Abstract
Neural progenitor cells (NPCs) have the capacity to proliferate and give rise to all major central nervous system cell types and represent a possible cell of origin in gliomagenesis. Deletion of the tumor suppressor gene Tp53 (p53) results in increased proliferation and self-renewal of NPCs and is a common genetic mutation found in glioma. We have identified Inhibitor of DNA Binding 2 (Id2) as a novel target gene directly repressed by p53 to maintain normal NPC proliferation. p53(-/-) NPCs express elevated levels of Id2 and suppression of Id2 expression is sufficient to inhibit the increased proliferation and self-renewal which results from p53 loss. Elevated expression of Id2 in wild type NPCs phenocopies the behavior of p53(-/-) NPCs by enhancing NPC proliferation and self-renewal. Interestingly, p53 directly binds to a conserved site within the Id2 promoter to mediate these effects. Finally, we have identified elevated ID2 expression in glioma cell lines with mutated P53 and demonstrated that constitutive expression of ID2 plays a key role in the proliferation of glioma stem-like cells. These findings indicate that Id2 functions as a pro-proliferative gene that antagonizes p53 mediated cell cycle regulation in NPCs and may contribute to the malignant proliferation of glioma-derived tumor stem cells.
Keywords: Genes, p53; Genes, Id2; Stem cell; Cell Proliferation
Introduction
Neural progenitor cells (NPCs) residing in the subventricular zone (SVZ) of adult mammals have the ability to give rise to all three major central nervous system (CNS) lineages throughout adulthood1, 2. The hypothesis that NPCs have the potential to give rise to brain tumors is underscored by the fact that brain tumors, especially gliomas, are often made up of tumor cells with a variety of characteristics representing multiple cell lineages3, 4. Importantly, tumor suppressor genes frequently found to be altered in human glioma, including PTEN, RB1, NF1 and TP53 (P53), function to regulate normal NPC homeostasis in mice5-9. The multipotentiality of NPCs and their ability to continually divide throughout the life span of the organism2 suggests the possibility that NPCs may be the cell of origin in glioma, and a number of mouse models of glioma provide strong evidence for this possibility7, 10, 11. In several mouse models of glioma, activation of the initiating oncogenic mutation results in altered differentiation, hyperplastic growth and enhanced self-renewal within NPCs in the SVZ7, 8, 10. However, very little is known about how genetic lesions common in brain tumors could drive the oncogenic transformation of multipotent adult NPCs.
We have studied the effect of TP53 (P53) pathway disruption in NPCs. P53 inactivation is thought to be an early event in gliomagenesis12, and is estimated to occur in 87% of such tumors13. Inherited germline mutations within the P53 gene, as seen in patients with Li-Fraumeni Syndrome, results in an increased incidence of glioma with an early onset14. Deletion of p53 in mice results in numerous pre-neoplastic phenotypes in NPCs including enhanced self-renewal, accelerated growth and an expanded SVZ8, 9 suggesting that P53 functions not only as a tumor suppressor, but also as an essential NPC regulatory gene. Targeted inactivation of the p53 DNA binding domain in cells expressing glial fibrillary acidic protein (GFAP+ cells) is sufficient to initiate glioma arising in SVZ progenitor cells, although many other GFAP+ cell types, including astrocytes, contain the initiating p53 mutation11. These studies support the notion that gliomas frequently arise from SVZ NPCs which have lost P53 function. Understanding how P53 functions to regulate the biology of NPCs should identify critical NPC regulatory pathways that when deregulated could promote brain tumorigenesis.
Id genes are known to promote many key aspects of NPC biology including proliferation and self-renewal15-17, two cellular processes critical for tumor growth. Four ID genes (ID1-ID4) exist within the mammalian genome and are thought to exert their function most often by hetero-dimerizing with basic helix-loop-helix (bHLH) transcription factors through the helix-loop-helix domain18, 19. ID proteins lack a basic DNA binding domain and, as a result, function as dominant-negative inhibitors of bHLH DNA binding. Most frequently this results in the inhibition of differentiation15, 20 and promotion of cell cycle progression15, 21. Recently our laboratory has identified a dramatic decrease in adult neurogenesis and NPC proliferation after targeted deletion of Id215, which appears to be critical for embryonic NPC growth and self-renewal17.
We have discovered a molecular mechanism that mediates, at least in part, p53 dependent hyperplasia of NPCs8, 9. We identified elevated expression of Id2 in p53(-/-) NPCs and demonstrated that Id2 expression alone is sufficient for maintaining the increased growth and self-renewal of p53(-/-) NPCs. We also found evolutionarily conserved p53 binding sites within the Id2 promoter and established that p53 binding to one of these directly represses Id2 transcription. Finally, we have identified elevated ID2 expression in glioma cell lines with mutant P53 and demonstrated that constitutive expression of ID2 plays a key role in the proliferation of glioma derived stem-like cells. These data indicate that p53 functions to regulate both NPC and glioma stem-like cell proliferation and self-renewal by directly modulating Id2, a known regulator of proliferation in NPCs15. Our data support a model by which loss of p53 function, common in the majority of gliomas, drives cell cycle transit by increasing Id2 expression thus potentiating the malignant transformation of NPCs.
Materials and Methods
Tissue Harvest and Culture
Primary NPCs were isolated from P0 mouse forebrains and cultured in DMEM/F12 containing 10ng/mL EGF and B27 as previously described22. To prepare glioma stem-like cells, tumor cell lines were replated in NPC proliferation media without EGF.
Proliferation and Apoptosis Analyses
2×104 NPCs growing in 24 well plates were counted using trypan blue exclusion, except for the experiment shown in Fig.1A, in which 5000 cells were plated. To determine cell cycle distribution NPCs stained with propidium iodide (PI) were examined by flow cytometry using Modfit. Carboxyfluorescein diacetate succinimidyl ester (CFSE, Invitrogen) labeling was performed as described previously23. Apoptosis was quantified by live cell flow cytometry using Annexin-V/PI (BD Biosciences) and analyzed using FlowJo. TUNEL staining was performed after plating on laminin-coated chamber slides.
Figure 1.
Aberrant Id2 expression observed in p53(-/-) NPCs. (A): Growth curve demonstrating the proliferation of NPCs after the loss of p53. Average of two experiments plated in triplicate, *p<0.05 (B): Cell cycle distribution of WT and p53(-/-) NPCs. Total of 3 individual experiments, *p<0.05 (C): Western blot of Id2 from WT and p53(-/-) NPCs. (D): Quantitative PCR for Id2 in WT and p53(-/-) NPCs. Average of three individual experiments plated in triplicate, *p<0.05. (E) Western blot for p53 after 2 Gy of irradiation. (F): Western blot for Id2 in WT and p53(-/-) NPCs after 2 Gy of irradiation. All error bars represent SEM. Abbreviations: WT, wild type. h, hour. Pre-Rad, before irradiation.
Cell Irradiation
NPCs were dissociated, plated in proliferation media for at least 24 hours, and treated with the indicated amounts of gamma radiation at 10 Gy/min in a 137Cs Irradiator.
Immunocytochemistry
To differentiate NPCs, cells were plated on laminin coated chamber slides and placed in DMEM/F12 containing L-glutamine, 10% fetal bovine serum and B27(with vitamin A, Invitrogen) for 14 days and immunostained with the indicated antibodies. For cytospins, 5×104 dissociated NPCs were centrifuged onto poly-L-lysine slides at 600 RPM and immunostained with mouse anti-Ki-67 antibody (Novacastra), detected by fluorescence, and quantified.
