Abstract
C/EBP and GCN4 are basic region-leucine zipper (bZIP) DNA-binding proteins that recognize the dyad-symmetric sequences ATTGCGCAAT and ATGAGTCAT, respectively. The sequence specificities of these and other bZIP proteins are determined by their alpha-helical basic regions, which are related at the primary sequence level. To identify amino acids that are responsible for the different DNA sequence specificities of C/EBP and GCN4, two kinds of hybrid proteins were constructed: GCN4-C/EBP chimeras fused at various positions in the basic region and substitution mutants in which GCN4 basic region amino acids were replaced by the corresponding residues from C/EBP. On the basis of the DNA-binding characteristics of these hybrid proteins, three residues that contribute significantly to the differences in C/EBP and GCN4 binding specificity were defined. These residues are clustered along one face of the basic region alpha helix. Two of these specificity residues were not identified as DNA-contacting amino acids in a recently reported crystal structure of a GCN4-DNA complex, suggesting that the residues used by C/EBP and GCN4 to make base contacts are not identical. A random binding site selection procedure also was used to define the optimal recognition sequences for three of the GCN4-C/EBP fusion proteins. These experiments identify an element spanning the hinge region between the basic region and leucine zipper domains that dictates optimal half-site spacing (either directly abutted for C/EBP or overlapping by one base pair for GCN4) in high-affinity binding sites for these two proteins.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abel T., Bhatt R., Maniatis T. A Drosophila CREB/ATF transcriptional activator binds to both fat body- and liver-specific regulatory elements. Genes Dev. 1992 Mar;6(3):466–480. doi: 10.1101/gad.6.3.466. [DOI] [PubMed] [Google Scholar]
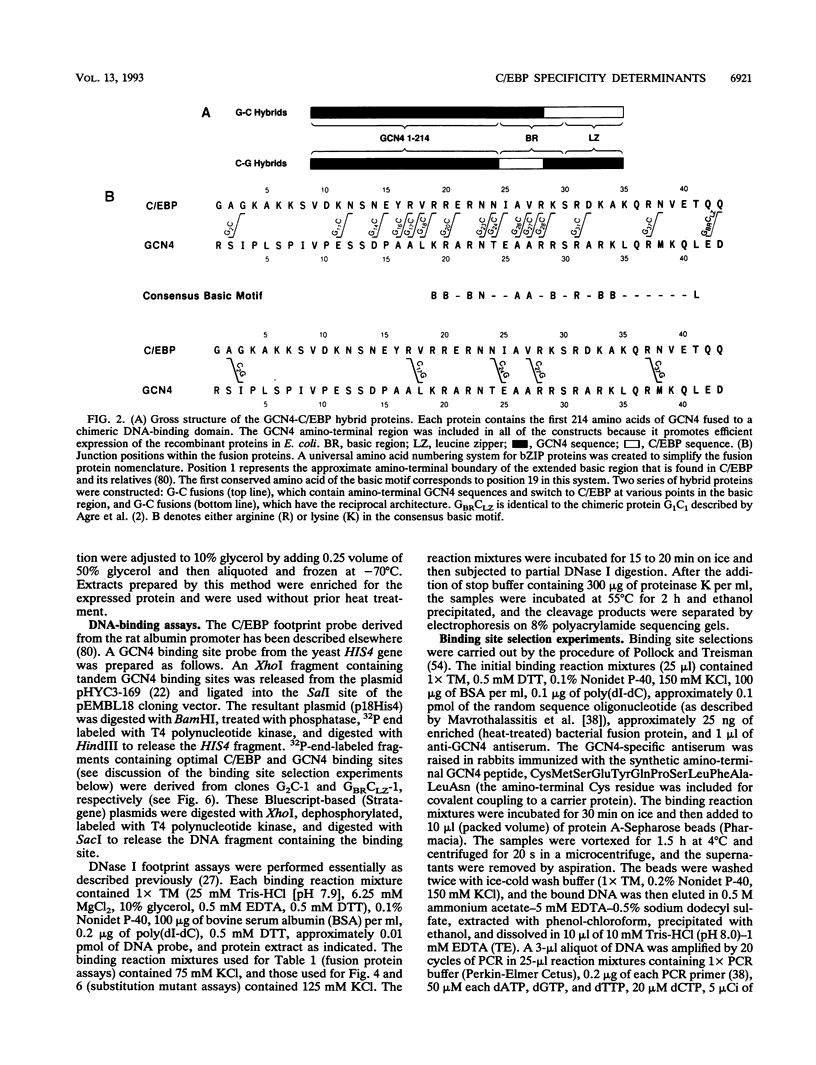
- Agre P., Johnson P. F., McKnight S. L. Cognate DNA binding specificity retained after leucine zipper exchange between GCN4 and C/EBP. Science. 1989 Nov 17;246(4932):922–926. doi: 10.1126/science.2530632. [DOI] [PubMed] [Google Scholar]
- Angel P., Imagawa M., Chiu R., Stein B., Imbra R. J., Rahmsdorf H. J., Jonat C., Herrlich P., Karin M. Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell. 1987 Jun 19;49(6):729–739. doi: 10.1016/0092-8674(87)90611-8. [DOI] [PubMed] [Google Scholar]
- Blackwell T. K., Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990 Nov 23;250(4984):1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Bohmann D., Bos T. J., Admon A., Nishimura T., Vogt P. K., Tjian R. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science. 1987 Dec 4;238(4832):1386–1392. doi: 10.1126/science.2825349. [DOI] [PubMed] [Google Scholar]
- Bohmann D., Tjian R. Biochemical analysis of transcriptional activation by Jun: differential activity of c- and v-Jun. Cell. 1989 Nov 17;59(4):709–717. doi: 10.1016/0092-8674(89)90017-2. [DOI] [PubMed] [Google Scholar]
- Cao Z., Umek R. M., McKnight S. L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991 Sep;5(9):1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- Cereghini S., Raymondjean M., Carranca A. G., Herbomel P., Yaniv M. Factors involved in control of tissue-specific expression of albumin gene. Cell. 1987 Aug 14;50(4):627–638. doi: 10.1016/0092-8674(87)90036-5. [DOI] [PubMed] [Google Scholar]
