Abstract
Salmonella enterica serotype Paratyphi A is a human-restricted pathogen and the cause of paratyphoid A fever. Using a high-throughput immunoscreening technique, in vivo-induced antigen technology (IVIAT), we identified 20 immunogenic bacterial proteins expressed in humans who were bacteremic with S. Paratyphi A but not those expressed in S. Paratyphi A grown under standard laboratory conditions. The majority of these proteins have known or potential roles in the pathogenesis of S. enterica. These include proteins implicated in cell adhesion, fimbrial structure, adaptation to atypical conditions, oxidoreductase activity, proteolysis, antimicrobial resistance, and ion transport. Of particular interest among these in vivo-expressed proteins were S. Paratyphi A (SPA)2397, SPA2612, and SPA1604. SPA2397 and SPA2612 are prophage related, and SPA1604 is in Salmonella pathogenicity island 11 (SPI-11). Using real-time quantitative PCR (RT-qPCR), we confirmed increased levels of mRNA expressed by genes identified by IVIAT in a comparison of mRNA levels in organisms in the blood of bacteremic patients to those in in vitro cultures. Comparing convalescent- to acute-phase samples, we also detected a significant increase in the reaction of convalescent-phase antibodies with two proteins identified by IVIAT: SPA2397 and SPA0489. SPA2397 is a phage-related lysozyme, Gp19, and SPA0489 encodes a protein containing NlpC/P60 and cysteine, histidine-dependent amidohydrolase/peptidase (CHAP) domains. In a previous study utilizing a different approach, we found that transcripts for 11 and 7 of the genes identified by IVIAT were detectable in organisms in the blood of humans in Bangladesh who were bacteremic with S. Paratyphi A and Salmonella enterica serovar Typhi, respectively. S. Paratyphi A antigens identified by IVIAT warrant further evaluation for their contributions to pathogenesis and might have diagnostic, therapeutic, or preventive relevance.
INTRODUCTION
There are >2,000 serotypes of Salmonella, which are non-lactose-fermenting Gram-negative bacteria. Salmonella infection usually manifests as gastroenteritis or a systemic infection. Systemic infection that includes persistent fever, hepatosplenomegaly, and persistent bacteremia is referred to as enteric fever. Enteric fever can be caused by S. enterica serotype Typhi, the cause of typhoid fever, or S. enterica serovar Paratyphi A, B, or C, the causes of paratyphoid fever (1). S. Typhi and S. Paratyphi infect 25 million individuals each year and are the cause of death in approximately 200,000 of those individuals (2). Recently, S. Paratyphi A has been isolated from patients at an increasing frequency in Asian countries, such as Bangladesh, India, Pakistan, Nepal, and Indonesia (3). S. Paratyphi A infection now accounts for approximately one-fifth of enteric fever cases in areas of South Asia (3), and existing typhoid vaccines do not protect against S. Paratyphi A infection. Multidrug-resistant S. Paratyphi A strains that do not respond to commonly used antibiotics are also increasingly being identified (4).
S. Typhi and S. Paratyphi A are human-restricted pathogens, making optimal animal models unavailable for studying host-pathogen interactions in these globally important infections (5). Data on S. Paratyphi A are particularly limited (6). Although they are in development, there currently is no commercially available S. Paratyphi A vaccine and no accurate rapid diagnostic assay to identify individuals with paratyphoid A fever (7). Most of our current understanding of S. Paratyphi A is based on extrapolations from murine models using S. enterica serovar Typhimurium and from studying humans infected with S. Typhi. To further our understanding of bacterial events occurring during human paratyphoid A infection, we applied an immunoscreening technique, in vivo-induced antigen technology (IVIAT), to identify immunogenic S. Paratyphi A proteins expressed in humans who are bacteremic with S. Paratyphi A that are not expressed in S. Paratyphi A organisms grown under standard laboratory conditions. IVIAT has previously been applied successfully to other human pathogens, including Vibrio cholerae, the cause of cholera, Bacillus anthracis, the cause of anthrax, and S. Typhi, the cause of typhoid fever (5, 8, 9).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Genomic DNA from Salmonella enterica serotype Paratyphi A ATCC 9150 (Salmonella Genetic Stock Center, Calgary, Alberta, Canada) was used to construct an inducible genomic expression library in an Escherichia coli BL21(DE3) host strain (New England BioLabs, Ipswich, MA). All strains were grown in Luria-Bertani (LB) broth at 37°C with aeration. Clones containing pET30c constructs (New England BioLabs) were grown in LB broth and solid agar containing 50 μg/ml kanamycin. Glycerol stocks were maintained at −80°C in LB medium supplemented with 15% glycerol (Sigma-Aldrich, St. Louis, MO).
Patient and control serum.
Paired acute- (days 0 to 2) and convalescent-phase (days 14 to 28) serum samples were obtained from eight individuals with S. Paratyphi A bacteremic infection presenting to the Mirpur or Kamalapur field sites of the International Centre for Diarrheal Disease Research, Bangladesh (ICDDR,B) in Dhaka, Bangladesh. In this study, control serum samples were also collected from cholera patients in Bangladesh at the acute and convalescent phases of illness. This study was approved by the institutional review boards at Massachusetts General Hospital, Boston, MA, and the ICDDR,B, Dhaka, Bangladesh.
Adsorption of serum.
