Abstract
There is widespread interest in characterizing the organization of human genetic variation around the world from a population perspective. Related to this are attempts to describe the pattern of genetic variation in the human species generally, including “recreational” genomics, the genome-based estimation of the ancestry of individuals. These approaches rest on subtle concepts of variation, time, and ancestry that are perhaps not widely appreciated. They share the idea that there are, or were, discrete panmictic human populations such that every person is either a member of such a population or is an admixed descendant of them. Ancestry fraction estimation is biased by assumptions about past and present human population structure, as when we trace ancestry to hypothetical unmixed ancestral populations, or assign an individual's ancestry to continental populations that are indistinguishable from classical “races.” Attempts to identify even individuals' local subpopulations are less precise than most (geneticists included) expect, because that is usually based on a small portion of a person's ancestry, relative to the much larger pool of comparably related ancestors. It is easier to show that two people have some relationship than to show who or where the actual ancestor was. There is an important distinction between individuals' demographic ancestry and the ancestry of their genes. Despite superficial appearances, these interpretations of genetic data are often based on typological rather than Darwinian thinking, raising important issues about the questions that are actually being asked.
Human genetic data are becoming available from unprecedented global sampling, larger sample sizes, and many loci assayed per individual. Rapidly growing databases have fueled interest in reconstructing the detailed history of our globally dispersed species at a level of detail that was not previously possible. The large amounts of genotyping that can be done inexpensively from small amounts of DNA has led to widespread anthropological interest in constructing the detailed ancestry even of single individuals, an interest that has quickly moved from the research lab to the commercial, recreational, and sociological domains (Shriver and Kittles 2004).
But what exactly is the question being asked? Strangely, this is far less clear even among experts. It is only individuals who inherit, carry, and transmit copies of the human genome, but the relationships between individuals, populations, and genes are the products of complex past evolutionary histories that we infer by making comparisons among contemporaries. Our inferences require simplifying models and assumptions that may bias what appear to be objective results (Long and Kittles 2003).
Human settlement history and population concepts
The underlying rationale for ancestry analysis is evolutionary, based on the idea of a common origin both for the DNA sequences at each nucleotide and for our species as a whole. Evidence from genomic variation suggests that anatomically modern humans arose in some single region somewhere in northeast Africa about 100,000–200,000 yr ago (Jobling et al. 2004). A diversity of genetic, archeological, and paleontological data suggests that this population expanded within Africa and subsequently, around 100,000 or fewer years ago, out of Africa into Europe, Asia, and Australia, followed by further expansion into the New World somewhere around 20,000–30,000 yr ago, and finally into some isolated areas, such as the Pacific islands, less than 10,000 yr ago.
This currently favored scenario envisions the process of settlement and expansion as irregular, affected by culture, terrain, chance and other factors including natural selection. At the frontier of human habitation small founder groups carrying a subset of genetic variation from the edge would expand into the next unoccupied territory (by modern humans, though probably occupied by some closely related hominin species). Ultimately, the genealogical ancestry of people in all regions of the world traces back to Africa through a series of these founder events and expanding populations. After the settlement of the major geographic regions there was exogamy practiced at the local level that created stochastic gene flow among adjacent small hunter–gatherer populations. There are irregularities due, for example, to divergent expansion routes related to geographic barriers.
Overall, today people indigenous to widely separated regions—that is, whose ancestors we believe to have resided continually in their respective regions since the distant past—are generally genetically more divergent than indigenes living in close quarters. This pattern of divergence relates to both the serial founder effects and the subsequent smoothing by local mate exchange patterns (Cavalli-Sforza et al. 1993; Ramachandran et al. 2005; Liu et al. 2006; Witherspoon et al. 2007; Jakobsson et al. 2008; Li et al. 2008; Novembre and Stephens 2008; Hunley et al. 2009). However, many large contemporary populations arose from major colonial or conquest migrations in more recent historical times, such as the Americas in the last 500 yr. Geneticists refer to mate exchanges between members of distantly located populations that were previously isolated from each other as admixture. The genetic structure of contemporary humans relates to both the deep evolutionary history of our species and the more recent mate exchanges.
People are often described in terms of their “populations,” but the meaning of that term is not as transparent as it seems. Large populations contain many levels of subdivision. At the more inclusive levels of subdivision are ethnic groups, language groups, nations, shared colonial history, and inter- and intracontinental migrations. The more exclusive levels of subdivision include moieties such as local clans and lineages. With the possible exception of some very isolated island or remote indigenous areas, most human populations exchange mates (often by rule) with other populations, at least their neighbors, and this obscures population boundaries and membership. Population boundaries, mating choices, and movements of people are also highly stochastic. All of these factors make human population strata and boundaries multilayered, porous, ephemeral, and difficult to identify. Samples for genetic analysis are collected using operational criteria imposed by investigators and may be more representative of these operational criteria than actual breeding groups and gene pools. Nevertheless, correctly identifying populations is important because, if population structure is not taken into account, it can produce false positive genetic associations with disease risk in genome-wide mapping studies (Freedman et al. 2004; Marchini et al. 2004; Voight and Pritchard 2005; Witherspoon et al. 2007). Population structure and dynamics are also of anthropological interest.
For these and other reasons, it has become common to couch analysis of human variation as the pursuit to find genetic subdivisions within large regions and communities, or even the whole world. Lately geneticists have been applying the term “population structure” to mean these kinds of subdivisions. However, some care is warranted because evolutionary population genetics uses the term “population structure” in a much broader sense that can encompass both the local mating pools of interest in recent disease association studies, as well as longer term history of our species including intercontinental migrations and founder events (see Wright 1969).