- Cohen D. R., Curran T. fra-1: a serum-inducible, cellular immediate-early gene that encodes a fos-related antigen. Mol Cell Biol. 1988 May;8(5):2063–2069. doi: 10.1128/mcb.8.5.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwarki V. J., Montminy M., Verma I. M. Both the basic region and the 'leucine zipper' domain of the cyclic AMP response element binding (CREB) protein are essential for transcriptional activation. EMBO J. 1990 Jan;9(1):225–232. doi: 10.1002/j.1460-2075.1990.tb08099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright R. H., Cossart P., Gicquel-Sanzey B., Beckwith J. Molecular basis of DNA sequence recognition by the catabolite gene activator protein: detailed inferences from three mutations that alter DNA sequence specificity. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7274–7278. doi: 10.1073/pnas.81.23.7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright R. H., Cossart P., Gicquel-Sanzey B., Beckwith J. Mutations that alter the DNA sequence specificity of the catabolite gene activator protein of E. coli. Nature. 1984 Sep 20;311(5983):232–235. doi: 10.1038/311232a0. [DOI] [PubMed] [Google Scholar]
- Ellenberger T. E., Brandl C. J., Struhl K., Harrison S. C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992 Dec 24;71(7):1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- Gaire M., Chatton B., Kedinger C. Isolation and characterization of two novel, closely related ATF cDNA clones from HeLa cells. Nucleic Acids Res. 1990 Jun 25;18(12):3467–3473. doi: 10.1093/nar/18.12.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentz R., Rauscher F. J., 3rd, Abate C., Curran T. Parallel association of Fos and Jun leucine zippers juxtaposes DNA binding domains. Science. 1989 Mar 31;243(4899):1695–1699. doi: 10.1126/science.2494702. [DOI] [PubMed] [Google Scholar]
- Guarente L., Bermingham-McDonogh O. Conservation and evolution of transcriptional mechanisms in eukaryotes. Trends Genet. 1992 Jan;8(1):27–32. doi: 10.1016/0168-9525(92)90021-u. [DOI] [PubMed] [Google Scholar]
- Guiltinan M. J., Marcotte W. R., Jr, Quatrano R. S. A plant leucine zipper protein that recognizes an abscisic acid response element. Science. 1990 Oct 12;250(4978):267–271. doi: 10.1126/science.2145628. [DOI] [PubMed] [Google Scholar]
- Hai T. W., Liu F., Coukos W. J., Green M. R. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989 Dec;3(12B):2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- Hartings H., Maddaloni M., Lazzaroni N., Di Fonzo N., Motto M., Salamini F., Thompson R. The O2 gene which regulates zein deposition in maize endosperm encodes a protein with structural homologies to transcriptional activators. EMBO J. 1989 Oct;8(10):2795–2801. doi: 10.1002/j.1460-2075.1989.tb08425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R., Krummel B., Saiki R. K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988 Aug 11;16(15):7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Lucchini G., Fink G. R. A synthetic HIS4 regulatory element confers general amino acid control on the cytochrome c gene (CYC1) of yeast. Proc Natl Acad Sci U S A. 1985 Jan;82(2):498–502. doi: 10.1073/pnas.82.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989 Apr 15;77(1):51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hoeffler J. P., Meyer T. E., Yun Y., Jameson J. L., Habener J. F. Cyclic AMP-responsive DNA-binding protein: structure based on a cloned placental cDNA. Science. 1988 Dec 9;242(4884):1430–1433. doi: 10.1126/science.2974179. [DOI] [PubMed] [Google Scholar]
