Abstract
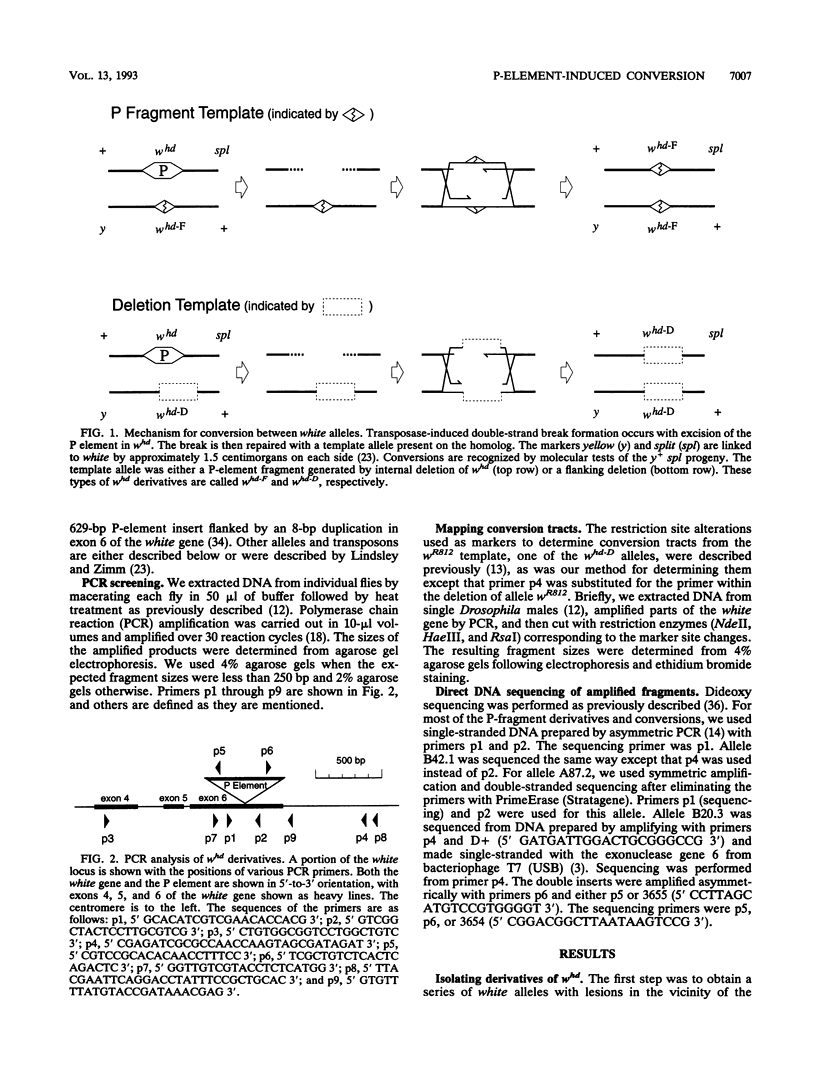
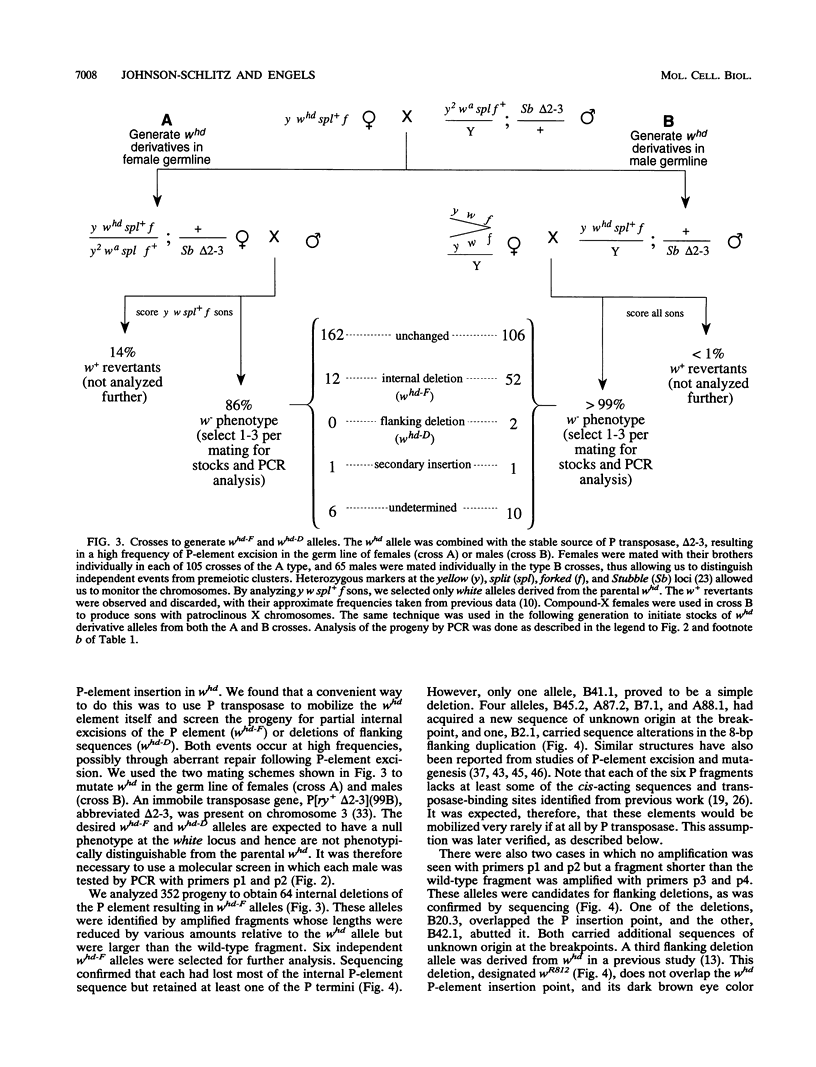
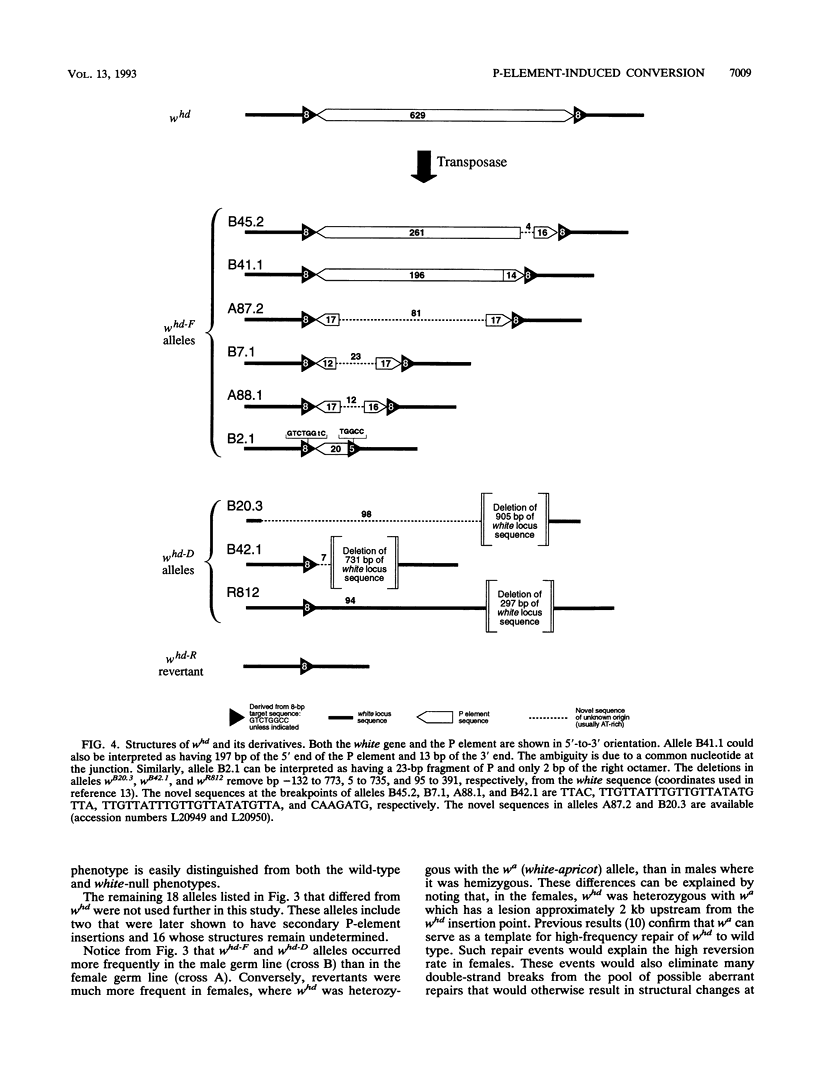
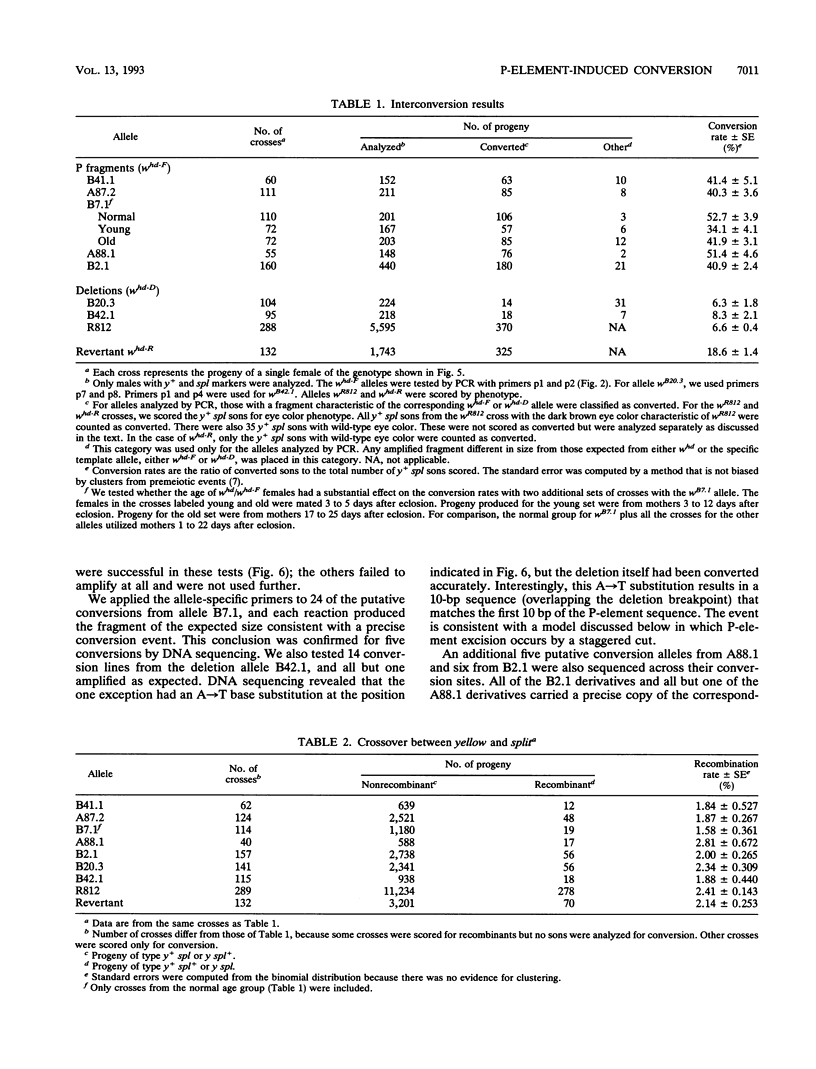
We studied the process by which whd, a P-element insertion allele of the Drosophila melanogaster white locus, is replaced by its homolog in the presence of transposase. These events are interpreted as the result of double-strand gap repair following excision of the P transposon in whd. We used a series of alleles derived from whd through P-element mobility as templates for this repair. One group of alleles, referred to collectively as whd-F, carried fragments of the P element that had lost some of the sequences needed in cis for mobility. The other group, whd-D, had lost all of the P insert and had some of the flanking DNA from white deleted. The average replacement frequencies were 43% for whd-F alleles and 7% for the whd-D alleles. Some of the former were converted at frequencies exceeding 50%. Our data suggest that the high conversion frequencies for the whd-F templates can be attributed at least in part to an elevated efficiency of repair of unexpanded gaps that is possibly caused by the closer match between whd-F sequences and the unexpanded gap endpoints. In addition, we found that the gene substitutions were almost exclusively in the direction of whd being replaced by the whd-F or whd-D allele rather than the reverse. The template alleles were usually unaltered in the process. This asymmetry implies that the conversion process is unidirectional and that the P fragments are not good substrates for P-element transposase. Our results help elucidate a highly efficient double-strand gap repair mechanism in D. melanogaster that can also be used for gene replacement procedures involving insertions and deletions. They also help explain the rapid spread of P elements in populations.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahn B. Y., Dornfeld K. J., Fagrelius T. J., Livingston D. M. Effect of limited homology on gene conversion in a Saccharomyces cerevisiae plasmid recombination system. Mol Cell Biol. 1988 Jun;8(6):2442–2448. doi: 10.1128/mcb.8.6.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banga S. S., Boyd J. B. Oligonucleotide-directed site-specific mutagenesis in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1735–1739. doi: 10.1073/pnas.89.5.