Darwinian and platonic concepts of variation
There are many ways to portray the degrees of genetic similarity and relationships among a set of global human samples. The samples can be displayed in tree diagrams constructed from genotype frequencies at multiple loci. Even if not intended, such trees may give the false impression that the populations represent phylogenetically evolving units whose differences arose independently since the time of a common ancestral population. This can be an important source of bias: Apparent separation times estimated from genetic data without accounting for gene flow can be much shorter than the real age of the populations (Weiss and Maruyama 1976).
Alternatively, multilocus genotypes of sampled individuals can be plotted, using methods like principal components analysis, often coded by symbol or color to show the clustering of individuals from the same sample relative to those from other samples. However, investigators identify the clusters in these diagrams post hoc by using subjective methods that are mainly of heuristic value. Clusters in these diagrams may reify divisions between populations that researchers create by carving discrete samples from an underlying more continuous distribution of local breeding populations. This problem has led some investigators to interpolate the pattern of genetic diversity over unsampled space for presentation in map form geographically as continuous, and more realistic, frequency isoclines (Cavalli-Sforza et al. 1993).
All of these approaches embrace a Darwinian view of relating differentiation to shared ancestry filtered through historical demography. However, they differ on the relative emphasis they give to four evolutionary processes (drift, mutation, migration, and natural selection), and as to whether they view the human gene pool as occupying an equilibrium or nonequilibrium state. Assuming equilibrium makes some analysis easier, but that can be at the expense of introducing bias.
Because of the difficulty in deriving explicit hypotheses from complex evolutionary processes, and developing valid statistical tools to test these hypotheses, there is a natural tendency for researchers to rely on readily available user-friendly computer packages and conduct analyses in an off-the-shelf fashion. Recent analyses have used a Bayesian K-populations cluster analysis as the tool of choice for analyzing the large-scale human population genetic data that are now available. These applications involve evolutionary and historical as well as quasitaxonomic concepts. A Bayesian K-populations cluster approach to variation treats our species, or some specified area of the world, in admixture terms, as if populated by people who either are members of discrete populations or are admixed descendants of such populations. Users of the Bayesian K-populations cluster approach refer to the discrete populations in various kinds of ancestry terms, such as by referring to them as “parental.” For example, samples of African-Americans collectively reflect a majority of ancestors from African and smaller fractions from European or other parental populations (Parra et al. 1998; McKeigue et al. 2000; Shriver et al. 2003). The mixing proportions are estimated by statistical analysis based on the admixed sample and donor genotype frequencies taken from samples of presumed parental populations, and may take into account the temporal dynamics of the admixture process (Pfaff et al. 2001).
This kind of admixture approach to human variation has been done most frequently by using the program structure, or programs implementing modifications of the same or a similar conceptual approach (Pritchard et al. 2000; Falush et al. 2003; Hoggart et al. 2004; McKeigue 2005; Tang et al. 2005, 2006; Zhu et al. 2006; Montana and Hoggart 2007). Here, we will use the phrase “structure-like analysis” to refer generically to this approach, regardless of the specific program used in any given paper. The popularity and ease of use of structure-like programs have fueled a recent trend to use the term ”population structure” in the limited admixture sense, and indeed to view history from this rather platonic view, as comprised of parental entities and their offspring. The architects of structure and related programs are well aware of the limitations of the method and state them clearly in their papers (Pritchard et al. 2000; Falush et al. 2003; Tang et al. 2005). However, applications of such programs are often made without heeding caveats or recognizing the limitations of the underlying models with respect to the questions and data at hand.
In structure-like analysis, typical input data consist of globally distributed polymorphisms (STRs, SNPs, indels, etc.) that are genotyped in a sample of individuals. Depending on the purpose and specific program, these may be from a series of intracontinental or global samples. The program user can optionally either specify the number of parental populations and provide their allele frequencies from external data, or can specify that number and have the program statistically group the sample and optimize their allele frequencies, or can have the program estimate both the optimized number of parental populations (K) and their allele frequencies. A parental population is assumed to be randomly mating with Hardy–Weinberg equilibrium genotype proportions, and the program uses likelihood ratio or other similar significance-testing criteria to identify such internally statistically homogeneous populations, and minimize any linkage disequilibrium between them, that is, to determine the statistically optimal population number and allele frequencies represented by the supplied data.
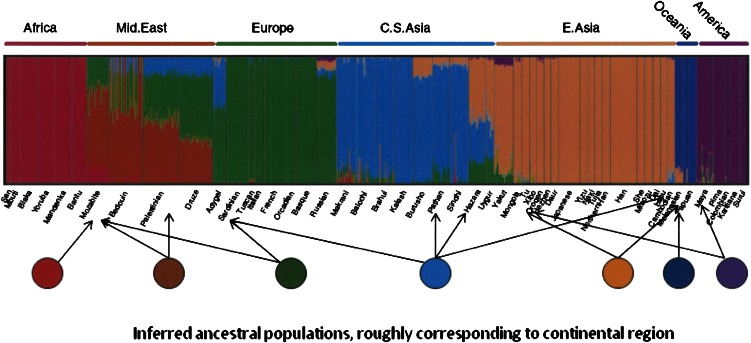
Once the parental populations have been characterized in terms of their allele frequencies, each individual in the data is assigned an estimated fraction of ancestry from each of the parental populations (which can be 1.0 for a member of a parental population). The analysis is graphically presented, usually by arranging the populations by their location, such as a west-to-east, Africa to Americas axis, as shown in Figure 1. The output analysis is shown across the top, where each individual in the sample is represented by a narrow vertical line divided into color segments proportional to that individual's fraction of admixture from the program-optimized K color-coded ancestral populations, of which there are seven in this example. Below, we have shown those assumed ancestral populations as discrete circles of homogenous corresponding colors, with arrows suggesting a few of their contributions to individuals in the sample.