DNA constructs
DNA corresponding to the human ID2 promoter was purchased from Switchgear Genomics (Menlo Park, CA). Mutagenesis of Id2 luciferase reporters was performed using the Stratagene Quik Change kit. Each of the two hID2-luc constructs deleting different binding sites also lacked a 10bp region of high G/C content 57bp upstream from the human ID2 transcriptional start site. pBMN-ID2 encoded an N-terminal Flag-tagged human ID2 cDNA. All constructs were verified by DNA sequencing.
Luciferase Assays
Luciferase reporter constructs, 800ng, were cotransfected with 20ng of pRL-TK Renilla and 200ng of pMAX-GFP into 293T cells using LipoD293 (SignaGen). Following 24 hours of incubation, cells were infected with recombinant adenovirus encoding LacZ or P53. Luciferase activity was assayed using Dual-Luciferase Assay (Promega).
Chromatin Immunoprecipitation
Chromatin was prepared from NPCs fixed with 1% formaldehyde, sheared by sonication (average 500bp), immunoprecipated with anti-p53 antibody (5ug, FL-393, Santa-Cruz) and processed using a chromatin immuno-precipitation kit (Millipore). PCR primer sequences included: p21(Cdkn1a): 5′CCAGAGGATACCTTGCAAGGC(sense), 5′TCTCTGTCTCCATTCATGCTCTCC(anti-sense). p53 site 1: 5′TCGCATCACTTTGCCACCTACACT(sense), 5′CGAATTGTAGGAACACTGTGCGGT(anti-sense). p53 site 2: 5′TGCTCCAAGTTGCAAAGCTTCACG(sense), 5′GGCAAATTGAGTACAGTGTGCGCT(anti-sense). p53 site 3: 5′ACCAATGGGAGAATTCGCCTGGTA(sense), 5′ACTGAAGGCTTTCATGCTGCTCGT(anti-sense).
Quantitative PCR
RNA was reverse-transcribed using iScript (Bio-Rad). Controls were incubated without reverse transcriptase. cDNA was quantified using MyiQ Sybr Green qPCR (Bio-Rad). Fold change was determined after normalization to β-actin (ΔΔ-ct). Primer sequences included: Mouse-Id2: 5′CCGCTGACCACCCTGAAC(sense), 5′CATTCGACATAAGCTCAGAATGGAATT(anti-sense). Human-Id2: 5′TCAGCCTGCATCACCAGAGA(sense), 5′CTGCAAGGACAGGATGCTGAT(anti-sense). p21(Cdkn1a): 5′GCAGATCCACAGCGATATCCA(sense), 5′GGTCGGACATCACCAGGATT(anti-sense).
Western Blotting
Lysates (100ug) of NPCs were analyzed by western blot using anti-Id2 1:500 (Santa Cruz, sc-489) or anti-Id2 1:500 (Cell-Signaling Technologies, D39E8), anti-p53 1:1000 (Santa-Cruz, sc-6243), anti-p21 1:500 (Santa-Cruz, sc-6246), anti-total PARP 1:2000 (Cell Signaling Technologies, 9542), or anti-β-actin 1:5000 (Sigma, A-1978) antibodies. For densitometry, images were quantified by relative pixel area using Photoshop.
NCI-60 microarray meta analysis
Microarray gene expression values24 were downloaded using the NCI cell miner webpage (http://discover.nci.nih.gov/cellminer), and the accompanying P53 mutational sequence data was obtained. Average gene expression was evaluated using a one-sided T test.
Experimental Animals
Experiments were performed using cells isolated from Trp53Tm-Tyj (p53(-/-)) C57BL/6 mice25. The Id2CAG-FID2 mouse was created within our laboratory (our unpublished data).
Results
Elevated and aberrant expression of Id2 in p53(-/-) NPCs
Multipotent NPCs can be cultured as neurospheres in vitro and give rise to all three CNS lineages upon differentiation (Supplemental Fig.1). Our laboratory and others have observed that targeted deletion of the tumor suppressor p53 is associated with increased proliferation of NPCs (Fig.1A)8, 9. The molecular mechanisms mediating the increased growth of p53(-/-) NPCs are largely unknown. Given the importance of p53 inactivation in brain tumor pathogenesis and the potential for NPCs to give rise to such tumors11, we sought to better understand the effect of p53 loss on NPC proliferation. We determined that p53 deletion altered the cell cycle distribution of NPCs in culture. p53(-/-) NPCs exhibited a significant decrease in the fraction of cells in the G0/G1 stage of the cell cycle and an increase in the percentage of cells in S-phase compared to WT NPCs (Fig.1B). This supports a role for p53 in restraining the proliferation of NPCs and is consistent with the observations of p53 dependent regulation of adult stem cells from neuronal8 and mammary tissues26.
Our laboratory has previously identified a dramatic decrease in the proliferation of Id2(-/-) NPCs cultured as neurospheres in vitro15. We noticed that the increased growth rate and percentage of cells in S phase in p53(-/-) NPCs contrasted sharply with the decreased proliferation of Id2(-/-) NPCs (unpublished data and reference 15). This led us to hypothesize that both Id2 and p53 may function to regulate NPC proliferation by modulating the G1-S transition. To investigate if altered Id2 expression occurs in p53(-/-) NPCs, we examined steady state Id2 levels in WT and p53(-/-) NPCs cultured as neurospheres. Western blot analysis revealed a substantial increase in Id2 protein expression in p53(-/-) neurospheres (Fig.1C) along with a significant increase in Id2 mRNA in p53(-/-) NPCs detected by quantitative PCR (Fig.1D). The increase in Id2 mRNA we observed under steady state conditions in p53(-/-) NPCs suggested that p53, either directly or indirectly, could modulate Id2 mRNA levels. To further investigate the extent which p53 might repress Id2, we treated WT and p53(-/-) NPCs with 2 Gy of gamma irradiation to increase p53 protein expression and transcriptional activity (Fig.1E) (for review see27). We observed a decrease in Id2 protein 4-12 hours after treatment with gamma irradiation (Fig.1F), which corresponds to the kinetics of p53 activation (Fig.1E). In contrast, p53(-/-) NPCs express elevated levels of Id2, and following irradiation and DNA damage, p53(-/-) NPCs fail to demonstrate decreased Id2. Taken together these data support a model in which p53 functions to inhibit Id2 expression and p53 loss leads to inappropriately high levels of Id2 expression.
To evaluate the possibility that Id2 expression is diminished because of loss of cells by apoptosis after DNA damage, we quantified the number of apoptotic cells following irradiation (2Gy) and observed that this dose does not induce apoptosis in WT or p53(-/-) NPCs (Supplemental Fig.2). To examine further the effect of p53 on Id2 expression, we introduced exogenous WT P53 into p53(-/-) NPCs using an adenovirus-derived vector. Adenoviral introduction of P53 suppressed Id2 protein to undetectable levels (Supplemental Fig.3) and is consistent with a role for p53 in inhibiting Id2 expression in NPCs.
