Abstract
The prevention of bacterial infections via immunization presents particular challenges. While outer membrane extracts are often protective, they are difficult and expensive to isolate and standardize and thus are often impractical for development and implementation in vaccination programs. In contrast, individual proteins, which are easily adapted for use in subunit vaccines, tend to be poorly protective. Consequently, identification of the specific characteristics of outer membrane-based immunogens, in terms of the antigen contents and contexts that are required for protective immunity, represents a major gap in the knowledge needed for bacterial vaccine development. Using as a model Anaplasma marginale, a persistent tick-borne bacterial pathogen of cattle, we tested two sets of immunogens to determine whether membrane context affected immunogenicity and the capacity to induce protection. The first immunogen was composed of a complex of outer membrane proteins linked by covalent bonds and known to be protective. The second immunogen was derived directly from the first one, but the proteins were individualized rather than linked. The antibody response induced by the linked immunogen was much greater than that induced by the unlinked immunogen. However, both immunogens induced protective immunity and an anamnestic response. These findings suggest that individual proteins or combinations of proteins can be successfully tested for the ability to induce protective immunity with less regard for overall membrane context. Once protective antigens are identified, immunogenicity could be enhanced by cross-linking to allow a reduced immunogen dose or fewer booster vaccinations.
INTRODUCTION
Surface-exposed protein complexes are essential both structurally and functionally for the success of bacterial pathogens (1). For example, major outer membrane proteins serve as structural components, while porins allow for the passive transport of molecules across the membrane, and secretion systems export molecules which co-opt the host cell (2, 3). Outer membrane proteins are not only essential for the bacterium but also serve as major targets of the innate and adaptive immune system and thus serve as particularly relevant targets for vaccine development.
The specific features of an immunogen that are required to induce protective immunity are largely unknown for a variety of bacterial pathogens. However, outer membrane extracts are often protective. For example, outer membrane extracts of Francisella tularensis provide significant protection from challenge (4), as does a fusion protein composed of B-cell epitopes from three Legionella pneumophila surface proteins (5). Similarly, a macromolecular complex composed of three Shigella flexneri virulence factors induces protective immunity (6). In contrast, individual recombinant or native outer membrane proteins tend to be poorly protective in the face of challenge (4, 5, 7). For example, the same three L. pneumophila proteins, when administered individually, induced partial to no protection despite a relatively robust antibody response (5). Similarly, Tul4 and FopA, outer membrane proteins of F. tularensis, when presented by attenuated Salmonella enterica serovar Typhimurium, provide overall poor protection (8, 9).
In the case of Anaplasma marginale, a rickettsial pathogen of cattle, an outer membrane protein immunogen, which is composed of more than 20 different proteins, has been shown to induce protection against anemia and bacteremia in nearly all animals and protection against infection in 40 to 70% of vaccinees (10–12). More recently, a cross-linked complex composed of 11 outer membrane proteins was identified that induces protective immunity similar to the outer membrane protein immunogen (12). In contrast, immunization with individual outer membrane proteins, either native or recombinant, generally results in poor or inconsistent protection (13–18).
Two common features of these effective immunogens are the presence of multiple antigens and the maintenance of spatial relationships among the antigens comprising the immunogen. It is unknown if the presence of a specific core set of antigens is sufficient to induce protective immunity or if the maintenance of the physical relationships between proteins is also required. We hypothesize that maintaining the physical association between outer membrane proteins will enhance their immunogenicity and that this close physical association is required for protection from challenge.
We used A. marginale immunogens to test the hypotheses. Animals were immunized with either the previously characterized cross-linked protein complex or the same protein complex separated into its individual components. The antibody response, level of protection, and recall response after challenge were then compared among the groups. Herein, we report the results and discuss the findings in the context of vaccine development for bacterial pathogens.
MATERIALS AND METHODS
Antigen preparation. (i) Anaplasma marginale isolation.
Intact A. marginale cells were isolated from infected cells as previously described (12), with the following modifications. Sonication (Branson digital sonifier 450; 400-W maximum output) was done at 40% of maximum for 3 min total in 30-s intervals. Isolated bacteria were resuspended in 500 μl of phosphate-buffered saline (PBS) and stored at −80°C.
(ii) Formulation and purification of the immunogen.
To create the linked immunogen, intact bacteria were treated with 3,3′-dithiobis[sulfosuccinimidyl propionate] (DTSSP), a membrane-impermeable cross-linking agent that reacts with primary amines and has a disulfide bond within the linking arm. The bacteria were lysed, and then gel electrophoresis was used to separate the resulting protein complexes from other cellular components, as described in detail previously (12). To create the unlinked immunogen, the linked immunogen was treated with 10 mM dithiothreitol (DTT) in 100 mM ammonium bicarbonate for 1 h at 56°C. After removal of the DTT, the reduced complexes were alkylated with 50 mM iodoacetamide in 100 mM ammonium bicarbonate for 45 min in the dark at room temperature. To ensure that any remaining complexes or partially reduced complexes were excluded from the immunogen, a second gel purification was done using a 4% polyacrylamide stacking gel as follows. The immunogen was boiled for 3 min in SDS-PAGE sample buffer containing β-mercaptoethanol and run on a 4% polyacrylamide stacking gel layered over a 0.8% agarose stacking gel, thus excluding large complexes and allowing for the recovery of all reduced components in one band at the dye front. A portion of the gel was stained with SYPRO Ruby (Invitrogen, Carlsbad, CA) to verify the presence of protein.