Figure 1.
Admixture structure analysis of a worldwide sample. The x-axis represents individuals from populations arrayed geographically roughly from west to east, as labeled above with sample sources identified below. There is a thin vertical bar for each individual, color-coded to represent his/her admixture proportions from “parental” populations. The parentals are shown schematically as circles below the diagram, with arrows indicating a few of their contributions to individuals in the sample (circles and arrows added by us for this paper). These parental representations are not part of the actual sample but are statistically abstracted from it, as if they actually exist (some individuals in some of the populations are statistically assigned 100% ancestry from one of the parental in this particular data set). This analysis was done using the structure-like program Frappe (that employs a different estimation procedure for similar objectives; Tang et al. 2005). Structure analysis figure reprinted with permission from Li et al. 2008, American Association for the Advancement of Science © 2008.
Whether the investigator uses external information or makes estimates from the samples at hand, the parental populations are abstractions that conform to only the simplest kind of genetic structure. This structure places heavy emphasis on the idea that the world once harbored distinct and independently evolved populations that have now undergone admixture of an unstated type (often seeming to connote admixture due to colonial era migrations). Regardless of the intent, this idea of population structure is unfortunately more in line with race concepts held by European explorers and traders than with the recent genetic evidence supporting the serial sampling model of human evolutionary history.
The ideal markers for this kind of analysis are private to, and in high frequency in, only one of the putative parental populations, or at least display major differences in frequency among the putative parental populations. Geneticists call markers with these characteristics ancestry informative markers (AIMs). However, not many documented single nucleotide polymorphisms (SNPs) are useful AIMs. Private variants at high frequency even within local demes are rather rare because common alleles are usually old and shared across populations, either by descent from the species' ancestor, or because they have spread by migrations and local gene flow. AIMs that have not reached complete fixation in one population (which is by far the typical case) provide some useful geographic information, but do not relieve the data of the need for probabilistic analysis. Therefore, most information in a genetic marker, even a putative AIM, is in frequency differences, and this makes it fail as a definitive tag for a particular region or population. In practice, the situation is even worse because most structure-like analyses use markers that were discovered in modest-size samples from only a few populations (mainly, Europe, West Sub-Saharan Africa, and East Asia), and registered in databases such as dbSNP (www.ncbi.nlm.nih.gov/projects/SNP/) or HapMap (www.hapmap.org) whose markers are intended primarily for gene mapping. How specific these markers are to a limited geographic region is often untested. For example, an AIM intended to reveal Native American ancestry may also be common in East Asians, and not private after all.
This raises the question as to what the evidence for the ancestral populations inferred by structure-like analysis actually represents. If the user has supplied their allele frequency definitions from other samples, the user has made decisions as to what and how many they are, their genetic definitions, and their “parental” status. Commonly, as in the case of Figure 1, the program has identified these populations statistically from the given data set, and a user-specified criterion for determining K, which is why we portray them as circles separated from the individual results. It is clear that in all cases these populations are statistical constructs that depend on the sampled data, not literal ancestors which is why we refer to them as platonic, and we should question how well, and even whether, they represent actual populations that existed in the past.
In structure-like analysis, individuals are ascribed ancestry as if panmictic (noninternally structured) parental populations actually existed in history, and it is possible to reconstruct their composition from the contemporary data. Some researchers treat the results implicitly or explicitly as a new fundamental truth, or even discovery of genetic relationships (Wilson et al. 2001), but there are often strange features that deserve further questioning. Is it plausible, as suggested in Figure 1, that the present Siberian Yakut population represents a three-way mix of East Asians, Europeans, and Native Americans? What do we make of contemporary Russians to whom the structure-like program attributes Native American ancestry? Such cases require geographically implausible mixes between distant populations.
The abstract nature of such results can be seen by the fact that sampled individuals had only two real parents, but are assigned multiple ancestral fractions from the presumed contemporaries. Of course, depending on how far back one wishes to go and the degree to which population ancestry is a valid concept, each of us has multiple distant ancestry. Thus, this ancestry approach not only uses contemporaries to represent assumed ancestral populations, but with implicit temporal depth that blurs gene pool fractions with ancestry fractions. The difference becomes especially important in the case of individual genetic ancestry estimation in recreational genomics (discussed below).
Although each individual is assigned fractions from the parentals, no individual in the sample needs to be a “pure” representative of a parental population. Thus, the data may define, but not include, the parental populations themselves. For example, in Figure 1 a “Middle East” parental is defined (colored brown), but few, if any, individuals in the data are assigned ancestry only from that population. We ask first whether it is possible to test such a model, second whether there are other evolutionary interpretations of the data, and third whether the alternatives are preferable to the seemingly simple admixture-based structure results.
The nature of abstraction in the K-populations analytic framework can perhaps be seen in yet another way, which is that large geographic areas, such as “Africa” or “Europe” are depicted as ancestral sources, as if those areas have no internal geographic stratification, which is not literally true. This is an artifact of the way that available discrete, spot samples, such as CEPH Europeans, are depicted as representing large geographic regions, such as “Europe,” and the width of each region depends on the number of individuals in the sample since each individual is represented by a vertical admixture bar. The artifactual nature of the result can be seen by the fact that when more populations or more markers are included, more parentals are usually inferred or admixture appears to be more complex, and large regions are themselves structured as recently shown in Europe (Bauchet et al. 2007; Novembre et al. 2008).