- Hunger S. P., Ohyashiki K., Toyama K., Cleary M. L. Hlf, a novel hepatic bZIP protein, shows altered DNA-binding properties following fusion to E2A in t(17;19) acute lymphoblastic leukemia. Genes Dev. 1992 Sep;6(9):1608–1620. doi: 10.1101/gad.6.9.1608. [DOI] [PubMed] [Google Scholar]
- Iyer S. V., Davis D. L., Seal S. N., Burch J. B. Chicken vitellogenin gene-binding protein, a leucine zipper transcription factor that binds to an important control element in the chicken vitellogenin II promoter, is related to rat DBP. Mol Cell Biol. 1991 Oct;11(10):4863–4875. doi: 10.1128/mcb.11.10.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. F., Landschulz W. H., Graves B. J., McKnight S. L. Identification of a rat liver nuclear protein that binds to the enhancer core element of three animal viruses. Genes Dev. 1987 Apr;1(2):133–146. doi: 10.1101/gad.1.2.133. [DOI] [PubMed] [Google Scholar]
- Johnson P. F., McKnight S. L. Eukaryotic transcriptional regulatory proteins. Annu Rev Biochem. 1989;58:799–839. doi: 10.1146/annurev.bi.58.070189.004055. [DOI] [PubMed] [Google Scholar]
- Katagiri F., Lam E., Chua N. H. Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature. 1989 Aug 31;340(6236):727–730. doi: 10.1038/340727a0. [DOI] [PubMed] [Google Scholar]
- Kouzarides T., Ziff E. Leucine zippers of fos, jun and GCN4 dictate dimerization specificity and thereby control DNA binding. Nature. 1989 Aug 17;340(6234):568–571. doi: 10.1038/340568a0. [DOI] [PubMed] [Google Scholar]
- Kouzarides T., Ziff E. The role of the leucine zipper in the fos-jun interaction. Nature. 1988 Dec 15;336(6200):646–651. doi: 10.1038/336646a0. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., Adashi E. Y., Graves B. J., McKnight S. L. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988 Jul;2(7):786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science. 1989 Mar 31;243(4899):1681–1688. doi: 10.1126/science.2494700. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Lee W., Haslinger A., Karin M., Tjian R. Activation of transcription by two factors that bind promoter and enhancer sequences of the human metallothionein gene and SV40. Nature. 1987 Jan 22;325(6102):368–372. doi: 10.1038/325368a0. [DOI] [PubMed] [Google Scholar]
- Lewis M., Jeffrey A., Wang J., Ladner R., Ptashne M., Pabo C. O. Structure of the operator-binding domain of bacteriophage lambda repressor: implications for DNA recognition and gene regulation. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):435–440. doi: 10.1101/sqb.1983.047.01.051. [DOI] [PubMed] [Google Scholar]
- Lichtsteiner S., Wuarin J., Schibler U. The interplay of DNA-binding proteins on the promoter of the mouse albumin gene. Cell. 1987 Dec 24;51(6):963–973. doi: 10.1016/0092-8674(87)90583-6. [DOI] [PubMed] [Google Scholar]