Convalescent-phase serum samples from eight individuals who were bacteremic with S. Paratyphi A were pooled. The serum samples were adsorbed extensively with in vitro-grown S. Paratyphi A strain ATCC 9150. The organisms were grown to late log phase under standard laboratory conditions (in vitro) in LB broth (37°C, aeration shaker); the cells were pelleted and resuspended in phosphate-buffered saline (PBS) with 0.05% sodium azide. To produce cell lysates, in vitro-grown cells were lysed with 0.1-mm zirconia-silica beads in a Mini-Beadbeater (BioSpec, Bartlesville, OK) per the manufacturer's instructions. Heat-denatured cell lysates were generated by boiling the cell lysates. Whole-cell S. Paratyphi A, nondenatured cell lysates, and heat-denatured cell lysates were separately immobilized on 0.5-μm polystyrene beads (Bangs Laboratories, Inc., Fishers, IN), and pooled convalescent-phase serum samples were serially adsorbed with whole-cell S. Paratyphi A and these beads. Adsorbed serum was aliquoted and stored at −80°C.
Construction of inducible genomic expression library of S. Paratyphi A.
Genomic DNA was extracted from S. Paratyphi A ATCC 9150 using a Qiagen genomic-tip 100/G and genomic buffer set (Qiagen, Valencia, CA). The DNA was sheared using a Covaris M220 AFA focused-ultrasonicator (Covaris, Woburn, MA) and settings were optimized to yield 500- to 1,500-bp fragments. The resulting fragments were gel purified and extracted using a QIAquick PCR purification kit (Qiagen, Valencia, CA). Blunt ends were created on the S. Paratyphi A fragments using the End-It DNA end-repair kit (Epicentre, Madison, WI), and the fragments were inserted into pET30c treated with EcoRV (New England BioLabs) and calf intestinal alkaline phosphatase (New England BioLabs). These products were electroporated into E. coli DH5α (Invitrogen, Carlsbad, CA) to generate libraries with all three possible open reading frames (ORFs). The resulting plasmids were pooled, and the library mixture was transformed into E. coli BL21(DE3) (New England BioLabs); we confirmed that >80% of the library contained inserts of 500- and 1,500-bp insertion sizes.
Screening the inducible genomic expression library for antigenic proteins expressed during S. Paratyphi A infection.
In the primary immunoscreening, aliquots of the genomic library were plated on LB plates with kanamycin overnight at 37°C to obtain approximately 500 colonies per plate. Colonies were transferred onto nitrocellulose membranes (Invitrogen, Carlsbad, CA), and the membranes were incubated on LB plates containing 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 37°C. Adherent colonies were lysed by placing the nitrocellulose membranes on chloroform-soaked blotting paper for 10 s. The membranes were air dried for 5 min and blocked with phosphate-buffered saline (PBS) containing 5% nonfat milk. The membranes were then incubated with adsorbed serum in PBS-0.25% Tween at a dilution of 1:10,000 overnight at 4°C. Clones reacting with adsorbed serum were detected using peroxidase-conjugated goat anti-human IgG antibody (MP Biomedicals Cappel, Aurora, OH) at a dilution of 1:3,000 in PBS-0.25% Tween. The immunoblots were developed using enhanced chemiluminescence (ECL) Western blotting detection reagents (GE Healthcare, Piscataway, NJ). Immunoreactive clones were recovered and glycerol stocks were frozen at −80°C.
A whole-colony immunoblot assay was used to confirm reactive clones by comparing their immunoreactivities to that of the control strain E. coli BL21(DE3) containing pET30 vector with no insert. Plasmids were purified from reactive clones using a QIAprep spin miniprep kit (Qiagen, Valencia, CA) and the inserts were sequenced with predesigned forward and reverse primers for the T7 promoter and termination sites. Since inserts might contain multiple open reading frames and fragments of genes, each identified gene was separately PCR amplified and cloned into pET30c as NdeI and NotI fragments. Whole-colony immunoblot assay was used in tertiary screening to confirm the immunoreactivities of individually cloned gene products by comparing their immunoreactivities to that of E. coli BL21(DE3) containing pET30 with no insert.
Prediction of function of antigens identified by IVIAT.
The functional classification of antigens identified by IVIAT was predicted using published studies of identified proteins in S. enterica and using the J. Craig Venter Institute (JCVI) Comprehensive Microbial Resource (http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi).
Confirmation of S. Paratyphi A gene expression in patients with S. Paratyphi A bacteremia by real-time quantitative PCR (RT-qPCR).
Bacterial mRNA expression levels of the genes encoding proteins identified by IVIAT were quantified to evaluate whether they were upregulated in the blood of infected humans compared to in vitro cultures. Previously collected blood samples were used that had been immediately stored in TRIzol LS (Invitrogen, Grand Island, NY) at the time of clinical presentation in febrile patients in Dhaka, Bangladesh. Samples from patients (n = 3) who were subsequently confirmed to have been bacteremic with S. Paratyphi A at the time of presentation were used. S. Paratyphi A cultures were grown to mid-logarithmic phase in LB at 37°C to measure mRNA expression levels in in vitro-grown bacteria. RNA was treated with an Ambion DNA-free kit (Austin, TX) and generated cDNA using random decamers and an Ambion RETROscript kit (Austin, TX), per the manufacturer's two-step RT-qPCR protocol. Real-time quantitative PCR analysis was performed using Bio-Rad iQ SYBR green supermix reagent, an MJ Research Chromo4 thermocycler (Bio-Rad), Bio-Rad Hard Shell 96-well BLK/WHT PCR plates, and Bio-Rad Microseal “B” film optical plate covers (Bio-Rad, Hercules, CA). Primers were digested with Beacon Design (Premier Biosoft, Palo Alto, CA), primer3, and BLAST software to optimize reaction thermodynamics. Primers were designed to produce PCR fragments of 150 to 225 bases, and each quantification was performed at least twice. The calculated threshold cycle (CT) was set in the lower/linear portion of product curves. MJ Opticon Monitor software version 3.1 (Bio-Rad, Hercules, CA) was used to quantify gene copy numbers against the concentration curve of pET30 tertiary-screened clone plasmids. Gene copy numbers were normalized against cDNA copies of 16S rRNA.
Comparison of acute- to convalescent-phase immunoreactivities of proteins identified by IVIAT.