Ultimately, in conceptually proper use, structure-like analysis provides a statistically effective means to reduce the complexity and graphically present high dimensional data, and a way to visualize individuals in the context of variation within and between samples. It does so in terms of functions of the raw data. It can reveal substructure that could confound genetic inference in relation to disease. This analysis can provide insight into real patterns but the process underlying the displayed patterns is not necessarily the admixture between isolated and independent ancestral populations.
Perhaps an exaggerated version of these points can be seen in a recent paper analyzing the ancestry of various presumably admixed (“Mestizo”) populations in the Americas (Wang et al. 2008). Here, parental populations include diverse samples from putatively “pure” (unadmixed, in some assumed sense) Native American populations, along with approximately representative European and African samples. Admixed urban populations from Central and South America were analyzed by structure to estimate their fractions of admixture from these global parentals. But this means that a sample from Mexico City and one from southern Chile were both treated as if they were hybrids from all of the 13 Central and South American Native American sources, and a population near Tierra del Fuego was described as having some Central and Northern Amerindian ancestry. The analysis makes sense in showing evidence that the mixed populations had higher fractions of putative admixture from the more nearby indigenous populations, but in doing this by using the admixture approach, it fictionalizes much of the idea of ancestral and admixed populations.
Despite these issues, this kind of analysis seems to make intuitive sense. But what is it? Whether the parental populations are externally user-defined or internally statistically defined, we cannot distinguish the analysis from a search for ideal types. Such analysis may use modern genetic data, but the statistical output is not conceptually different from classical racial analysis based on morphology. Biologists since—and including—Darwin have known that there cannot be classical platonic essences in populations, otherwise evolution would be impossible. Succinctly, variation is the central reality. “Parental” populations are platonic because they are abstractions that never actually existed yet are used to infer reality as if they did. Reality has not stopped the long persistence of “structure” analysis to define disconnected “pure” types—races—of which everyone is either a member or a hybrid (Darwin 1871; Hooton 1926; Baur et al. 1931; Boyd 1950; Kittles and Weiss 2003; Weiss and Fullerton 2005; Morris-Reich 2006). Some of the most serious human abuses in history have been justified in such terms (Kevles 1995).
Investigators may be innocently unaware of this history, but misapplied structure analysis essentially replaces classical racial types based on multivariate metrical or morphological characteristics by types defined by multilocus allele frequencies without carefully thinking through the evolutionary implications. This is an interesting kind of typology in which every member of the type is actually different. In the genetic approach, there are Hardy–Weinberg proportions at many loci; everybody of a given parental type will have a different genotype, randomly drawn from the same type-specific allele frequency vector, yet at any given locus the same genotype can be found, if with reduced probability, in people of other types. Yet, these modern genetic constructions are produced by evolutionary biologists and superficially appear to be about evolutionary history.
Often there is no explicit accounting for the fact that at some point the parental populations (whether they could be real today, or at any time in the past) must share ancestry with each other, and that the different parental populations ultimately share varying degrees of ancestry. Even if one were to grant that contemporary data only provide estimates of, rather than actual, ancestral parental genotype frequencies, there is no reason to think that there ever were isolated, homogeneous parental populations at any point in our human past. Why do we, even in science, so uncritically accept admixture-based analyses of global samples that give the appearance that human variation is clustered into a few major populations, portrayed in much the same way as classical races? These are not pleasant thoughts, but it is important to learn from history, and sometimes it is valuable to be brought face to face with one's tacit assumptions or the nature of their underlying rationale.
Recent group and individual ancestry estimation and recreational genomics
Social concepts may in part be responsible for a reliance on admixture to explain human variation. Because we have become used to thinking of humans as living in populations identified as units by culture, language, location (e.g., towns, countries, and villages), and because most of our existing human data are from such discretely defined sampling units, ancestry questions are usually phrased in population-specific terms: What “tribe” or “country” did my ancestors come from? This nonscientific view of life has perhaps been reinforced by actual recent admixture in the United States and other former European colonies. In the age of sail, and to a lesser extent before that, it has been possible for individuals who were born in the most widely dispersed parts of the world, with very little direct gene flow between them, to meet and mate. Such mating clearly occurred in Viking, Mongol, European, Na Dene (northern North American), and Bantu expansions, but the migration of Europeans and Africans to the New World has contributed heavily to the importance of admixture in population structure. Indeed, for centuries laws that regulated the legitimacy of intermarriage defined and reinforced the races we perceive today.
An African American really may have identifiable ancestors who came to North America directly from Africa and Europe, where their recent prior ancestors were at least mainly localized to those respective continents. In this context an admixture way of describing human variation certainly has some heuristic value. Natural curiosity has led to widespread interest in individuals tracing the history of immigration of their own genealogical ancestors into the New World, and many who cannot do that directly (e.g., African Americans whose New World ancestors were slaves with little in the way of adequate documentary records) have shown great interest in estimates of their genetic ancestry. Serving this interest are a fluid landscape of ∼30 companies that offer “recreational” genetic ancestry analysis, each using these various concepts in varying ways.
This is recreational in that it is for edification rather than for practical purposes, and because it is known (at least to the vendors, whether or not to their customers) to be, at best, approximate. But it is likely to become much more serious as ethics meets science, when results are interpreted in inheritance, tribal, legal, or disease-related terms. The reasons have been cogently assessed from bioethical and societal points of view (Bolnick et al. 2007), though defenders of genomic ancestry services have a differing point of view (Wagner and Shriver 2007). The area is controversial because different companies have provided different ancestry results for the same person because the results depend on each company's set of parental populations, genetic markers, assumptions, and interpretations.