Inhibition of Id2 expression is sufficient to prevent the hyperplastic growth and the enhanced self-renewal of p53(-/-) NPCs
We hypothesized that Id2 was a key mediator of the accelerated proliferation of NPCs which occurs after loss of p538, 9. To test this hypothesis, we inhibited the expression of Id2 in p53(-/-) NPCs by retroviral infection of shRNAs targeting Id2 (Origene, HuSH system) and examined the proliferation of these NPCs. Western blot analysis of neurosphere cell cultures stably expressing Id2 shRNAs (p53(-/-)/sh-Id2) demonstrated decreased expression of Id2 compared to levels detected in cells infected with a scrambled, non-specific control shRNA (p53(-/-)/sh-Scramble, Fig.2A). Early passage p53(-/-)/sh-Id2 NPCs routinely formed significantly smaller neurospheres than p53(-/-)/sh-Scramble NPCs (Fig.2B), suggesting a decrease in proliferation after inhibition of Id2. To characterize the observed decrease in proliferation, p53(-/-)/sh-Scramble and p53(-/-)/sh-Id2 neurospheres were dissociated, fixed to glass slides, and immunostained using an antibody against the proliferation marker Ki6728. We observed a significant decrease in the frequency of Ki67+ cells in p53(-/-)/sh-Id2 NPCs compared to control cells (Fig.2C). To further quantify the alterations in growth kinetics following Id2 inhibition in p53(-/-) NPCs, we examined early passage p53(-/-)/sh-Id2 NPCs in a growth assay. After one week of growth, each independent p53(-/-)/sh-Id2 culture grew significantly slower than control p53(-/-)/sh-Scramble NPCs (Fig.2D). This decreased level of proliferation was indistinguishable from the proliferation rate observed in WT NPCs. The inhibition of Id2 expression did not alter the expression of the cyclin-dependant kinase inhibitor, p21, in either p53(-/-)/sh-Id2 or Id2(-/-) NPCs (Supplemental Fig.4). Similarly we did not observe changes in the percentage of cells positive for nestin expression (Supplemental Fig.5). To determine if decreased Id2 expression alters the differentiation of p53(-/-) NPCs, we differentiated p53(-/-)/sh-Scramble and p53(-/-)/sh-Id2 cultures and found no indication that Id2 levels affected the capacity of these cells to form neurons, astrocytes or oligodendrocytes (Supplemental Fig.6).
Figure 2.
Reduction of Id2 expression is sufficient to inhibit the enhanced proliferation of p53(-/-) NPCs. (A): Id2 western blot after infection with Id2 shRNAs and stable selection. (B): Representative phase contrast micrograph and quantification of p53(-/-)/shId2, control, and WT neurospheres two days after passage, scale bar=100 um, 4× magnification, *p<0.001 (C): Ki67 staining of control (left panel) and p53(-/-)/shId2 (center panel) dissociated NPCs, 10× magnification. (Right panel) Comparison of Ki67+ cells in p53(-/-)/shScramble and p53(-/-)/shId2 NPCs. Average of three cytospins, 50,000 cells per slide, counted in triplicate, *p<0.05. (D): Growth curve of WT, p53(-/-)/shScramble and two independent p53(-/-)/shId2 NPC cultures. Representative experiment plated in triplicate, *p<0.01 for each p53(-/-)/shId2 culture vs. p53(-/-)/shScramble. WT vs p53(-/-)/shId2 cultures p>0.05 at day 7. (E): Secondary sphere formation assay. Average of three independent experiments plated in duplicate, *p<0.01. (F) Id2 western blot 48 hours following retroviral infection. (G) TUNEL stain and quantification. 20× magnification, scale bar =25 um. Average of two experiments plated in duplicate, *p<0.05. All error bars represent SEM. Abbreviations: WT, wild type.
The ability of NPCs to self-renew, as measured in an assay for secondary spheres, is critical to the expansion of neurospheres in vitro and evidence suggests that loss of p53 increases the rate of NPC self-renewal8. Recently, Id proteins were implicated in regulating the self-renewal rate of NPCs17. To determine if Id2 can affect the self-renewal of p53(-/-) NPCs, we plated p53(-/-)/sh-Id2 and p53(-/-)/sh-Scramble NPCs in secondary sphere formation assays. NPCs were dissociated and plated at a limiting dilution of approximately 1 cell per well in 96 well plates to determine the frequency with which individual cells would re-form neurospheres. We observed that suppression of Id2 expression reduced the self-renewal rate of p53(-/-) NPCs to levels comparable to WT (Fig.2E).
We next evaluated the effect of Id2 inhibition on the rate of p53(-/-) apoptosis. Infection of Id2 shRNAs with recombinant retrovirus resulted in decreased Id2 levels but inhibition was not below the levels observed in WT NPCs (Fig.2F). Decreased expression of Id2 resulted in a significant increase in the frequency of TUNEL positive cells 48 hours after treatment (Fig.2G). These data suggest that Id2 expression, increased as the result of p53 inactivation, contributes to maintain enhanced growth and increased self-renewal of p53(-/-) NPCs as a result of both increased proliferation and decreased apoptosis.
Elevated expression of ID2 is sufficient to increase the proliferation and self-renewal of WT NPCs
The identification of Id2 as a critical mediator of p53 dependent NPC proliferation implies that Id2 functions to modulate cell cycle progression. Increased expression of Id2 in cells which possess WT p53 should phenocopy the hyperplastic growth and enhanced self-renewal of p53(-/-) NPCs described by others8, 9. To examine this possibility, we used a transgenic mouse from our laboratory in which GFP tagged human ID2 was placed under the conditional regulation of the chicken actin gene (CAG) promoter (unpublished data). Under basal conditions, the CAG promoter transcribes the chloramphenicol acetyltransferase (CAT) gene which is flanked by loxP sites (Fig.3A). Upon Cre mediated recombination, the floxed CAT gene is excised from the genome placing the ID2 transgene downstream of the constitutively active CAG promoter (Id2CAG-FID2).
Figure 3.
Generation and characterization of Id2CAG-FID2 transgenic NPCs. (A): Genetic map of the Id2CAG-FID2 transgene, bold left pointing arrows are LoxP sites, small arrows indicate primer locations. (B): Genomic recombination of the transgenic allele after Cre recombination. PCR of genomic DNA from neurospheres after infection with Cre adenovirus using the indicated primers. (C): Dose dependent activation of the Id2CAG-FID2 transgene by Ad-Cre. Total Id2 expression was quantified by qPCR (top). Corresponding Cre expression as determined by semi-quantitative RT-PCR (bottom). (D): Western blot for Id2 in Id2CAG-ID2 NPCs following recombinant adenovirus infection. Abbreviations: CAG, chicken actin gene promoter; CAT, chloramphenicol acetyl-transferase. pA; polyadenylation signal; hId2, human Id2; GFP, green fluorescent protein; AG2, actin gene primer 2; CAT2, chloramphenicol acetyl-transferase primer 2; CAT3 chloramphenicol acetyl-transferase primer 3; Id2R, human Id2 reverse primer; Ad-LacZ, beta galactosidase expressing adenovirus; Ad-Cre, Cre recombinase expressing adenovirus.
