Abstract
Fungi, plants, and bacteria accumulate potassium via two distinct molecular machines not directly coupled to ATP hydrolysis. The first, designated TRK, HKT, or KTR, has eight transmembrane helices and is folded like known potassium channels, while the second, designated HAK, KT, or KUP, has 12 transmembrane helices and resembles MFS class proteins. One of each type functions in the model organism Neurospora crassa, where both are readily accessible for biochemical, genetic, and electrophysiological characterization. We have now determined the operating balance between Trk1p and Hak1p under several important conditions, including potassium limitation and carbon starvation. Growth measurements, epitope tagging, and quantitative Western blotting have shown the gene HAK1 to be much more highly regulated than is TRK1. This conclusion follows from three experimental results: (i) Trk1p is expressed constitutively but at low levels, and it is barely sensitive to extracellular [K+] and/or the coexpression of HAK1; (ii) Hak1p is abundant but is markedly depressed by elevated extracellular concentrations of K+ and by coexpression of TRK1; and (iii) Carbon starvation slowly enhances Hak1p expression and depresses Trk1p expression, yielding steady-state Hak1p:Trk1p ratios of ∼500:1, viz., 10- to 50-fold larger than that in K+- and carbon-replete cells. Additionally, it appears that both potassium transporters can adjust kinetically to sustained low-K+ stress by means of progressively increasing transporter affinity for extracellular K+. The underlying observations are (iv) that K+ influx via Trk1p remains nearly constant at ∼9 mM/h when extracellular K+ is progressively depleted below 0.05 mM and (v) that K+ influx via Hak1p remains at ∼3 mM/h when extracellular K+ is depleted below 0.1 mM.
INTRODUCTION
Essentially all living cells are metabolically dependent on high (∼150 mM) intracellular concentrations of potassium ions, but two fundamentally different strategies are used to maintain high internal [K+] in the face of generally low external concentrations (<10 mM). In animal cells, concentrative uptake of K+ is chemically coupled to the ATP-driven extrusion of sodium ions, and the diffusional efflux of K+ ions into extracellular concentrations ranging from nearly 1 to 10 mM determines ordinary resting membrane voltages (Vm) of −70 to −90 mV. In contrast, most plant cells, fungi, and bacteria possess a strongly electrogenic proton extrusion pump, and concentrative K+ influxes are driven by the membrane voltage, which can range beyond −300 mV (1, 2).
At least four modes of K+ uptake occur in these cells: (i) diffusive, fast influx via K+-specific channels (3, 4); (ii) simple facilitation, a protein-mediated slow flux formerly termed facilitated diffusion; (iii) H+- or Na+-coupled symport of potassium ions, formerly termed secondary active transport (5–9); and (iv) in some species, ATP-coupled K+ influx pumping (10, 11).
Modes ii and iii have both been attributed to two distinct and large classes of transport proteins, generally dubbed Trk and Kup in bacteria and Trk (or HKT) and HAK in plants and fungi. For the Kup-HAK proteins, no crystal structures have yet been reported, but available sequences define 12 transmembrane helices. This indicates general similarity to the MFS class of membrane transporters and suggests a mechanism of rocker-switched access for transfer of K+ ions (12–15). For the Trk-HKT proteins, early sequence data revealed strong similarity to K+ channels and led to a theoretical model (16, 17) based on the bacterial potassium channel, KcsA (18). That model, which varied slightly among the bacterial, fungal, and plant homologues, has been substantiated by a variety of point mutation and sidedness studies (19–22). More recently, VpTrkH, a bacterial homologue from Vibrio parahaemolyticus, has been crystallized (23), confirming the major features of the Durell and Guy model and pointing to a fundamentally channel-like permeation process for K+ ions.
Independent results from a variety of plant, fungal, and bacterial preparations have suggested that representatives of both classes of protein mediate K+ transport in mode ii and, separately, in mode iii. With a few exceptions, however, defining the actual mode of transport has proven elusive. That issue has arisen pointedly in the model organism Neurospora crassa, where potassium uptake coupled to proton influx (symport) was first demonstrated (2, 5), well ahead of cloning and sequencing techniques. Actual proof of K+-H+ symport emerged from direct voltage measurements on potassium-starved spherocytes of Neurospora, which displayed large, fast, and reversible depolarizations in response to pulses of 10 to 100 μM K+. Fifteen years later, two K+ transporter genes were cloned from Neurospora (24), coding for one protein in each class, viz., N. crassa Trk1p (NcTrk1p) and NcHak1p. These were partially characterized after expression in Saccharomyces, where K+ uptake measurements, made in the absence of extracellular sodium ions, indicated both to be high-affinity transporters. Northern blot analysis of Neurospora, following K+ starvation, failed to find mRNA for TRK1, but it did identify abundant mRNA for HAK1, which prompted the hypothesis that the HAK1 protein, not the TRK1 protein, was responsible for K+-H+ symport in the earlier experiments (24).
We have now undertaken a thorough test of the situation directly in Neurospora, making particular use of single-gene deletion strains recently provided by the Neurospora genome project (25). In normal K+-containing media, TRK1 protein (Trk1p) is expressed constitutively at a low level and is rather insensitive to growth conditions. HAK1 (Hak1p) is also expressed constitutively but is strongly suppressed by high extracellular potassium and also by the simultaneous presence of Trk1p. Other metabolic conditions impinge on both. For example, carbon starvation of Neurospora, superimposed on K+ starvation, slowly diminishes Trk1p while simultaneously enhancing Hak1p. One net result of these changes is that the expression ratio Hak1p:Trk1p, which is in the neighborhood of 10:1 under normal laboratory conditions, rises to more than 500:1 after 3 h of starvation for K+ and carbon.
MATERIALS AND METHODS
Strains of N. crassa used in this study are listed in Table 1. Knockout strains for single genes encoding the critical potassium transporters TRK1 and HAK1, for the apparently silent homologue TRK2, and for the K+ channel TOK1 were generated within the Neurospora functional genomics project (25) by homologous replacement with the hygromycin B resistance gene (hph). These four strains, along with two knockout strains for the DNA repair enzyme MUS51 and wild-type strains ORSa and 74-OR23-1VA, were obtained from the Fungal Genetics Stock Center (FGSC; Kansas City, KS). Knockout of the MUS51 gene greatly enhances the frequency of homologous recombination in Neurospora (26).
Table 1.
Strains used in this study
| Strain name | Laboratory file no. | Genotypea | Origin, cross, and/or transformationb |
|---|---|---|---|
| {+++}c | WT74 | TRK1 TRK2 HAK1 | FGSC 4200a/2489A (WT74-OR23-IVA) |
| {Δ++} | Nc06449 | trk1::hphd TRK2 HAK1 | FGSC 12678a/12679A (NCU06449) |
| {+Δ+} | Nc02456 | TRK1 trk2::hph HAK1 | FGSC 1612A (NCU02456) |
| {++Δ} | Nc00790 | TRK1 TRK2 hak1::hph | FGSC 13815a/13816A(NCU00790) |
| {+++ tok1Δ} | Nc12045 | TRK1 TRK2 HAK1 tok1::hph | FGSC 12045A |
| {+ΔΔ} | KAR6 | TRK1 trk2::hph hak1::hph | From {++Δ} × {ΔΔ+} |
| {Δ+Δ} | KAR12 | trk1::hph TRK2 hak1::hph | From {++Δ} × {ΔΔ+} |
| {ΔΔ+} | KAR8 | trk1::hph trk2::hph HAK1 | From {Δ++} × {+Δ+} |
| {ΔΔΔ} | KAR5 | trk1::hph trk2::hph hak1::hph | From {++Δ} × {ΔΔ+} |
| {ΔΔΔ tok1Δ} | KAR117-1-28 | trk1::hph trk2::hph hak1::hph tok1::hph | From {ΔΔΔ} × {+++ tok1Δ} |
| {+++ mus}e | 9718 | TRK1 TRK2 HAK1 | FGSC 9718a |
| {+Δ+ mus} | KAR106 | TRK1 trk2::hph HAK1 | From {ΔΔΔ} × {+++ mus} |
| {+ΔΔ mus} | KAR15 | TRK1 trk2::hph hak1::hph | From {ΔΔΔ} × {+++ mus} |
| {ΔΔ+ mus} | KAR81 | trk1::hph trk2::hph HAK1 | From {ΔΔΔ} × {+++ mus} |
| {ΔΔΔ mus} | KAR75 | trk1::hph trk2::hph hak1::hph | From {ΔΔΔ} × {+++ mus} |
| {+++ mus his-3} | 9717 | TRK1 TRK2 HAK1 his3 | FGSC 9717A |
| {ΔΔΔ mus his-3} | KAR79 | trk1::hph trk2::hph hak1::hph his3 | From {ΔΔΔ mus} × {+++ mus his-3} |
| {TRK1-HA} | KAR77 | TRK1-HA trk2::hph hak1::hph | Transform {ΔΔΔ mus} |
| {HA-HAK1} | KAR105 | trk1::hph trk2::hph HA-HAK1 | Transform {ΔΔΔ mus} |
| {TRK1-HA HA-HAK1} | KAR108 | TRK1-HA trk2::hph HA-HAK1 | From {TRK1-HA} × {HA-HAK1} |
| {TRK1-HA HAK1} | KAR109 | TRK1-HA trk2::hph HAK1 | From {ΔΔ+ mus} × {TRK1-HA} |
| {TRK1 HA-HAK1} | KAR110 | TRK1 trk2::hph HA-HAK1 | From {+ΔΔ mus} × {HA-HAK1} |
| {NcppTRK1}f | KAR101 | trk1::hph trk2::hph hak1::hph his-3::pPMA1-TRK1 | Transform {ΔΔΔ mus his-3} |
| {NcppHAK1} | KAR100 | trk1::hph trk2::hph hak1::hph his3::pPMA1-HAK1 | Transform {ΔΔΔ mus his-3} |
| {NcppTRK1-HA} | NTH3 | trk1::hph trk2::hph hak1::hph his3::pPMA1-TRK1-HA | Transform {ΔΔΔ mus his-3} |
| {NcppHA-HAK1} | KAR107 | trk1::hph trk2::hph hak1::hph his3::pPMA1-HA-HAK1 | Transform {ΔΔΔ mus his-3} |
| {NcppTRK1 HA-HAK1} | KAR115 | trk1::hph trk2::hph HA-HAK1 his3::pPMA1-TRK1 | From {NcppTRK1} × {HA-HAK1} |
| {NcppHAK1 TRK1-HA} | KAR114 | TRK1-HA trk2::hph hak1::hph his3::pPMA1-HAK1 | From {TRK-HA} × {NcppHAK1} |
Potassium transporter genes were under the control of their respective native promoters, except in the six strains denoted by Ncpp.