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastia D., Germino J., Crosa J. H., Ram J. The nucleotide sequence surrounding the replication terminus of R6K. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2095–2099. doi: 10.1073/pnas.78.4.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels S. B., Chovnick A. P element transposition in Drosophila melanogaster: an analysis of sister-chromatid pairs and the formation of intragenic secondary insertions during meiosis. Genetics. 1993 Mar;133(3):623–636. doi: 10.1093/genetics/133.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels S. B., Peterson K. R., Strausbaugh L. D., Kidwell M. G., Chovnick A. Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics. 1990 Feb;124(2):339–355. doi: 10.1093/genetics/124.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C., Capecchi M. R. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol Cell Biol. 1992 Aug;12(8):3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels W. R., Johnson-Schlitz D. M., Eggleston W. B., Sved J. High-frequency P element loss in Drosophila is homolog dependent. Cell. 1990 Aug 10;62(3):515–525. doi: 10.1016/0092-8674(90)90016-8. [DOI] [PubMed] [Google Scholar]
- Engels W. R. The estimation of mutation rates when premeiotic events are involved. Environ Mutagen. 1979;1(1):37–43. doi: 10.1002/em.2860010110. [DOI] [PubMed] [Google Scholar]
- Engels W. R. The origin of P elements in Drosophila melanogaster. Bioessays. 1992 Oct;14(10):681–686. doi: 10.1002/bies.950141007. [DOI] [PubMed] [Google Scholar]
- Fishman-Lobell J., Haber J. E. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992 Oct 16;258(5081):480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- Gloor G. B., Nassif N. A., Johnson-Schlitz D. M., Preston C. R., Engels W. R. Targeted gene replacement in Drosophila via P element-induced gap repair. Science. 1991 Sep 6;253(5024):1110–1117. doi: 10.1126/science.1653452. [DOI] [PubMed] [Google Scholar]
- Gyllensten U. B., Erlich H. A. Generation of single-stranded DNA by the polymerase chain reaction and its application to direct sequencing of the HLA-DQA locus. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7652–7656. doi: 10.1073/pnas.85.20.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber J. E. Exploring the pathways of homologous recombination. Curr Opin Cell Biol. 1992 Jun;4(3):401–412. doi: 10.1016/0955-0674(92)90005-w. [DOI] [PubMed] [Google Scholar]