To compare the immunoreactivities of S. Paratyphi A proteins identified by IVIAT in acute- versus convalescent-phase serum samples of patients with bacteremia, in vitro transcription-translation-based protein expression and enzyme-linked immunosorbent assay (ELISA) methods were used. S. Paratyphi A proteins were expressed using the Promega E. coli T7 S30 extract system (Promega, Madison, WI), and the resulting products were bound to 96-well ELISA plates (Nunc, Rochester, NY). Wells were immune probed with pooled acute- or convalescent-phase serum from four S. Paratyphi A bacteremic patients (1:150 dilution in PBS with 5% milk) that was distinct from serum in the screening pool. Pooled acute (day 2) or convalescent (day 21) sera were also used from four Bangladeshi patients with cholera caused by Vibrio cholerae O1 who presented to the same care facility in Bangladesh as did the paratyphoid patients. Immunoreactivity was detected with peroxidase-conjugated goat anti-human immunoglobulin (IgG) antibody (MP Biomedicals Cappel, Aurora, OH) at a dilution of 1:1,000 in PBS with 5% milk and was developed using SuperSignal West Femto maximum sensitivity substrate (Thermo Scientific, Rockford, IL). Reactions were read using a SpectraMax Gemini XPS (Molecular Devices, Sunnyvale, CA), and results were expressed as fold-change (sample protein reactivity to empty vector reactivity) that equilibrated cholera immunoreactivity to 1.
Statistical analyses.
Comparisons of gene expression levels between in vivo and in vitro conditions were tested for significance using one-way analysis of variance (ANOVA) (Holm-Šídák). When we compared the differences in the fold-increase of immune responses, we used unpaired t tests. Analyses were performed using GraphPad Prism 5.0 and SigmaStat 3.1.
RESULTS
Identification of S. Paratyphi A antigens by IVIAT.
We screened 100,000 clones from the S. Paratyphi A inducible genomic expression library and identified 505 immunoreactive clones in our primary screening. Forty-seven of these clones had clear and consistent immunoreactivity on repetitive (secondary) screening. Plasmids of these 47 clones contained 87 whole or partial S. Paratyphi A open reading frame fragments. We subcloned each identified open reading frame (ORF) in its entirety into pET30, electroporated resultant plasmids into E. coli BL21(DE3), and confirmed consistent immunoreactivity for 20 resultant clones (Table 1).
Table 1.
Proteins identified by applying IVIAT to S. enterica serotype Paratyphi A
| Functional category | SPA locusa | SPA genea | Description |
|---|---|---|---|
| Cell envelope and adhesion | SPA0021 | bcfA | Fimbrial subunit |
| SPA0181 | stkE | Putative fimbrial protein | |
| SPA0201 | stfA | Putative fimbrial subunit | |
| SPA0703 | stcD | Putative fimbrial protein | |
| SPA1533 | osmE | Osmotically inducible lipoprotein E precursor | |
| SPA1604 | envE | Putative lipoprotein | |
| SPA1645 | ycfL | Putative lipoprotein | |
| SPA3787 | yifL | Putative lipoprotein | |
| SPA4148 | yjeI | Putative membrane protein | |
| Cell wall and antimicrobial resistance | SPA1644 | ycfM | Penicillin binding protein activator, putative lipoprotein |
| Energy metabolism, nutrient acquisition, cellular processes | SPA0489 | NlpC/P60 superfamily of enzymes and CHAP domain | |
| SPA0608 | napB | Cytochrome c type protein, NapB precursor | |
| SPA0652 | cirA | Colicin I receptor precursor, iron acquisition | |
| SPA1239 | ldhA | d-Lactate dehydrogenase | |
| SPA1734 | scsD | Suppressor for copper sensitivity D, secreted protein | |
| SPA2362 | Probable terminal oxidase subunit II | ||
| SPA2948 | yggG | Putative metalloendopeptidase | |
| SPA4117 | melB | Melibiose carrier protein | |
| Mobile and extrachromosomal element functions | SPA2397 | gp19 | Phage lysozyme |
| SPA2612 | Putative phage gene |
SPA, S. Paratyphi A.
Functional classification of antigens identified by IVIAT.
We were able to assign a functional classification to the 20 proteins expressed from genes identified by IVIAT (Table 1). The majority of the proteins have known or potential roles in the pathogenesis of S. enterica infection. These included proteins that are implicated in fimbrial structure (S. Paratyphi A [SPA]0021, SPA0181, SPA0201, and SPA0703), the cell envelope and membrane (SPA1533, SPA1604, SPA1645, SPA3787, and SPA4148), energy metabolism and cellular processes (SPA0489, SPA0608, SPA0652, SPA1239, SPA1734, SPA2362, SPA2948, and SPA4117), and antimicrobial resistance (SPA1644). Two genes were phage related (SPA2397 and SPA2612) (10).
Comparison of S. Paratyphi A genes identified by IVIAT to homologs in other Enterobacteriaceae.
Homologs of genes encoding 10 of the 20 S. Paratyphi A proteins identified by IVIAT are not found in E. coli (SPA0021, SPA0181, SPA0201, SPA0489, SPA0703, SPA1604, SPA1734, SPA2397, SPA2362, and SPA2612). Two of these genes, SPA2397 and SPA2612, reside in the prophage regions of the S. Paratyphi A genome designated SPA-1 and SPA-3-P2, respectively. Ten of the genes (SPA0608, SPA0652, SPA1239, SPA1533, SPA1644, SPA1645, SPA2948, SPA3787, SPA4117, and SPA4148) have homologs in E. coli as well as in various Salmonella spp., including S. Typhi, S. Paratyphi B and C, and nontyphoidal Salmonella spp. Homologs of 5 of the 20 S. Paratyphi A genes identified by IVIAT are not found in S. Typhi (or E. coli): SPA0181, SPA0201, SPA0703, SPA2397, and SPA2612. One of these, the stkE (SPA0181) gene that encodes a putative fimbrial protein, is specific to S. Paratyphi A and S. enterica serotype Heidelberg, and it is not found in other bacteria.
