Abstract
The leptospiral immunoglobulin-like (Lig) proteins LigA and LigB possess immunoglobulin-like domains with 90-amino-acid repeats and are adhesion molecules involved in pathogenicity. They are conserved in pathogenic Leptospira spp. and thus are of interest for use as serodiagnostic antigens and in recombinant vaccine formulations. The N-terminal amino acid sequences of the LigA and LigB proteins are identical, but the C-terminal sequences vary. In this study, we evaluated the protective potential of five truncated forms of LigA and LigB proteins from Leptospira interrogans serovar Canicola as DNA vaccines using the pTARGET mammalian expression vector. Hamsters immunized with the DNA vaccines were subjected to a heterologous challenge with L. interrogans serovar Copenhageni strain Spool via the intraperitoneal route. Immunization with a DNA vaccine encoding LigBrep resulted in the survival of 5/8 (62.5%) hamsters against lethal infection (P < 0.05). None of the control hamsters or animals immunized with the other vaccine preparations survived. The vaccine induced an IgG antibody response and, additionally, conferred sterilizing immunity in 80% of the surviving animals. Our results indicate that the LigBrep DNA vaccine is a promising candidate for inclusion in a protective leptospiral vaccine.
INTRODUCTION
Leptospirosis is a neglected infectious disease that is caused by pathogenic spirochetes of the genus Leptospira (1, 2). Around 350,000 to 500,000 cases of severe human infection are reported annually, but it is believed that this number is an underestimate of the true number of cases, due to a combination of factors, including a lack of surveillance, diagnostic tests, and notification in countries with large disease burdens (3). This constitutes a public health problem in developing countries, with outcomes that range from subclinical infections to severe pulmonary hemorrhage or Weil's syndrome and fatality rates of up to 20 to 50% (4, 5).
Reservoir hosts are typically asymptomatic and often serologically negative. The risk of acquiring leptospirosis is associated with contact with animals (6). Leptospires colonize the renal tubules of reservoir animals and are shed into the urine. Thus, direct contact with animal tissues or urine can cause individuals to become infected (2). Additionally, the bacteria can survive for several months in the external environment (3, 7). Most rodent species are natural carriers and contribute to the dissemination of pathogenic leptospires (2, 6). Leptospirosis is an important occupational disease and in particular affects farmers, slaughterhouse workers, pet traders, veterinarians, rodent catchers, and sewer workers (8). In livestock, infection by leptospires is associated with abortion, stillbirth, milk drop syndrome, and occasionally death (9).
Pathogenic leptospires enter the body via skin abrasions and mucous membranes and successfully infect the individual by binding to extracellular matrix compounds and host cells. The interaction of leptospires with pathogen recognition receptors is a fundamental issue in leptospiral immunity as well as in immunopathology. Since leptospirosis is a zoonotic disease that affects humans and animals, disease treatment strategies and prophylactic means, such as effective vaccines, are needed, but their development remains challenging. The currently available vaccines consist of inactivated whole-cell leptospires that confer short-lived immunity, fail to provide cross-protection against the large number of pathogenic Leptospira serovars (>200), and require boosters (10, 11). The greatest difficulty in developing a vaccine against leptospirosis is finding an antigen that elicits long-lasting, cross-protective, and sterilizing immunity. Surface-exposed outer membrane proteins (OMPs) are attractive for use as vaccines because they are relatively well conserved and, if exposed on the cell surface, constitute targets for interactions with host immune mediators (12–14).
Leptospiral immunoglobulin-like (Lig) proteins A, B, and C contain domains of 90-amino-acid repeats that were identified in adhesion molecules such as intimin in Escherichia coli and invasin in Yersinia pseudotuberculosis (15, 16). A special interest in Lig proteins has arisen because of their involvement in pathogenic mechanisms. Several studies have been conducted to evaluate these antigens as recombinant vaccines (reviewed in reference 17). In addition, they are present only in pathogenic leptospires, are virulence determinants regulated by osmolality, and mediate interactions between multiple host extracellular matrix proteins, including fibronectin, fibrinogen, collagen, and laminin (18–20). They also interact with the immune system by binding to regulatory protein factor H and C4b-binding protein (C4BP) (21) and to the complement proteins C3b and C4b (22). The amino acid sequences of Lig proteins are highly conserved (70 to 99% identity) (23). The N-terminal portions of the LigA and LigB proteins are identical (LigBrep), but the other regions of the proteins vary (LigAni, LigBni, and LigBct) (23, 24). While LigA is found in some pathogenic Leptospira spp., LigB is found in all species (23), which makes it a promising antigen for use in vaccine formulations and in diagnostic tests for leptospirosis.
A vaccine against Leptospira interrogans that used plasmid DNA was first tested in 2005 (25). Gerbils were immunized with a mammalian expression vector that carried the full gene of hemolysin-associated protein 1 (Hap-1 [also called LipL32]) under the control of a cytomegalovirus enhancer-promoter, which resulted in protection of 60% of the vaccinated animals. However, the survival rate of the control group was 35%. Subsequently, the LigA protein was evaluated, and DNA vaccines were produced with two truncated regions, i.e., the conserved portion (amino acids 32 to 626) and the variable portion (amino acids 631 to 1225), which were coadministered to hamsters (26). The vaccine afforded 100% protection in a homologous challenge; however, the survival rates of the control group were 50 to 75%, which indicated the low virulence of the L. interrogans serovar Pomona strain that was used in the challenge. In the present study, fragments that contain domains of LigA and LigB were cloned in a mammalian expression vector and evaluated as DNA vaccines in a heterologous challenge, using the hamster model.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The L. interrogans serovar Canicola strain Hond Utrecht IV and the virulent L. interrogans serogroup Icterohaemorrhagiae serovar Copenhageni strain Spool were cultivated at 30°C in Ellinghausen-McCullough-Johnson-Harris (EMJH) medium (Difco) supplemented with Leptospira enrichment EMJH medium. The E. coli TOP10 strain was grown at 37°C in Luria-Bertani (LB) medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl, and 2% agar) to which 100 μg/ml ampicillin had been added.