- Heslip T. R., Williams J. A., Bell J. B., Hodgetts R. B. A P element chimera containing captured genomic sequences was recovered at the vestigial locus in Drosophila following targeted transposition. Genetics. 1992 Aug;131(4):917–927. doi: 10.1093/genetics/131.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houck M. A., Clark J. B., Peterson K. R., Kidwell M. G. Possible horizontal transfer of Drosophila genes by the mite Proctolaelaps regalis. Science. 1991 Sep 6;253(5024):1125–1128. doi: 10.1126/science.1653453. [DOI] [PubMed] [Google Scholar]
- Kaufman P. D., Doll R. F., Rio D. C. Drosophila P element transposase recognizes internal P element DNA sequences. Cell. 1989 Oct 20;59(2):359–371. doi: 10.1016/0092-8674(89)90297-3. [DOI] [PubMed] [Google Scholar]
- Kaufman P. D., Rio D. C. P element transposition in vitro proceeds by a cut-and-paste mechanism and uses GTP as a cofactor. Cell. 1992 Apr 3;69(1):27–39. doi: 10.1016/0092-8674(92)90116-t. [DOI] [PubMed] [Google Scholar]
- Kidwell M. G. Evolution of hybrid dysgenesis determinants in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1655–1659. doi: 10.1073/pnas.80.6.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liskay R. M., Letsou A., Stachelek J. L. Homology requirement for efficient gene conversion between duplicated chromosomal sequences in mammalian cells. Genetics. 1987 Jan;115(1):161–167. doi: 10.1093/genetics/115.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins M. C., Rio D. C., Rubin G. M. cis-acting DNA sequence requirements for P-element transposition. Genes Dev. 1989 May;3(5):729–738. doi: 10.1101/gad.3.5.729. [DOI] [PubMed] [Google Scholar]
- Mézard C., Pompon D., Nicolas A. Recombination between similar but not identical DNA sequences during yeast transformation occurs within short stretches of identity. Cell. 1992 Aug 21;70(4):659–670. doi: 10.1016/0092-8674(92)90434-e. [DOI] [PubMed] [Google Scholar]
- Nassif N., Engels W. DNA homology requirements for mitotic gap repair in Drosophila. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1262–1266. doi: 10.1073/pnas.90.4.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brochta D. A., Gomez S. P., Handler A. M. P element excision in Drosophila melanogaster and related drosophilids. Mol Gen Genet. 1991 Mar;225(3):387–394. doi: 10.1007/BF00261678. [DOI] [PubMed] [Google Scholar]
