Abstract
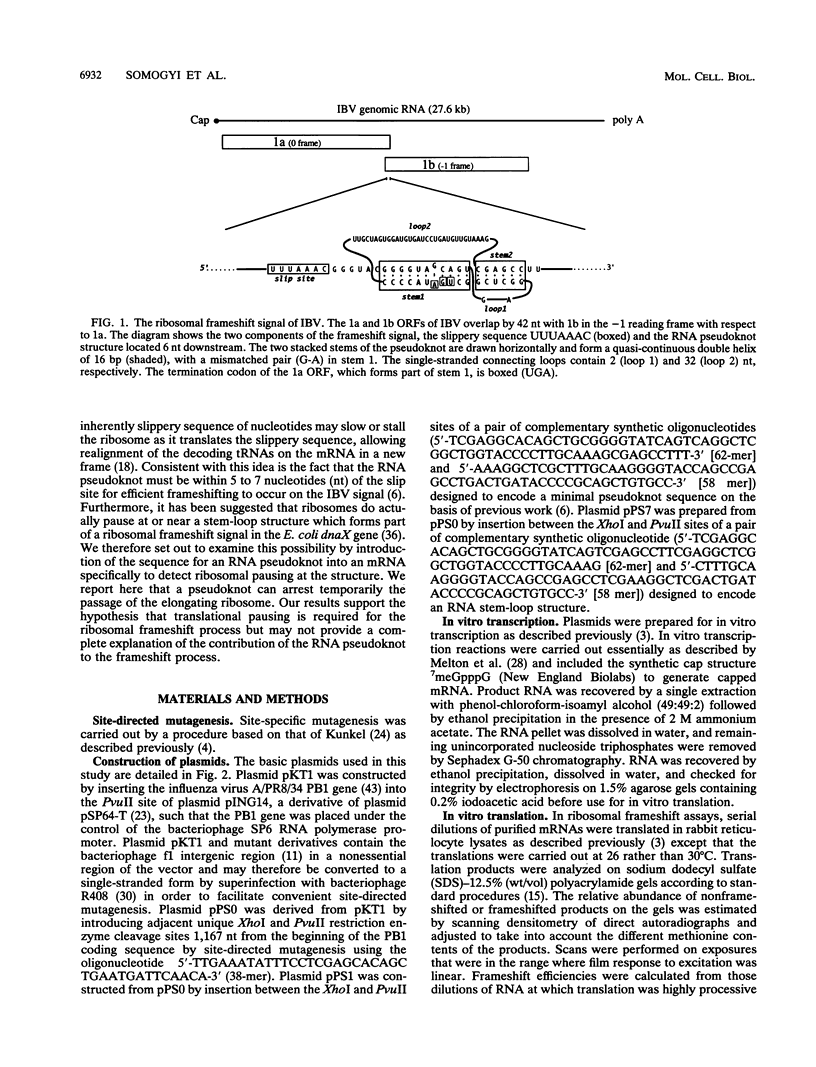
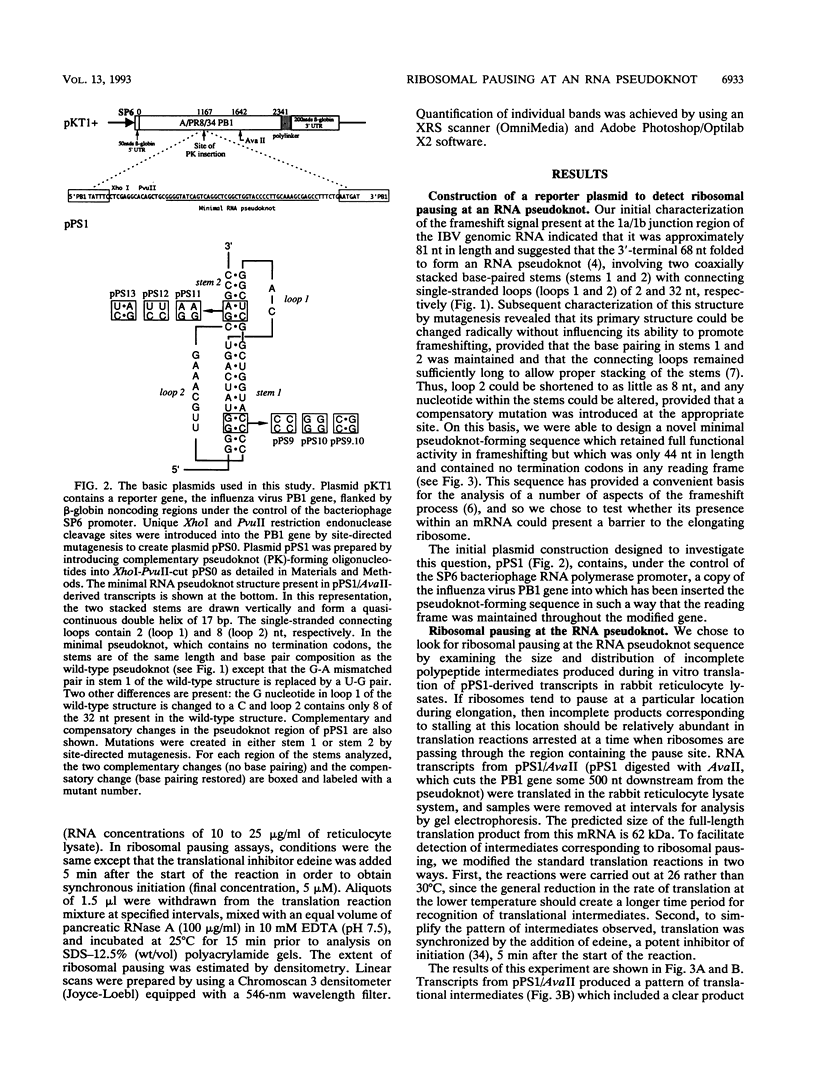
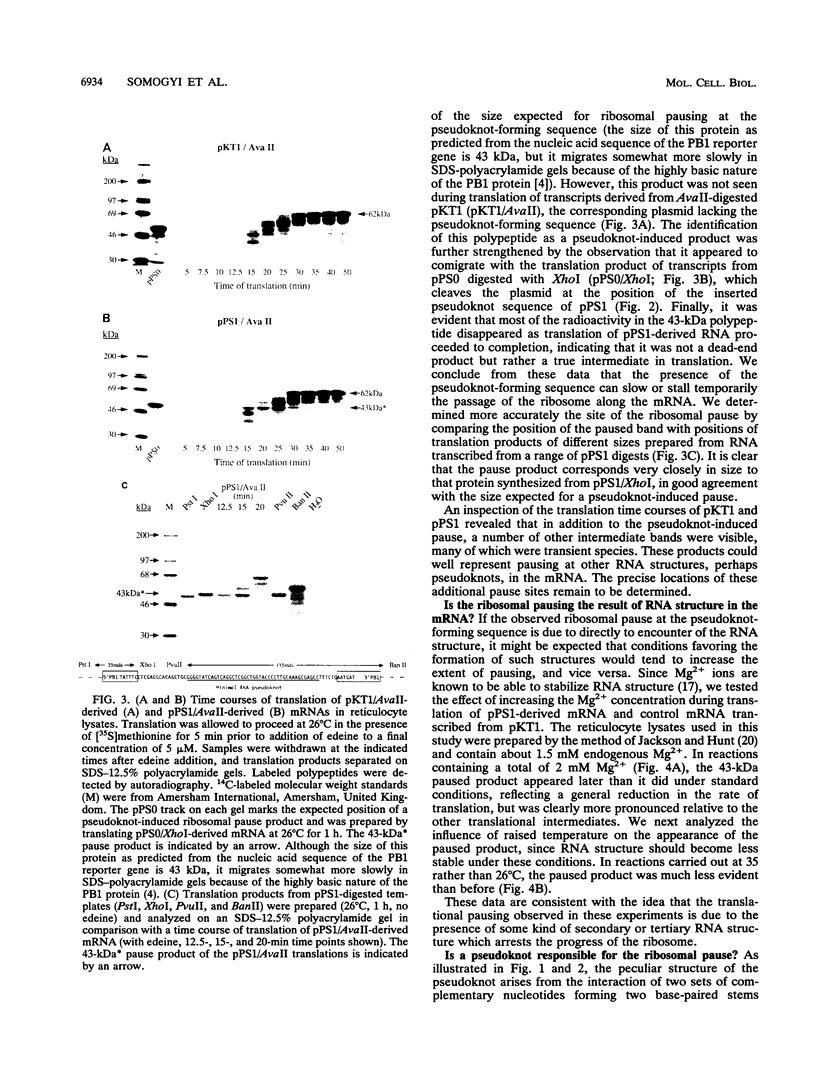
The genomic RNA of the coronavirus infectious bronchitis virus contains an efficient ribosomal frameshift signal which comprises a heptanucleotide slippery sequence followed by an RNA pseudoknot structure. The presence of the pseudoknot is essential for high-efficiency frameshifting, and it has been suggested that its function may be to slow or stall the ribosome in the vicinity of the slippery sequence. To test this possibility, we have studied translational elongation in vitro on mRNAs engineered to contain a well-defined pseudoknot-forming sequence. Insertion of the pseudoknot at a specific location within the influenza virus PB1 mRNA resulted in the production of a new translational intermediate corresponding to the size expected for ribosomal arrest at the pseudoknot. The appearance of this protein was transient, indicating that it was a true paused intermediate rather than a dead-end product, and mutational analysis confirmed that its appearance was dependent on the presence of a pseudoknot structure within the mRNA. These observations raise the possibility that a pause is required for the frameshift process. The extent of pausing at the pseudoknot was compared with that observed at a sequence designed to form a simple stem-loop structure with the same base pairs as the pseudoknot. This structure proved to be a less effective barrier to the elongating ribosome than the pseudoknot and in addition was unable to direct efficient ribosomal frameshifting, as would be expected if pausing plays an important role in frameshifting. However, the stem-loop was still able to induce significant pausing, and so this effect alone may be insufficient to account for the contribution of the pseudoknot to frameshifting.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkins J. F., Weiss R. B., Gesteland R. F. Ribosome gymnastics--degree of difficulty 9.5, style 10.0. Cell. 1990 Aug 10;62(3):413–423. doi: 10.1016/0092-8674(90)90007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenbeek P. J., Pachuk C. J., Noten A. F., Charité J., Luytjes W., Weiss S. R., Spaan W. J. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 1990 Apr 11;18(7):1825–1832. doi: 10.1093/nar/18.7.