Evaluation of immunogen.
Using previously published methods, SDS-PAGE gel electrophoresis was done in nonreducing conditions followed by either staining for total protein using SYPRO Ruby or Western blotting to demonstrate effective reduction of the disulfide bonds in the unlinked immunogen. For Western blotting, monoclonal antibodies to detect Omp9 (121/1055, 4 μg/ml) were used as previously described (12).
Quantitative Western blotting was done to verify that approximately equal amounts of a given protein were present in the doses of linked and unlinked immunogen. We boiled 9.4 μg of each immunogen for 3 min in sample buffer containing β-mercaptoethanol, followed by treatment with 0.1 M iodoacetamide for 15 min in the dark. Following transblotting, Western blotting was done as described previously (19) using 4 μg/ml of a monoclonal antibody (121/1055) directed against Omp9. Densitometry was done using Quantity One 4.6.9 one-dimensional (1-D) analysis software (Bio-Rad), and a Student's t test was used to determine if differences between groups were statistically significant (JMP software version 9; JMP, Cary, NC).
Immunization and challenge. (i) Animals.
The bovine lymphocyte antigen-DRB3 alleles of 15 Holstein steers were determined by the PCR restriction fragment length polymorphism method and sequencing exon 2 of the DRB3 gene (20–22). The animals were allocated into three groups of five animals per group such that haplotypes were matched or half-matched among all groups (Table 1).
Table 1.
Immunization with linked proteins results in higher titers than immunization with unlinked proteins
| Immunization group and animal no. | DRB3 haplotype | Postimmunization titer |
|
|---|---|---|---|
| IgG | IgG2 | ||
| Linked proteins | |||
| 66 | *0101/*0101 | ≥10,000 | ≥10,000 |
| 124 | *1101/*1201 | ≥10,000 | ≥10,000 |
| 130 | *0101/*1501 | ≥10,000 | ≥10,000 |
| 1252 | *2703/*1201 | ≥10,000 | ≥10,000 |
| 100 | *1501/*1501 | ≥10,000 | ≥10,000 |
| Unlinked proteins | |||
| 95 | *0101/*0101 | 300 | <100 |
| 142 | *1101/*1201 | 300 | 1,000 |
| 150 | *1101/*1501 | 300 | 1,000 |
| 149 | *2703/*2703 | 300 | 1,000 |
| 164 | *0101/*1501 | 300 | 100 |
| Adjuvant only | |||
| 107 | *1010/*1010 | <100 | <100 |
| 102 | *1101/*0201 | <100 | <100 |
| 123 | *1010/*1501 | <100 | <100 |
| 131 | *2703/*2703 | <100 | <100 |
| 103 | *1501/*1501 | <100 | <100 |
The animals were immunized subcutaneously at 3-week intervals with 40 μg of either linked proteins or unlinked proteins suspended in 1 mg of saponin in a total volume of 1 ml. The third group of calves was similarly immunized on the same schedule with 1 mg of saponin only.
(ii) Challenge.
Animals were challenged 2 months after the final immunization by intravenous inoculation of 1 × 104 A. marginale (St. Maries strain) cells, as previously described (12).
Evaluation of immunization. (i) Determination of titers.
Titers were determined using SDS-PAGE and immunoblotting as described previously (12). The equivalent of 6 × 108 A. marginale cells were loaded in each well, electrophoresed at 70 to 80 V, and transferred to nitrocellulose. To determine the total immunoglobulin G (IgG) and IgG2 titers, serum was diluted from 1:100 to 1:100,000. Antibody binding was detected with horseradish peroxidase-labeled goat anti-bovine IgG (Kirkegaard & Perry Laboratories, Gaithersburg, MD) or horseradish peroxidase-labeled mouse anti-bovine IgG2 (Serotec, Oxford, United Kingdom) at a 1:4,000 dilution and developed with an enhanced chemiluminescence (ECL) Western blotting detection system (Pierce, Rockford, IL). Statistical significance of the differences between postimmunization titers was determined using the chi-square test (JMP software version 9). The statistical significance of differences in the antibody response following challenge was determined using the Kruskal-Wallis test based on data ranks blocked by days postinfection. Statistical differences between individual groups were determined using the Wilcoxon rank-sum test (JMP software version 9).
(ii) Measurement of protection.