Six strains (marked FGSC) were obtained from the Fungal Genetics Stock Center (Kansas City, MO), for which the three single-gene deletion strains (NCU06449, NCU02456, and NCU00790) were constructed by the Neurospora Genome consortium (25). All other strains were made in this laboratory, either by transformations or by crosses going back to the FGSC strains.
Curly brackets denote strain names by the critical gene constructs.
hph is a gene for hygromycin B phosphotransferase, conferring resistance to hygromycin B.
All strains from here downward also carry mus51::bar (bar is a gene conferring resistance to the herbicide bialaphos).
Ncpp indicates pPMA1, a strong promoter that is native to the Neurospora plasma membrane H+-pumping ATPase.
Gene locus numbers for TRK1, TRK2, HAK1, TOK1, and MUS51 are NCU06449, NCU02456, NCU00790, NCU04065, and NCU08290, respectively. All other strains in this study were constructed using standard genetic and molecular biological techniques (27). Strain names, detailed genotypes, and sources/constructs for all strains used in these experiments are listed in Table 1. In order to make the genetic construct(s) in each experiment as clear and direct as possible while minimizing the need for cross-referencing, we have named each strain according to its critical genes and have placed each strain name in curly brackets, e.g., {ΔΔΔ}, {TRK1-HA}, and {NcppHA-HAK1}. Otherwise, gene names are written in italics, uppercase for wild-type genes and lowercase plus “Δ” for deleted genes, viz., TRK1 and trk1Δ. Protein names are written in mixed-case nonitalic font and usually end with “p,” such as Trk1p. These conventions are followed throughout the text and figures.
Measurements of growth and intracellular potassium.
Drop tests and batch cultures were both used to assess growth. Strains were maintained (25°C for 7 to 10 days) on 2% agar slants or flasks of Vogel's minimal medium: 36.8 mM KH2PO4, 8.9 mM Na3 citrate (pH 5.8), 25 mM NH4NO3, 0.8 mM MgSO4, 0.7 mM CaCl2, and trace elements (28), made up in distilled water plus 2% sucrose. In K+-free Vogel's medium, potassium phosphate was replaced by NaH2PO4, thus elevating extracellular concentrations of Na+ ([Na+]o) to 63.5 mM. For all experiments, harvested conidia were washed and resuspended in glass-distilled water prior to inoculation.
Drop tests were devised to compare strains bearing seven combinations of single, double, and triple deletions of TRK1, TRK2, and HAK1 genes (see Fig. 2), as well as two combinations with TOK1 deleted. Seven-μl aliquots of conidial suspensions (at 106 conidia/ml) were pipetted onto 2% agar plates containing K+-free Vogel's salts, 0.2% glucose, 3% sorbose (to enforce colonial growth), and KCl at 0.03, 0.1, 1, 3, or 10 mM. Plates were incubated at 30°C for 3 days and scanned on a CanoScan 8400F digital scanner (Canon U.S.A., Inc., Lake Success, NY) at 600 dpi.
Fig 2.
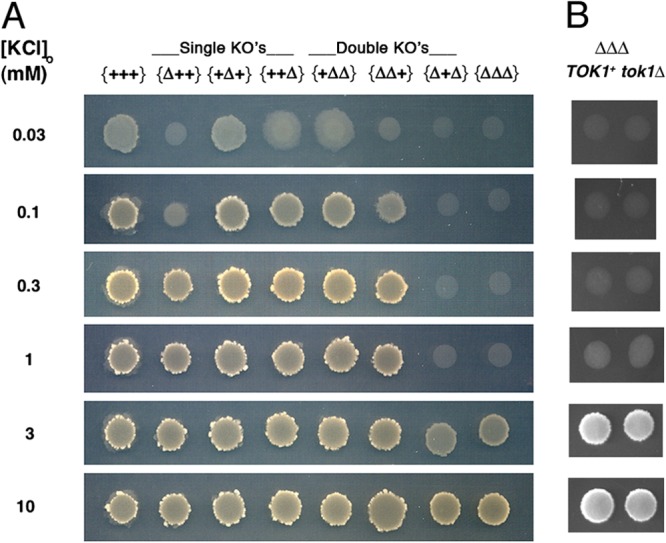
Trk1p and/or Hak1p supports growth of Neurospora on low-potassium medium. Three-day drop colonies, grown at 30°C, were used. Column headers designate the presence of the wild type (+) or the knockout (Δ) for each gene, in the order (left to right) TRK1, TRK2, HAK1; viz., the relevant genotype of strain {+++} is TRK1 TRK2 HAK1, that of strain {Δ++} is trk1Δ TRK2 HAK1, that of strain {++Δ} is TRK1 TRK2 hak1Δ, etc. The potassium channel TOK1 was wild type in panel A, but deleted in the right-hand column of panel B. Suspensions of conidia (7 μl at 106 cells/ml) were pipetted onto agar plates containing K+-free Vogel's minimal medium plus KCl, as indicated, with 3% sorbose to enforce colonial morphology (see Materials and Methods). The bright, scabrous ring surrounding each robust colony represents profuse conidiophores.
Batch cultures were used to quantitate growth and the specific potassium and protein contents of strains bearing the hemagglutinin (HA)-tagged genes (see below), all constructed in the mus51Δ background. Washed conidia were resuspended in distilled water at ∼108/ml and then inoculated at 2 × 106/ml into 500-ml Erlenmeyer flasks containing 150 ml K+-free Vogel's salts plus 2% sucrose and KCl at 0.1, 0.3, 1, or 37 mM. These suspensions were incubated at 25°C on an orbital shaker (150 rpm), and germinating conidia were filter harvested at roughly 2-h intervals. The cells in 40-ml aliquots of suspension were collected on predried, preweighed Millipore filters (DAWP; 5-μm pores; Millipore Corp., Bedford MA), rinsed twice in 10 ml of distilled water, dried (37°C) overnight, weighed, and then extracted in 0.5 ml of 0.1 M HNO3 (30 min at 98°C). Debris was spun out, and potassium in the supernatant was assayed on an atomic absorption photometer (Model 943; Instrumentation Laboratory, Lexington, MA). Intracellular K+ concentrations ([K+]i) were calculated as mmol/kg cell water (mM), using an intracellular wet/dry weight ratio (ICW) of 2.54 (29). Semilog plots of cell dry weights versus time proved to be linear between 4 and 12 h (designated 0 to 8 h in Fig. 3, 8, and 9), yielding specific growth rates and doubling times for each strain.
Fig 3.
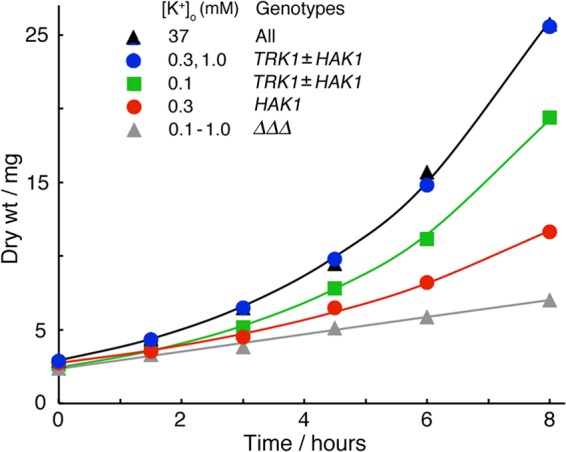
Limitation of exponential growth (batch cultures) by low extracellular K+ in strains deleted of all three K+ transporter genes and then transformed with HA-tagged TRK1 and/or HAK1. Measured dry weights were grouped and averaged for similar plots. Under all conditions, growth became stably exponential for at least 8 h irrespective of the initial extracellular K+ concentration. Smooth curves are least-square-fitted exponentials; specific rate constants (top to bottom) were 0.274, 0.259, 0.182, and ∼0.08 h−1, corresponding to doubling times of 2.53, 2.68, 3.81, and >8 h, respectively. Batch cultures of 150 ml were inoculated with 2 × 106 conidia/ml in K+-free Vogel's salts plus 2% sucrose plus KCl as indicated, shaken at 25°C, filter harvested (40-ml aliquots) at 1.5- or 2-h intervals, and rinsed 3× in distilled water. Dry weights and cell potassium were measured as described in Materials and Methods. Experiments were run in duplicate and repeated 2 to 4 times for each strain and [K+]o. The scatter of measured dry weights (±1 standard deviation [SD]) was ∼15% of the mean value at most points. The initial growth of such cultures, following harvesting and rinsing of conidia, is always slow and created a lag phase (∼4 h) prior to the plotted zero time. For simplicity, only the K+ transporter genes are indicated here. Table 1 lists full genotypes and strain names.