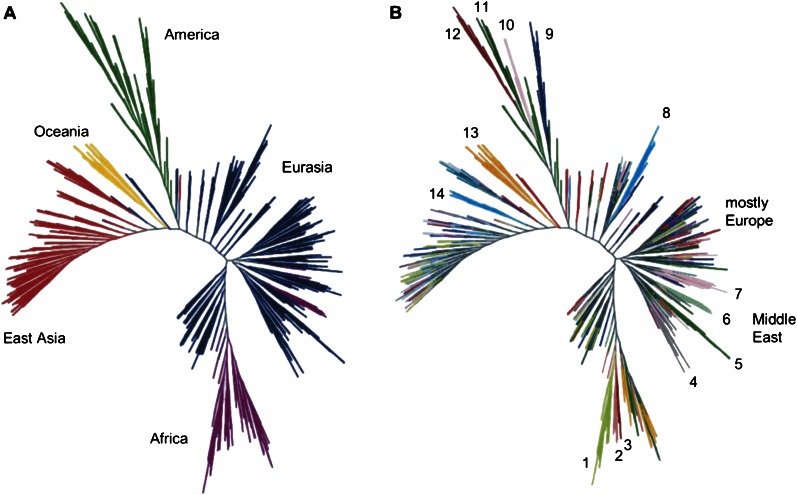
Will these problems go away with more data and more refined estimates? The answer is unclear and to some degree depends on the quantity that one wishes to estimate. There are alternatives to the structure-like analysis of genetic diversity and relatedness at the level of individuals. For example, a recent analysis that used essentially the same set of individuals as in Figure 1, without considering individuals in terms of population constructs (Nievergelt et al. 2007). Instead, the researchers used the neighbor-joining algorithm to build an unrooted similarity tree linking all individuals on the basis of pairwise distances, as shown in Figure 2. In this tree, individuals from the same general geographic region sit near each other (Fig. 2A) (the geographic axis is reversed from that given in Figure 1) and there are clusters of people from the same geographic region. However, the impression of typological orderliness of human variation disappears. Notice that people from the major geographic regions do not always appear in ‘monophyletic’ clusters. For example, Eurasians populate the most inclusive group that includes all Africans. Africans, Pacific Islanders, and Native Americans appear in the most inclusive group that includes all Eurasians. Moreover, Figure 2B uses a different color-coding of the leaves on the same tree to show that people do not cluster neatly by local population within continental regions. Any person's closest genomic match is likely to trace back to the same broad geographic area, but not necessarily in the same local group. This again shows that it is the use and interpretation of results, not the programs themselves that are at issue. Sampling that was not dependent on prior population definitions, would provide even less cluster-like results.
Figure 2.
Illusions can be generated by population-based structure analysis. Individual genotype relationships from the CEPH-diversity 51 global population samples; neighbor-joining similarity trees were constructed from the matrix of pairwise differences (reprinted from Nievergelt et al. 2007). Panel A color codes the major world regions. The analysis correctly grouped individuals on this broad criterion. However, in panel B, the color code identifies the population source (number and text annotation represents different populations), showing intermingling of similarity within geographic regions. For details see the original paper (Nievergelt et al. 2007).
The data analyzed in this study consisted solely of indigenous people; however, in a similar kind of analysis, recently admixed individuals, such as African-Americans, would be placed proportionately in between the Africans and Europeans in the sample, as has been done before with individual-specific ancestry analysis (e.g., Shriver and Kittles 2004; Shriver et al. 2005). Clearly, even relative to this sparse population-based sample, an individual in the sample could not identify a home “village,” much less a single, ancestral village from centuries past, as is a common interpretation in today's recreational ancestry testing arena, where the tested individuals are not part of the background data set.
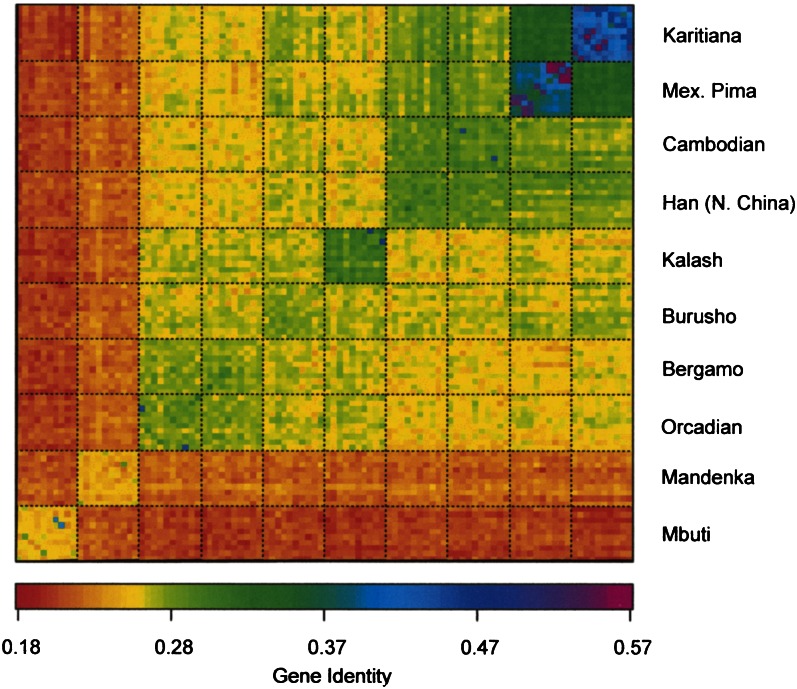
Curiously, both the structure-like and tree-building approaches gloss over one of the most basic questions that an individual might ask about their genetic similarities and differences compared to others—How much do I share genetically with others? For example, how likely is someone from East Asia to have the same genotype at a locus as someone from Europe? Does the probability change if the person's recent ancestors migrated from one continent to another? Figure 3 provides answers to this sort of question using publicly available data from Noah Rosenberg's Website (http://rosenberglab.bioinformatics.med.umich.edu/datasets.html). Here we have selected 100 individuals (10 individuals from each of 10 populations) from the CEPH diversity panel. We calculated the average of Nei's unbiased gene identity statistic over 678 loci within each individual, and between all pairs of individuals (Nei 1987). We chose this statistic because of its interpretability—gene identity within individuals is simply the proportion of loci at which the individual is homozygous; gene identity between a pair of individuals is the expected homozygosity if the pair was to produce a child. Figure 3 presents the entire 100 × 100 matrix with color-coding to visualize the numerical results.