Real-time quantitative PCR analysis of expression of IVIAT-identified antigens.
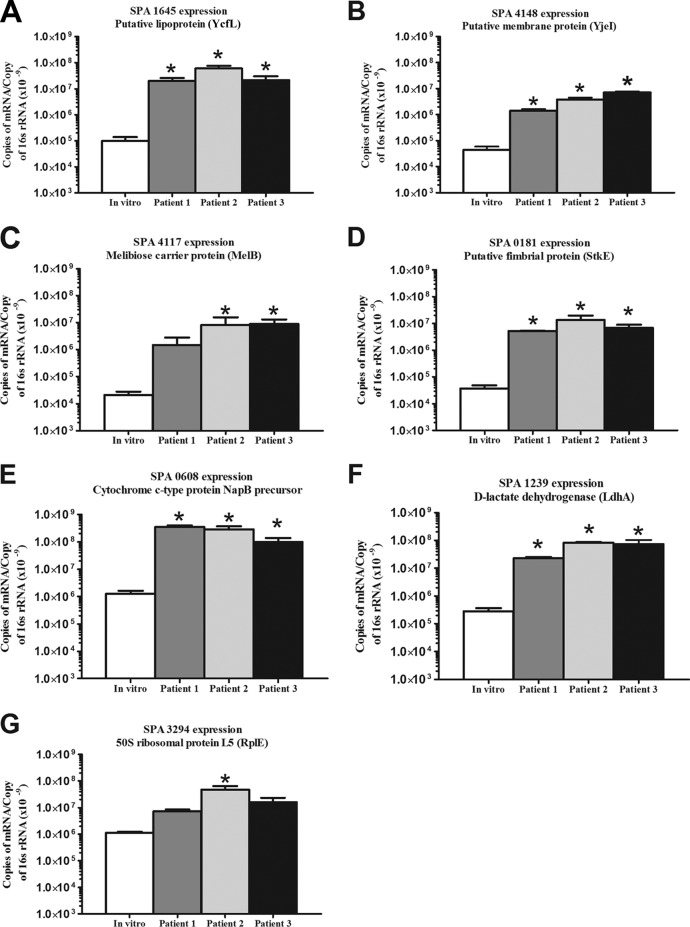
To confirm the IVIAT results, we quantified the expression of six of the identified S. Paratyphi A genes (SPA0181, SPA0608, SPA1239, SPA1645, SPA4117, and SPA4148) in the blood of all three S. Paratyphi A-infected patients. We were unable to detect a sufficient mRNA signal for the remaining genes in TRIzol-preserved in vivo samples, perhaps reflecting the low organism CFU/ml load that is common in enteric fever (11). For the genes whose mRNA we were able to detect in vivo, we compared the expression levels in bacteremic humans to those in in vitro-grown bacterial samples, normalizing by 16S rRNA. We also assessed in vivo versus in vitro expression levels for the S. Paratyphi A housekeeping gene SPA3294 (rplE) that encodes the 50S ribosomal protein L5 (12). We found that the genes whose proteins were identified by IVIAT were upregulated in the blood of bacteremic patients compared to the in vitro-grown cultures (Fig. 1). The fold-changes of the expression of these genes between in vivo and in vitro samples were 125- to 2,103-fold.
Fig 1.
Real-time quantitative PCR analysis of S. Paratyphi A genes identified by IVIAT (A to G) comparing RNA recovered from blood of bacteremic patients to in vitro culture of S. Paratyphi A. Analysis also includes a housekeeping gene, SPA3294 (G), which encodes the 50S ribosomal protein L5 (RplE). The mean number of copies of mRNA per copy of 16S rRNA and the standard errors of the mean are presented. *, P < 0.05.
In vitro transcription-translation-based ELISA.
Comparing convalescent- to acute-phase serum samples, we detected a significant increase in the reaction of convalescent-phase IgG antibodies with two genes identified by IVIAT, SPA0489 and SPA2397 (P ≤ 0.05; Fig. 2), while control patients with cholera did not have any significant changes in immunoreactivity between samples. SPA2397 encodes Gp19, a lysozyme, and SPA0489 encodes a protein that contains NlpC/P60 and cysteine, histidine-dependent amidohydrolase/peptidase (CHAP) domains.
Fig 2.
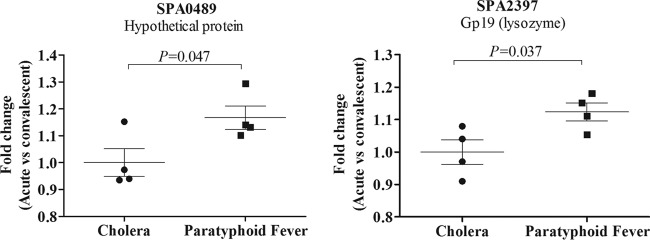
Fold-change immunoreactivity (reactivity in convalescent-phase compared to that in acute-phase serum samples) to Gp19 (SPA2397) and SPA0489 (protein of unknown function) in samples obtained from patients infected with S. enterica serotype Paratyphi A or V. cholerae (controls). Acute- and convalescent-phase samples were collected at days 0 and 14 to 28, respectively. Columns represent means, and error bars represent standard errors of the mean.