Fig 8.
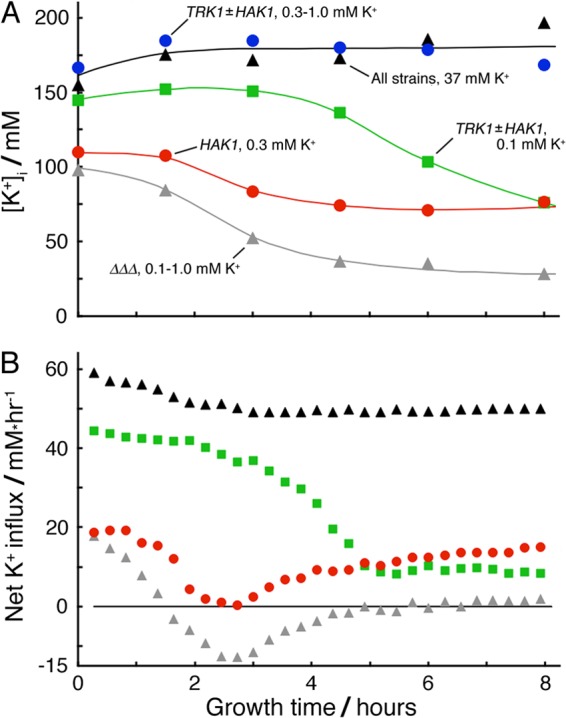
Time courses of intracellular K+ concentration and net flux during exponential growth of Neurospora on limiting potassium. The experiment and measurements were the same as those described for Fig. 3. (A) Cell-associated potassium measured and [K+]i calculated as described in Materials and Methods. Smoothed curves were drawn by hand. Different starting values of [K+]i reflect net loss of cellular potassium during the lag phase (−4 to 0 h). Scatter of measured [K+]i values (±1 SD) was ∼20% of the mean value. (B) Implied net fluxes were calculated from the increment of total cellular potassium over each successive 16-min interval along the appropriate smooth curve in panel A and then divided by the average cell mass during the same interval along the corresponding fitted curve in Fig. 3. Positive values represent net influx.
Fig 9.
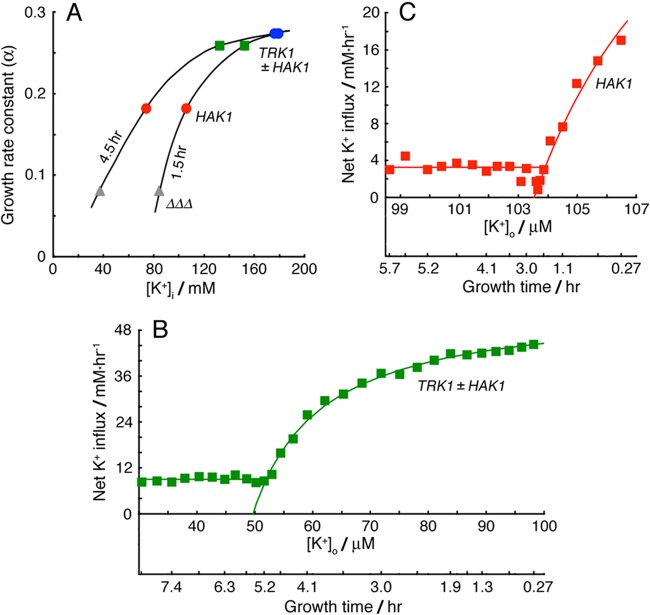
Neurospora can modulate the K+ concentration dependence of growth rate and the substrate affinities of both K+ transporters. (A) Apparent dependence of specific growth rate upon the intracellular K+ concentration, early (1.5 h) and midway (4.5 h) in exponential growth. Parametric plot of data from Fig. 3; smooth curves were drawn by hand. (B and C) Net K+ fluxes plotted against extracellular K+ concentrations for cells growing exponentially after inoculation into Vogel's medium (K+-free) supplemented with 0.1 mM KCl. Initial values of [K+]o at time 0 were greater than 0.1 mM due to loss of cellular K+ during the lag phase. [K+]o was then gradually depleted by net uptake. Smooth curves were fitted to a Michaelis function having an offset concentration, 49.7 and 103.5 μM, respectively. Below the offset concentrations, steady fluxes were calculated as the average values out to 8 h: 9.04 and 3.26 mM/h, respectively.
For analysis of membrane proteins, 75-ml batch cultures were harvested from late exponential phase (16 h at 25°C) by filtration through 5-μm Millipore filters, washed with 50 ml distilled water, and extracted/processed as described below. For starvation studies, batch cultures were harvested at 12 h and transferred to potassium-free Vogel's salts plus 1% glucose. Individual cultures were subsequently reharvested for assays of protein and intracellular K+ at intervals of ∼1 h. In one group of experiments (see Fig. 7), 100 mM KCl was added to the culture medium at 5 h. In several other groups (see Fig. 10), batch cultures were again harvested and transferred to K+-free and sugar-free Vogel's salts.
Fig 7.
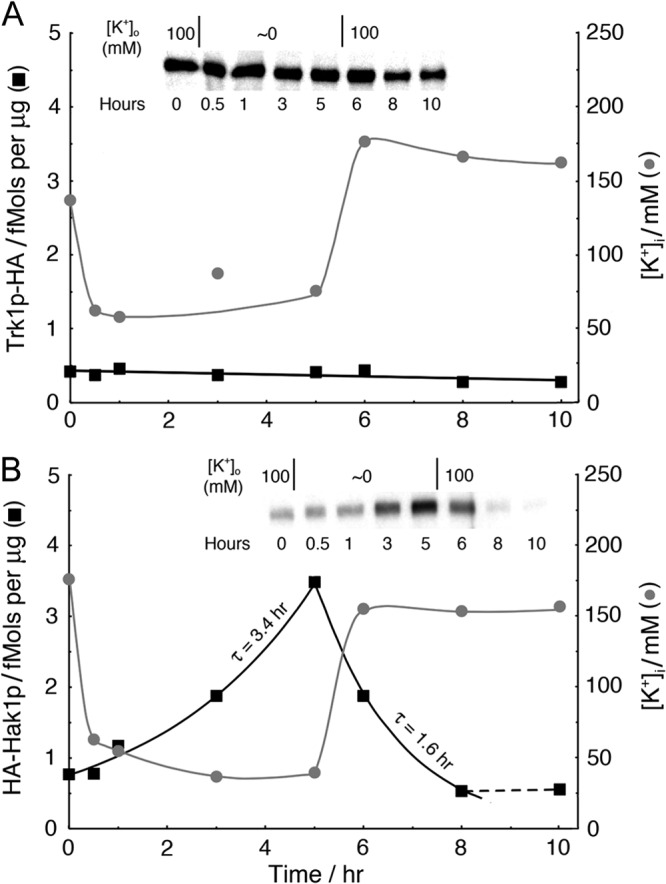
Rapid shifts of potassium-ion concentration affect Hak1p expression only slowly and do not affect Trk1p expression at all. Time courses of Trk1p-HA (A, ■) and HA-Hak1p (B, ■) and of intracellular K+ ( ) during potassium starvation (0 to 5 h), followed by potassium repletion (5 to 10 h). Removal of [K+]o (harvest, rinse, and resuspend) required less than 1 min, which was very fast compared to the decline of [K+]i. For strains {TRK1-HA} and {HA-HAK1}, the preparative growth medium was K+-free Vogel's salts plus 100 mM KCl and 2% sucrose. For the insets, the load was 5 μg membrane protein in each lane. High-sensitivity ECL reagent was used for panel A.
) during potassium starvation (0 to 5 h), followed by potassium repletion (5 to 10 h). Removal of [K+]o (harvest, rinse, and resuspend) required less than 1 min, which was very fast compared to the decline of [K+]i. For strains {TRK1-HA} and {HA-HAK1}, the preparative growth medium was K+-free Vogel's salts plus 100 mM KCl and 2% sucrose. For the insets, the load was 5 μg membrane protein in each lane. High-sensitivity ECL reagent was used for panel A.
Fig 10.