To prepare NPCs that overexpressed ID2, we established neurosphere cultures from the brains of P0 Id2CAG-FID2 mice. Dispersed neurospheres were infected with adenovirus expressing either beta-galactosidase or Cre recombinase (Ad-LacZ or Ad-Cre respectively). To determine if genomic recombination occurred at the Id2CAG-FID2 locus after Ad-Cre infection, we used primers which specifically amplify the recombined allele by priming from the CAG promoter and the ID2 gene (primers AG2 and ID2R, Fig.3A) or which amplify the CAT gene (CAT2 and CAT3 primers). Genomic PCR analysis of adenoviral-infected neurospheres indicated that transgene recombination occurred only in NPCs which received Ad-Cre (Fig.3B). However the persistence of CAT gene PCR products in Ad-Cre infected cultures indicated that recombination was not 100% efficient and suggests that these cultures also contained cells which have not undergone recombination and do not express the ID2 transgene (Fig.3B). Next we performed quantitative PCR for total Id2 transcript in Id2CAG-FID2 infected cells and semi-quantitative RT-PCR for Cre expression. We observed a dose dependent increase in total Id2 mRNA expression dependent on the amount of Ad-Cre virus used for infection (Fig.3C) and western blot analysis of Id2 from Id2CAG-FID2/Ad-Cre neurospheres demonstrated an enhanced expression of ID2 (Fig.3D) at the predicted weight, 42 kDa (molecular weight ID2: 15kDa, GFP: 27 kDa).
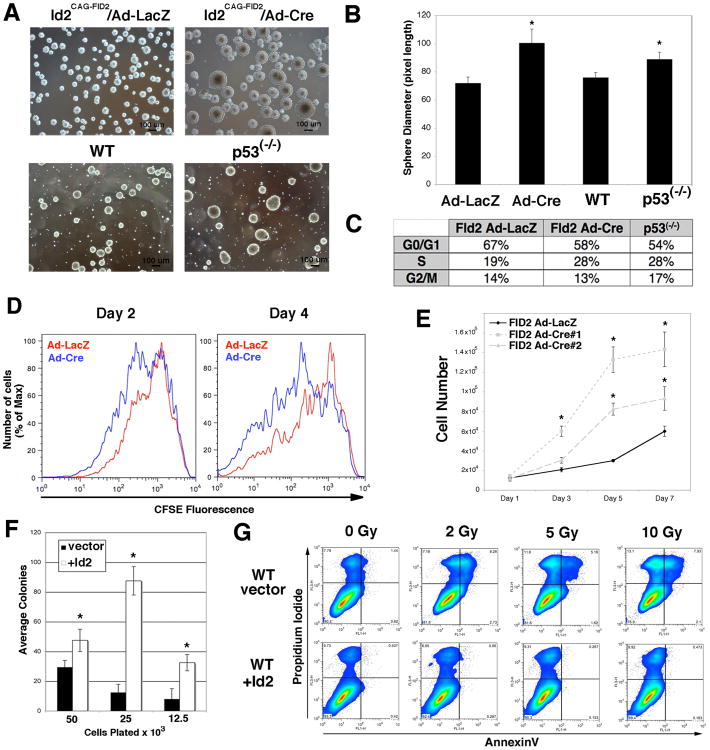
We noted that Id2CAG-FID2/Ad-Cre NPCs routinely formed larger neurospheres than control Ad-LacZ cells with a diameter similar to p53(-/-) NPCs (Fig.4A&B). We therefore examined the cell cycle distribution of Id2CAG-FID2 NPCs, and found that proliferating Id2CAG-FID2/Ad-Cre NPCs mimic the cell cycle distribution of p53(-/-) NPCs (Fig.4C). These data indicate that both Id2 gain of function and p53 loss of function result in NPCs with similar cell cycle distributions. To more rigorously evaluate the growth of Id2CAG-FID2 NPCs, we employed the use of CFSE staining. We identified a decrease in CFSE fluorescence intensity in Id2CAG-FID2/Ad-Cre infected cells at 2 and 4 days post labeling (Fig.4D), suggesting that enhanced expression of ID2 is sufficient to increase the proliferation of NPCs expressing p53 (median fluorescence at day 4: Id2CAG-FID2/Ad-LacZ=474, Id2CAG-FID2/Ad-Cre=169).
Figure 4.
Enhanced expression of ID2 increases the growth and self-renewal of NPCs. (A): Phase contrast micrograph of Id2CAG-FID2/Ad-LacZ, Id2CAG-FID2/Ad-Cre, WT and p53(-/-) neurospheres 2 days after plating, 4× magnification. (B): Quantification of the diameter of neurosphere cultures, *p<0.05. (C): Cell cycle distributions of NPCs growing in culture determined by propidium iodide staining. Representative experiment. (D) CFSE fluorescence histograms from Id2CAG-FID2/Ad-Cre NPCs. Representative histograms from three individual experiments. (E): Growth curve of control Id2CAG-FID2/Ad-LacZ and two independent Id2CAG-FID2/Ad-Cre NPC cultures, *p<0.05. (F): Secondary sphere formation of Id2 expressing NPCs immobilized in soft agar. Representative experiment plated in triplicate *p<0.05. (G) Evaluation of apoptosis by AnnexinV/PI flow cytometry after irradiation, pseudo-color density plot. Abbreviations: WT, wild type. Ad-LacZ, Beta-galactosidase adenovirus. Ad-Cre, Cre adenovirus. Gy, Gray.
To further examine alterations in cellular proliferation, we assayed the proliferation of Id2CAG-FID2/Ad-Cre NPCs and observed that Id2CAG-FID2/Ad-Cre NPCs proliferated more rapidly than Id2CAG-FID2/Ad-LacZ cells (Fig.4E). However increased Id2 expression did not alter the expression of p21 (Supplemental Fig.7). To examine the effect of Id2 on NPC self-renewal, we infected WT NPCs with retrovirus encoding flag-tagged human ID2. Following infection of NPCs with recombinant retrovirus, we used antibiotic selection to prepare polyclonal cultures expressing elevated levels of ID2 (Supplemental Fig.8). After two weeks of growth as single cells suspended in soft agar29, 30, we observed a significantly increased number of secondary neurospheres formed from NPCs expressing ID2 (Fig.4F). We interpret these results to indicate that ID2 can function to increase NPC self-renewal.
We next investigated the effect of Id2 expression on apoptosis 24 hours after radiation (2-10Gy) induced DNA damage by AnnexinV/PI flow cytometry. Under these conditions, ID2 expression resulted in decreased propidium iodide-positive and AnnexinV/propidium iodide double positive cells (Fig.4G). These data suggest that Id2 expression may function to inhibit programmed cell death and are consistent with the observation that inhibition of Id2 expression results in increased apoptosis in p53(-/-) NPCs (Fig.2G).
Binding of P53 to the Id2 promoter is required for P53 repression
Recently, multiple mechanisms of p53 mediated transcriptional repression have been characterized31. To determine if p53 could physically associate with the Id2 promoter, we first identified regions of the Id2 promoter evolutionarily conserved between mouse and human by pair-wise sequence analysis 1 kb upstream from the transcriptional start site using VISTA32 (http://genome.lbl.gov/vista/index.shtml, Fig.5A top). We identified three potential p53 binding sites which occurred in evolutionarily conserved regions shared by the human and mouse promoter regions (Fig.5A, center). These binding sites contain deviations of no more than 3 bp from the 20 nucleotide consensus p53 binding motif (RRRCWWGYYY(0-13 bp spacer)RRRCWWGYYY)33 and these deviations are highly conserved between species (Fig.5A, bottom). The sequences of each of the p53 binding sites contain nucleotide spacers between the decameric half sites (Fig.5A, bottom). The presence of nucleotide spacer regions occur almost exclusively within binding sites of p53 target genes that are transcriptionally repressed by p5334-36, supporting our observations that p53 functions as a transcriptional repressor of Id2.