DISCUSSION
In this study, we applied the IVIAT technique to S. Paratyphi A and identified 20 immunogenic proteins expressed uniquely in vivo. IVIAT is a technique that identifies bacterial antigens expressed in infected humans and not in bacteria grown under standard laboratory conditions. Identifying such a subgroup of antigens can assist in focusing analysis efforts, since antigens expressed uniquely in humans might play significant or critical roles in in vivo survival or pathogenesis. For 11 of the S. Paratyphi A genes encoding proteins identified by IVIAT, we have previously demonstrated by a different capture and amplification transcriptional profiling technique, selective capture of transcribed sequences (SCOTS), mRNA expression in the blood of humans in Bangladesh who were bacteremic with S. Paratyphi A (12). Using SCOTS, we have similarly identified seven of the corresponding 20 genes in the blood of humans who were bacteremic with S. Typhi in Bangladesh (13). We identified mRNA for four of the genes in both S. Paratyphi A- and S. Typhi-infected patients (12, 13). Our identification by transcriptional analysis of the majority of the genes detected by IVIAT suggests the complementary nature of these approaches and might support the potential importance of these genes in infected humans.
The 20 genes identified in this IVIAT study can be categorized into several functional groups. These include proteins involved in cell adhesion, fimbrial structure, adaptation to atypical conditions, energy metabolism, oxidoreductase activity, proteolysis, antimicrobial resistance, and ion transport. Our results suggest that S. Paratyphi A expresses genes in vivo that might be involved in recognizing and responding to the in vivo environment, including genes involved in energy metabolism, use of alternate energy sources, and membrane attachment and signaling. These observations are in agreement with those from our previous transcriptional-based analyses (12, 13).
Previous studies have shown that the fimbrial proteins of S. Typhi (5) and S. Typhimurium (14) are upregulated under in vivo compared to in vitro conditions. In this study, we also identified a number of fimbrial genes that were upregulated, including bcfA, stfA, stkE, and stcD, and whose proteins were immunogenic. BcfA and StfA are detected on the surfaces of bacterial cells after oral infection of mice with S. Typhimurium and are not expressed when the bacteria are grown in vitro (14, 15). Additionally, infected mice that were seroconverted to these fimbrial proteins (14), and mice immunized with BcfA-glutathione S-transferase (GST) and StfA-GST fusion proteins had reduced fecal shedding after challenge with S. Typhimurium (14). Homologs of bcfA and stfA are found in nontyphoidal Salmonella spp. and S. Paratyphi B and C, but not in E. coli. S. Typhi has a homolog of bcfA, but not stfA. stkE and stcD were also identified in our screen, as both are putative fimbrial genes involved in cell adherence. In mice infected with S. Typhimurium, StcD appears to be involved in gastrointestinal and long-term systemic infections (16, 17). Homologs of stcD are found in nontyphoidal Salmonella, but not in S. Typhi, S. Paratyphi B or C, or E. coli. The stk operon was reported at first to be specific for S. Paratyphi A (18, 19); however, a homolog of stkE appears to be present in the S. enterica serotype Heidelberg strain. Using RT-qPCR, we confirmed the increased in vivo expression of the transcript of stkE in the blood of patients with S. Paratyphi A bacteremia, suggesting that StkE may play some role in adherence in infected humans.
In addition to fimbrial antigens, we identified a number of cell envelope proteins in our analyses. YjeI, a putative membrane protein, has homologs in S. Typhi, S. Typhimurium, and E. coli. In E. coli, yjeI is upregulated by external acidification (20). In our study, we confirmed upregulation of yjeI in the blood of patients infected with S. Paratyphi A. YifL is a putative lipoprotein with homologs in various organisms, including S. Typhi, nontyphoidal Salmonella spp., E. coli, and also S. Paratyphi B and C. EnvE is a putative lipoprotein expressed by envE (SPA1604) located within Salmonella pathogenicity island 11 (SPI-11). SPI-11 contains a number of genes involved in S. enterica pathogenesis, including pagC, whose expression is regulated in vivo by the PhoP/Q regulon (21, 22). The PhoP/Q regulatory system controls a network of S. enterica genes involved in virulence and survival within macrophages (21, 22). Of note, envE is present in a variety of Salmonella spp. and is transcribed in the opposite direction of pagC. osmE is also found in S. Typhi, S. Typhimurium, and E. coli, and encodes an outer membrane protein. osmE is induced by elevated osmolarity and might be useful for adaptation to atypical conditions that typically lead to poor bacterial growth (23, 24). In a previous study, the gene expression profiles of S. Typhi-infected macrophages demonstrated upregulation of osmE compared to in vitro conditions (25). Of the 20 proteins identified by IVIAT, two were in the same operon: ycfL and ycfM. The ycf operon harbors six genes (encoding a hypothetical protein, two putative lipoproteins, beta-hexosaminidase, thiamine kinase, and purine nucleoside phosphoramidase). YcfL is a putative lipoprotein that is potentially involved in cell adhesion. YcfM-LpoB is a penicillin-binding protein (PBP) activator essential for peptidoglycan synthesis. A previous study showed that the outer membrane lipoprotein LpoB is an essential cofactor for PBP action, and it promotes peptidoglycan synthesis in E. coli (26). Homologs of both ycfL and ycfM are present in a wide variety of organisms, including both typhoidal and nontyphoidal Salmonella spp. and E. coli.
Using IVIAT, we identified a number of S. Paratyphi A genes that encode proteins involved in energy metabolism, nutrient acquisition, and transport and binding (melB, napB, ldhA, scsD, cirA, yggG, SPA0489, and SPA2362), and we confirmed the increased expression of three of these genes (melB, ldhA, and napB) by RT-qPCR in the blood of S. Paratyphi A-infected patients. melB is present in a variety of organisms, including E. coli and different Salmonella species, and encodes a melibiose permease (27). In S. Typhimurium, melibiose permease is a symporter that couples melibiose transport with the transport of Na+, Li+, or H+ cations (28). scsD encodes the secreted protein ScsD, a suppressor of copper sensitivity that has homologs in a number of bacteria (29). S. Typhimurium ScsD restores copper tolerance in copper-sensitive E. coli mutants (29). Another metabolism- and nutrient-related gene identified by IVIAT is cirA, which encodes a colicin I receptor. During infection, S. enterica requires iron for survival, metabolism, and pathogenesis (10). CirA is an outer membrane receptor that controls catecholate-mediated iron uptake in Salmonella (30). CirA is vital for norepinephrine-enhanced growth and motility (31, 32), and cirA mutants colonize mice poorly. napB is a cytochrome c-type protein involved in anaerobic respiration in S. Typhimurium (33), suggesting a potential role in the survival of S. Paratyphi A in the in vivo environment.