Animals.
Female golden Syrian hamsters and BALB/c mice were housed at the animal facility of the Biotechnology Unit of the Federal University of Pelotas. During the experiment, all animals were maintained in accordance with the recommendations of the Brazilian College of Animal Experimentation and the Ethics Committee for Animal Experimentation of the Federal University of Pelotas.
Genomic DNA extraction.
Genomic DNA from L. interrogans serovar Canicola strain Hond Utrecht IV was extracted using the GFX genomic DNA purification kit (GE Healthcare), following the protocol for Gram-negative bacteria. The extracted DNA was analyzed by agarose gel electrophoresis, in order to evaluate its integrity and quality, and was stored at −20°C.
DNA vaccine construction.
DNA sequences encoding fragments of LigA and LigB proteins were amplified using oligonucleotides designed according to the L. interrogans serovar Copenhageni strain FIOCRUZ L1-130 genome sequence (GenBank accession no. AE016823; region 533414.0.537088 and region 526395.0.532067, respectively), using L. interrogans serovar Canicola strain Hond Utrecht IV genomic DNA (GenBank accession no. EU289225) as the template. The oligonucleotides used for gene amplification and the corresponding regions are shown in Table 1. PCR was carried out under the following conditions: 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 7 min. The reactions were performed in a final volume of 25 μl that contained 2.5 μl of 10× buffer, 0.5 μl of 10 mM deoxynucleoside triphosphates, 150 nM each primer, 0.5 μl (1 unit) of Taq DNA polymerase (Invitrogen), 0.5 μl of 50 mM MgCl2, 18 μl of Milli-Q water, and 2 μl (50 ng) of template DNA, in a Mastercycler thermocycler (Eppendorf). PCR-amplified products were loaded onto a 1% agarose gel for electrophoresis and then visualized by GelRed (Invitrogen) staining.
Table 1.
Oligonucleotides used to amplify the target lig sequences
| Antigen used for vaccine preparation | Plasmid and oligonucleotidea | Amino acidsb | Molecular mass (kDa) |
|---|---|---|---|
| LigAni | pAE_F, 5′-CCGCTCGAGGATATTCTTACCGTTTC-3′ | 629–1224 | 64 |
| pAE_R, 5′-CGGGAATTCTGGCTCCGTTTTA-3′ | |||
| pTARGET_F, 5′-ATGGATATTCTTACCGTTTC-3′ | |||
| LigBni | pAE_F, 5′-CCGCTCGAGGATATTGCTGAAATT-3′ | 629–1112 | 52 |
| pAE_R, 5′-CGGGAATTCTCCAGATACTGAACC-3′ | |||
| pTARGET_F, 5′-ATGCTCGAGGATATTGCTGAAATT-3′ | |||
| LigBrep | pAE_F, 5′-CCGCTCGAGGTGTTTATGAAGAAA-3′ | 1–628 | 72 |
| pAE_R, 5′-ACTCTCGAGCGTATTAGAGGAAT-3′ | |||
| pTARGET_F, 5′-ATGGTGTTTATGAAGAAAATATTTTTG-3′ | |||
| LigBct1 | pAE_F, 5′-CCGCTCGAGAATACAATCCTCACA-3′ | 1113–1501 | 45 |
| pAE_R, 5′-GGGAAGCTTTATAAAAATATGAGA-3′ | |||
| pTARGET_F, 5′-ATGAATACAATCCTCACA-3′ | |||
| LigBct2 | pAE_F, 5′-CCGCTCGAGGCAGGTTCAAGACC-3′ | 1502–1891 | 45 |
| pAE_R, 5′-GGGAAGCTTTTATTGATTCTGTT-3′ | |||
| pTARGET_F, 5′-ATGGCAGGTTCAAGACC-3′ |
In the oligonucleotide sequences, the underlined letters denote the restriction sites.
Amino acid coordinates in the LigA and LigB proteins of L. interrogans serovar Copenhageni strain FIOCRUZ L1-130.
The gene regions encoding LigAni, LigBni, LigBrep, LigBct1, and LigBct2 were amplified by PCR and cloned into the pTARGET mammalian expression vector (Promega), which presents a multiple cloning site, neomycin-selectable marker, lacZα region, T7 RNA polymerase promoter, simian virus 40 (SV40) late polyadenylation signal, and human cytomegalovirus (CMV) promoter. Electrocompetent E. coli TOP10 cells were transformed with the plasmids and cultured in LB broth containing ampicillin. The recombinant plasmids were verified by restriction digestion and DNA sequencing using the DYEnamic ET dye terminator cycle sequencing kit for MegaBACE DNA analysis systems, MegaBACE 500 (GE Healthcare). DNA extraction was performed using the NucleoBond Xtra maxikit for plasmid DNA purification (Macherey-Nagel). The DNA was quantified in a Qubit fluorometer (Invitrogen), according to the manufacturer's recommendations.