Starting at 10 days postchallenge, all animals were bled daily, and the packed cell volume and bacteremia were determined by capillary tube centrifugation and counting of the percentage of infected erythrocytes in Giemsa-stained blood smears, respectively. These parameters were tracked until the percentage of infected erythrocytes returned to below 1%. Data analysis started 27 days postchallenge, which was the first day infected erythrocytes were microscopically detectable. An analysis of variance on data ranks (SAS software version 9.1; SAS, Cary, NC) was done to determine if differences in bacteremia among the groups were statistically significant.
RESULTS
Evaluation of immunogen.
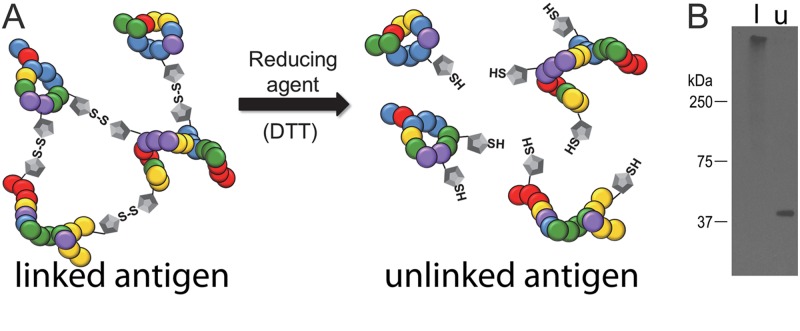
The immunogen in which the proteins were covalently linked (linked immunogen) was made by isolating and treating intact A. marginale with the cross-linking agent DTSSP (12). The resulting high-molecular-weight complexes were then isolated based on size. We derived the second immunogen (unlinked immunogen) from the first by reducing the disulfide bonds within the linking arm of DTSSP, thus separating the complex into individual proteins (Fig. 1).
Fig 1.
Illustration and definition of the linked and unlinked immunogens. (A) A cartoon illustrating the disulfide bonds between adjacent outer membrane proteins formed by the cross-linking agent 3,3′-dithiobis[sulfosuccinimidyl propionate] (DTSSP) in the linked immunogen. The disulfide bonds were reduced through treatment with dithiothreitol (DTT) to form the unlinked immunogen. (B) To determine if reduction of disulfide bonds in the linked immunogen was successful, SDS-PAGE gel electrophoresis was done under nonreducing conditions followed by Western blotting. Omp9 is confined to the stacking gel of the lane containing the linked immunogen (l). The migration of Omp9 to its expected molecular mass of 40 kDa and the lack of additional bands in the unlinked immunogen (u) demonstrate that in the linked immunogen the reduction of the disulfide bonds to produce the unlinked immunogen was successful.
Gel electrophoresis followed by staining for total protein and Western blotting was done to confirm that (i) cross-linking resulted in formation of the expected high-molecular-weight protein complex, (ii) treatment of the linked complexes with DTT was effective in reducing the disulfide bonds to produce the unlinked immunogen, and (iii) any remaining high-molecular-weight complexes were excluded from the unlinked proteins. After formulation, electrophoresis of both immunogens was done under nonreducing conditions to maintain the cross-links in the linked immunogen and to ensure that the treatment and purification of the unlinked immunogen were effective. Findings were similar for the total protein stain and Western blotting to detect Omp9. After we stained the linked proteins for total protein, it was apparent that all detectable protein was maintained in the stacking gel, as expected. As a marker, Omp9 was exclusively identified in the stacking gel, indicating that cross-linking with DTSSP was successful and that individualized proteins were excluded from the immunogen (Fig. 1). For the unlinked proteins, multiple protein bands between approximately 20 and 250 kDa were apparent after staining for total protein, as expected (data not shown) (12). Omp9 was detected only at the expected molecular mass of 40 kDa, indicating that the reduction of the cross-linked immunogen resulted in the individualization of intact proteins (Fig. 1). Additionally, protein staining and Omp9 were absent in the stacking gel containing the unlinked immunogen, indicating exclusion of the large protein complexes in the unlinked immunogen (Fig. 1).
The total amount of protein in each dose of each immunogen was the same; however, the treatment of the linked complexes with DTT to reduce the disulfide bonds may have resulted in protein degradation in the unlinked proteins. To verify that protein degradation was not a significant factor, quantitative Western blotting was done to compare the amounts of Omp9 in both the linked and unlinked immunogens. The mean optical densities of the triplicate bands representing Omp9 were 3,786 ± 7.5 and 3,807 ± 15.4 in the linked and unlinked immunogens, respectively (Fig. 2). There were no statistically significant differences between groups (P > 0.05).
Fig 2.
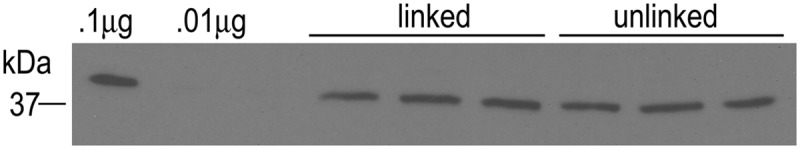
Quantification of proteins within the immunogens using Omp9 as a marker. To verify that approximately equal amounts of a given protein were present in the doses of linked and unlinked immunogen, quantitative Western blotting was done. We electrophoresed 9.4 μg of each immunogen under reducing conditions in triplicate and used 0.1 μg and 0.01 μg of recombinant Omp9 for comparison. Western blotting was done using an anti-Omp9 monoclonal antibody. Bands were evaluated using densitometry. The mean optical densities were 3,786 ±7.5 and 3,807 ± 15.4 in the linked and unlinked immunogens, respectively. The difference was not statistically significant.