- O'Hare K., Rubin G. M. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell. 1983 Aug;34(1):25–35. doi: 10.1016/0092-8674(83)90133-2. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W. Fungal recombination. Microbiol Rev. 1985 Mar;49(1):33–58. doi: 10.1128/mr.49.1.33-58.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H., Groenen J. T. Targeted alterations of the Caenorhabditis elegans genome by transgene instructed DNA double strand break repair following Tc1 excision. EMBO J. 1992 Jan;11(1):287–290. doi: 10.1002/j.1460-2075.1992.tb05051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick M. A. The repair of double-strand breaks in DNA; a model involving recombination. J Theor Biol. 1976 Jun;59(1):97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- Robertson H. M., Preston C. R., Phillis R. W., Johnson-Schlitz D. M., Benz W. K., Engels W. R. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988 Mar;118(3):461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M., Kidwell M. G., Bingham P. M. The molecular basis of P-M hybrid dysgenesis: the nature of induced mutations. Cell. 1982 Jul;29(3):987–994. doi: 10.1016/0092-8674(82)90462-7. [DOI] [PubMed] [Google Scholar]
- Rubnitz J., Subramani S. The minimum amount of homology required for homologous recombination in mammalian cells. Mol Cell Biol. 1984 Nov;4(11):2253–2258. doi: 10.1128/mcb.4.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searles L. L., Greenleaf A. L., Kemp W. E., Voelker R. A. Sites of P element insertion and structures of P element deletions in the 5' region of Drosophila melanogaster RpII215. Mol Cell Biol. 1986 Oct;6(10):3312–3319. doi: 10.1128/mcb.6.10.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentry J. W., Kaiser K. P element transposition and targeted manipulation of the Drosophila genome. Trends Genet. 1992 Oct;8(10):329–331. doi: 10.1016/0168-9525(92)90267-8. [DOI] [PubMed] [Google Scholar]
- Shen P., Huang H. V. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics. 1986 Mar;112(3):441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer B. S., Gold L., Gauss P., Doherty D. H. Determination of the amount of homology required for recombination in bacteriophage T4. Cell. 1982 Nov;31(1):25–33. doi: 10.1016/0092-8674(82)90401-9. [DOI] [PubMed] [Google Scholar]