Figure 5.
p53 interacts directly with the Id2 promoter. (A): Pairwise sequence analysis and identification of potential p53 binding sites in the mouse and human Id2 promoters using VISTA. All Id2 promoter and gene regions with greater than 70% conservation between species are shaded within the plot as determined using a 100bp window. Numbers represent distance from the Id2 transcriptional start site. (B): Chromatin immuno-precipitation for p53 interaction with the Id2 promoter. P21 promoter was used as a positive control. P53 ChIP from p53(-/-) extracts were used as a negative control. Representative image from three independent experiments. (C): Schematic of the human ID2 promoter driven luciferase reporter obtained from Switchgear Genomics (Menlo Park, CA). hID2-luc contained from -704 bp to +353 bp of the ID2 promoter. Numbers indicated are in relation to the Id2 transcriptional start site. (D): Effect of dose dependent expression of P53 on ID2 promoter driven luciferase and p53 binding site mutant constructs. Average of two experiments plated in triplicate, *p<0.05. Error bars represent SEM. Abbreviations: kb, kilobase. UTR, untranslated region. Ad-P53, P53 expressing adenovirus. WT, wild type. IP, immuno-precipitation. Ab, antibody.
To determine if p53 can physically associate with these three p53 binding sites, we performed p53 chromatin immuno-precipitation (ChIP). Using primers flanking each of the three p53 binding sites to probe for the presence of the corresponding DNA fragments, we found that Id2 promoter DNA corresponding to p53 site 3 immuno-precipitated with p53, but not to sites 1 or 2 (Fig.5B). We verified the specificity of this immuno-precipitation by examining a known p53 binding site in the promoter of the p53 target gene p2137, as well as immunoprecipitating from p53(-/-) extracts. These data indicated that p53 can physically interact with the Id2 promoter under normal proliferative conditions in vitro.
To determine if the p53 binding sites located in the Id2 promoter are required for p53-mediated repression, we utilized a commercially available human ID2 luciferase construct (hID2-luc, SwitchGear Genomics) that contained p53 binding sites 2 and 3 (Fig.5C). To assess the effect of P53 on the hID2-luc reporter we transfected 293T cells with the hID2-luc reporter and titrated in increasing amounts of Ad-P53. To avoid the potentially confounding effects of apoptosis, we assayed 293T cells for luciferase activity 16 hours post infection before any evidence of cell death was discernable visually and PARP cleavage was minimal38 (unpublished data). We observed that increasing amounts of P53 expression resulted in a dose dependent decrease of hID2-luc reporter expression (Fig.5D). This result indicates that P53 can act as a transcriptional repressor of ID2 and that this repression occurs within the cloned promoter region containing p53 sites 2 and 3. We then created additional luciferase reporter constructs in which individual sites were ablated using site directed mutagenesis. We began these experiments by deleting either site 2 (hID2-lucΔ site 2) or site 3 (hID2-lucΔ site 3). Addition of Ad-P53 virus led to the repression of hID2-lucΔ site 2 (Fig.5D) indicating that p53 site 2 does not function to repress ID2 expression. In contrast, hID2-lucΔ site 3 did not demonstrate P53 mediated repression (Fig.5D) thereby identifying a putative conserved site within the Id2 promoter that functions to repress transcription in a p53 dependent manner. The identification of endogenous p53 physically associated with binding site 3 by ChIP, coupled with the observation that deletion of p53 site 3 abolished ID2 repression (Fig.5D), provides strong evidence that p53 binding site 3 is a critical cis-acting element mediating p53 repression of ID2 expression.
Identification of ID2 as a bona fide target of p53 repression in human cancer cell lines
The loss of P53 function occurs commonly in a multitude of tumor types27. This fact prompted us to determine if WT P53 repressed ID2 in human cancer cell lines. We sought to determine if high levels of ID2 expression correlated with the loss of P53 function and reduced ID2 expression with WT P53. We therefore conducted a meta-analysis of previously compiled microarray data from the NCI-60 panel of cell lines evaluating gene expression changes after DNA damage, a condition that is known to activate P53 activity (Fig.1E)24, 27. We stratified the NCI-60 panel into two groups based on P53 mutational status. This assignment was predictive of elevated expression of the established P53 response gene, P21, after irradiation (Fig.6A). Next we evaluated changes in ID2 expression, and the only other ID family member, ID3, reported in this dataset. We discovered a significant decrease in ID2 gene expression after irradiation only in tumors that retained WT P53 and could not identify P53 dependent repression of ID3 expression after DNA damage (Fig.6A). Based on these data we conclude P53 repression of ID2 is likely to occur in human cancer cell lines bearing a WT P53 gene and this repression was not seen in all members of the Id gene family.
Figure 6.
Characterization of ID2 as a bona fide P53 repression target in human cancer cell lines. (A): Meta-analysis of microarray gene expression profiling of the NCI-60 panel of human cancer cell lines after induction of DNA damage by irradiation. Left: examination of the expression of the canonical P53 target gene, P21, after DNA damage induced p53 activation. Center: examination of the expression of ID2 after DNA damage induced P53 activation. Right: examination of the expression of ID3, another member of the ID gene family, after DNA damage induced p53 activation. *p<0.05. (B): Survey of ID2 expression in human glioma cell lines by Western blot. Induction of P21 expression after DNA damage was used as a control for P53 function. All error bars represent SEM. Abbreviations: WT, wild type; MT, mutant.
Based on the similarities between NPCs and glioma cells39 we chose to evaluate further the relationship between ID2 and P53 within human glioma cell lines. We selected four well-characterized glioma lines. One of these, U87, has WT P53 and three are P53 mutant lines: U251, SW1088 and A17240. U251 was amongst the six glioma cell lines in the NCI-60 panel. To verify WT P53 functionality, we treated each cell line with 2Gy gamma irradiation and screened for P21 induction. As expected, the P53 WT U87 glioma line expresses P21 after DNA damage, while P21 expression was not induced in cell lines with mutated P53 (Figure 6B). Consistent with our previous findings, we were unable to detect ID2 in the P53 WT U87 cell line, but we did find high levels of ID2 in each of the three P53 mutant glioma lines (Figure 6B). This suggests that WT P53 can repress ID2 in glioma-derived tumor cell lines, but mutant P53 cannot.
Increased expression of ID2 enhances the proliferation of glioma stem cells
Our current experiments demonstrate that P53 can function to repress ID2 in some human tumor cell lines (Fig.6) and glioma cell lines with mutated P53 express higher levels of ID2 than the P53 WT U87 glioma line (Fig.6B). Therefore we tested whether ID2 over-expression could promote the growth of glioma cells in a manner similar to that observed in p53(-/-) NPCs (Figs. 2 & 4). Glioma cell lines cultured in defined media known to support the growth of NPCs become enriched for cells with characteristics of tumor stem cells41 and mirror phenotypes of primary tumors39, 42. We therefore employed this culture method to study the effects of ID2 expression in glioma stem-like cells. We prepared U251 glioma cells that expressed elevated levels of ID2 using recombinant retrovirus encoding ID2 (U251 +Id2, Fig.7A). We observed larger sphere size and increased growth of U251 +Id2 glioma stem-like cells compared to U251 vector control cells (Figs.7B&C). Similar to our observations in NPCs, increased expression of ID2 in glioma stem-like cells results in an increase in proliferation, suggesting that ID2 is a key driver of cell cycle progression in both normal and transformed cells of the CNS.