Breadth of antibody response to immunization.
Groups of five calves each were immunized with the linked proteins, unlinked proteins, or adjuvant only. The breadth of the antibody response directed against A. marginale antigens isolated from erythrocytes was measured by Western blotting using postimmunization serum. In both groups of animals, the primary antibody response was directed against Msp2 (36 kDa) and Msp3 (74 kDa), the major immunodominant proteins of A. marginale. As previously reported, two other bands were also identified, at 100 kDa and 25 kDa. These findings indicate that a similar complement of antigens was present in each immunogen (Fig. 3), despite the differences in formulation. There was some variation among individual animals; however, an antibody response similar to this was seen previously when using the linked immunogen (12).
Fig 3.
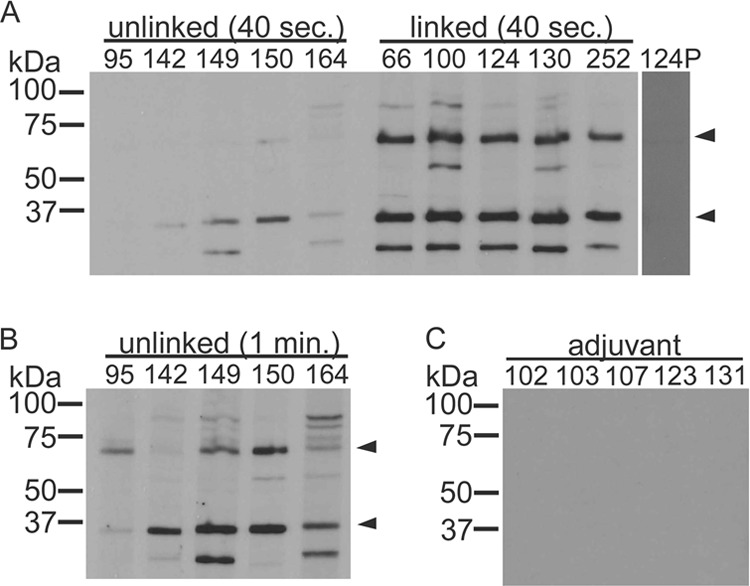
Western blots demonstrating that the breadth of the antibody responses induced by immunization were similar in both groups of immunized animals. An immunoblot was exposed to film for 40 s for both immunized groups (A) and for 1 min for the unlinked group in order to more clearly demonstrate bands (B). Antibodies in both groups were predominantly directed toward Msp2 (36 kDa) and Msp3 (74 kDa), which are known immunodominant A. marginale proteins (arrowheads). The unlinked group received cross-linked outer membrane protein complexes in which the cross-links had been reduced. The linked group received intact cross-linked outer membrane protein complexes. Animal numbers are listed across the top. No specific antibodies were detected in animals prior to the immunization shown for animal 124 (124P) or in animals inoculated with (C) adjuvant alone. The serum dilution was 1:100 for all groups.
Postimmunization antibody titers.
After the third immunization, the A. marginale-specific titers were determined using Western blotting. Overall, the antibody responses were weaker in the animals that received the unlinked immunogen than in the animals that received the linked immunogen. Thus, all animals were immunized two more times to test whether this finding was attributable to an intrinsic difference in immunogenicity that could be overcome with additional booster immunizations. However, after the final immunization, all animals in the group receiving the linked immunogen had total IgG titers that were significantly higher than those in the animals that received the unlinked immunogen (P = 0.0002) (Table 1). IgG2 titers were also done, as IgG2 directed toward outer membrane proteins is associated with protective immunity (11). Similar to the total IgG titers, the IgG2 titers in the animals receiving the linked immunogen were significantly higher (P = 0.001) than in the animals receiving the unlinked immunogen (Table 1).