Evaluation of DNA vaccine functionality by transfection of mammalian cells.
To confirm that the DNA vaccine constructs were functional and expressed the target proteins, Vero cells were transfected with the plasmids (pTARGET/LigAni, pTARGET/LigBni, pTARGET/LigBrep, pTARGET/LigBct1, and pTARGET/LigBct2) using Lipofectin (Invitrogen), according to the manufacturer's protocol. Briefly, Vero cells were maintained in flasks in Dulbecco's modified Eagle's medium (DMEM) supplemented with 100 IU/ml penicillin, 100 μg/ml streptomycin, and 10% (vol/vol) heat-inactivated fetal bovine serum (FBS). When a minimum of 50% confluence was reached, the cells were transfected with 2 μg of plasmid DNA precondensed with Lipofectin, in serum-free DMEM. Transfection with an empty control vector (pTARGET) was performed as a negative control. Forty-eight hours after transfection, the cells were collected and their protein expression was analyzed by indirect immunofluorescence. For that, the transfected cells were washed 3 times with phosphate-buffered saline (PBS) (pH 7.4) and then fixed with methanol for 10 min at 4°C. PBS plus 10% FBS was added, and the slides were incubated for 30 min at 30°C in a dark humid chamber. Another washing with PBS plus FBS was performed, antibodies (anti-LigAni, anti-LigBrep, anti-LigBni, or anti-LigBct) diluted in PBS at a ratio of 1:100 were added, and the samples were incubated for 2 h at 4°C in a dark humid chamber. After washing with PBS plus FBS, anti-mouse IgG-fluorescein isothiocyanate conjugate (Sigma) diluted 1:80 was added, and the cell suspension was incubated for 1 h at 30°C. Then, another round of PBS plus FBS washing was performed, 10 μl of mounting medium was added, and the coverslips were sealed. Staining was visualized by fluorescence microscopy (Olympus), at an excitation wavelength of 450 nm.
Expression and purification of His6-LigA and -B proteins.
The target sequences were amplified by PCR using the purified genomic DNA of L. interrogans serovar Canicola strain Hond Utrecht with the oligonucleotides detailed in Table 1. The resulting gene products, LigAni, LigBni, LigBrep, LigBct1, and LigBct2, were inserted into the plasmid pAE (27), and the ligation products were used to transform electrocompetent E. coli TOP10 cells. Recombinant clones were selected, and the target gene presence was confirmed by restriction enzyme digestion and sequencing using the DYEnamic ET dye terminator cycle sequencing kit for MegaBACE DNA analysis systems, MegaBACE 500 (GE Healthcare). The resulting recombinant plasmids were used to transform E. coli BL21(DE3) Star. The cells were cultured at 37°C under agitation (200 rpm) for ∼3.5 h (optical density at 600 nm, 0.7) and then were treated with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h. The cells were harvested (7,000 × g for 20 min at 4°C), and the pellet was suspended in solubilization buffer (8 M urea, 200 mM NaH2PO4, 0.5 M NaCl, and 5 mM imidazole at pH 8.0). The cells were sonicated (3 times, 45 s) and then centrifuged (10,000 × g for 60 min at 4°C), and the supernatant was collected and filtered through a 0.8-μm filter (Millipore). The recombinant proteins were purified by immobilized metal ion affinity chromatography using Ni2+-Sepharose HisTrap columns (GE Healthcare). Fractions that contained the eluted proteins were identified by SDS-PAGE and Western blotting. The relevant fractions were dialyzed at 4°C in PBS (pH 7.4) that contained decreasing concentrations of urea in each step. The protein concentrations were determined using a bicinchoninic acid protein assay kit (Pierce).
Antisera against recombinant LigA and LigB proteins.
The fractions of recombinant LigA and LigB proteins were adsorbed onto 15% Alhydrogel (Invitrogen) as adjuvant. Female BALB/c mice (4 to 6 weeks old) were injected intramuscularly with 100 μg recombinant proteins, with two subsequent booster injections at 2-week intervals. Negative-control mice were injected with PBS/Alhydrogel. Two weeks after each immunization, the mice were bled from the retro-orbital plexus, and the antibodies present in the pooled serum samples were monitored by enzyme-linked immunosorbent assay (ELISA), for use in the indirect immunofluorescence assay.
Hamster immunization.
Vaccines were prepared with 100 μg of DNA and 15% Alhydrogel. Fifty female hamsters at 6 weeks of age were allocated into 7 groups, i.e., 5 DNA vaccine groups (LigAni, LigBni, LigBrep, LigBct1, and LigBct2), which contained 8 animals each, and 2 control groups, a killed whole leptospires group and an empty pTARGET vector group, which contained 4 and 6 animals, respectively. The hamsters were vaccinated with two intramuscular injections in the hind leg, at 21-day intervals. Forty-two days after the first dose, all of the hamsters were challenged intraperitoneally with 10 leptospires, which was equivalent to 5× the 50% lethal dose (LD50) of the L. interrogans serovar Copenhageni strain Spool. The negative-control group was inoculated with an empty pTARGET plasmid (100 μg) plus 15% Alhydrogel, and the positive-control group was immunized with a bacterin vaccine consisting of 109 heat-killed whole leptospires, as described previously (28). Hamsters were monitored daily for clinical signs of leptospirosis and were euthanized when clinical signs of terminal disease appeared.