Figure 7.

Increased expression of ID2 enhances the proliferation of glioma stem-like cells. (A): Western blot for ID2 in stable U251 glioma stem-like cells. (B): Phase contrast micrographs of control and ID2 expressing glioma stem-like cells propagated in NPC media. 4× magnification. (C): Growth curve of control and ID2 expressing glioma stem-like cells. Representative experiment plated in triplicate, *p<0.05. Error bars represent SEM.
Discussion
Identification of the molecular basis of neural stem and progenitor cell homeostasis is of critical importance for understanding normal CNS development and the potential of these cells to initiate tumorigenesis7, 10, 11. P53 is known to control the size of the neural stem cell compartment in vivo under normal physiologic conditions8. We have discovered that p53 directly represses Id2 and modulates NPC proliferation. This previously unrecognized regulatory activity implicates Id2 as a key functional element mediating p53 function in NPCs. While Id2 has been well established to promote cell cycle progression, our data suggests that Id2 can also function to inhibit apoptosis in NPCs.
We have evaluated in NPCs previously established mechanisms of Id2 mediated growth promotion through antagonism of the p21 or Rb pathways43, 44. Although displaying pronounced alterations in proliferation, neither p53(-/-)/shId2 nor Id2CAG-Cre/Ad-Cre NPCs exhibit changes in p21 expression (Supplemental Figs.4&7). Similarly we have not observed decreased expression of E2F1 target genes including Ccnd1, Cdc6 and Cdc25a in Id2(-/-) NPCs as would be predicted if increased Rb activity was responsible for the Id2(-/-) NPC growth defect (unpublished data). This could imply that a novel and previously unrecognized mechanism exists for Id2 in promoting the growth of NPCs.
Our findings demonstrate that p53 functions by repressing Id2 through binding to the conserved p53 binding site 3 within the Id2 promoter (Fig.5). The context in which p53 inhibits target gene expression can be determined, in part, by the composition of the p53 response element itself31, 45. While the exact mechanism of p53 repression of Id2 has not been determined, the structure of the p53 response elements within the Id2 promoter suggests the recruitment of chromatin remodeling complexes, rather than steric hindrance through binding site overlap, as a putative mechanism35. Two key characteristics found in the majority of p53 repressed target genes that distinguish these from sites of activation are: (i) The presence of a spacer region between the two decameric half sites34, 36, 46, and (ii) substitutions within the core “CWWG” motif45. Both of these attributes occur in the p53 binding site 3 present within the Id2 promoter and are highly conserved between mouse and human. Substitutions in the core motif may affect DNA flexibility and tetrameric p53 binding resulting in conformational changes that may contribute to differential cofactor recruitment45, 47. Further elucidation of the mechanisms determining p53-mediated repression, including defining the genetic elements required for this repression, will lead to an enhanced understanding of the coordinated gene expression required for p53 pathway function.
Our observation of increased Id2 expression in p53 deficient NPCs (Fig.1) may also be of significance for cancer, since inactivation of the P53 pathway has been reported to occur in 87% of glioblastomas13, tumors which may arise in NPCs11. Because many biological characteristics and molecular pathways are shared by normal NPCs and glioma stem-like cells, the identification of key effector genes downstream of p53 in both of these cell types is of great interest. We found that ID2 was highly expressed in glioma-derived cell lines that lacked a functional P53 (Fig. 6), and ID2 can drive the proliferation of glioma-derived tumor stem cells (Fig. 7). These findings complement our observation of the importance of Id2 expression in NPCs and suggest that ID2 is a critical effector gene driving the proliferation of P53 deficient brain tumor stem-like cells and thereby contributing to oncogenesis (Fig.7).
WT P53 has not previously been recognized as a regulator of ID2, and has not been examined in normal tissue. Fontemaggi et. al.48, have reported that mutant P53 can activate ID2 expression in the colon cancer cell line, SW480, and in comparable experiments using this same cell line, Yan et al.49 have reported repression of ID2 by mutant p53, but not the wildtype allele. Importantly, these authors did not evaluate ID2 expression in reporter assays which contained our newly identified p53 binding site 3. The finding of ID2 activation by mutant P53 is consistent with our observation that WT P53 is associated with low levels of ID2 expression in the diverse NCI-60 panel of cancer cell lines (Fig.6A), including cell lines derived from colon tumors. We specifically examined two glioma lines containing hotspot mutations in the same amino acid, R273, as was examined by Fontemaggi et. al.48 and found that these exhibited high levels of ID2 expression (Fig.6B). Importantly, these mutations are known to be gain-of-function mutations50 suggesting that some mutations within the P53 gene may elevate ID2 expression by converting P53 from a transcriptional repressor to an activator.
Finally, a recent molecular sub-classification of gliomas from The Cancer Genome Atlas Project has associated P53 mutations most closely with gene expression signatures similar to oligodendrocyte progenitor cells51. Many cellular processes altered in P53 inactivated tumors strikingly coincide with Id2 mediated processes including the promotion of NPC proliferation (this publication,15, 17, 52) and inhibition of oligodendrocyte maturation53, 54. These studies and our data strongly suggest a role for Id2 in regulating the malignant characteristics of P53 deficient glioma stem-like cells. The identification of key molecular changes that drive NPC and tumor stem cell growth, such as enhanced Id2 expression, have the potential to inform future targeted therapies.
Summary
p53 directly represses Id2 transcription to inhibit the proliferation of NPCs and glioma stem-like cells.
Supplementary Material
Supplemental Figure 1: NPCs cultured as neurospheres are multipotent upon differentiation. NPC cultures give rise to GFAP+ astrocytes, Tuj1+ neurons and O4+ oligodendrocytes 14 days after the addition of serum (20× magnification for GFAP and Tuj1, 10× for O4). Representative experiment.
Supplemental Figure 2: 2 Gy irradiation of NPCs does not increase the amount of apoptosis (12 hours after treatment). AnnexinV/PI flow cytometery for apoptosis after irradiation of WT and p53(-/-) NPCs. Representative experiment. Abbreviations: Gy, gray.
Supplemental Figure 3: Expression of exogenous P53 in p53(-/-) NPCs represses Id2. Western blot for p53 and Id2 in adenoviral infected p53(-/-) NPCs. Lysates were collected 16 hours post infection. Abbreviations: Ad-LacZ, beta-galactosidase expressing adenovirus. Ad-p53, P53 expressing adenovirus.
Supplemental Figure 4: Inhibition of Id2 does not alter p21 expression. (A) Quantitative PCR analysis of Id2 and p21 expression in p53(-/-) NPCs after Id2 inhibition. Average of cDNAs from three individual sh-Id2 cultures plated in triplicate. (B) p21 western blot of lysates from WT and Id2-/- NPCs. Representative image of results from three independent experiments. (C) Quantification of p21 western analysis by densitometry, n=3. Error bars represent standard deviation.
Supplemental Figure 5: Inhibition of Id2 expression does not alter the percentage of nestin+ NPCs. p53(-/-)/shScramble and p53(-/-)/sh-Id2 NPCs immunostained with anti-nestin antibody. 10× magnification. Representative images from two experiments plated in duplicate.