- Mavrothalassitis G., Beal G., Papas T. S. Defining target sequences of DNA-binding proteins by random selection and PCR: determination of the GCN4 binding sequence repertoire. DNA Cell Biol. 1990 Dec;9(10):783–788. doi: 10.1089/dna.1990.9.783. [DOI] [PubMed] [Google Scholar]
- Moye-Rowley W. S., Harshman K. D., Parker C. S. Yeast YAP1 encodes a novel form of the jun family of transcriptional activator proteins. Genes Dev. 1989 Mar;3(3):283–292. doi: 10.1101/gad.3.3.283. [DOI] [PubMed] [Google Scholar]
- Mueller C. R., Maire P., Schibler U. DBP, a liver-enriched transcriptional activator, is expressed late in ontogeny and its tissue specificity is determined posttranscriptionally. Cell. 1990 Apr 20;61(2):279–291. doi: 10.1016/0092-8674(90)90808-r. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y., Nathans D. The basic region of Fos mediates specific DNA binding. EMBO J. 1989 Dec 1;8(12):3833–3841. doi: 10.1002/j.1460-2075.1989.tb08561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberg M., Schuermann M., Hunter J. B., Müller R. Two functionally different regions in Fos are required for the sequence-specific DNA interaction of the Fos/Jun protein complex. Nature. 1989 Apr 13;338(6216):589–590. doi: 10.1038/338589a0. [DOI] [PubMed] [Google Scholar]
- Nishina H., Sato H., Suzuki T., Sato M., Iba H. Isolation and characterization of fra-2, an additional member of the fos gene family. Proc Natl Acad Sci U S A. 1990 May;87(9):3619–3623. doi: 10.1073/pnas.87.9.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neil K. T., Hoess R. H., DeGrado W. F. Design of DNA-binding peptides based on the leucine zipper motif. Science. 1990 Aug 17;249(4970):774–778. doi: 10.1126/science.2389143. [DOI] [PubMed] [Google Scholar]
- O'Neil K. T., Shuman J. D., Ampe C., DeGrado W. F. DNA-induced increase in the alpha-helical content of C/EBP and GCN4. Biochemistry. 1991 Sep 17;30(37):9030–9034. doi: 10.1021/bi00101a017. [DOI] [PubMed] [Google Scholar]
- O'Shea E. K., Klemm J. D., Kim P. S., Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991 Oct 25;254(5031):539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- O'Shea E. K., Rutkowski R., Kim P. S. Evidence that the leucine zipper is a coiled coil. Science. 1989 Jan 27;243(4890):538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- Oakley M. G., Dervan P. B. Structural motif of the GCN4 DNA binding domain characterized by affinity cleaving. Science. 1990 May 18;248(4957):847–850. doi: 10.1126/science.2111578. [DOI] [PubMed] [Google Scholar]
- Oeda K., Salinas J., Chua N. H. A tobacco bZip transcription activator (TAF-1) binds to a G-box-like motif conserved in plant genes. EMBO J. 1991 Jul;10(7):1793–1802. doi: 10.1002/j.1460-2075.1991.tb07704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant A. R., Brandl C. J., Struhl K. Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: analysis of yeast GCN4 protein. Mol Cell Biol. 1989 Jul;9(7):2944–2949. doi: 10.1128/mcb.9.7.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- Patel L., Abate C., Curran T. Altered protein conformation on DNA binding by Fos and Jun. Nature. 1990 Oct 11;347(6293):572–575. doi: 10.1038/347572a0. [DOI] [PubMed] [Google Scholar]
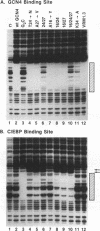