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Boursnell M. E., Binns M. M., Bilimoria B., Blok V. C., Brown T. D., Inglis S. C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 1987 Dec 1;6(12):3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S. C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989 May 19;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Jenner A. J., Inglis S. C. Mutational analysis of the "slippery-sequence" component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1992 Sep 20;227(2):463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Rolley N. J., Jenner A. J., Inglis S. C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1991 Aug 20;220(4):889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamorro M., Parkin N., Varmus H. E. An RNA pseudoknot and an optimal heptameric shift site are required for highly efficient ribosomal frameshifting on a retroviral messenger RNA. Proc Natl Acad Sci U S A. 1992 Jan 15;89(2):713–717. doi: 10.1073/pnas.89.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinman J. D., Icho T., Wickner R. B. A -1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto G. P., Enea V., Zinder N. D. Functional analysis of bacteriophage f1 intergenic region. Virology. 1981 Oct 30;114(2):463–473. doi: 10.1016/0042-6822(81)90226-9. [DOI] [PubMed] [Google Scholar]
- Feng Y. X., Yuan H., Rein A., Levin J. G. Bipartite signal for read-through suppression in murine leukemia virus mRNA: an eight-nucleotide purine-rich sequence immediately downstream of the gag termination codon followed by an RNA pseudoknot. J Virol. 1992 Aug;66(8):5127–5132. doi: 10.1128/jvi.66.8.5127-5132.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- Hatfield D. L., Levin J. G., Rein A., Oroszlan S. Translational suppression in retroviral gene expression. Adv Virus Res. 1992;41:193–239. doi: 10.1016/S0065-3527(08)60037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack A., Ladner J. E., Rhodes D., Brown R. S., Klug A. A crystallographic study of metal-binding to yeast phenylalanine transfer RNA. J Mol Biol. 1977 Apr 15;111(3):315–328. doi: 10.1016/s0022-2836(77)80054-5. [DOI] [PubMed] [Google Scholar]
- Jacks T., Madhani H. D., Masiarz F. R., Varmus H. E. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988 Nov 4;55(3):447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R. J., Howell M. T., Kaminski A. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem Sci. 1990 Dec;15(12):477–483. doi: 10.1016/0968-0004(90)90302-r. [DOI] [PubMed] [Google Scholar]