- Sugawara N., Haber J. E. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992 Feb;12(2):563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Takasu-Ishikawa E., Yoshihara M., Hotta Y. Extra sequences found at P element excision sites in Drosophila melanogaster. Mol Gen Genet. 1992 Mar;232(1):17–23. doi: 10.1007/BF00299132. [DOI] [PubMed] [Google Scholar]
- Thomas K. R., Deng C., Capecchi M. R. High-fidelity gene targeting in embryonic stem cells by using sequence replacement vectors. Mol Cell Biol. 1992 Jul;12(7):2919–2923. doi: 10.1128/mcb.12.7.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubota S. I., Huong D. V. Capture of flanking DNA by a P element in Drosophila melanogaster: creation of a transposable element. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):693–697. doi: 10.1073/pnas.88.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubota S., Ashburner M., Schedl P. P-element-induced control mutations at the r gene of Drosophila melanogaster. Mol Cell Biol. 1985 Oct;5(10):2567–2574. doi: 10.1128/mcb.5.10.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubota S., Schedl P. Hybrid dysgenesis-induced revertants of insertions at the 5' end of the rudimentary gene in Drosophila melanogaster: transposon-induced control mutations. Genetics. 1986 Sep;114(1):165–182. doi: 10.1093/genetics/114.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman A. S., Liskay R. M. Differential effects of base-pair mismatch on intrachromosomal versus extrachromosomal recombination in mouse cells. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5340–5344. doi: 10.1073/pnas.84.15.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt V. M., Ingles C. J., Urdea M. S., Rutter W. J. Homology requirements for recombination in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4768–4772. doi: 10.1073/pnas.82.14.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley M., Noguchi P. D., Sensabaugh S. M., Odenwald W. F., Kassis J. A. The Drosophila gene escargot encodes a zinc finger motif found in snail-related genes. Mech Dev. 1992 Feb;36(3):117–127. doi: 10.1016/0925-4773(92)90063-p. [DOI] [PubMed] [Google Scholar]