Figure 3.
Gene identity within and between 100 individuals selected from populations throughout the world. Africa (Mbuti, Mandenka), Europe (Orcadians, Bergamo), South Asia (Burusho, Kalash), East Asia (Han, Cambodian), Americans (Pima, Karitiana).
This depiction illustrates several features. First, within several populations there exist pairs of individuals with high gene identity. These individuals are likely close relatives. Second, some individuals have about the same gene identity with others that belong to different populations, as they do with others in their own population. This is particularly true for Orcadians and Bergamo, and for Han from North China and Cambodians. Thousands of kilometers separate both pairs of populations. Third, the well-known trend for increasing homozygosity as geographic distance increases from Africa is immediately evident. Fourth, there is substantial gene identity between any pair of individuals throughout the world. Fifth, there is high gene identity within and between Kalash individuals but in comparison to people in different geographic regions the Kalash present a pattern that is similar to Europeans and South Asians. This result is somewhat surprising because the structure program output depicts the Kalash as a unique ancestral population.
The point has been shown quantitatively in a somewhat different way by others (Bamshad et al. 2004; Witherspoon et al. 2007), and is acknowledged in some structure-like papers (Rosenberg et al. 2002). Even between distant continents, there is a substantial probability that, if one looks only at a modest number of loci, the closest genetic match to a person may be on another continent, and as in Figure 2B, even with genomewide markers, this nonspecificity applies even more on an intracontinental basis. The greater continental than local predictability of genetic similarities is entirely predictable from the stochastic, small-demic, locally restricted nature of human evolutionary population dynamics until recent centuries.
Conclusions
Currently the genetic ancestry service landscape is quite fluid, different companies offering many different kinds of ancestry service, based on different data, criteria, names, and degrees of specificity. However, they generally share the basic idea of using a series of population-specific samples as parental and estimating in some probabilistic way the distribution of overall genomic ancestry a testee has from them. And, except for clear-cut recent examples, these approaches are vague about accuracy, and are nonevolutionary in concept in ways we have described.
Although DNA data have the aura of providing definitive answers to population and individual ancestry questions, they require careful interpretation in terms of both the laws of inheritance and the evolutionary process. Untrained individuals, and even some professionals, will have a difficult time reconciling the nuances of interpretation with the bottom-line aura that DNA carries. This places private companies and public not-for-profit services in a difficult position because they must convince their customers of both the value and limitations of the product. At present, each such service uses different data and assumptions, and they each generate differing results. We provide here a list of points that the informed customer should understand in order to insure that the product is both safe and fairly represented.
All people are relatives of varying degree. Any meaningful statement about shared ancestry must specify a frame of reference. There are different reference frames for analyzing genetic data, but these frames are not equally encompassing, nor are they neutral with respect to social ideology. The intercontinental migration era that mainly began 500 yr ago is one frame of reference, but it carries the baggage of centuries of biologically flawed assertions by conquerors and slave owners about the essential nature of the subordinate “races.”
No gene or segment of DNA carries the complete, or even a large fraction of, information about the people who are an individual's ancestors. This is because while all of the copies of a gene in the human gene pool coalesce to an ancestral sequence at some point in history, different genes, indeed individual SNPs, whether within the same individual or between two individuals, coalesce independently, often at widely separated times and geographic locations.
An individual cannot conclude that they have a close affinity to a particular ethnic group or local geographic population simply because their genome holds a DNA sequence some of whose parts have matches in that population. Such a conclusion would require demonstrating that the DNA sequence is not present in other places, it would require demonstrating that the gene pool of that ethnic group or local population had been closed and immobile for centuries or millennia, and it would require that that DNA sequence is in linkage disequilibrium with so many other portions of the genome that it is a good proxy for the genome as a whole.
Genetic methods assess the similarity relationship between individuals on the basis of sharing alleles at polymorphic loci. However, the results from different loci will vary widely because the vast majority of common polymorphisms are shared widely throughout world populations, which is evolutionarily why they are common. Recent studies show that by assaying an insufficient number of genetic loci, individuals from different populations can appear genetically more similar than individuals from the same population. The frequency of alleles affects the results, and the frequency spectrum is affected by the size and nature of the sample from which alleles are identified. Nevertheless, there will always be substantially wide range local geographic populations that are compatible with the “genetic” ancestry of any individual, no matter how many loci are included.