- Jackson R. J., Hunt T. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 1983;96:50–74. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- Klausner R. D., Rouault T. A., Harford J. B. Regulating the fate of mRNA: the control of cellular iron metabolism. Cell. 1993 Jan 15;72(1):19–28. doi: 10.1016/0092-8674(93)90046-s. [DOI] [PubMed] [Google Scholar]
- Kozak M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc Natl Acad Sci U S A. 1986 May;83(9):2850–2854. doi: 10.1073/pnas.83.9.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984 Sep 25;12(18):7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Shieh C. K., Gorbalenya A. E., Koonin E. V., La Monica N., Tuler J., Bagdzhadzhyan A., Lai M. M. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991 Feb;180(2):567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingelbach K., Dobberstein B. An extended RNA/RNA duplex structure within the coding region of mRNA does not block translational elongation. Nucleic Acids Res. 1988 Apr 25;16(8):3405–3414. doi: 10.1093/nar/16.8.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans R. M., Van Steeg M. H., Verlaan P. W., Pleij C. W., Bosch L. Mutational analysis of the pseudoknot in the tRNA-like structure of turnip yellow mosaic virus RNA. Aminoacylation efficiency and RNA pseudoknot stability. J Mol Biol. 1992 Jan 5;223(1):221–232. doi: 10.1016/0022-2836(92)90727-2. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa S., Bishop D. H. Identification and analysis of the gag-pol ribosomal frameshift site of feline immunodeficiency virus. Virology. 1992 Feb;186(2):389–397. doi: 10.1016/0042-6822(92)90004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russel M., Kidd S., Kelley M. R. An improved filamentous helper phage for generating single-stranded plasmid DNA. Gene. 1986;45(3):333–338. doi: 10.1016/0378-1119(86)90032-6. [DOI] [PubMed] [Google Scholar]
- Snijder E. J., den Boon J. A., Bredenbeek P. J., Horzinek M. C., Rijnbrand R., Spaan W. J. The carboxyl-terminal part of the putative Berne virus polymerase is expressed by ribosomal frameshifting and contains sequence motifs which indicate that toro- and coronaviruses are evolutionarily related. Nucleic Acids Res. 1990 Aug 11;18(15):4535–4542. doi: 10.1093/nar/18.15.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan W., Cavanagh D., Horzinek M. C. Coronaviruses: structure and genome expression. J Gen Virol. 1988 Dec;69(Pt 12):2939–2952. doi: 10.1099/0022-1317-69-12-2939. [DOI] [PubMed] [Google Scholar]