The ldhA gene, encoding d-lactate dehydrogenase, is involved in pyruvate metabolism. In E. coli, this enzyme converts pyruvate to d-lactate under anaerobic and low-pH conditions (34). In an acidic pH environment, E. coli upregulates the expression of this enzyme by 10-fold (34), and the deletion of ldhA in E. coli results in reduced glucose consumption and bacterial growth (35). Active transport of metabolites via membrane-bound vesicles is also energetically coupled to d-lactate dehydrogenase activity in E. coli (36).
YggG is a zinc-dependent metalloprotease with homologs in various pathogenic organisms, including S. Typhi, nontyphoidal Salmonella spp., and E. coli (37, 38). YggG is a membrane-associated heat shock protein that binds to the Era protein in stress response (38). Era is a membrane-associated GTPase and is important for the viability of E. coli (39). In E. coli under normal conditions, the expression of yggG is upregulated by heat shock and UV irradiation (39).
Using IVIAT, we also identified SPA0489; this gene encodes a protein with NlpC/P60 and CHAP domains (40, 41). NlpC/P60 proteins are a family of cell wall peptidases found in several bacterial lineages (40), with CHAP domains being associated with amidase function (41). SPA0489 is only present in Salmonella spp. We confirmed convalescent-phase reactivity to SPA0489 in humans recovering from paratyphoid fever, supporting the expression of this protein in vivo.
The S. Paratyphi A genome harbors three prophages, SPA-1, -2, and -3 (10). SPA-1 is a lambdoid prophage and encodes 47 genes (SPA2385 to SPA2431). SPA-2 and -3 are P2-type prophages designated SPA-2-SopE (SPA2554 to SPA2600) and SPA-3-P2 (SPA2601 to SPA2625) (10). We identified Gp19 (SPA2397), which is encoded in SPA-1, and SPA2612, which is encoded in SPA-3-P2, in our analysis. We confirmed convalescent-phase reactivity to Gp19 in humans recovering from paratyphoid fever, supporting the expression of GP19 in vivo.
Our study has a number of limitations. IVIAT does not identify antigens expressed both in vitro and in vivo that might play important roles in pathogenesis and host-pathogen interactions. IVIAT also only identifies antigens that stimulate humoral immunity and does not identify antigens that induce only innate or cellular responses (5). Also, IVIAT does not predict whether any of the identified immune responses are involved in clearing infection or mediating host protection. It simply uses a comparative immunoscreening technique to identify antigens expressed in vivo that might warrant further evaluation. During IVIAT, we also adsorbed convalescent-phase serum using bacteria grown in LB broth at 37°C with aeration. It is very possible that protein expression profiles might vary under a range of in vitro conditions, but once again, the purpose of IVIAT is to identify antigens that might warrant additional analysis. Also, in our study, we were only able to confirm the specific upregulation of gene expression for 6 of the 20 identified genes using RT-qPCR. We suspect that this might reflect the very low median copy number of S. enterica organisms in the blood of infected humans, <1 CFU/ml, a reality that impedes mRNA profiling and quantification in human typhoid and paratyphoid fever (42).
In summary, we have identified 20 S. Paratyphi A proteins expressed in vivo during human infection that are immunogenic for humoral immunity. Some identified antigens are only present in Salmonella strains producing enteric fever and are not found in other Enterobacteriaceae. We confirmed the upregulated expression of IVIAT-identified genes in the blood of patients with S. Paratyphi A bacteremia. IVIAT-identified antigens warrant a focused evaluation to further understand their roles in S. Paratyphi A pathogenesis and potentially in diagnosis and disease control.
ACKNOWLEDGMENTS
This work was supported by the ICDDR,B and by grants from the National Institutes of Health, including the National Institute of Allergy and Infectious Diseases (R21 AI100023 and U01 AI077883 to E.T.R. and F.Q., U01 AI058935 to S.B.C., E.T.R., and F.Q., Training Grant in Vaccine Development and Public Health (D43 TW005572 to M.M.A., F.K., A.S., T.S., E.T.R., and F.Q.), Career Development Awards (K01 TW07409 to J.B.H., K08 AI089721 to R.C.C., and K08 AI100923 to D.T.L.), an American Recovery and Reinvestment Act (ARRA) postdoctoral fellowship in Global Infectious Diseases (TW005572 to D.T.L. and R.C.C.), a Clinical Research Scholars Award (R24 TW007988 to S.B.C.) from the Fogarty International Center, a Physician Scientist Early Career Award from the Howard Hughes Medical Institute (to R.C.L.), a Massachusetts General Hospital Physician Scientist Development Award from Massachusetts General Hospital (to R.C.C.), a Burroughs Wellcome Fund/American Society of Tropical Medicine and Hygiene Postdoctoral Fellowship in Tropical Infectious Diseases (to D.T.L.), a Harvard College Program for Research in Science and Engineering Fellowship (to L.L.T.), and an iSURF Award from the Harvard Global Health Institute (to L.L.T.).