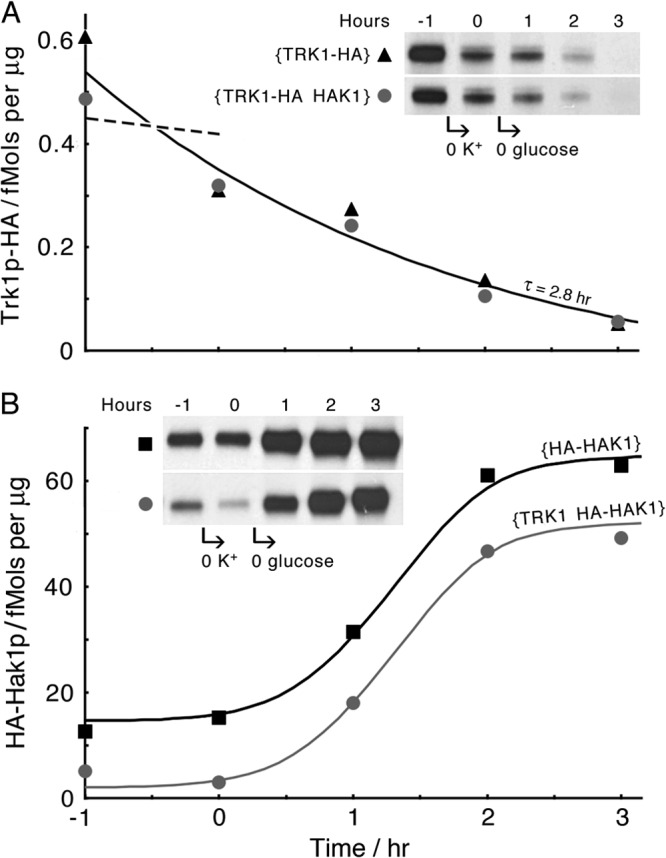
Carbon starvation demonstrates further involvement of HAK1-Hak1p in the metabolic program of N. crassa. Time courses of TRK1p-HA depression (A) and HA-Hak1p enhancement (B) during 1 h of starvation for K+, followed by 3 h of starvation for sugar and K+. Note that simultaneous expression of both genes, TRK1 and HAK1, appeared irrelevant to the effect of carbon starvation on the expression time course of either protein, even though the steady-state prestarvation level of Hak1p was significantly reduced by coexpression of TRK1. The general protocol was similar to that described for Fig. 7, starting with cells grown for 16 h in Vogel's medium plus 2% sucrose. The smooth curve shown in panel A is least-square fitted to all 10 data points; the dashed line represents the expected time course of Trk1p-HA during the hour of starvation for K+ alone, based on data from Fig. 7A. Smooth curves shown in panel B were drawn by hand and are identical, except for the vertical separation of 12.6 fmol. For the panel A inset, the load is 15 μg membrane protein in each lane; for the inset in panel B, the load is 1 μg in each lane.
Construction of transformation cassettes.
Since deletion of all three K+ transporter genes (strains {ΔΔΔ} and {ΔΔΔ mus}) essentially abolishes growth of Neurospora on media containing ≤1 mM KCl (see Fig. 2), growth complementation at low extracellular concentrations of potassium ([K+]o) could be used to select for transformation by TRK1, HAK1, or their tagged variants in the ΔΔΔ background. Transformation cassettes were designed for exact homologous replacement of the native open reading frames (ORFs). The 5′ and 3′ untranslated regions (UTRs) of ∼1 kb for each gene were incorporated into the transformation cassettes, such that the introduced tagged genes would be flanked by the intact native 5′ and 3′ UTRs and thus would be under the control of the native promoters and termination sequences. Two different tags, the HA epitope (YPYDVPDYA) and the FLAG epitope (DYKDDDDK), were tested at a variety of insertion sites. The HA epitope yielded much clearer Western blots than the FLAG epitope for both transporter genes, and the C terminus of TRK1 and N terminus of HAK1 proved the best insert sites, so the tagged genes are designated TRK1-HA and HA-HAK1, respectively.
In order to assemble the TRK1-HA cassette, a segment including the 5′ UTR, the TRK1 ORF, and HA was constructed by PCR using wild-type genomic DNA as a template; likewise, a segment of the 3′ UTR was amplified by PCR. These segments, bearing an MluI site at the 5′ end and also at the 3′ end, were spliced, cut with MluI restriction endonuclease, and ligated into the pIBI25 vector. The list and explanation of primers (K1-f to K4-r) used in these constructs is given in Table 2. The HA-HAK1 cassette was assembled similarly, using genomic DNA and the primers (K5-f to K8-r) also shown in Table 2. For both completed constructs, the ligation mixtures were transformed into E. coli electrocompetent cells (DH10B; 18290-015; Life Technologies/Invitrogen Corp., Grand Island, NY), and the amplified plasmids, sequenced to ensure fidelity, were linearized by incubation with the appropriate restriction enzymes, MluI for TRK1-HA and KpnI for HA-HAK1. A homemade high-fidelity DNA polymerase (generous gift of Efim Golub, Department of Genetics, Yale University) was used to carry out all of the PCRs; T4 DNA ligase and restriction endonucleases were purchased from New England BioLabs Inc., Beverly, MA.
Table 2.
Primers used to construct the transformation cassettes
| Namea | Sequenceb (5′→3′) | Region |
|---|---|---|
| K1-f | cacgtcACGCGTcttgcatacaagttctggtgggcgtg | TRK1 5′ UTR + TRK1 ORF (from −1694 to 2978) |
| K2-r | tcaggcgtagtcggggacgtcgtaggggtactcgctaaaatcagtaggagtggacgggaaac | |
| K3-f | cgacgtccccgactacgcctgatggatgtttcttgttcctatggcg | TRK1 3′ UTR (from 2984 to 4261) |
| K4-r | cactagACGCGTctaaccagagccaacatcaccatcctc | |
| K5-f | cacgtcGGTACCctcgagtccagtacgagaagatggaagttgag | HAK1 5′ UTR (from −1325 to 0) |
| K6-r | ggcgtagtcggggacgtcgtaggggtagttgtccatctttttttttatggaagtg | |
| K7-f | gacgtccccgactacgccgacactacagcagcccccaaac | HAK1 ORF + HAK1 3′ UTR (from 1 to 4199) |
| K8-r | cactagGGTACCgatgggtgtacattctgtacactgggtgttcc | |
| PP1-f | cacgtaGCGGCCGCgttgtggttccgtagaggtag | PMA1 5′ UTR (from −943 to 0) |
| PP2-r | gtacctTCTAGAtggcgttatggtttgcag |
f, forward primer; r, reverse primer.
Uppercase letters designate the restriction sites used for cloning. Italics designate start and stop codons. Coordinates for the DNA fragments are defined from 1 = A of the start codons. Underlining designates the coding sequence for the HA epitope.
Transformation and genotyping.
The TRK1-HA and HA-HAK1 genes were transformed into strains {ΔΔΔ mus} and {ΔΔΔ mus his-3} by electroporation. Stock cultures of the recipient strains were grown on Vogel's medium plus 2% sucrose with or without 0.5 mg/ml histidine. Conidia from 5- to 7-day-old slants were harvested in sterile distilled water, washed, transferred to fresh medium, and germinated for 2 h at 30°C (50 ml of medium containing ∼5 × 107 conidia/ml on an orbital shaker at 250 rpm). The germinated conidia were harvested by centrifugation (3,000 × g for 5 min), washed twice with 10 ml 1 M sorbitol, and resuspended at a density of ∼108 conidia/ml. In each transformation, a 40-μl aliquot was mixed with 1 to 3 μg of linearized DNA, transferred into an iced 0.2-cm electroporation cuvette, and electropulsed via a Bio-Rad Gene Pulser at 1.5 kV (600 Ω and 25 μF; Bio-Rad Laboratories, Hercules, CA).
For transforming TRK1-HA and HA-HAK1 into their homologous loci, the electropulsed conidial suspensions were spread on K+-limited plates made with 2% purified agar and K+-free Vogel's medium plus 0.2% glucose, 3% sorbose, and 1 mM KCl. Transformants were identified by robust growth on 1 mM KCl. For transforming TRK1, HAK1, or their HA-tagged variants into the his3 locus, electropulsed conidia were spread on histidine-free Vogel's medium. Because these inserts were designed to reconstitute the HIS3 wild-type gene, transformants were identified by growth on histidine-free plates. After 3 to 5 days (30°C in the dark), colonies were transferred to slants of the appropriate selective medium (lacking sorbose to allow normal filamentous growth). Homokaryons were obtained by backcrossing, as described by Davis and de Serres (27).
Genotypes of the homokaryotic progeny were verified by PCR via one primer from inside the transformation cassette and one primer outside and using template genomic DNA extracted by the method of Rose et al. (30). Mycelium from an overnight standing culture (1 ml Vogel's minimal medium at 30°C, ∼10 mg wet weight) was removed with a sterile hook, suspended in lysis mix, and broken by a 3-min shock in an SI-D236 cell disruptor (Scientific Industries, Bohemia, NY). The complete lysis mix contained 0.1 ml acid-washed glass beads (G-8772; Sigma-Aldrich Inc., St. Louis MO), 0.1 ml lysing buffer (2% Triton X-100, 1% sodium dodecyl sulfate, 100 mM NaCl, 1 mM EDTA, 10 mM Tris base, pH 8.0, and NaOH), and 0.1 ml phenol-chloroform (1:1). After brief mixing with 0.1 ml Tris-EDTA (TE; 10 mM Tris, pH 8, 1 mM EDTA), the slurry was spun down in a microcentrifuge (16,300 rpm, 5 min), and 0.2 ml of the aqueous phase was mixed with 0.5 ml 100% ethanol. The resulting precipitated DNA was collected by centrifugation, washed in 70% ethanol, and vacuum dried. Dry genomic DNA was resuspended in 50 μl TE, and ∼0.2 μg of DNA (volume, ∼0.5 μl) was used as the template for PCR (PicoMaxx PCR system 600422; Stratagene, La Jolla, CA).