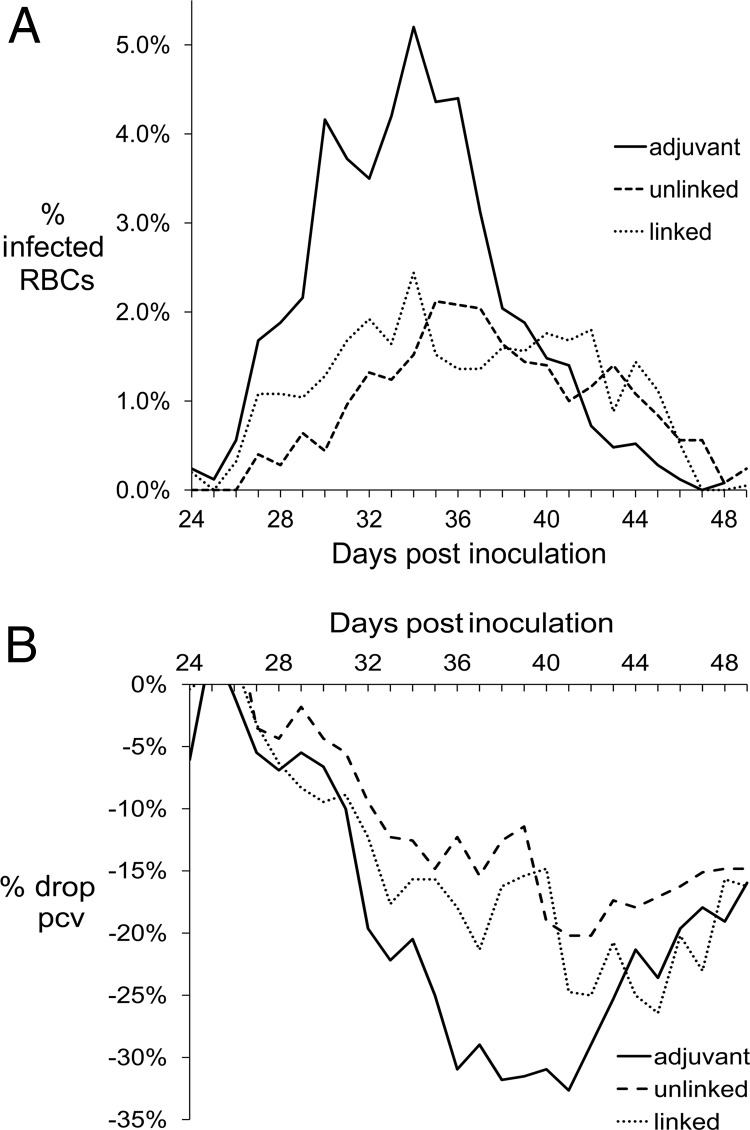
Protection from challenge.
Upon challenge with A. marginale (St. Maries strain), animals immunized with either linked or unlinked proteins were similarly and significantly protected from bacteremia compared to the animals that received only adjuvant (P ≤ 0.0001) (Fig. 4). The means of the maximum percent infected erythrocytes were 3.0% and 3.3%, respectively, for the group that received the linked and unlinked immunogens and 8.0% for the group that received only adjuvant. Both groups of immunized animals developed less severe anemia than the animals that received only adjuvant. Specifically, the group that received the linked proteins and the group that received the unlinked proteins had 35% and 27% drops in packed cell volumes, respectively. The group that received only adjuvant had a 39% drop in packed cell volume. These differences were not statistically significant.
Fig 4.
Protection against A. marginale upon challenge. The two immunogens induced similar levels of protection from challenge. The means of the percent infected erythrocytes (A) or percent drop in packed cell volume (B) are plotted by days postinoculation. The adjuvant group received only adjuvant and served as the positive control. The linked group was immunized with cross-linked outer membrane proteins. The unlinked group was immunized with the outer membrane protein complexes, in which the cross-links had been reduced. The immunized groups were significantly (P ≤ 0.0001) protected from bacteremia, but not anemia, compared to the control group.
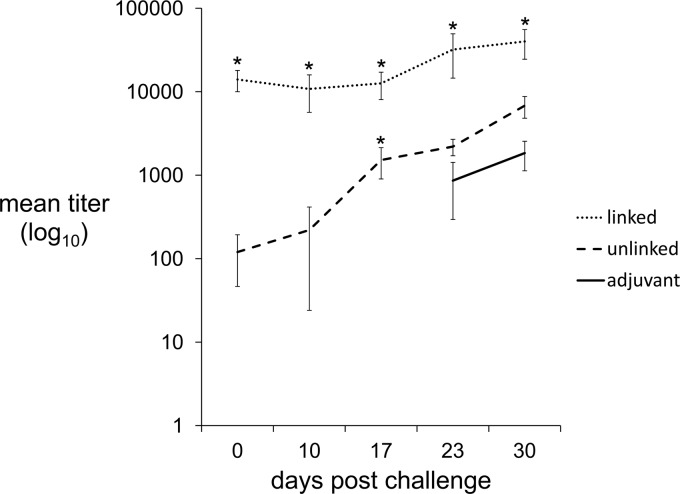
Antibody recall response postchallenge.
The induction of a memory response is essential for effective immunization. The antibody titers induced by the unlinked immunogen were low compared to those induced by the linked immunogen. However, levels of protection from challenge were similar between these groups; thus, we hypothesized that immunization with the unlinked proteins was sufficient to induce a recall response. To test this hypothesis, we used Western blotting to perform weekly measurements of the change in total IgG titers from the time of challenge to 30 days postchallenge (Fig. 5). The rise in titers in both immunized groups was exponential, while the rise in titers of the group that received the adjuvant was linear. In the group immunized with the unlinked proteins, a rise in titer occurred between 10 and 17 days postinoculation. In comparison, titers started to rise between days 17 and 23 in the group that received only adjuvant. This difference was statistically significant (P = 0.007); thus, the hypothesis was accepted.