Sharing ancestors does not equate to sharing the genes that these ancestors carried. This is true even for close relatives. For example, full siblings have exactly the same set of ancestors, but do not have the same multiple locus genotypes.
The ancestors that contribute to genetic kinship existed historically, but information collected from contemporaries is the raw materials of genetic ancestry testing. Because of this, deducing relationships requires us to apply genetic principles and make assumptions about the breeding structure, evolution, and existence of populations that no longer exist. Deductions are bound to assumptions, which often embody preconceived notions, such as racial taxonomies that have no validity. Their impact on the accuracy, even of vaguely specified ancestry questions, may be substantial. Genotypic affinity is related to, but not identical with, genetic or demographic ancestry. Genotypes may predict an individual's broad geographic ancestral homeland(s), but the homeland does not predict his genotype. Above all, a present-day population is not a literal ancestor!
Acknowledgments
We thank Anne Buchanan, Kari Schroeder, and three reviewers for helpful comments on this manuscript. This work was supported by funding from the National Institutes of Health (MH063749) and the Penn State Evan Pugh Professors research fund (K.M.W.), and funding from the National Science Foundation (NSF 0321610) and the University of Michigan Medical School (J.C.L.). We claim full responsibility for all interpretations and any errors of omission or commission.
Footnotes
Article is online at http://www.genome.org/cgi/doi/10.1101/gr.076539.108.
References
- Bamshad M., Wooding S., Salisbury B.A., Stephens J.C. Deconstructing the relationship between genetics and race. Nat. Rev. Genet. 2004;5:598–609. doi: 10.1038/nrg1401. [DOI] [PubMed] [Google Scholar]
- Bauchet M., McEvoy B., Pearson L.N., Quillen E.E., Sarkisian T., Hovhannesyan K., Deka R., Bradley D.G., Shriver M.D. Measuring European population stratification with microarray genotype data. Am. J. Hum. Genet. 2007;80:948–956. doi: 10.1086/513477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baur E., Fischer E., Lenz F. Human heredity. Macmillan; New York: 1931. [Google Scholar]
- Bolnick D.A., Fullwiley D., Duster T., Cooper R.S., Fujimura J.H., Kahn J., Kaufman J.S., Marks J., Morning A., Nelson A., et al. Genetics. The science and business of genetic ancestry testing. Science. 2007;318:399–400. doi: 10.1126/science.1150098. [DOI] [PubMed] [Google Scholar]
- Boyd W.C. Genetics and the races of man: An introduction to modern physical anthropology. Little Brown; Boston: 1950. [Google Scholar]
- Cavalli-Sforza L.L., Menozzi P., Piazza A. Demic expansions and human evolution. Science. 1993;259:639–646. doi: 10.1126/science.8430313. [DOI] [PubMed] [Google Scholar]
- Darwin C. The descent of man and selection in relation to sex. J. Murray; London: 1871. [Google Scholar]
- Falush D., Stephens M., Pritchard J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman M.L., Reich D., Penney K.L., McDonald G.J., Mignault A.A., Patterson N., Gabriel S.B., Topol E.J., Smoller J.W., Pato C.N., et al. Assessing the impact of population stratification on genetic association studies. Nat. Genet. 2004;36:388–393. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- Hoggart C.J., Shriver M.D., Kittles R.A., Clayton D.G., McKeigue P.M. Design and analysis of admixture mapping studies. Am. J. Hum. Genet. 2004;74:965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooton E.A. Methods of racial analysis. Science. 1926;63:75–81. doi: 10.1126/science.63.1621.75. [DOI] [PubMed] [Google Scholar]
- Hunley K.L., Heale M.E., Long J.C. The global pattern of gene identity variation reveals a history of long-range migrations, founder effects and local mate exchange: Implications for biological race. Am. J. Phys. Anthropol. 2009 doi: 10.1002/ajpa.20932. [DOI] [PubMed] [Google Scholar]
- Jakobsson M., Scholz S.W., Scheet P., Gibbs J.R., VanLiere J.M., Fung H.C., Szpiech Z.A., Degnan J.H., Wang K., Guerreiro R., et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- Jobling M., Hurles M., Tyler-Smith C. Human evolutionary genetics: Origins, peoples & disease. Garland; New York: 2004. [Google Scholar]
- Kevles D.J. In the name of eugenics: Genetics and the uses of human heredity. Harvard University Press; Cambridge, MA: 1995. [Google Scholar]
- Kittles R.A., Weiss K.M. Race, ancestry, and genes: Implications for defining disease risk. Annu. Rev. Genomics Hum. Genet. 2003;4:33–67. doi: 10.1146/annurev.genom.4.070802.110356. [DOI] [PubMed] [Google Scholar]
- Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Liu H., Prugnolle F., Manica A., Balloux F. A geographically explicit genetic model of worldwide human-settlement history. Am. J. Hum. Genet. 2006;79:230–237. doi: 10.1086/505436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.C., Kittles R.A. Human genetic diversity and the nonexistence of biological races. Hum. Biol. 2003;75:449–471. doi: 10.1353/hub.2003.0058. [DOI] [PubMed] [Google Scholar]