Isolation of plasma membranes.
Neurospora plasma membranes were obtained by the method of Bowman et al. (31), with slight modifications. The filtered and rinsed cell pad from each batch culture (see above) was lifted, resuspended in 1.5 ml of digesting buffer (0.59 M sucrose, 5 mM EDTA, 50 mM NaH2PO4 at pH 5.8, 1 mM β-mercaptoethanol [β-ME] plus 4,600 U of snail gut enzyme [G3060-04; U.S. Biological]), and incubated for 1 h at 30°C, with occasional mixing. Digestion was terminated by addition of 6 ml of cold buffer, the mixture was spun at 5,000 × g for 5 min, and the pellet was resuspended in 7 ml of stabilizing buffer (0.33 M sucrose, 1 mM EGTA, 0.3% bovine serum albumin fraction V, titrated to pH 7.1 with NaOH; see reference 28) plus protease inhibitor cocktail (PIC; 10 μg/ml each of aprotinin, leupeptin, pepstatin, and chymostatin). The suspension was then homogenized in a glass-and-Teflon tissue grinder and spun at 5,000 × g for 5 min. This pellet extraction cycle was repeated three more times, and the resulting supernatants were pooled and then respun twice, first (at 10,000 × g for 10 min) to remove mitochondria and then (18,500 × g for 20 min) to collect the plasma membranes (PMs). The PM pellet was rinsed in 15 ml of washing buffer (1 mM EGTA-Tris at pH 7.5) plus PIC, by resuspension with the glass-Teflon homogenizer, and was spun down again (18,500 × g, 20 min). The final pellet was resuspended in 100 μl of washing buffer, sampled for quality testing, frozen, and stored at −80°C. (Our standard assay for quality of N. crassa membrane preparations is hydrolytic activity of the plasma-membrane ATPase, NcPma1p [31].)
Quantitative Western blot analysis.
Samples were first assayed for total protein content by the method of Lowry et al. (32) and then separated by SDS-PAGE (33). Electrophoresis was carried out on precast polyacrylamide gradients (4–15% TGX precast gel 456-1086; Bio-Rad) run for 1.5 h at 100 V, and the resulting gels were electroblotted to polyvinylidene fluoride (PVDF) membranes (IPVH 15150; Millipore). These were blocked by 1 h of incubation with 5% fat-free milk in TS buffer (10 mM Trizma base titrated to pH 7.4 with HCl, 0.9% NaCl, and 0.1% Tween 20) and stained for 1 h (25°C) with primary anti-HA monoclonal antibody (MMS-101P; Covance Corp., Berkeley, CA) and for a second hour (25°C) with secondary anti-mouse antibody coupled to horseradish peroxidase (HRP; 4021; Promega Corp., Madison, WI). (MMS-101P proved to be the only non-cross-reacting antibody commercially available. Control tests on wild-type [nontagged] membrane samples found several other commercial preparations to cross-react with an abundant protein running near the mass of NcTrk1p or NcHak1p [∼100 kDa].) Blots were visualized using Denville HyGlo ECL reagent (E2400; Denville Scientific, Metuchen NJ) exposed to Kodak XAR autoradiograph film and were imaged on the CanoScan 8400 at 600 dpi. Each row image was analyzed via ImageQuant software (Molecular Dynamics Inc., Sunnyvale, CA), measuring band intensities with respect to background levels.
The actual membrane content of HA epitope, whether attached to Trk1p or to Hak1p, was quantitated via reference samples of HA-tagged Pma1p (the plasma membrane ATPase proton pump) from Saccharomyces cerevisiae. A membrane-protein suspension in which HA-tagged S. cerevisiae Pma1p (ScPma1p) had been established as 20% of the total protein (Coomassie staining; A. B. Mason and K. E. Allen, unpublished data) was loaded onto four lanes of each gel (2, 3, 4, and 5 ng of S. cerevisiae membrane protein), interspersed with the test samples of Neurospora membrane protein. After antibody (ECL) staining of the HA epitope, band intensities for HA-ScPma1p were converted to fmol of HA epitope via the known molecular mass of HA-ScPma1p, 100.8 kDa. The overall procedure is demonstrated in Fig. 1, which plots the calibrating samples as red circles and the test samples as black circles. For each separate gel, the resulting calibration curve was fitted by a simple logarithmic expression: ECL band intensity = A + B · ln(nanograms of HA-Pma1), where A and B were the fitting parameters. The inverse expression was then used to calculate the likely fmol of HA epitope as TRK-HA or HA-Hak1 in each sample of plasma membrane protein from Neurospora. Although resulting standard curves varied somewhat from gel to gel, repeated measurements of HA-Hak1 in a single Neurospora plasma membrane source agreed within 5%.
Fig 1.
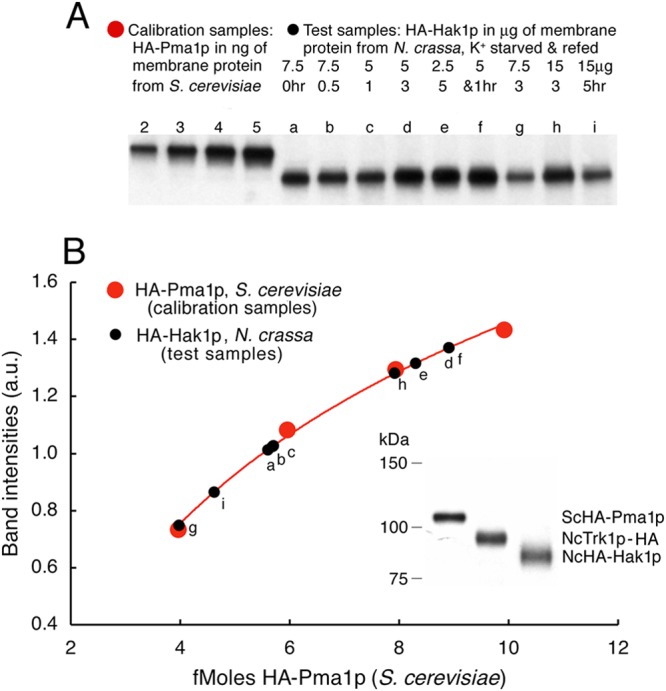
Quantitative calibration of Western blots using Saccharomyces HA-tagged Pma1p. HA-tagged proteins were electrophoresed on precast polyacrylamide gradients blotted onto PVDF membranes, blocked with milk, stained first with anti-HA monoclonal antibody and then with anti-mouse antibody, and visualized by means of ECL reagent (see Materials and Methods). Each gel was run with four lanes containing ScPma1p N terminally tagged with the HA epitope (red circles) and 2, 3, 4, and 5 ng of membrane protein at 20% purity (A. B. Mason and K.E. Allen, unpublished data), representing 4, 6, 8, and 10 fmol of HA, respectively, attached to Pma1p. The test Neurospora membrane preparations contained 2.5 to 15 μg of membrane protein (black circles), including HA-tagged Hak1p. (A) Blots showing the four calibrating loads of Saccharomyces membrane protein and several test loads of Neurospora membrane protein. Samples of Neurospora were starved of K+ for 0.5 to 5 h and then refed 10 mM K+ for 1 to 5 h. (B) Plotted points indicate digitization of the bands shown in panel A. The smooth curve indicates the least-square fit of a simple log function to the calibrating points (red). Test points (black) are plotted as per-band intensity (ordinate) and are measured in fmol (abscissa). (Inset, lower right) Single blot comparing Saccharomyces HA-Pma1p to the two Neurospora proteins of interest here.
Essential note about the graphs: representative equal-load gels were used for the display of the Western blots [see Fig. 4 and insets in Fig. 6, 7, and 10], but these could not be used for quantitation because of saturation in some lanes. Therefore, the graphs shown in Fig. 5 to 7 and 10 were obtained from separate gels prepared with loads estimated to fall within the range of the calibrating samples.
Fig 4.