Fig 5.
Linked and unlinked immunogens induce an antibody recall response. The total IgG titers were measured weekly after challenge. Titers in the group that received the unlinked immunogen started to rise between days 10 and 17 postchallenge. Titers in the control group started to rise in response to challenge between days 17 and 23. Asterisks indicate a statistically significant difference between immunized (linked or unlinked) groups and the control (adjuvant) group. In the group receiving only adjuvant, titers were less than 100 in all animals between days 0 and 17 and thus are not plotted on the log10 scale.
DISCUSSION
The known immunogens that confer protective immunity to A. marginale are (i) a complete outer membrane preparation and (ii) a cross-linked complex of 11 outer membrane proteins (10–12). The complete outer membrane preparation is composed of fragments of outer membranes, as visualized by electron microscopy, and includes more than 20 proteins, as determined by two-dimensional gel electrophoresis (10, 23). Similarly, in the protein complex, the intermolecular relationship between proteins is maintained by using chemically induced covalent bonds rather than membrane components. While both of these immunogens are protective, the specific antigens and, importantly, the requirement for the maintenance of intermolecular relationships between proteins in the induction of protective immunity are unknown. In this study, we demonstrated that the maintenance of these intermolecular relationships enhanced the immunogenicity of the preparation. Thus, we accept the first hypothesis that the maintenance of a physical association between proteins enhances immunogenicity. However, both formulations of the immunogen were similarly and significantly protective in the face of challenge. Thus, the second hypothesis, that the physical linkage between proteins is necessary for protection from challenge, is rejected. Differences between major histocompatibility complex (MHC) class II haplotypes are unlikely to account for the observed differences in antibody responses, because animals were distributed among the groups based on their MHC class II haplotypes such that animals with matched and half-matched haplotypes were equally distributed among the three groups (Table 1).
There are several possible explanations for the enhanced antibody response to the linked proteins compared to the individualized proteins. Differences in protein content of the linked and unlinked immunogens are unlikely to play a significant role in the observed differences, as equal quantities of total protein were administered to each animal. However, differences in the quantities of individual proteins may vary between the immunogens due to protein breakdown during reduction of the disulfide bonds to produce the unlinked immunogen. To test for this possibility, we used Omp9 as a marker. The optical densities of Omp9 were similar in the linked and unlinked immunogens. Thus, it is unlikely that the observed differences in titers were due to variations in the amounts of individual proteins within each immunogen. Additionally, in the group receiving the unlinked immunogen, there was little change in the titers between the third immunization and the fifth immunization (data not shown), indicating that the unlinked immunogen had inherently low immunogenicity and increased exposure to the immunogen had little effect on the antibody response.
Due to linked recognition, the linked proteins may have elicited an enhanced antibody response compared to the unlinked proteins. Linked recognition is the principle whereby a given B cell can be activated only by helper T cells that respond to the same antigen. By the linking of B-cell and T-cell epitopes in close proximity to one another, the antibody response to poorly immunogenic molecules can be enhanced. This method is used in the Haemophilus influenzae type b childhood vaccine to prevent meningitis (24, 25). In this vaccine, the poorly immunogenic, but protective, B-cell epitope (capsular polysaccharide) is conjugated to the strongly antigenic tetanus toxoid. CD4+ T cells primed during recognition of tetanus toxoid antigen can then provide help to B cells, which produce antibody targeting the capsular polysaccharide. In the case of A. marginale, linkage between molecules has been shown to enhance antibody production and may also increase the repertoire of antibodies that target the type IV secretion system proteins (26, 27).
A second but not mutually exclusive possibility is that the physical properties of the immunogen, such as particulate size and charge, maintenance or partial maintenance of conformational epitopes or repetitive elements, or an increased depot effect of the linked protein immunogen, led to more efficient antigen uptake, processing, and presentation compared to the unlinked protein immunogen (28–31).
Regardless of the formulation, both groups of animals were similarly and significantly protected from challenge, indicating that the protective epitope or epitopes were present in both antigens. The antibody response to the unlinked immunogen was low overall. However, by day 17 postchallenge all animals in this group had a >3-fold rise in titers. The mean rise in titers from challenge to 30 days postchallenge in this group was approximately 3,000-fold. In comparison, it was not until day 30 that all animals that received adjuvant demonstrated a >3-fold rise in titers. In the animals that received the linked proteins, titers were high at the time of challenge (mean titer, 14,000), dropped in two animals during the first 10 days after challenge, and then increased (mean titer at 30 days postchallenge, 40,000), demonstrating a strong response both to immunization and to challenge; thus both immunogens induced a memory response capable of being recalled upon challenge.