Evaluation of antibody responses.
IgG antibodies against the antigens LigAni, LigBrep, LigBni, LigBct1, and LigBct2 in the animal sera were monitored through ELISA. ELISA plates (PolySorp surface; Nunc) were coated with 200 ng per well of each purified protein for evaluation of hamsters immunized with Lig DNA vaccines, 200 ng of a pool of these proteins for evaluation of hamsters immunized with the empty pTARGET plasmid, or 5 × 107 cells/ml of whole L. interrogans used as antigen for evaluation of hamsters immunized with the killed whole leptospires. The antigens were diluted in carbonate-bicarbonate buffer (pH 9.6) and incubated at 4°C for 16 to 18 h. Then, they were washed 3 times with PBS (pH 7.6) containing 0.05% Tween 20 (PBST) and were blocked with 5% (dried skim milk) blocking buffer for 1 h at 37°C. Following 3 washes with PBST, the sera of the immunized animals, diluted 1:50 in PBS, were added and the plates were incubated at 37°C for 1 h. After 3 washes with PBST, goat anti-IgG-peroxidase conjugate (Serotec, USA) diluted 1:8,000 was added and the plates were incubated at 37°C for 1 h. Finally, the plates were washed 5 times with PBST, and the reaction products were visualized with o-phenylenediamine dihydrochloride (Sigma-Aldrich) and hydrogen peroxide. The reactions were stopped by adding 0.1 M sulfuric acid, and absorbances were determined at 492 nm using a Multiskan MCC/340 ELISA reader (Titertek Instruments, USA). Mean values were calculated from serum samples assayed in triplicate.
Culturing and histopathological analysis.
The hamsters that survived to day 30 postchallenge were euthanized. Kidney and lung tissues were then harvested and studied with respect to histopathology. Sterilizing immunity was determined by culture isolation of leptospires from kidney samples. From each organ, 1 to 2 g of tissue was aseptically removed, processed, and transferred to 5 ml of EMJH medium (pH 7.2). Dark-field microscopy was performed during an 8-week incubation period, to identify positive cultures. For histopathological studies, kidney and lung tissue samples were fixed in 10% formalin (pH 7.0) and embedded in paraffin. Six sections, 5 to 6 μm thick, were obtained from the organs, stained with hematoxylin and eosin, and examined by a qualified pathologist.
Statistical analysis.
Variance analysis was used to determine significant differences between the assay results. The Bonferroni test was used to determine significant differences in the serological assay results. The Fisher exact test and the Wilcoxon log rank test were used to determine significant differences in mortality and survival rates, respectively, using Prism 5 (GraphPad, USA). Differences were considered to be statistically significant at a P value of <0.05.
RESULTS
Lig protein expression in mammalian cells.
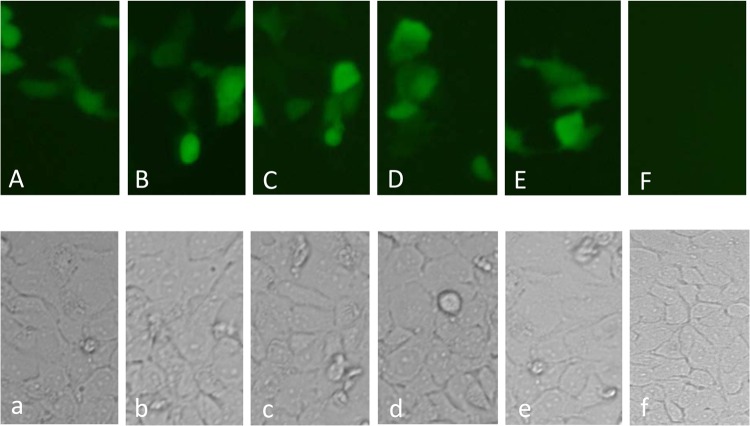
The lig genes were amplified from genomic DNA from L. interrogans serovar Canicola strain Hond Utrecht IV and cloned into the pTARGET mammalian expression vector. The functionality of the constructed plasmids was evaluated in mammalian cells (Vero), and expression of the recombinant proteins was visualized by indirect immunofluorescence. Our results clearly revealed the expression of LigAni, LigBni, LigBrep, LigBct1, and LigBct2 (Fig. 1). No reaction was detected after incubation with polyclonal anti-LigA and anti-LigB sera for the Vero control cells, which had been transfected with empty pTARGET plasmid.
Fig 1.
(A to F) Indirect immunofluorescence detection of LigAni (A), LigBni (B), LigBrep (C), LigBct1 (D), and LigBct2 (E) proteins expressed in Vero cells, using anti-Lig polyclonal sera; the negative control (F) corresponds to Vero cells that were transfected with the empty pTARGET plasmid and incubated with anti-Lig polyclonal sera. Visualization was performed with a 40× objective on an Olympus BX71 fluorescence microscope. (a to f) Corresponding bright-field microscopic views of the Vero cells.
Expression and purification of Lig proteins.
The recombinant proteins LigAni, LigBni, LigBrep, LigBct1, and LigBct2 were expressed and purified from E. coli BL21(DE3) Star and showed the expected sizes of 64, 52, 72, 45, and 45 kDa, respectively (data not shown). All of the proteins were insoluble, and they were resolubilized in urea. The yields of the recombinant proteins were 88 mg/liter LigAni, 2.58 mg/liter LigBni, 4.05 mg/liter LigBrep, 5.42 mg/liter LigBct1, and 1.52 mg/liter LigBct2.
