.7.2605-2613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Ong MS, Richmond TJ, Davey CA. DNA stretching and extreme kinking in the nucleosome core. J Mol Biol. 2007;368:1067–1074. doi: 10.1016/j.jmb.2007.02.062. [DOI] [PubMed] [Google Scholar]
- Park J, Myong S, Niedziela-Majka A, Lee KS, Yu J, Lohman TM, Ha T. PcrA Helicase Dismantles RecA Filaments by Reeling in DNA in Uniform Steps. Cell. 2010;142:544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad TK, Robertson RB, Visnapuu ML, Chi P, Sung P, Greene EC. A DNA-translocating Snf2 molecular motor: Saccharomyces cerevisiae Rdh54 displays processive translocation and extrudes DNA loops. J Mol Biol. 2007;369:940–953. doi: 10.1016/j.jmb.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat Methods. 2006;3:891–893. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- Schwanbeck R, Xiao H, Wu C. Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex. J Biol Chem. 2004;279:39933–39941. doi: 10.1074/jbc.M406060200. [DOI] [PubMed] [Google Scholar]
- Shundrovsky A, Smith CL, Lis JT, Peterson CL, Wang MD. Probing SWI/SNF remodeling of the nucleosome by unzipping single DNA molecules. Nat Struct Mol Biol. 2006;13:549–554. doi: 10.1038/nsmb1102. [DOI] [PubMed] [Google Scholar]
- Sirinakis G, Clapier CR, Gao Y, Viswanathan R, Cairns BR, Zhang Y. The RSC chromatin remodelling ATPase translocates DNA with high force and small step size. EMBO J. 2011;30:2364–2372. doi: 10.1038/emboj.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- Stockdale C, Flaus A, Ferreira H, Owen-Hughes T. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J Biol Chem. 2006;281:16279–16288. doi: 10.1074/jbc.M600682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohner R, Wachsmuth M, Dachauer K, Mazurkiewicz J, Hochstatter J, Rippe K, Langst G. A ‘loop recapture’ mechanism for ACF-dependent nucleosome remodeling. Nat Struct Mol Biol. 2005;12:683–690. doi: 10.1038/nsmb966. [DOI] [PubMed] [Google Scholar]
- Stryer L, Haugland RP. Energy Transfer - a Spectroscopic Ruler. Proc Natl Acad Sci USA. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto RK, Edayathumangalam RS, White CL, Melander C, Gottesfeld JM, Dervan PB, Luger K. Crystal structures of nucleosome core particles in complex with minor groove DNA-binding ligands. J Mol Biol. 2003;326:371–380. doi: 10.1016/s0022-2836(02)01407-9. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. Characterization of the Imitation Switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Holde K, Yager T. Models for chromatin remodeling: a critical comparison. Biochem Cell Biol. 2003;81:169–172. doi: 10.1139/o03-038. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- Vary JC, Jr., Gangaraju VK, Qin J, Landel CC, Kooperberg C, Bartholomew B, Tsukiyama T. Yeast Isw1p forms two separable complexes in vivo. Mol Cell Biol. 2003;23:80–91. doi: 10.1128/MCB.23.1.80-91.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan D, Chua EY, Davey CA. Crystal structures of nucleosome core particles containing the ‘601’ strong positioning sequence. J Mol Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Stockdale C, Flaus A, Szczelkun MD, Owen-Hughes T. Evidence for DNA translocation by the ISWI chromatin-remodeling enzyme. Mol Cell Biol. 2003;23:1935–1945. doi: 10.1128/MCB.23.6.1935-1945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widom J. Role of DNA sequence in nucleosome stability and dynamics. Q Rev Biophys. 2001;34:269–324. doi: 10.1017/s0033583501003699. [DOI] [PubMed] [Google Scholar]
- Yamada K, Frouws TD, Angst B, Fitzgerald DJ, DeLuca C, Schimmele K, Sargent DF, Richmond TJ. Structure and mechanism of the chromatin remodelling factor ISW1a. Nature. 2011;472:448–453. doi: 10.1038/nature09947. [DOI] [PubMed] [Google Scholar]
- Yang JG, Madrid TS, Sevastopoulos E, Narlikar GJ. The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat Struct Mol Biol. 2006;13:1078–1083. doi: 10.1038/nsmb1170. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns BR, Peterson CL, Bustamante C. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol Cell. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.