Several possible mechanisms are involved in models that may explain the lack of correlation between antibody titer and protective immunity. The first is that overall antibody titer is not measuring antibody directed against the key neutralization-sensitive epitopes. This is supported by evidence that the predominant antibody response (and thus the titer) to A. marginale is directed against two immunodominant, abundant, hypervariable surface proteins, Msp2 and Msp3. However, results of recent studies suggest that the immune response directed toward these surface proteins does not play a major role in vaccine-induced immunity. Thus, the lack of correlation between protection and titer could be attributed to the discordance between the overall titer and neutralizing antibody. The second is that there may be effector mechanisms, including complement-mediated opsonization, which are triggered by a threshold level of antibody. Antibody levels above the threshold would not add to the response, and thus the observed lack of correlation would result. The third is that protection is antibody independent. While the current model suggests that the effective immune response directed against A. marginale is a Th1-type response characterized by secretion of gamma interferon (IFN-γ) and interleukin-2 (IL-2), the mechanism has not been definitively identified and there are several known (and undoubtedly yet undiscovered) mechanisms that may be involved in protective immunity. It is unlikely that classical MHC-restricted CD8+ cytotoxic T cells play a significant role in immunity, as A. marginale resides in erythrocytes, which lack MHC molecules. However, there may be roles for nonclassical CD8+ T cells or gamma-delta T cells in immunity. The data in the present article provide both the rationale and an experimental model to identify these mechanisms.
Overall these findings suggest that future studies to identify protective antigens can rely on testing of individual or groups of candidate proteins with less emphasis on the maintenance of intermolecular relationships between proteins, thus simplifying the process of antigen discovery. Once the protective antigen or antigens have been identified, linking those antigens into high-molecular-weight complexes, which is readily accomplished, may be a useful tool in enhancing their immunogenicity in order to minimize the number of doses required for the induction of protective immunity.
ACKNOWLEDGMENTS
We thank Emma Karel, Ralph Horn, and James Allison for their expertise and assistance in animal care. We are grateful for the technical assistance of Shelly Whidbee, Beverly Hunter, Debbie Alperin, Eric Sutten, and Kathleen White. Marc Evans assisted with the statistical analysis.
This research was supported by USDA-ARS grant 5348-32000-033-00D, USDA-ARS cooperative agreement 5348-3200-033-03S, and National Institutes of Health grants AI044005, AI007025, and AI053692.
Footnotes
Published ahead of print 27 February 2013
REFERENCES
- 1. Silhavy TJ, D Kahne, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2:a000414 doi:10.1101/cshperspect.a000414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bishop RE. 2008. Structural biology of membrane-intrinsic beta-barrel enzymes: sentinels of the bacterial outer membrane. Biochim. Biophys. Acta 1778:1881–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hatch TP. 1996. Disulfide cross-linked envelope proteins: the functional equivalent of peptidoglycan in chlamydiae? J. Bacteriol. 178:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huntley JF, Conley PG, Rasko DA, Hagman KE, Apicella MA, Norgard MV. 2008. Native outer membrane proteins protect mice against pulmonary challenge with virulent type A Francisella tularensis. Infect. Immun. 76:3664–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xu Y, Guan W, Xu JN, Cao DP, Yang BB, Chen DL, Chen JP. 2011. Evaluation of the protective immunity of the Legionella pneumophila recombinant protein FlaA/MompS/PilE in an A/J. mouse model. Vaccine 29:4051–4057 [DOI] [PubMed] [Google Scholar]
- 6. Turbyfill KR, Hartman AB, Oaks EV. 2000. Isolation and characterization of a Shigella flexneri invasin complex subunit vaccine. Infect. Immun. 68:6624–6632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fulop M, Manchee R, Titball R. 1996. Role of two outer membrane antigens in the induction of protective immunity against Francisella tularensis strains of different virulence. FEMS Immunol. Med. Microbiol. 13:245–247 [DOI] [PubMed] [Google Scholar]