IgG immune responses.
The antibody responses induced by the LigA and LigB constructs were analyzed at time points 0 (preimmune), 21, and 42 days postimmunization (DPI). Our results demonstrated that IgG antibodies were generated following immunization. The hamsters that were immunized with pTARGET/LigAni and pTARGET/LigBrep presented significant antibody titers at 21 DPI (P < 0.05), which increased at 42 DPI. At this time point, pTARGET/LigBni and pTARGET/LigBct2 also induced significant humoral immune responses (P < 0.05). As shown in Fig. 2, the antibody titers increased after the booster immunization (21 days), and the highest titers were observed at 42 days for hamsters that had been immunized with pTARGET/LigBrep, followed by pTARGET/LigAni. Specific antibodies were not detected in the groups that received pTARGET or pTARGET/LigBct2. A significant IgG immune response against the whole L. interrogans used as the antigen in the ELISA was observed after the boost (42 days) with the killed whole leptospires.
Fig 2.
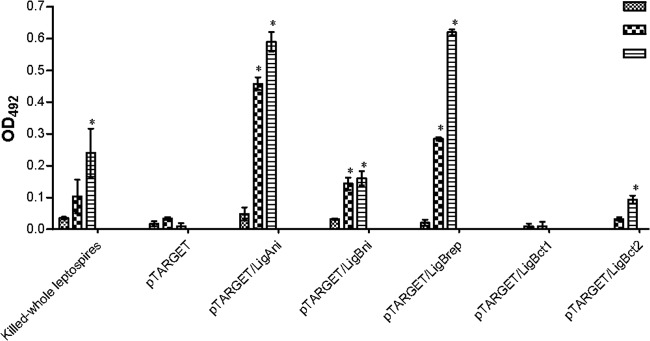
Antibody responses of hamsters immunized with Lig DNA vaccines, as measured by ELISA. Recombinant LigAni, LigBni, LigBrep, LigBct1, and LigBct2 proteins were used as antigens in the ELISA. The results are expressed as the mean absorbances for all of the animals in each group. OD492, optical density at 492 nm. *, P < 0.05 compared to the group that received the empty pTARGET plasmid.
Prophylactic effects of DNA vaccines in hamster challenge.
The protective efficacies of the DNA vaccines were evaluated in terms of survival times, histopathological findings for vital organs, and leptospiral loads in the kidneys. The survival data (Wilcoxon log rank test) showed that animals immunized with pTARGET/LigBrep, followed by a heterologous challenge, were significantly protected (survival rate, 62.5%; P < 0.05) (Table 2). In contrast, none of the other DNA vaccines had a protective effect, and the median length of survival was 10 days (Fig. 3). The animals in the positive-control group, which had been immunized with killed whole leptospires (Table 2 and Fig. 3), were protected against death (100%; P < 0.05). None of the animals that received the empty pTARGET vector survived (median survival time, 11 days), confirming the virulence of the challenge strain. The hamsters that died during the experiment showed severe external hemorrhaging, swelling, and complete closure of the eyes, which is a characteristic of the challenge strain, in addition to commonly described symptoms such as dyspnea, hemorrhage, and lethargy.
Table 2.
Immunoprotective potential of LigA and LigB proteins as DNA vaccines
| Vaccine | No. of survivors/total no. of animals | Times of death (days) | Survival rate (%)a |
|---|---|---|---|
| Killed whole leptospires | 4/4 | 100b | |
| pTARGET | 0/6 | 6, 10, 11, 11, 11, 11 | 0 |
| pTARGET/LigAni | 0/8 | 10, 10, 10, 10, 11, 12, 12, 12 | 0 |
| pTARGET/LigBni | 0/8 | 10, 10, 10, 10, 10, 12, 12, 12 | 0 |
| pTARGET/LigBrep | 5/8 | 10, 12, 20 | 62.5b |
| pTARGET/LigBct1 | 0/8 | 10, 10, 10, 10, 11, 12, 12, 12 | 0 |
| pTARGET/LigBct2 | 0/8 | 5, 7, 9, 10, 10, 10, 10, 10 | 0 |
Surviving animals were observed for 30 days.
P < 0.05.
Fig 3.
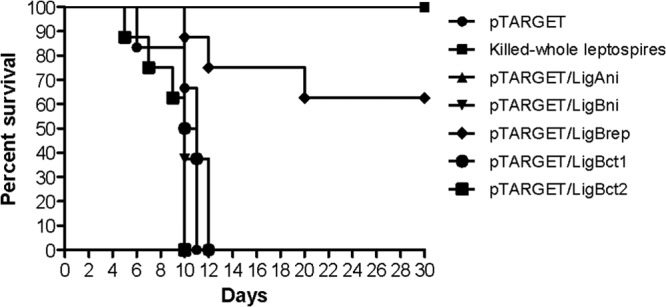
Survival times of hamsters immunized with Lig DNA vaccines or with killed whole leptospires after heterologous challenge with 101 leptospires of the virulent L. interrogans serovar Copenhageni strain Spool. The Wilcoxon log rank test was used to determine significant survival differences between the immunized groups and the control (empty pTARGET plasmid) group. The groups immunized with killed whole leptospires and LigBrep show P values of <0.05.