Supplemental Figure 6: Id2 inhibition does not affect the differentiation capacity of p53(-/-) NPCs. (A) Immunocytochemistry of p53(-/-)/shScramble and p53(-/-)/sh-Id2 NPCs after 14 days of differentiation for markers of astroglial, neuronal and oligodendroglial lineages. Representative images from two biological replicates. 10× magnification. (B) Quantification of the percentages of cell types formed in (A).
Supplemental Figure 7: Increased expression of ID2 in NPCs does not alter p21 expression. Quantitative PCR for Id2 and p21 expression in Id2CAG-FID2/Ad-LacZ and Id2CAG-FID2/Ad-Cre after recombinant adenoviral infection. Average of three experiments plated in triplicate.
Supplemental Figure 8: Stable retroviral expression of flag-tagged ID2 in NPCs. Id2 western blot from stable polyclonal cultures. Id2(-/-) NPC lysate included as a control.
Acknowledgments
We would like to thank Sarah Gilman for expert handing of all animals used in this study. We also would like to acknowledge the help of Dr. Jiang Gui with statistical analysis.
Research support: This study was supported by an NIH fellowship F31NS064634 (BRP), the Theodora B. Betz Foundation (MAI) and the Jordon and Kyra Memorial Foundation (MAI).
Footnotes
Disclosures of Potential Conflicts of Interest: The authors have no potential conflicts of interest.
Author contribution summary: Brenton R. Paolella: Conception and design, Financial support, Collection of data, Data analysis and interpretation, Manuscript writing.
Matthew C. Havrda: Conception and design, Collection of data, Data analysis and interpretation, Manuscript writing.
Akio Mantani: Provision of study material.
Christina M. Wray: Collection of Data.
Zhonghua Zhang: Provision of study material.
Mark A. Israel: Conception and design, Financial support, Data analysis and interpretation, Manuscript writing, Final approval of manuscript.
Publisher's Disclaimer: Disclaimers: none
References
- 1.Lois C, Garcia-Verdugo JM, Alvarez-Buylla A. Chain migration of neuronal precursors. Science. 1996;271:978–981. doi: 10.1126/science.271.5251.978. [DOI] [PubMed] [Google Scholar]
- 2.Doetsch F, Caille I, Lim DA, et al. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 3.Chen R, Nishimura MC, Bumbaca SM, et al. A Hierarchy of Self-Renewing Tumor-Initiating Cell Types in Glioblastoma. Cancer Cell. 2011;17:362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 4.Pollard SM, Yoshikawa K, Clarke ID, et al. Glioma Stem Cell Lines Expanded in Adherent Culture Have Tumor-Specific Phenotypes and Are Suitable for Chemical and Genetic Screens. Cell Stem Cell. 2009;4:568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 5.Zheng H, Ying H, Yan H, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferguson KL, Callaghan SM, O'Hare MJ, et al. The Rb-CDK4/6 signaling pathway is critical in neural precursor cell cycle regulation. J Biol Chem. 2000;275:33593–33600. doi: 10.1074/jbc.M004879200. [DOI] [PubMed] [Google Scholar]
- 7.Alcantarallaguno S, Chen J, Kwon C, et al. Malignant Astrocytomas Originate from Neural Stem/Progenitor Cells in a Somatic Tumor Suppressor Mouse Model. Cancer Cell. 2009;15:45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gil-Perotin S. Loss of p53 Induces Changes in the Behavior of Subventricular Zone Cells: Implication for the Genesis of Glial Tumors. Journal of Neuroscience. 2006;26:1107–1116. doi: 10.1523/JNEUROSCI.3970-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meletis K. p53 suppresses the self-renewal of adult neural stem cells. Development. 2005;133:363–369. doi: 10.1242/dev.02208. [DOI] [PubMed] [Google Scholar]
- 10.Dai C. PDGF autocrine stimulation dedifferentiates cultured astrocytes and induces oligodendrogliomas and oligoastrocytomas from neural progenitors and astrocytes in vivo. Genes & Development. 2001;15:1913–1925. doi: 10.1101/gad.903001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Yang J, Zheng H, et al. Expression of Mutant p53 Proteins Implicates a Lineage Relationship between Neural Stem Cells and Malignant Astrocytic Glioma in a Murine Model. Cancer Cell. 2009;15:514–526. doi: 10.1016/j.ccr.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hesselager G, Uhrbom L, Westermark B, et al. Complementary effects of platelet-derived growth factor autocrine stimulation and p53 or Ink4a-Arf deletion in a mouse glioma model. Cancer Research. 2003;63:4305–4309. [PubMed] [Google Scholar]
- 13.The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li FP, Fraumeni JF, Jr, Mulvihill JJ, et al. A cancer family syndrome in twenty-four kindreds. Cancer Res. 1988;48:5358–5362. [PubMed] [Google Scholar]
- 15.Havrda MC, Harris BT, Mantani A, et al. Id2 Is Required for Specification of Dopaminergic Neurons during Adult Olfactory Neurogenesis. Journal of Neuroscience. 2008;28:14074–14087. doi: 10.1523/JNEUROSCI.3188-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yun K. Id4 regulates neural progenitor proliferation and differentiation in vivo. Development. 2004;131:5441–5448. doi: 10.1242/dev.01430. [DOI] [PubMed] [Google Scholar]
- 17.Jung S, Park R, Kim S, et al. Id proteins facilitate self renewal and proliferation of neural stem cells. Stem Cells Dev. 2009 doi: 10.1089/scd.2009.0093. [DOI] [PubMed] [Google Scholar]
- 18.Benezra R, Davis RL, Lockshon D, et al. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-y. [DOI] [PubMed] [Google Scholar]
- 19.Jogi A, Persson P, Grynfeld A, et al. Modulation of basic helix-loop-helix transcription complex formation by Id proteins during neuronal differentiation. J Biol Chem. 2002;277:9118–9126. doi: 10.1074/jbc.M107713200. [DOI] [PubMed] [Google Scholar]
- 20.Cai L, Morrow EM, Cepko CL. Misexpression of basic helix-loop-helix genes in the murine cerebral cortex affects cell fate choices and neuronal survival. Development. 2000;127:3021–3030. doi: 10.1242/dev.127.14.3021. [DOI] [PubMed] [Google Scholar]
- 21.Iavarone A, Garg P, Lasorella A, et al. The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev. 1994;8:1270–1284. doi: 10.1101/gad.8.11.1270. [DOI] [PubMed] [Google Scholar]
- 22.Reynolds BA, Weiss S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev Biol. 1996;175:1–13. doi: 10.1006/dbio.1996.0090. [DOI] [PubMed] [Google Scholar]
- 23.Groszer M, Erickson R, Scripture-Adams DD, et al. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- 24.Amundson SA, Do KT, Vinikoor LC, et al. Integrating Global Gene Expression and Radiation Survival Parameters across the 60 Cell Lines of the National Cancer Institute Anticancer Drug Screen. Cancer Research. 2008;68:415–424. doi: 10.1158/0008-5472.CAN-07-2120. [DOI] [PubMed] [Google Scholar]
- 25.Jacks T, Remington L, Williams BO, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 26.Cicalese A, Bonizzi G, Pasi CE, et al. The Tumor Suppressor p53 Regulates Polarity of Self-Renewing Divisions in Mammary Stem Cells. Cell. 2009;138:1083–1095. doi: 10.1016/j.cell.2009.06.048. [DOI] [PubMed] [Google Scholar]