- 8. Sjostedt A, Sandstrom G, Tarnvik A. 1992. Humoral and cell-mediated immunity in mice to a 17-kilodalton lipoprotein of Francisella tularensis expressed by Salmonella typhimurium. Infect. Immun. 60:2855–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mann BJ, Ark NM. 2009. Rationally designed tularemia vaccines. Expert Rev. Vaccines 8:877–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tebele N, McGuire TC, Palmer GH. 1991. Induction of protective immunity by using Anaplasma marginale initial body membranes. Infect. Immun. 59:3199–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brown WC, Shkap V, Zhu D, McGuire TC, Tuo W, McElwain TF, Palmer GH. 1998. CD4(+) T-lymphocyte and immunoglobulin G2 responses in calves immunized with Anaplasma marginale outer membranes and protected against homologous challenge. Infect. Immun. 66:5406–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Noh SM, Brayton KA, Brown WC, Norimine J, Munske GR, Davitt CM, Palmer GH. 2008. Composition of the surface proteome of Anaplasma marginale and its role in protective immunity induced by outer membrane immunization. Infect. Immun. 76:2219–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Palmer GH, Barbet AF, Cantor GH, McGuire TC. 1989. Immunization of cattle with the MSP-1 surface protein complex induces protection against a structurally variant Anaplasma marginale isolate. Infect. Immun. 57:3666–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Palmer GH, Barbet AF, Davis WC, McGuire TC. 1986. Immunization with an isolate-common surface protein protects cattle against anaplasmosis. Science 231:1299–1302 [DOI] [PubMed] [Google Scholar]
- 15. Palmer GH, McElwain TF. 1995. Molecular basis for vaccine development against anaplasmosis and babesiosis. Vet. Parasitol. 57:233–253 [DOI] [PubMed] [Google Scholar]
- 16. Palmer GH, McGuire TC. 1984. Immune serum against Anaplasma marginale initial bodies neutralizes infectivity for cattle. J. Immunol. 133:1010–1015 [PubMed] [Google Scholar]
- 17. Palmer GH, Oberle SM, Barbet AF, Goff WL, Davis WC, McGuire TC. 1988. Immunization of cattle with a 36-kilodalton surface protein induces protection against homologous and heterologous Anaplasma marginale challenge. Infect. Immun. 56:1526–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abbott JR, Palmer GH, Kegerreis KA, Hetrick PF, Howard CJ, Hope JC, Brown WC. 2005. Rapid and long-term disappearance of CD4+ T lymphocyte responses specific for Anaplasma marginale major surface protein-2 (MSP2) in MSP2 vaccinates following challenge with live A. marginale. J. Immunol. 174:6702–6715 [DOI] [PubMed] [Google Scholar]
- 19. Noh SM, Brayton KA, Knowles DP, Agnes JT, Dark MJ, Brown WC, Baszler TV, Palmer GH. 2006. Differential expression and sequence conservation of the Anaplasma marginale msp2 gene superfamily outer membrane proteins. Infect. Immun. 74:3471–3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Norimine J, Brown WC. 2005. Intrahaplotype and interhaplotype pairing of bovine leukocyte antigen DQA and DQB molecules generate functional DQ molecules important for priming CD4(+) T-lymphocyte responses. Immunogenetics 57:750–762 [DOI] [PubMed] [Google Scholar]
- 21. Park YH, Joo YS, Park JY, Moon JS, Kim SH, Kwon NH, Ahn JS, Davis WC, Davies CJ. 2004. Characterization of lymphocyte subpopulations and major histocompatibility complex haplotypes of mastitis-resistant and susceptible cows. J. Vet. Sci. 5:29–39 [PubMed] [Google Scholar]
- 22. Van Eijk MJ, Stewart-Haynes JA, Lewin HA. 1992. Extensive polymorphism of the BoLA-DRB3 gene distinguished by PCR-RFLP. Anim. Genet. 23:483–496 [DOI] [PubMed] [Google Scholar]
- 23. Lopez JE, Siems WF, Palmer GH, Brayton KA, McGuire TC, Norimine J, Brown WC. 2005. Identification of novel antigenic proteins in a complex Anaplasma marginale outer membrane immunogen by mass spectrometry and genomic mapping. Infect. Immun. 73:8109–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murphy K, Travers P, Walport M. 2008. The humoral immune response, p 379–495 Murphy KM, Travers P, Walport M. (ed), Janeway's immunobiology, 7th ed. Garland Science, New York, NY [Google Scholar]
- 25. Zepp F. 2010. Principles of vaccine design—lessons from nature. Vaccine 28(Suppl 3):C14–C24 [DOI] [PubMed] [Google Scholar]
- 26. Macmillan H, Norimine J, Brayton KA, Palmer GH, Brown WC. 2008. Physical linkage of naturally complexed bacterial outer membrane proteins enhances immunogenicity. Infect. Immun. 76:1223–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morse K, Norimine J, Palmer GH, Sutten EL, Baszler TV, Brown WC. 2012. Association and evidence for linked recognition of type IV secretion system proteins VirB9-1, VirB9-2, and VirB10 in Anaplasma marginale. Infect. Immun. 80:215–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bachmann MF, Jennings GT. 2010. Vaccine delivery: a matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 10:787–796 [DOI] [PubMed] [Google Scholar]
- 29. Jennings GT, Bachmann MF. 2007. Designing recombinant vaccines with viral properties: a rational approach to more effective vaccines. Curr. Mol. Med. 7:143–155 [DOI] [PubMed] [Google Scholar]
- 30. Fifis T, Gamvrellis A, Crimeen-Irwin B, Pietersz GA, Li J, Mottram PL, McKenzie IF, Plebanski M. 2004. Size-dependent immunogenicity: therapeutic and protective properties of nano-vaccines against tumors. J. Immunol. 173:3148–3154 [DOI] [PubMed] [Google Scholar]
- 31. Storni T, Ruedl C, Renner WA, Bachmann MF. 2003. Innate immunity together with duration of antigen persistence regulate effector T cell induction. J. Immunol. 171:795–801 [DOI] [PubMed] [Google Scholar]