Necropsy of the immunized hamsters that survived 30 days postchallenge did not reveal the presence of macroscopic lesions in the organs, pulmonary hemorrhaging, or jaundice. An exception was one animal (1/5 [20%]) in the pTARGET/LigBrep group that presented with pulmonary hemorrhaging, apparent renal injury, and jaundice. Histopathological changes in the lung of this animal included alveolar hemorrhage, edema, capillary congestion, and leukocyte infiltration. The predominant lesions in the kidneys included interstitial nephritis, renal tubular failure, and necrosis.
The leptospiral loads in the kidneys of the surviving hamsters were evaluated by culturing. All of the animals that had been immunized with killed whole leptospires were culture negative, which indicated the prophylactic efficacy in preventing infection. Only 1 of the 5 hamsters that received the DNA vaccine (pTARGET/LigBrep) and survived upon challenge had a positive kidney culture. The other four animals (80%) were culture negative, which indicated that sterilizing immunity had been induced.
DISCUSSION
In our study, we used DNA vaccines that encoded five portions of the LigA and LigB proteins, coadministered with Alhydrogel. We found that the plasmid construct expressing the region that is identical in the LigA and LigB (LigBrep) proteins of L. interrogans serovar Canicola protected 62.5% (5/8) of hamsters in a heterologous challenge with L. interrogans serovar Copenhageni. Additionally, the vaccine induced an IgG antibody response and conferred sterilizing immunity in 80% of the surviving hamsters. Other portions of the LigA and LigB proteins that were evaluated in the present study (LigAni, LigBni, LigBct1, and LigBct2) failed to protect the hamsters from lethal infection.
Compared to the LigBrep DNA vaccine, the LigAni fraction induced similar antibody levels; however, they were not protective. The other Lig fractions did not induce high antibody levels or afford protection in hamsters. The importance of the humoral immune response induced by recombinant Lig proteins is unclear. Several authors have associated high antibody titers with protection against lethal infection (26, 29–31). In contrast, a recent study suggested that protection by LigA was not due solely to the humoral immune response induced by the protein or its fractions (32). DNA vaccines often induce lower antibody titers than conventional vaccines, and several reports have demonstrated that DNA vaccines formulated with Alhydrogel have increased protective efficacy, with antibody titers increased 10- to 100-fold (33, 34). Highly potent DNA vaccines may allow reductions in the quantity of DNA that is required for protective efficacy, and therefore they might be beneficial with respect to vaccine costs and manufacturing. In DNA vaccination strategies, the biosynthetic machinery of the host cells is responsible for producing the antigen (35). We believe that the difference in the levels of antibody responses induced by Lig fractions is due to antigenic characteristics or to a possible failure in correct protein expression by the cell machinery of the hamsters immunized with the DNA vaccine constructs. It has been reported that the protective epitope of LigA protein is conformational rather than linear and that specific Lig domains are required for proper conformation (32). The Lig proteins are of great interest, and several studies have demonstrated their potential uses in diagnostic assays and vaccine development (reviewed in reference 17). Lig proteins are surface-exposed OMPs in pathogenic leptospires and therefore are potential targets of a protective immune response (36). However, the mechanism of Lig protein-mediated protection has not been elucidated. Previous studies examined the ability of Lig proteins to confer protection (24, 26, 29–32, 37), but the results were inconclusive in several cases because of the animal models that were used and the low virulence of the challenge strains. In our study, we used the classic animal model of leptospirosis, the golden Syrian hamster, and 101 leptospires (5 × LD50) of a highly virulent strain in our heterologous challenge experiment. This resulted in lethal infections in all of the negative-control animals and the animals from groups vaccinated with constructs containing nonprotective portions of Lig A and LigB proteins (Table 2 and Fig. 3).
This is the first study to evaluate five portions of the Lig A and LigB proteins in a heterologous challenge. A previous study evaluated three portions of Lig proteins, i.e., LigAni (amino acids 625 to 1224), LigBni (amino acids 625 to 1259), and LigBrep (amino acids 131 to 645), and only LigAni administered as purified recombinant protein afforded protection of hamsters against a homologous challenge (67 to 100%) (24). Our results demonstrate that LigBrep, when presented as a DNA vaccine, has a prophylactic effect against lethal infection. Since LigB is a surface-exposed protein, anti-LigB antibodies might act as opsonins and block the interaction of leptospires with host cells during the initial stage of infection (29). The LigBrep fraction contains Lig domains that are highly conserved among pathogenic Leptospira spp. (23); therefore, it has the greatest potential as a universal vaccine against leptospirosis.
The presence of ligB in all pathogenic Leptospira spp. (23) is important when cross-protection against multiple serovars of Leptospira spp. is sought. In addition, although the identity of the ligB gene in the L. interrogans serovars Canicola and Copenhageni is only 96.7% (23), the region encoding LigBrep is more conserved, with >98% identity among serovars of L. interrogans. The lack of protection with the other lig fragments might have been due to the lower level of identity between the strain used as the source of DNA and the challenge strain. The LigA homology between the Canicola and Copenhageni serovars is only 87% (23).
Interestingly, the kidneys of 80% (4/5) of the hamsters that survived the heterologous challenge were culture negative, and no histopathological lesions were found. Therefore, the DNA vaccine that contained the LigBrep-encoding portion from L. interrogans serovar Canicola was able to protect against a L. interrogans serovar Copenhageni challenge. However, the vaccination protocol might be able to be improved to increase the DNA vaccine potency. Among the strategies that have been evaluated are the use of more-efficient promoters, codon optimization, addition of traditional or genetic adjuvants, electroporation, intradermal delivery, and prime-boost strategies (38). Additionally, an effective vaccine against leptospirosis is likely not to be based on a single antigen but rather to be composed of several antigens that would be able to afford complete protection against different Leptospira spp. and serovars. In conclusion, LigBrep is a promising candidate to compose an effective vaccine for leptospirosis control.