Footnotes
Published ahead of print 13 March 2013
REFERENCES
- 1.Fangtham M, Wilde H. 2008. Emergence of Salmonella paratyphi A as a major cause of enteric fever: need for early detection, preventive measures, and effective vaccines. J. Travel Med. 15:344–350 [DOI] [PubMed] [Google Scholar]
- 2.Crump JA, Luby SP, Mintz ED. 2004. The global burden of typhoid fever. Bull. World Health Organ. 82:346–353 [PMC free article] [PubMed] [Google Scholar]
- 3.Maskey AP, Day JN, Phung QT, Thwaites GE, Campbell JI, Zimmerman M, Farrar JJ, Basnyat B. 2006. Salmonella enterica serovar Paratyphi A and S. enterica serovar Typhi cause indistinguishable clinical syndromes in Kathmandu, Nepal. Clin. Infect. Dis. 42:1247–1253 [DOI] [PubMed] [Google Scholar]
- 4.Tankhiwale SS, Agrawal G, Jalgaonkar SV. 2003. An unusually high occurrence of Salmonella enterica serotype Paratyphi A in patients with enteric fever. Indian J. Med. Res. 117:10–12 [PubMed] [Google Scholar]
- 5.Harris JB, Baresch-Bernal A, Rollins SM, Alam A, LaRocque RC, Bikowski M, Peppercorn AF, Handfield M, Hillman JD, Qadri F, Calderwood SB, Hohmann E, Breiman RF, Brooks WA, Ryan ET. 2006. Identification of in vivo-induced bacterial protein antigens during human infection with Salmonella enterica serovar Typhi. Infect. Immun. 74:5161–5168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santander J, Curtiss R., III 2010. Salmonella enterica serovars Typhi and Paratyphi A are avirulent in newborn and infant mice even when expressing virulence plasmid genes of Salmonella Typhimurium. J. Infect. Dev. Ctries. 4:723–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raffatellu M, Wilson RP, Winter SE, Bäumler AJ. 2008. Clinical pathogenesis of typhoid fever. J. Infect. Dev. Ctries. 2:260–266 [DOI] [PubMed] [Google Scholar]
- 8.Hang L, John M, Asaduzzaman M, Bridges EA, Vanderspurt C, Kirn TJ, Taylor RK, Hillman JD, Progulske-Fox A, Handfield M, Ryan ET, Calderwood SB. 2003. Use of in vivo-induced antigen technology (IVIAT) to identify genes uniquely expressed during human infection with Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 100:8508–8513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rollins SM, Peppercorn A, Young JS, Drysdale M, Baresch A, Bikowski MV, Ashford DA, Quinn CP, Handfield M, Hillman JD, Lyons CR, Koehler TM, Calderwood SB, Ryan ET. 2008. Application of in vivo induced antigen technology (IVIAT) to Bacillus anthracis. PLoS One 3:e1824 doi:10.1371/journal.pone.0001824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McClelland M, Sanderson KE, Clifton SW, Latreille P, Porwollik S, Sabo A, Meyer R, Bieri T, Ozersky P, McLellan M, Harkins CR, Wang C, Nguyen C, Berghoff A, Elliott G, Kohlberg S, Strong C, Du F, Carter J, Kremizki C, Layman D, Leonard S, Sun H, Fulton L, Nash W, Miner T, Minx P, Delehaunty K, Fronick C, Magrini V, Nhan M, Warren W, Florea L, Spieth J, Wilson RK. 2004. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 36:1268–1274 [DOI] [PubMed] [Google Scholar]
- 11.Wain J, Hosoglu S. 2008. The laboratory diagnosis of enteric fever. J. Infect. Dev. Ctries. 2:421–425 [DOI] [PubMed] [Google Scholar]
- 12.Sheikh A, Charles RC, Rollins SM, Harris JB, Bhuiyan MS, Khanam F, Bukka A, Kalsy A, Porwollik S, Brooks WA, LaRocque RC, Hohmann EL, Cravioto A, Logvinenko T, Calderwood SB, McClelland M, Graham JE, Qadri F, Ryan ET. 2010. Analysis of Salmonella enterica serotype Paratyphi A gene expression in the blood of bacteremic patients in Bangladesh. PLoS Negl. Trop. Dis. 4:e908 doi:10.1371/journal.pntd.0000908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sheikh A, Charles RC, Sharmeen N, Rollins SM, Harris JB, Bhuiyan MS, Arifuzzaman M, Khanam F, Bukka A, Kalsy A, Porwollik S, Leung DT, Brooks WA, LaRocque RC, Hohmann EL, Cravioto A, Logvinenko T, Calderwood SB, McClelland M, Graham JE, Qadri F, Ryan ET. 2011. In vivo expression of Salmonella enterica serotype Typhi genes in the blood of patients with typhoid fever in Bangladesh. PLoS Negl. Trop. Dis. 5:e1419 doi:10.1371/journal.pntd.0001419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphries A, Deridder S, Bäumler AJ. 2005. Salmonella enterica serotype Typhimurium fimbrial proteins serve as antigens during infection of mice. Infect. Immun. 73:5329–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Humphries AD, Raffatellu M, Winter S, Weening EH, Kingsley RA, Droleskey R, Zhang S, Figueiredo J, Khare S, Nunes J, Adams LG, Tsolis RM, Bäumler AJ. 2003. The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol. Microbiol. 48:1357–1376 [DOI] [PubMed] [Google Scholar]
- 16.Weening EH, Barker JD, Laarakker MC, Humphries AD, Tsolis RM, Bäumler AJ. 2005. The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect. Immun. 73:3358–3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawley TD, Chan K, Thompson LJ, Kim CC, Govoni GR, Monack DM. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2:e11 doi:10.1371/journal.ppat.0020011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards RA, Olsen GJ, Maloy SR. 2002. Comparative genomics of closely related salmonellae. Trends Microbiol. 10:94–99 [DOI] [PubMed] [Google Scholar]
- 19.Bronowski C, Winstanley C. 2009. Identification and distribution of accessory genome DNA sequences from an invasive African isolate of Salmonella Heidelberg. FEMS Microbiol. Lett. 298:29–36 [DOI] [PubMed] [Google Scholar]