- Marchini J., Cardon L.R., Phillips M.S., Donnelly P. The effects of human population structure on large genetic association studies. Nat. Genet. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- McKeigue P.M. Prospects for admixture mapping of complex traits. Am. J. Hum. Genet. 2005;76:1–7. doi: 10.1086/426949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeigue P.M., Carpenter J.R., Parra E.J., Shriver M.D. Estimation of admixture and detection of linkage in admixed populations by a Bayesian approach: Application to African-American populations. Ann. Hum. Genet. 2000;64:171–186. doi: 10.1017/S0003480000008022. [DOI] [PubMed] [Google Scholar]
- Montana G., Hoggart C. Statistical software for gene mapping by admixture linkage disequilibrium. Brief. Bioinform. 2007;8:393–395. doi: 10.1093/bib/bbm035. [DOI] [PubMed] [Google Scholar]
- Morris-Reich A. Race, ideas, and ideals: A comparison of Franz Boas and Hans F.K. Gunther. Hist. Eur. Ideas. 2006;32:313–332. [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. Columbia University Press; New York: 1987. [Google Scholar]
- Nievergelt C.M., Libiger O., Schork N.J. Generalized analysis of molecular variance. PLoS Genet. 2007;3:e51. doi: 10.1371/journal.pgen.0030051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J., Stephens M. Interpreting principal component analyses of spatial population genetic variation. Nat. Genet. 2008;40:646–649. doi: 10.1038/ng.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J., Johnson T., Bryc K., Kutalik Z., Boyko A.R., Auton A., Indap A., King K.S., Bergmann S., Nelson M.R., et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra E.J., Marcini A., Akey J., Martinson J., Batzer M.A., Cooper R., Forrester T., Allison D.B., Deka R., Ferrell R.E., et al. Estimating African American admixture proportions by use of population-specific alleles. Am. J. Hum. Genet. 1998;63:1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff C.L., Parra E.J., Bonilla C., Hiester K., McKeigue P.M., Kamboh M.I., Hutchinson R.G., Ferrell R.E., Boerwinkle E., Shriver M.D. Population structure in admixed populations: Effect of admixture dynamics on the pattern of linkage disequilibrium. Am. J. Hum. Genet. 2001;68:198–207. doi: 10.1086/316935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J.K., Stephens M., Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran S., Deshpande O., Roseman C.C., Rosenberg N.A., Feldman M.W., Cavalli-Sforza L.L. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl. Acad. Sci. 2005;102:15942–15947. doi: 10.1073/pnas.0507611102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N.A., Pritchard J.K., Weber J.L., Cann H.M., Kidd K.K., Zhivotovsky L.A., Feldman M.W. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Shriver M.D., Kittles R.A. Genetic ancestry and the search for personalized genetic histories. Nat. Rev. Genet. 2004;5:611–618. doi: 10.1038/nrg1405. [DOI] [PubMed] [Google Scholar]
- Shriver M.D., Parra E.J., Dios S., Bonilla C., Norton H., Jovel C., Pfaff C., Jones C., Massac A., Cameron N., et al. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum. Genet. 2003;112:387–399. doi: 10.1007/s00439-002-0896-y. [DOI] [PubMed] [Google Scholar]
- Shriver M.D., Mei R., Parra E.J., Sonpar V., Halder I., Tishkoff S.A., Schurr T.G., Zhadanov S.I., Osipova L.P., Brutsaert T.D., et al. Large-scale SNP analysis reveals clustered and continuous patterns of human genetic variation. Hum. Genomics. 2005;2:81–89. doi: 10.1186/1479-7364-2-2-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Peng J., Wang P., Risch N.J. Estimation of individual admixture: Analytical and study design considerations. Genet. Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- Tang H., Coram M., Wang P., Zhu X., Risch N. Reconstructing genetic ancestry blocks in admixed individuals. Am. J. Hum. Genet. 2006;79:1–12. doi: 10.1086/504302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight B.F., Pritchard J.K. Confounding from cryptic relatedness in case-control association studies. PLoS Genet. 2005;1:e32. doi: 10.1371/journal.pgen.0010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J., Shriver M.D. Misinformation, social construction and genomic ancestry testing. Science. 2007. http://www.sciencemag.org/cgi/eletters/318/5849/399.
- Wang S., Ray N., Rojas W., Parra M.V., Bedoya G., Gallo C., Poletti G., Mazzotti G., Hill K., Hurtado A.M., et al. Geographic patterns of genome admixture in Latin American Mestizos. PLoS Genet. 2008;4:e1000037. doi: 10.1371/journal.pgen.1000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss K.M., Maruyama T. Archeology, population genetics and studies of human racial ancestry. Am. J. Phys. Anthropol. 1976;44:31–49. doi: 10.1002/ajpa.1330440106. [DOI] [PubMed] [Google Scholar]
- Weiss K.M., Fullerton S.M. Racing around, getting nowhere. Evol. Anthropol. 2005;14:165–169. [Google Scholar]
- Wilson J.F., Weale M.E., Smith A.C., Gratrix F., Fletcher B., Thomas M.G., Bradman N., Goldstein D.B. Population genetic structure of variable drug response. Nat. Genet. 2001;29:265–269. doi: 10.1038/ng761. [DOI] [PubMed] [Google Scholar]
- Witherspoon D.J., Wooding S., Rogers A.R., Marchani E.E., Watkins W.S., Batzer M.A., Jorde L.B. Genetic similarities within and between human populations. Genetics. 2007;176:351–359. doi: 10.1534/genetics.106.067355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Evolution and the genetics of populations. Vol. 2. The theory of gene frequencies. University of Chicago Press; Chicago: 1969. pp. 290–291. [Google Scholar]
- Zhu X., Zhang S., Tang H., Cooper R. A classical likelihood based approach for admixture mapping using EM algorithm. Hum. Genet. 2006;120:431–445. doi: 10.1007/s00439-006-0224-z. [DOI] [PubMed] [Google Scholar]