ACKNOWLEDGMENTS
This work was supported by grants from FAPERGS and CNPq (Brazilian government).
We are grateful to Michele dos Santos for technical assistance and to Alan McBride for reviewing the English of the manuscript.
Footnotes
Published ahead of print 13 March 2013
REFERENCES
- 1.Faine SB, Adler B, Bolin C, Perolat P. 1999. Leptospira and leptospirosis, 2nd ed MediSci, Melbourne, Australia [Google Scholar]
- 2.Levett PN. 2001. Leptospirosis. Clin. Microbiol. Rev. 14:296–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reis RB, Ribeiro GS, Felzemburgh RD, Santana FS, Mohr S, Melendez AX, Queiroz A, Santos AC, Ravines RR, Tassinari WS, Carvalho MS, Reis MG, Ko AI. 2008. Impact of environment and social gradient on Leptospira infection in urban slums. PLoS Negl. Trop. Dis. 2:e228 doi:10.1371/journal.pntd.0000228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartskeerl RA. 2006. Leptospirosis: current status and future trends. Indian J. Med. Microbiol. 24:309. [DOI] [PubMed] [Google Scholar]
- 5.McBride AJ, Athanazio DA, Reis MG, Ko AI. 2005. Leptospirosis. Curr. Opin. Infect. Dis. 18:376–386 [DOI] [PubMed] [Google Scholar]
- 6.Bharti AR, Nally JE, Ricaldi JN, Matthias MA, Diaz MM, Lovett MA, Levett PN, Gilman RH, Willig MR, Gotuzzo E, Vinetz JM. 2003. Leptospirosis: a zoonotic disease of global importance. Lancet Infect. Dis. 3:757–771 [DOI] [PubMed] [Google Scholar]
- 7.Lau C, Smythe L, Weinstein P. 2010. Leptospirosis: an emerging disease in travellers. Travel Med. Infect. Dis. 8:33–39 [DOI] [PubMed] [Google Scholar]
- 8.Hartskeerl RA, Collares-Pereira M, Ellis WA. 2011. Emergence, control and re-emerging leptospirosis: dynamics of infection in the changing world. Clin. Microbiol. Infect. 17:494–501 [DOI] [PubMed] [Google Scholar]
- 9.Guitian J, Thurmond MC, Hietala SK. 1999. Infertility and abortion among first-lactation dairy cows seropositive or seronegative for Leptospira interrogans serovar hardjo. J. Am. Vet. Med. Assoc. 215:515–518 [PubMed] [Google Scholar]
- 10.Ko AI, Goarant C, Picardeau M. 2009. Leptospira: the dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat. Rev. Microbiol. 7:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sonrier C, Branger C, Michel V, Ruvoen-Clouet N, Ganiere JP, Andre-Fontaine G. 2000. Evidence of cross-protection within Leptospira interrogans in an experimental model. Vaccine 19:86–94 [DOI] [PubMed] [Google Scholar]
- 12.Koizumi N, Watanabe H. 2005. Leptospirosis vaccines: past, present, and future. J. Postgrad. Med. 51:210–214 [PubMed] [Google Scholar]
- 13.Cullen PA, Haake DA, Adler B. 2004. Outer membrane proteins of pathogenic spirochetes. FEMS Microbiol. Rev. 28:291–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haake DA, Mazel MK, McCoy AM, Milward F, Chao G, Matsunaga J, Wagar EA. 1999. Leptospiral outer membrane proteins OmpL1 and LipL41 exhibit synergistic immunoprotection. Infect. Immun. 67:6572–6582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isberg RR, Voorhis DL, Falkow S. 1987. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell 50:769–778 [DOI] [PubMed] [Google Scholar]
- 16.Jerse AE, Yu J, Tall BD, Kaper JB. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. U. S. A. 87:7839–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dellagostin OA, Grassmann AA, Hartwig DD, Felix SR, da Silva EF, McBride AJ. 2011. Recombinant vaccines against leptospirosis. Hum. Vaccin. 7:1215–1224 [DOI] [PubMed] [Google Scholar]
- 18.Choy HA, Kelley MM, Chen TL, Moller AK, Matsunaga J, Haake DA. 2007. Physiological osmotic induction of Leptospira interrogans adhesion: LigA and LigB bind extracellular matrix proteins and fibrinogen. Infect. Immun. 75:2441–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsunaga J, Medeiros MA, Sanchez Y, Werneid KF, Ko AI. 2007. Osmotic regulation of expression of two extracellular matrix-binding proteins and a haemolysin of Leptospira interrogans: differential effects on LigA and Sph2 extracellular release. Microbiology 153:3390–3398 [DOI] [PubMed] [Google Scholar]
- 20.Matsunaga J, Sanchez Y, Xu X, Haake DA. 2005. Osmolarity, a key environmental signal controlling expression of leptospiral proteins LigA and LigB and the extracellular release of LigA. Infect. Immun. 73:70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castiblanco-Valencia MM, Fraga TR, Silva LB, Monaris D, Abreu PA, Strobel S, Jozsi M, Isaac L, Barbosa AS. 2012. Leptospiral immunoglobulin-like proteins interact with human complement regulators factor H, FHL-1, FHR-1, and C4BP. J. Infect. Dis. 205:995–1004 [DOI] [PubMed] [Google Scholar]