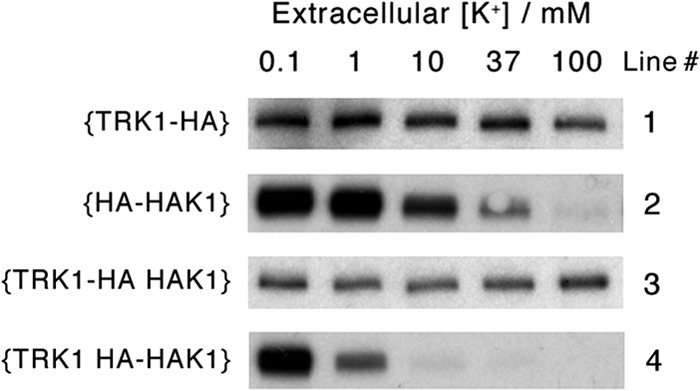
Elevated [K+]o and/or coexpression of TRK1 strongly suppresses HAK1p, but high [K+]o and coexpression of HAK1 only weakly affect expression of TRK1p. Raw Western blot data for plasma membranes isolated from four different strains of N. crassa, {TRK1-HA}, {HA-HAK1}, {TRK1-HA HAK1}, and {TRK1 HA-HAK1}, are shown. The parent strain {ΔΔΔ mus} had been deleted of all three K+ transporters and also of MUS51 in order to suppress heterologous recombination. Data are from a single representative experiment of 75-ml batch cultures (n = 3); all lanes were loaded with equal plasma membrane protein (5 μg/lane) despite saturation of the signal intensity in some lanes. Methods and calibration were the same as those described for Fig. 1.
Fig 6.
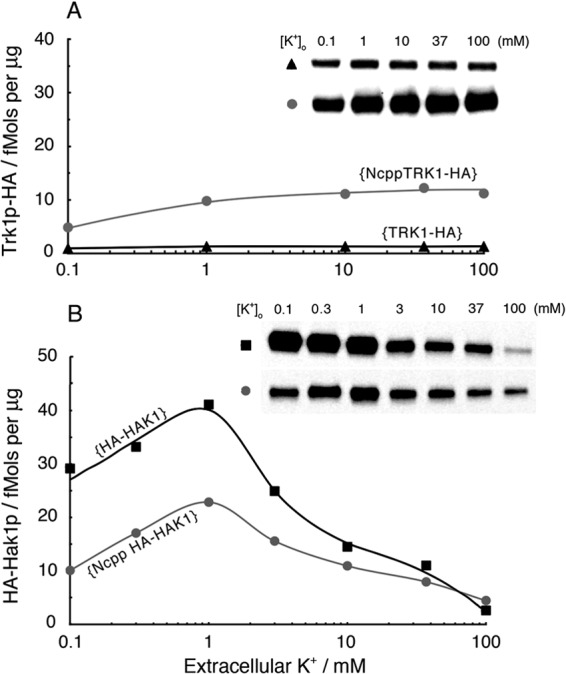
A strong promoter (Ncpp) enhances expression of TRK1 protein but depresses expression of HAK1 protein. (A) Western blot analysis of HA-tagged Trk1p produced via the native NcTRK1 promoter (▲, strain {TRK1-HA}) or via the NcPMA1 promoter (Ncpp; strain {NcppTRK1-HA};  ). Steady-state enhancement by Ncpp was ∼5-fold at low [K+]o and ∼10-fold at high [K+]o. For the inset, the load is 20 μg membrane protein in each lane. (B) Western blot analysis of HA-tagged Hak1p produced via the native NcHAK1 promoter (strain {HA-HAK}; ■) or via the NcPMA1 promoter (Ncpp; strain {NcppHA-HAK1};
). Steady-state enhancement by Ncpp was ∼5-fold at low [K+]o and ∼10-fold at high [K+]o. For the inset, the load is 20 μg membrane protein in each lane. (B) Western blot analysis of HA-tagged Hak1p produced via the native NcHAK1 promoter (strain {HA-HAK}; ■) or via the NcPMA1 promoter (Ncpp; strain {NcppHA-HAK1};  ). Quantitative gels from two separate experiments were scaled (to values at [K+]o of 1 mM) and averaged. Note that steady-state depression by Ncpp, about 2.5-fold at low [K+]o, was released at high [K+]o. Methods were the same as those described for Fig. 1, 4, and 5. For the inset, the load is 1.5 μg for each lane.
). Quantitative gels from two separate experiments were scaled (to values at [K+]o of 1 mM) and averaged. Note that steady-state depression by Ncpp, about 2.5-fold at low [K+]o, was released at high [K+]o. Methods were the same as those described for Fig. 1, 4, and 5. For the inset, the load is 1.5 μg for each lane.
Fig 5.
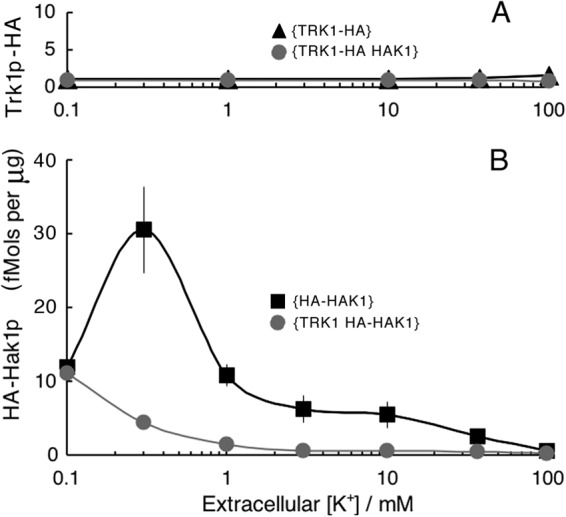
Quantitative summary of TRK1-HAK1 interactions, averaged for four separate experiments, as described for Fig. 4. Methods and calibration were the same as those described for Fig. 1. Scatter bars (±1 standard error) are mostly within the plotting symbols.
Overexpressing HA-tagged TRK1 and HAK1 genes.
The promoter for the essential gene PMA1, which encodes the major proton pump in fungal plasma membranes (34–36), is useful as a strong, constitutive promoter (37, 38). A plasmid bearing the PMA1 promoter was constructed from plasmid pMF272 (originally designed to target the his3 locus [39]) by modification as follows. A segment containing the ccg-1 promoter, the multiple cloning site (MCS), and SGFP (encoding s-green fluorescent protein) was excised via NotI/EcoRI restriction and replaced with a short fragment containing the same MCS flanked by NotI and EcoRI sites, thus making plasmid pAR282. Separately, a 1-kb segment of DNA located upstream of the NcPMA1 start codon, and designated here Ncpp, was amplified by PCR using genomic DNA from wild-type strain 2489A as the template, along with the PP1-f/PP2-r primer pair (Table 2). After restriction with NotI/XbaI, this was cloned into pAR282 to give plasmid pAR283. Untagged and HA-tagged versions of both TRK1 and HAK1 were cloned into pAR 283 following XbaI/XmaI cuts for TRK1 and following XbaI/EcoRI cuts for HAK1. The resulting four plasmids were linearized with DraI endonuclease before transformation of the {ΔΔΔ mus51 his-3} strain (Table 1).
RESULTS
Growth experiments.
Expression cloning of Neurospora DNA in a K+-deficient strain of Saccharomyces cerevisiae (24, 40) and homology searching in the completed N. crassa genome sequence (25, 41–43) have together revealed three K+ transport genes in Neurospora: TRK1, TRK2, and HAK1. To gain an initial idea of the physiological roles of the corresponding transport proteins, combination knockout strains were constructed and tested in drop colony growth experiments over a range of extracellular K+ concentrations (0.03 to 10 mM) (Fig. 2A). TRK1, expressed in strains {++Δ} and {+ΔΔ}, supported robust growth down to 0.1 mM [K+]o, while HAK1, expressed in strains {Δ++} and {ΔΔ+}, was significantly less effective, yielding robust growth only down to 0.3 mM [K+]o. Deletion of TRK1 from HAK1+ strains reduced both colony size (diameter) and mycelial density (reflectance) compared to the control colonies (cf. {Δ++} with {+++} or {ΔΔ+} with {+Δ+} for [K+]o of ≤0.3 mM). The reciprocal deletions of HAK1 had no such effects on TRK1+ colonies. TRK2, expressed alone in strain {Δ+Δ}, was unable to support growth on low K+ and also failed to augment growth due to either TRK1 or HAK1 (cf. {++Δ} with {+ΔΔ} or {Δ++} with {ΔΔ+}). For this reason, all further experiments (see Fig. 4 to 10) focused on TRK1 and HAK1 and were carried out on strains deleted of the TRK2 gene.
Two other K+ transport mechanisms did not play a significant role under the conditions used below and were not considered further: (i) a separate low-affinity system for potassium uptake, indicated by the sharp increase of colony sizes and densities at high [K+]o (≥3 mM), especially in strains {Δ+Δ} and {ΔΔΔ), and presumed to be analogous to the NSC pathway previously described in patch-clamp studies of S. cerevisiae (44, 45), and also (ii) a Neurospora Tok1 K+ channel, expected to be a strong outward rectifier and not to allow significant K+ influx, again analogously to its homologue in yeast (46–48). Confirmation of the lack of effect of these channels on K+-dependent growth was obtained from experiments on several strains deleted of TOK1, as illustrated, for example, in Fig. 2B.