- Szer W., Kurylo-Borowska Z. Effect of edeine on aminoacyl-tRNA binding to ribosomes and its relationship to ribosomal binding sites. Biochim Biophys Acta. 1970 Dec 14;224(2):477–486. doi: 10.1016/0005-2787(70)90580-0. [DOI] [PubMed] [Google Scholar]
- Sørensen M. A., Kurland C. G., Pedersen S. Codon usage determines translation rate in Escherichia coli. J Mol Biol. 1989 May 20;207(2):365–377. doi: 10.1016/0022-2836(89)90260-x. [DOI] [PubMed] [Google Scholar]
- Tang C. K., Draper D. E. Unusual mRNA pseudoknot structure is recognized by a protein translational repressor. Cell. 1989 May 19;57(4):531–536. doi: 10.1016/0092-8674(89)90123-2. [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z. Translational frameshifting in the Escherichia coli dnaX gene in vitro. Nucleic Acids Res. 1991 May 11;19(9):2457–2462. doi: 10.1093/nar/19.9.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C., Tzeng T. H., Bruenn J. A. Ribosomal movement impeded at a pseudoknot required for frameshifting. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8636–8640. doi: 10.1073/pnas.89.18.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D. H., Sugimoto N., Freier S. M. RNA structure prediction. Annu Rev Biophys Biophys Chem. 1988;17:167–192. doi: 10.1146/annurev.bb.17.060188.001123. [DOI] [PubMed] [Google Scholar]
- Tzeng T. H., Tu C. L., Bruenn J. A. Ribosomal frameshifting requires a pseudoknot in the Saccharomyces cerevisiae double-stranded RNA virus. J Virol. 1992 Feb;66(2):999–1006. doi: 10.1128/jvi.66.2.999-1006.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin S. L., Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988 Nov;7(11):3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt J. R., Puglisi J. D., Tinoco I., Jr RNA pseudoknots. Stability and loop size requirements. J Mol Biol. 1990 Jul 20;214(2):455–470. doi: 10.1016/0022-2836(90)90193-P. [DOI] [PubMed] [Google Scholar]
- den Boon J. A., Snijder E. J., Chirnside E. D., de Vries A. A., Horzinek M. C., Spaan W. J. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J Virol. 1991 Jun;65(6):2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]