- 22.Choy HA. 2012. Multiple activities of LigB potentiate virulence of Leptospira interrogans: inhibition of alternative and classical pathways of complement. PLoS One 7:e41566 doi:10.1371/journal.pone.0041566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McBride AJ, Cerqueira GM, Suchard MA, Moreira AN, Zuerner RL, Reis MG, Haake DA, Ko AI, Dellagostin OA. 2009. Genetic diversity of the leptospiral immunoglobulin-like (Lig) genes in pathogenic Leptospira spp. Infect. Genet. Evol. 9:196–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva EF, Medeiros MA, McBride AJ, Matsunaga J, Esteves GS, Ramos JG, Santos CS, Croda J, Homma A, Dellagostin OA, Haake DA, Reis MG, Ko AI. 2007. The terminal portion of leptospiral immunoglobulin-like protein LigA confers protective immunity against lethal infection in the hamster model of leptospirosis. Vaccine 25:6277–6286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Branger C, Chatrenet B, Gauvrit A, Aviat F, Aubert A, Bach JM, Andre-Fontaine G. 2005. Protection against Leptospira interrogans sensu lato challenge by DNA immunization with the gene encoding hemolysin-associated protein 1. Infect. Immun. 73:4062–4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faisal SM, Yan W, Chen CS, Palaniappan RU, McDonough SP, Chang YF. 2008. Evaluation of protective immunity of Leptospira immunoglobulin like protein A (LigA) DNA vaccine against challenge in hamsters. Vaccine 26:277–287 [DOI] [PubMed] [Google Scholar]
- 27.Ramos CR, Abreu PA, Nascimento AL, Ho PL. 2004. A high-copy T7 Escherichia coli expression vector for the production of recombinant proteins with a minimal N-terminal His-tagged fusion peptide. Braz. J. Med. Biol. Res. 37:1103–1109 [DOI] [PubMed] [Google Scholar]
- 28.Seixas FK, da Silva EF, Hartwig DD, Cerqueira GM, Amaral M, Fagundes MQ, Dossa RG, Dellagostin OA. 2007. Recombinant Mycobacterium bovis BCG expressing the LipL32 antigen of Leptospira interrogans protects hamsters from challenge. Vaccine 26:88–95 [DOI] [PubMed] [Google Scholar]
- 29.Palaniappan RU, McDonough SP, Divers TJ, Chen CS, Pan MJ, Matsumoto M, Chang YF. 2006. Immunoprotection of recombinant leptospiral immunoglobulin-like protein A against Leptospira interrogans serovar Pomona infection. Infect. Immun. 74:1745–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan W, Faisal SM, McDonough SP, Divers TJ, Barr SC, Chang CF, Pan MJ, Chang YF. 2009. Immunogenicity and protective efficacy of recombinant Leptospira immunoglobulin-like protein B (rLigB) in a hamster challenge model. Microbes Infect. 11:230–237 [DOI] [PubMed] [Google Scholar]
- 31.Cao Y, Faisal SM, Yan W, Chang YC, McDonough SP, Zhang N, Akey BL, Chang YF. 2011. Evaluation of novel fusion proteins derived from extracellular matrix binding domains of LigB as vaccine candidates against leptospirosis in a hamster model. Vaccine 29:7379–7386 [DOI] [PubMed] [Google Scholar]
- 32.Coutinho ML, Choy HA, Kelley MM, Matsunaga J, Babbitt JT, Lewis MS, Aleixo JA, Haake DA. 2011. A LigA three-domain region protects hamsters from lethal infection by Leptospira interrogans. PLoS Negl. Trop. Dis. 5:e1422 doi:10.1371/journal.pntd.0001422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwissa M, Lindblad EB, Schirmbeck R, Reimann J. 2003. Codelivery of a DNA vaccine and a protein vaccine with aluminum phosphate stimulates a potent and multivalent immune response. J. Mol. Med. (Berl.) 81:502–510 [DOI] [PubMed] [Google Scholar]
- 34.Wang S, Liu X, Fisher K, Smith JG, Chen F, Tobery TW, Ulmer JB, Evans RK, Caulfield MJ. 2000. Enhanced type I immune response to a hepatitis B DNA vaccine by formulation with calcium- or aluminum phosphate. Vaccine 18:1227–1235 [DOI] [PubMed] [Google Scholar]
- 35.Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, Felgner PL. 1990. Direct gene transfer into mouse muscle in vivo. Science 247:1465–1468 [DOI] [PubMed] [Google Scholar]
- 36.Matsunaga J, Barocchi MA, Croda J, Young TA, Sanchez Y, Siqueira I, Bolin CA, Reis MG, Riley LW, Haake DA, Ko AI. 2003. Pathogenic Leptospira species express surface-exposed proteins belonging to the bacterial immunoglobulin superfamily. Mol. Microbiol. 49:929–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koizumi N, Watanabe H. 2004. Leptospiral immunoglobulin-like proteins elicit protective immunity. Vaccine 22:1545–1552 [DOI] [PubMed] [Google Scholar]
- 38.Saade F, Petrovsky N. 2012. Technologies for enhanced efficacy of DNA vaccines. Expert Rev. Vaccines 11:189–209 [DOI] [PMC free article] [PubMed] [Google Scholar]