- Pollock R., Treisman R. A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res. 1990 Nov 11;18(21):6197–6204. doi: 10.1093/nar/18.21.6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu W. T., Struhl K. Highly conserved residues in the bZIP domain of yeast GCN4 are not essential for DNA binding. Mol Cell Biol. 1991 Oct;11(10):4918–4926. doi: 10.1128/mcb.11.10.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu W. T., Struhl K. The leucine zipper symmetrically positions the adjacent basic regions for specific DNA binding. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):6901–6905. doi: 10.1073/pnas.88.16.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransone L. J., Wamsley P., Morley K. L., Verma I. M. Domain swapping reveals the modular nature of Fos, Jun, and CREB proteins. Mol Cell Biol. 1990 Sep;10(9):4565–4573. doi: 10.1128/mcb.10.9.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson J. S., Richardson D. C. Amino acid preferences for specific locations at the ends of alpha helices. Science. 1988 Jun 17;240(4859):1648–1652. doi: 10.1126/science.3381086. [DOI] [PubMed] [Google Scholar]
- Roman C., Platero J. S., Shuman J., Calame K. Ig/EBP-1: a ubiquitously expressed immunoglobulin enhancer binding protein that is similar to C/EBP and heterodimerizes with C/EBP. Genes Dev. 1990 Aug;4(8):1404–1415. doi: 10.1101/gad.4.8.1404. [DOI] [PubMed] [Google Scholar]
- Ryden T. A., Beemon K. Avian retroviral long terminal repeats bind CCAAT/enhancer-binding protein. Mol Cell Biol. 1989 Mar;9(3):1155–1164. doi: 10.1128/mcb.9.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder K., Lanahan A., Perez-Albuerne E., Nathans D. jun-D: a third member of the jun gene family. Proc Natl Acad Sci U S A. 1989 Mar;86(5):1500–1503. doi: 10.1073/pnas.86.5.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder K., Lau L. F., Nathans D. A gene activated by growth factors is related to the oncogene v-jun. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1487–1491. doi: 10.1073/pnas.85.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi P., Ransone L. J., Lamph W. W., Verma I. M. Direct interaction between fos and jun nuclear oncoproteins: role of the 'leucine zipper' domain. Nature. 1988 Dec 15;336(6200):692–695. doi: 10.1038/336692a0. [DOI] [PubMed] [Google Scholar]
- Sauer R. T. Transcriptional control. Scissors and helical forks. Nature. 1990 Oct 11;347(6293):514–515. doi: 10.1038/347514b0. [DOI] [PubMed] [Google Scholar]
- Schindler U., Menkens A. E., Beckmann H., Ecker J. R., Cashmore A. R. Heterodimerization between light-regulated and ubiquitously expressed Arabidopsis GBF bZIP proteins. EMBO J. 1992 Apr;11(4):1261–1273. doi: 10.1002/j.1460-2075.1992.tb05170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers J. W., Struhl K. Changing fos oncoprotein to a jun-independent DNA binding protein with GCN4 dimerization specificity by swapping "leucine zippers". Nature. 1989 Sep 7;341(6237):74–76. doi: 10.1038/341074a0. [DOI] [PubMed] [Google Scholar]
- Sellers J. W., Vincent A. C., Struhl K. Mutations that define the optimal half-site for binding yeast GCN4 activator protein and identify an ATF/CREB-like repressor that recognizes similar DNA sites. Mol Cell Biol. 1990 Oct;10(10):5077–5086. doi: 10.1128/mcb.10.10.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman J. D., Vinson C. R., McKnight S. L. Evidence of changes in protease sensitivity and subunit exchange rate on DNA binding by C/EBP. Science. 1990 Aug 17;249(4970):771–774. doi: 10.1126/science.2202050. [DOI] [PubMed] [Google Scholar]