For a quantitative study of cellular dependence on the two main K+ transporters, strains carrying hemagglutinin (HA)-tagged versions of the TRK1 and HAK1 genes were constructed (Table 1) and tested for the ability to grow exponentially in liquid suspension cultures. In the experiment shown in Fig. 3, freshly harvested conidia of N. crassa were rinsed and resuspended in minimal medium at 25°C, where they displayed a typical 3- to 4-h lag before germinating. Cells expressing only TRK1 then grew exponentially at the same specific rate (α) of 0.274 h−1 (doubling time of 2.53 h) at starting [K+]o as low as 0.3 mM (blue circles), falling slightly to 0.259 h−1 at 0.1 mM [K+]o (green squares). In contrast, cells expressing only HAK1 slowed to a specific growth rate of 0.182 h−1 at 0.3 mM [K+]o (red circles) and 0.144 h−1 at 0.1 mM [K+]o (data not shown). For cells deleted of both TRK1 and HAK1 and inoculated into [K+]o at or below 1 mM (grey triangles), growth was barely detectable and could be described as either quasiexponential, α < 0.08 h−1, or apparently linear, ∼0.6 mg dry weight/h. These results quantitatively confirm the advantage of Trk1p over Hak1p at low [K+]o. Further implications of the growth results, including the time courses of intracellular K+ concentration ([K+]i) and net fluxes, are considered later (see Fig. 8).
Regulation of HAK1 and TRK1 expression.
Most of the Neurospora strains listed in Table 1, including those used for the liquid growth experiments shown in Fig. 3, were constructed explicitly to allow expression analysis via Western blotting, making use of the HA epitope. Four clear results emerged, which are qualitatively demonstrated in the blots of Fig. 4: (i) expression of Trk1p was only weakly affected by [K+]o in the growing cell suspension, whereas (ii) expression of Hak1p was strongly inhibited by 10 mM [K+]o and almost completely suppressed by 100 mM [K+]o. Furthermore, (iii) coexpression of HAK1 had no significant effect on the cellular level of Trk1p, but (iv) coexpression of TRK1 clearly accentuated the inhibitory effect on Hak1p by elevated [K+]o. In other words, regulation of these two K+ transport proteins in Neurospora is in no way reciprocal; when the TRK1 gene is present behind its native promoter, it is dominant and sufficient down to the lowest tolerable K+ levels (Fig. 3). The HAK1 system is more closely tied to Neurospora's metabolic regulatory program, but it becomes useful mainly when Trk1p is absent or blocked or when other metabolic limitations are imposed, e.g., carbon starvation (see below).
Quantitative analysis of these results required careful calibration of the Western blots, and for this purpose, HA-tagged proton pump protein (Pma1p) from Saccharomyces cerevisiae was used as the standard (see Materials and Methods). Data averaged from three sets of gels similar to those of Fig. 4 yielded the plots of Fig. 5, showing not only that Trk1p is dominant and insensitive to the presence of the HAK1 gene or protein but also that it is scarce, being present at only 1 to 2 fmol per μg of membrane protein (Fig. 5A), at any value of [K+]o, compared to more than 10-fold that level of Hak1p (Fig. 5B) for low values of [K+]o. However, as expected from the results shown in Fig. 4, Hak1p was suppressed to very low levels by either elevated [K+]o or expressed TRK1 gene/protein.
These results were further emphasized by several experiments with Ncpp, the promoter that normally controls expression of the gene encoding Neurospora's plasma membrane proton pump (NcPMA1). Ncpp itself is not sensitive to [K+], whether intracellular or extracellular, but it is a routine strong promoter (37, 38). Western blot analysis of HA-tagged Trk1p and Hak1p, expressed behind Ncpp rather than their native promoters, found Ncpp to increase Trk1p approximately 10-fold in isolated plasma membranes, regardless of the potassium level (Fig. 6A). But, Ncpp not only failed to increase Hak1p in membrane preparations but actually decreased expression (Fig. 6B) at all [K+]o values below 100 mM; evidently, the native Hak1 promoter is more effective than Ncpp, especially with low extracellular potassium. Cross-expression experiments were fully consistent with these findings. That is, elevating the level of Trk1p ∼10-fold by means of expression behind Ncpp further depressed the expression levels of Hak1p (data not shown).
Figures 4 to 6 demonstrate steady-state regulation of Hak1p versus Trk1p over a time scale of hours during exponential growth, but those data say nothing about how rapidly regulatory shifting occurs when the extracellular potassium is suddenly raised or lowered. Figure 7 addresses that question for both transporter proteins. Cells of strain {HA-HAK1}, which had been pregrown in 100 mM K+ to be optimally loaded with potassium, were quickly rinsed and resuspended in K+-free medium (Fig. 7B). Intracellular [K+] had declined by ∼65% when the first measurements were made, 30-min postrinse, and rose almost equally fast when [K+]o was restored to 100 mM at 5 h. However, the corresponding upregulation and downregulation of HA-tagged Hak1p required several hours, rising with an apparent time constant (τ) of 3.44 h (half-time of 2.37 h) and falling with a τ of 1.60 h (half-time of 1.10 h). These numbers can be compared to time constants of ∼20 min for activating or deactivating high-affinity transport of either glucose or ammonium ions, induced by the appropriate starvation or replenishment (49, 50). In the obvious control experiment on {TRK1-HA}, HA-tagged Trk1p barely responded to potassium depletion, as is shown in Fig. 7A.
Modulation of cellular K+ content, fluxes, and affinities.
Apart from questions about molecular details, there are two principal modes for regulating protein function: (i) raising or lowering the amount of the specified protein present, and (ii) modulating the kinetic properties of that protein. Mode-i changes for Trk1p and Hak1p are described in Fig. 4 to 7, especially in response to different potassium regimens. Mode-ii changes can be extracted, in part, from additional properties of exponentially growing cells, e.g., from the time courses of intracellular potassium and of the fluxes implied by the measured cell mass and [K+]i. Cells sustaining the maximal growth rate (α = 0.274 h−1) had stabilized [K+]i within the control range, 165 to 190 mM, by 90 min after the onset of exponential growth (Fig. 8A, triangles and blue circles) and were taking up net K+ at ∼50 mM/h (Fig. 8B), or at approximately 1 mM/min, the previously established requirement for optimal growth of Neurospora (51, 52). However, potassium-limited cells (green squares, red circles, and grey triangles) were characterized by lowered [K+]i values at the onset of exponential growth (viz., by extra leakage of potassium from the conidia during the lag phase), by progressive decline of [K+]i as growth continued, and by falling net influx (Fig. 8B) as extracellular [K+] declined due to continued uptake. Cells with the most severe potassium limitation, lacking both K+ transporters and started at [K+]o of 1 mM or lower ({ΔΔΔ mus}; grey triangles in Fig. 8B), sank into several hours of actual K+ loss (net efflux; negative values in Fig. 8B) before stabilizing the flux at nearly zero with [K+]i below 40 mM (Fig. 8A).
Important quantitative comparisons emerged. For example, although the specific growth rate for strain {TRK1-HA} started in 0.1 mM K+ was 60% higher than that for strain {HA-HAK1} started in 0.3 mM K+ (α = 0.259 and 0.182 h−1, respectively), both the net influxes of K+ and the actual [K+]i values were nearly the same by 8 h (Fig. 3 and 8A and B). This means, more explicitly, that the apparent dependence of α upon [K+]i was steep near the outset of exponential growth (Fig. 9A, 1.5 h), but relaxed at later times (e.g., Fig. 9A, 4.5 h). Thus, within limits, growing Neurospora cells could adapt to the restricted availability of potassium without sacrificing growth rate.
Furthermore, this adaptation clearly involved increasing substrate affinity for both Trk1p and Hak1p, a phenomenon previously proposed for Neurospora and Saccharomyces (53, 54). It can readily be seen in the present experiments by plotting the net K+ influx versus extracellular potassium concentration, as [K+]o was depleted by growth. In Fig. 9B and C, strains bearing TRK1-HA and/or HA-HAK1 were inoculated into K+-free Vogel's medium plus 0.1 mM KCl. Loss of K+ from the conidia during the lag phase had slightly increased extracellular K+ at time zero, but the onset of exponential growth was accompanied by K+ influxes of ∼45 mM/h in the presence of Trk1p with or without Hak1p (Fig. 9B) and ∼18 mM/h in the presence of Hak1p alone (Fig. 9C). In both cases, the initial plots of flux versus [K+]o were hyperbolic but were offset from the origin (0 μM) by 49.7 μM for Trk1p and 103.6 μM for Hak1p, corresponding to projected growth times of ∼5.5 and ∼1.9 h, respectively. However, as [K+]o approached those offset values, the fluxes stabilized at ∼9 mM/h with Trk1p, with or without Hak1p, and at ∼3.2 mM/h with only Hak1p. The stabilized fluxes continued for several hours, to the end of exponential growth, despite continually declining extracellular potassium. Progressive, adaptive, increasing affinity of both transporters for substrate (K+) thus is a necessary inference. (HAK protein made no significant contribution either to mass expansion or to K+ net flux in the presence of TRK protein. That is, Trk1p-HA expression was equivalent to that of Trk1p-HA plus HA-Hak1p. It is not known whether Hak1p was deactivated or simply insignificant when present under these conditions.)
Quantitatively, these results reemphasize that growth of K+-limited (or K+-starved) cells of N. crassa is better supported via TRK protein than via HAK protein; the effective Km for K+ transport is lower and the effective Vmax is higher with Trk1p than with Hak1p when each is tested separately. Finally, it is clear that exponential growth of Neurospora is not rigidly dependent upon either the net K+ influx or the actual intracellular concentration.
Cross-pathway effects: carbon starvation.