- 27.Meek DW. Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer. 2009;9:714–723. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 28.Gerdes J, Schwab U, Lemke H, et al. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. International journal of cancer. 1983;31:13–20. doi: 10.1002/ijc.2910310104. [DOI] [PubMed] [Google Scholar]
- 29.Penuelas S, Anido J, Prieto-Sanchez RM, et al. TGF-beta increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell. 2009;15:315–327. doi: 10.1016/j.ccr.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 30.Sun P, Xia S, Lal B, et al. DNER, an epigenetically modulated gene, regulates glioblastoma-derived neurosphere cell differentiation and tumor propagation. Stem Cells. 2009;27:1473–1486. doi: 10.1002/stem.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang B, Xiao Z, Ko HL, et al. Cell cycle. Vol. 2010. Georgetown, Tex: The p53 response element and transcriptional repression; p. 9. [DOI] [PubMed] [Google Scholar]
- 32.Mayor C, Brudno M, Schwartz JR, et al. Bioinformatics. Vol. 16. Oxford, England: 2000. VISTA: visualizing global DNA sequence alignments of arbitrary length; pp. 1046–1047. [DOI] [PubMed] [Google Scholar]
- 33.el-Deiry WS, Kern SE, Pietenpol JA, et al. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 34.Hoffman WH, Biade S, Zilfou JT, et al. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem. 2002;277:3247–3257. doi: 10.1074/jbc.M106643200. [DOI] [PubMed] [Google Scholar]
- 35.Riley T, Sontag E, Chen P, et al. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 36.Tokino T, Thiagalingam S, el-Deiry WS, et al. p53 tagged sites from human genomic DNA. Hum Mol Genet. 1994;3:1537–1542. doi: 10.1093/hmg/3.9.1537. [DOI] [PubMed] [Google Scholar]
- 37.Bunz F. Requirement for p53 and p21 to Sustain G2 Arrest After DNA Damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 38.Oliver FJ, de la Rubia G, Rolli V, et al. Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis. Lesson from an uncleavable mutant. J Biol Chem. 1998;273:33533–33539. doi: 10.1074/jbc.273.50.33533. [DOI] [PubMed] [Google Scholar]
- 39.Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 40.Berglind H, Pawitan Y, Kato S, et al. Analysis of p53 mutation status in human cancer cell lines: a paradigm for cell line cross-contamination. Cancer Biol Ther. 2008;7:699–708. doi: 10.4161/cbt.7.5.5712. [DOI] [PubMed] [Google Scholar]
- 41.Günther HS, Schmidt NO, Phillips HS, et al. Glioblastoma-derived stem cell-enriched cultures form distinct subgroups according to molecular and phenotypic criteria. Oncogene. 2008;27:2897–2909. doi: 10.1038/sj.onc.1210949. [DOI] [PubMed] [Google Scholar]
- 42.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 43.Israel MA, Hernandez MC, Florio M, et al. Id gene expression as a key mediator of tumor cell biology. Cancer Research. 1999;59:1726s–1730s. [PubMed] [Google Scholar]
- 44.Lasorella A, Iavarone A, Israel MA. Id2 specifically alters regulation of the cell cycle by tumor suppressor proteins. Mol Cell Biol. 1996;16:2570–2578. doi: 10.1128/mcb.16.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang B, Xiao Z, Ren EC. Redefining the p53 response element. Proc Natl Acad Sci USA. 2009;106:14373–14378. doi: 10.1073/pnas.0903284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riley T, Sontag E, Chen P, et al. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 47.Balagurumoorthy P, Lindsay SM, Harrington RE. Atomic force microscopy reveals kinks in the p53 response element DNA. Biophysical chemistry. 2002;101-102:611–623. doi: 10.1016/s0301-4622(02)00169-2. [DOI] [PubMed] [Google Scholar]
- 48.Fontemaggi G, Dell'orso S, Muti P, et al. Id2 gene is a transcriptional target of the protein complex mutant p53/E2F1. Cell Cycle. 9 doi: 10.4161/cc.9.12.11915. [DOI] [PubMed] [Google Scholar]
- 49.Yan W, Liu G, Scoumanne A, et al. Suppression of Inhibitor of Differentiation 2, a Target of Mutant p53, Is Required for Gain-of-Function Mutations. Cancer Research. 2008;68:6789–6796. doi: 10.1158/0008-5472.CAN-08-0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harbor perspectives in biology. 2:a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verhaak RGW, Hoadley KA, Purdom E, et al. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5:603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- 53.Chen Y, Wu H, Wang S, et al. The oligodendrocyte-specific G protein–coupled receptor GPR17 is a cell-intrinsic timer of myelination. Nature Neuroscience. 2164:1–11. doi: 10.1038/nn.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samanta J. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–4142. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: NPCs cultured as neurospheres are multipotent upon differentiation. NPC cultures give rise to GFAP+ astrocytes, Tuj1+ neurons and O4+ oligodendrocytes 14 days after the addition of serum (20× magnification for GFAP and Tuj1, 10× for O4). Representative experiment.
Supplemental Figure 2: 2 Gy irradiation of NPCs does not increase the amount of apoptosis (12 hours after treatment). AnnexinV/PI flow cytometery for apoptosis after irradiation of WT and p53(-/-) NPCs. Representative experiment. Abbreviations: Gy, gray.
Supplemental Figure 3: Expression of exogenous P53 in p53(-/-) NPCs represses Id2. Western blot for p53 and Id2 in adenoviral infected p53(-/-) NPCs. Lysates were collected 16 hours post infection. Abbreviations: Ad-LacZ, beta-galactosidase expressing adenovirus. Ad-p53, P53 expressing adenovirus.
Supplemental Figure 4: Inhibition of Id2 does not alter p21 expression. (A) Quantitative PCR analysis of Id2 and p21 expression in p53(-/-) NPCs after Id2 inhibition. Average of cDNAs from three individual sh-Id2 cultures plated in triplicate. (B) p21 western blot of lysates from WT and Id2-/- NPCs. Representative image of results from three independent experiments. (C) Quantification of p21 western analysis by densitometry, n=3. Error bars represent standard deviation.
Supplemental Figure 5: Inhibition of Id2 expression does not alter the percentage of nestin+ NPCs. p53(-/-)/shScramble and p53(-/-)/sh-Id2 NPCs immunostained with anti-nestin antibody. 10× magnification. Representative images from two experiments plated in duplicate.
Supplemental Figure 6: Id2 inhibition does not affect the differentiation capacity of p53(-/-) NPCs. (A) Immunocytochemistry of p53(-/-)/shScramble and p53(-/-)/sh-Id2 NPCs after 14 days of differentiation for markers of astroglial, neuronal and oligodendroglial lineages. Representative images from two biological replicates. 10× magnification. (B) Quantification of the percentages of cell types formed in (A).
Supplemental Figure 7: Increased expression of ID2 in NPCs does not alter p21 expression. Quantitative PCR for Id2 and p21 expression in Id2CAG-FID2/Ad-LacZ and Id2CAG-FID2/Ad-Cre after recombinant adenoviral infection. Average of three experiments plated in triplicate.
Supplemental Figure 8: Stable retroviral expression of flag-tagged ID2 in NPCs. Id2 western blot from stable polyclonal cultures. Id2(-/-) NPC lysate included as a control.