- Singh K., Dennis E. S., Ellis J. G., Llewellyn D. J., Tokuhisa J. G., Wahleithner J. A., Peacock W. J. OCSBF-1, a maize ocs enhancer binding factor: isolation and expression during development. Plant Cell. 1990 Sep;2(9):891–903. doi: 10.1105/tpc.2.9.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. The DNA-binding domains of the jun oncoprotein and the yeast GCN4 transcriptional activator protein are functionally homologous. Cell. 1987 Sep 11;50(6):841–846. doi: 10.1016/0092-8674(87)90511-3. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
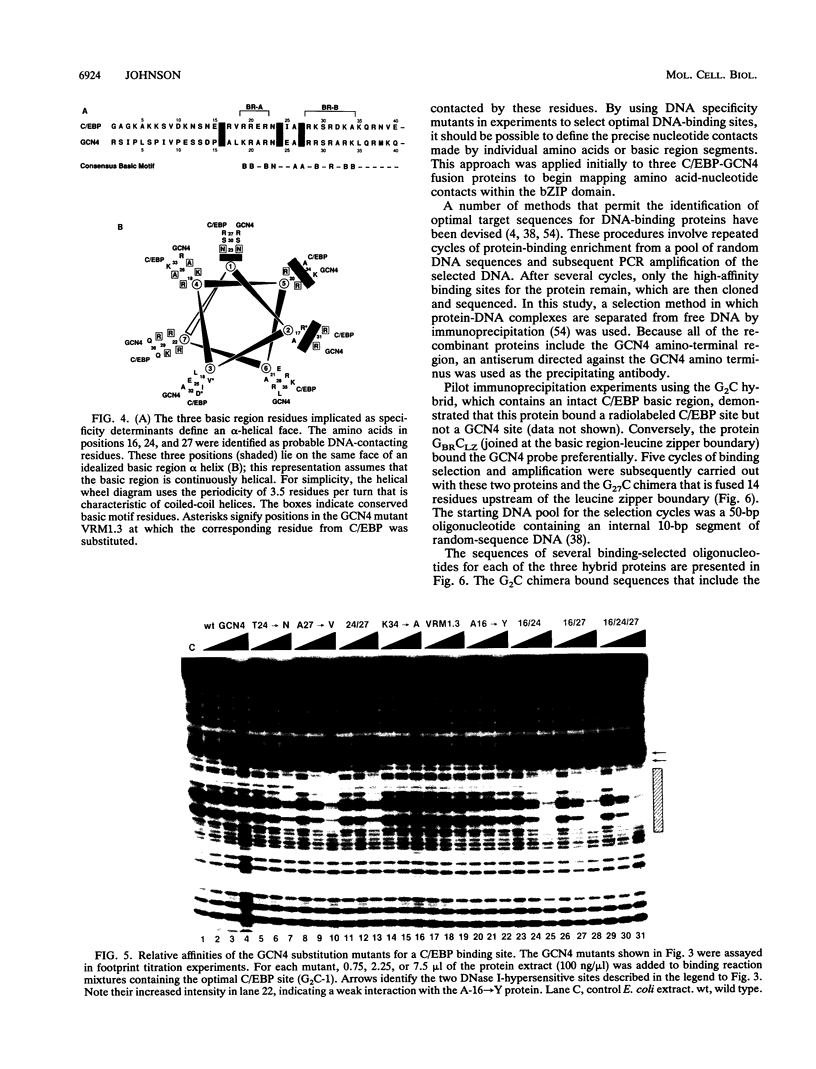
- Suckow M., von Wilcken-Bergmann B., Müller-Hill B. Identification of three residues in the basic regions of the bZIP proteins GCN4, C/EBP and TAF-1 that are involved in specific DNA binding. EMBO J. 1993 Mar;12(3):1193–1200. doi: 10.1002/j.1460-2075.1993.tb05760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata T., Nakayama T., Mikami K., Iwabuchi M. HBP-1a and HBP-1b: leucine zipper-type transcription factors of wheat. EMBO J. 1991 Jun;10(6):1459–1467. doi: 10.1002/j.1460-2075.1991.tb07666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata T., Takase H., Takayama S., Mikami K., Nakatsuka A., Kawata T., Nakayama T., Iwabuchi M. A protein that binds to a cis-acting element of wheat histone genes has a leucine zipper motif. Science. 1989 Sep 1;245(4921):965–967. doi: 10.1126/science.2772648. [DOI] [PubMed] [Google Scholar]
- Talanian R. V., McKnight C. J., Kim P. S. Sequence-specific DNA binding by a short peptide dimer. Science. 1990 Aug 17;249(4970):769–771. doi: 10.1126/science.2389142. [DOI] [PubMed] [Google Scholar]
- Turner R., Tjian R. Leucine repeats and an adjacent DNA binding domain mediate the formation of functional cFos-cJun heterodimers. Science. 1989 Mar 31;243(4899):1689–1694. doi: 10.1126/science.2494701. [DOI] [PubMed] [Google Scholar]
- Vinson C. R., Sigler P. B., McKnight S. L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989 Nov 17;246(4932):911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- Weisshaar B., Armstrong G. A., Block A., da Costa e Silva O., Hahlbrock K. Light-inducible and constitutively expressed DNA-binding proteins recognizing a plant promoter element with functional relevance in light responsiveness. EMBO J. 1991 Jul;10(7):1777–1786. doi: 10.1002/j.1460-2075.1991.tb07702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S. C., Cantwell C. A., Johnson P. F. A family of C/EBP-related proteins capable of forming covalently linked leucine zipper dimers in vitro. Genes Dev. 1991 Sep;5(9):1553–1567. doi: 10.1101/gad.5.9.1553. [DOI] [PubMed] [Google Scholar]
- Yon J., Fried M. Precise gene fusion by PCR. Nucleic Acids Res. 1989 Jun 26;17(12):4895–4895. doi: 10.1093/nar/17.12.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Straaten F., Müller R., Curran T., Van Beveren C., Verma I. M. Complete nucleotide sequence of a human c-onc gene: deduced amino acid sequence of the human c-fos protein. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3183–3187. doi: 10.1073/pnas.80.11.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]