- 20.Kannan G, Wilks JC, Fitzgerald DM, Jones BD, Bondurant SS, Slonczewski JL. 2008. Rapid acid treatment of Escherichia coli: transcriptomic response and recovery. BMC Microbiol. 8:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charles RC, Harris JB, Chase MR, Lebrun LM, Sheikh A, LaRocque RC, Logvinenko T, Rollins SM, Tarique A, Hohmann EL, Rosenberg I, Krastins B, Sarracino DA, Qadri F, Calderwood SB, Ryan ET. 2009. Comparative proteomic analysis of the PhoP regulon in Salmonella enterica serovar Typhi versus Typhimurium. PLoS One 4:e6994 doi:10.1371/journal.pone.0006994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunn JS, Alpuche-Aranda CM, Loomis WP, Belden WJ, Miller SI. 1995. Characterization of the Salmonella Typhimurium pagC/pagD chromosomal region. J. Bacteriol. 177:5040–5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gutierrez C, Barondess J, Manoil C, Beckwith J. 1987. The use of transposon TnphoA to detect genes for cell envelope proteins subject to a common regulatory stimulus. Analysis of osmotically regulated genes in Escherichia coli. J. Mol. Biol. 195:289–297 [DOI] [PubMed] [Google Scholar]
- 24.Conter A, Menchon C, Gutierrez C. 1997. Role of DNA supercoiling and rpoS sigma factor in the osmotic and growth phase-dependent induction of the gene osmE of Escherichia coli K12. J. Mol. Biol. 273:75–83 [DOI] [PubMed] [Google Scholar]
- 25.Faucher SP, Porwollik S, Dozois CM, McClelland M, Daigle F. 2006. Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc. Natl. Acad. Sci. U. S. A. 103:1906–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. 2010. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143:1110–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizushima K, Awakihara S, Kuroda M, Ishikawa T, Tsuda M, Tsuchiya T. 1992. Cloning and sequencing of the melB gene encoding the melibiose permease of Salmonella Typhimurium LT2. Mol. Gen. Genet. 234:74–80 [DOI] [PubMed] [Google Scholar]
- 28.Guan L, Nurva S, Ankeshwarapu SP. 2011. Mechanism of melibiose/cation symport of the melibiose permease of Salmonella Typhimurium. J. Biol. Chem. 286:6367–6374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta SD, Wu HC, Rick PD. 1997. A Salmonella Typhimurium genetic locus which confers copper tolerance on copper-sensitive mutants of Escherichia coli. J. Bacteriol. 179:4977–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bearson BL, Bearson SM, Uthe JJ, Dowd SE, Houghton JO, Lee I, Toscano MJ, Lay DC., Jr 2008. Iron-regulated genes of Salmonella enterica serovar Typhimurium in response to norepinephrine and the requirement of fepDGC for norepinephrine-enhanced growth. Microbes Infect. 10:807–816 [DOI] [PubMed] [Google Scholar]
- 31.Williams PH, Rabsch W, Methner U, Voigt W, Tschäpe H, Reissbrodt R. 2006. Catecholate receptor proteins in Salmonella enterica: role in virulence and implications for vaccine development. Vaccine 24:3840–3844 [DOI] [PubMed] [Google Scholar]
- 32.Bearson BL, Bearson SM, Lee IS, Brunelle BW. 2010. The Salmonella enterica serovar Typhimurium QseB response regulator negatively regulates bacterial motility and swine colonization in the absence of the QseC sensor kinase. Microb. Pathog. 48:214–219 [DOI] [PubMed] [Google Scholar]
- 33.Wei Y, Miller CG. 1999. Characterization of a group of anaerobically induced, fnr-dependent genes of Salmonella Typhimurium. J. Bacteriol. 181:6092–6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bunch PK, Mat-Jan F, Lee N, Clark DP. 1997. The ldhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology 143(Pt 1):187–195 [DOI] [PubMed] [Google Scholar]
- 35.Kabir MM, Ho PY, Shimizu K. 2005. Effect of ldhA gene deletion on the metabolism of Escherichia coli based on gene expression, enzyme activities, intracellular metabolite concentrations, and metabolic flux distribution. Biochem. Eng. J. 26:1–11 [Google Scholar]
- 36.Hong JS, Kaback HR. 1972. Mutants of Salmonella Typhimurium and Escherichia coli pleiotropically defective in active transport. Proc. Natl. Acad. Sci. U. S. A. 69:3336–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Y, Dong K, Zhang X, Zhang B, Hou L, Chen N, Chen S. 2008. Expression and regulation of the yggG gene of Escherichia coli. Curr. Microbiol. 56:14–20 [DOI] [PubMed] [Google Scholar]
- 38.Kuan CS, Wong MT, Choi SB, Chang CC, Yee YH, Wahab HA, Normi YM, Too WCS, Few LL. 2011. Klebsiella pneumoniae yggG gene product: a zinc-dependent metalloprotease. Int. J. Mol. Sci. 12:4441–4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang Y, Zhang B, Dong K, Zhang X, Hou L, Wang T, Chen N, Chen S. 2007. Up-regulation of yggG promotes the survival of Escherichia coli cells containing Era-1 mutant protein. FEMS Microbiol. Lett. 275:8–15 [DOI] [PubMed] [Google Scholar]
- 40.Anantharaman V, Aravind L. 2003. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 4:R11 doi:10.1186/gb-2003-4-2-r11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bateman A, Rawlings ND. 2003. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 28:234–237 [DOI] [PubMed] [Google Scholar]
- 42.Wain J, Diep TS, Ho VA, Walsh AM, Nguyen TT, Parry CM, White NJ. 1998. Quantitation of bacteria in blood of typhoid fever patients and relationship between counts and clinical features, transmissibility, and antibiotic resistance. J. Clin. Microbiol. 36:1683–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]