The first demonstration that transport of sugars and other neutral molecules can be unequivocally electrophoretic (55) solidified the notion of membrane transport within the complex of metabolic reactions then emerging as targets for glucose regulation, especially in N. crassa (56–58). Glucose withdrawal leads to the upregulation of high-affinity glucose transport (55, 56), to proliferation of vacuoles (59), to enhancement of cytoplasmic buffer capacity (D. Sanders and C. L. Slayman, unpublished results), and probably, as in Saccharomyces (60), to gradual disassembly of the vacuolar ATPase (B. Bowman, unpublished results). In addition, full carbon starvation increases membrane resistivity (49, 50), thereby magnifying voltage changes associated with active transport of potassium ions (5). The latter fact affords a robust approach for quantitative study of the K+ transporters in Neurospora (A. Rivetta, K. E. Allen, and C. L. Slayman, unpublished data), but accurate analysis requires knowledge of the separate effects of starvation upon the two proteins Trk1p and Hak1p.
This information was obtained by a simple extension of the experiment shown in Fig. 7, removing extracellular sugar from the mycelial suspension after a suitable period of starvation for K+. As shown in Fig. 10A, tagged Trk1p declined with a time constant of 2.8 h, corresponding to a half-time of 1.9 h. Neither the rate nor the apparent endpoint was influenced by the presence (circles) or absence (triangles) of HAK1/Hak1p. However, the observed decline of Trk1p-HA during pure K+ starvation was unexpectedly fast (Fig. 10A, dashed line) and not distinguishable from that with sugar starvation.
Hak1p itself, on the other hand, barely responded to 1 h of pure K+ starvation (Fig. 10B) but then rose from control values around 15 fmol/μg membrane protein to steady values near 65 fmol/μg membrane protein over the 3 h of carbon starvation (glucose removal). Although the presence (circles) or absence (squares) of TRK1/Trk1p did not affect the increase of Hak1p associated with carbon starvation (∼50 fmol/μg membrane protein), TRK1-Trk1p did lower the control value of Hak1p at the onset of K+ starvation by approximately 70%. This result is qualitatively consistent with the results shown in Fig. 5.
Overall, the Hak1p:Trk1p ratio, which varied in the range of 10:1 to 30:1 (depending mainly on K+ repletion) in normally grown Neurospora mycelium (Fig. 5, 6, and 8), was increased to >500:1 by the regimen of serial starvation for K+ and carbon. This emphasizes again that HAK1-Hak1p is fundamentally incorporated into the metabolic program of Neurospora in a manner that TRK1-Trk1p is not.
DISCUSSION
Basic conclusions.
Both of the TRK and HAK families of transporter genes are distributed widely in the plant and fungal kingdoms (61–64), and they can be complemented by two families of cation-pumping ATPases (10, 11), although the latter are not present in Neurospora (65). In higher plants, regulation of K+ uptake involves not only several isoforms of the HAK and/or TRK proteins but also many specialized tissues and organs. Regulation in these systems therefore must be very complex. On the other hand, regulation in the best-known simple eukaryotic system, the yeast S. cerevisiae, has evolved mainly around two K+ transporters, both of the TRK family, while completely excluding the HAK family. N. crassa, in possessing K+ transporters from both families while having only moderate cytological specialization, affords intermediate complexity for characterizing and modeling transporter function and regulation. The principal results, presented in Fig. 4 to 7, are simple, clear-cut, and quite distinct from most previous reports on plants or fungi.
The important background physiological conditions are ample carbon source, moderated salts (especially 63.5 mM Na+ and 25 mM NH4+), and pHo buffered (citrate/phosphate) in the range of 5.8 to 5.3, well within the broad optimum for K+ transport in N. crassa (66). Under these conditions, Neurospora Trk1p is kinetically a more effective transporter than Hak1p, in having both a higher per-site turnover number and a lower Michaelis constant (K0.5) for substrate K+ (Fig. 2, rows 1 to 3 and columns 2 and 4). It also supports robust growth below 100 μM [K+]o, compared with ∼300 μM for Hak1p. Otherwise put, Trk1p provides for stable potassium accumulation in Neurospora while behaving like a classic constitutive enzyme: it is expressed at a nearly constant level over wide-ranging K+ concentrations and can even override sudden shifts of concentration (Fig. 7A), but it is susceptible to approximately 10-fold depletion during energy limitation (i.e., carbon starvation) (Fig. 10A). In absolute terms, Trk1p expression is low in Neurospora, in the range of 1 ± 0.5 fmol/μg membrane protein, which translates to ∼4 nmol Trk1p per liter of intracellular water (ICW). Since the net flux of ∼50 mmol/h · liter ICW (Fig. 8B, blue circles and triangles) equals 14 μmol/s · liter ICW, a per-site transport number of 3.5 × 103 K+ ions/site · s is implied, which is high for conventional ion transporters but very low for conventional ion channels.
Hak1p, on the other hand, behaves like a classic derepressible enzyme in Neurospora, present at low levels (1 to 5 fmol/μg membrane protein; Fig. 5B) when its substrate is available at high concentration but elevated 10-fold or more when substrate is scarce and when Trk1p is missing (Fig. 4 and 5B). Its implied per-site turnover number, in sustained support of exponential growth (as in Fig. 8B), lies in the range of 1 × 102 to 3 × 102 K+ ions/site · s, appropriate for a conventional ion transport protein. However, carbon starvation, on top of K+ limitation, can drive up the expression of Hak1p by another order of magnitude (Fig. 10B), and under that condition, Hak1p becomes the dominant K+ uptake protein in Neurospora, by a factor of >500, over Trk1p. Since carbon starvation was the practical background condition for discovery of membrane currents due to K+-H+ cotransport (2, 5), it is clear that Hak1p must have been the responsible protein. This inference was also reached earlier by Haro et al. (24), who compared Northern blot measurements in Neurospora to transport measurements on NcHak1p heterologously expressed in Saccharomyces. A similar relationship between Trk1p and Hak1p has also been described recently for the methylotrophic yeast Hansenula polymorpha (67). Present data do not rule out the possibility that Trk1p also carries out H+-K+ cotransport, but they do make that mode unnecessary. Detailed quantitative analysis of the underlying electrical data is needed to clarify the actual functioning of both transport proteins (Rivetta et al., unpublished).
Adaptations.
Mechanistically more mysterious is the fact that both Trk1p and Hak1p are able to increase their affinities (or decrease their Michaelis constants) for the transported substrate, as K+ limitation becomes increasingly severe. Such a process had been implied before (53, 54, 68, 69); here, it is demonstrated and quantitated (Fig. 9B and C). We have no explanation for why the velocity-versus-concentration plots (flux versus [K+]o) are offset from the origin, but before zero is actually reached the proteins appear to change, stabilizing their fluxes at low values that are still sufficient for exponential growth. This condition of Trk1p and Hak1p might be termed the sliding-affinity mode; it occurs without perturbing the effectiveness of Trk1p compared to that of Hak1p. The growth studies (Fig. 3) showed that Trk1p can take up potassium as fast from ∼0.1 mM [K+]o (10 to 11 mM/h at 5 h; Fig. 3C) as can Hak1p at ∼0.3 mM [K+]o, and in the sliding-affinity mode, Trk1p stabilizes at a 3-fold higher net flux than Hak1p, while it also starts the process at a lower value of [K+]o. The origin of these changes, viz., the binding protein(s) presumed to be coactively involved, remains to be identified.
However, elevation of membrane voltage, such as has been demonstrated to occur during carbon starvation (50, 55), could have exactly that effect on transporter kinetics. This possibility has been described at least twice in theory (70, 71) and has previously been argued to account for flux data in S. cerevisiae (72). Elevation of membrane voltage during carbon starvation results from increased membrane resistance, which in turn reflects metabolic downregulation of temporarily unneeded membrane transporters. Such behavior is an energy shift device to enhance the cells' ability to scavenge the depleted nutrient, namely, sugars in the case of carbon starvation and potassium, we suggest, in the present experiments.
Adaptation to sustained potassium limitation is certainly a property of the whole organism, not solely of the K+ transporters. Cells expressing Trk1p and germinated in 0.1 mM K+ sustained their starting exponential growth rate (α = 0.259 h−1) (Fig. 3, green squares) for more than 8 h despite the ∼50% decline of intracellular [K+] that accompanied cellular volume expansion and reduction of net influx (Fig. 8A and B) due to extracellular [K+] depletion. Similar adaptation took place in cells expressing only Hak1p (Fig. 3, red circles) but with lower absolute rates, viz., α = 0.188 h−1 started in 0.3 mM K+ (Fig. 8) and α = 0.144 h−1 in 0.1 mM K+ (data not shown). Even cells missing both defined K+ transporters ({ΔΔΔ mus}) grew exponentially when constrained on low K+ (Fig. 3, triangles), though very slowly, as [K+]i continued to fall. In this case, residual K+ flux necessarily occurred through a low-affinity/nonselective pathway, probably corresponding to that dubbed NSC1 in Saccharomyces (44, 45). That path would carry current under voltage clamp conditions, but probably also would mediate ion exchange: K+ for H+, Na+, amino cations generally, and NH4+ in particular (52, 72). It is likely that ammonium ions (with a crystal radius of 1.48 Å) substituted for potassium ions (crystal radius of 1.33 Å) in those enzymes whose activities normally make use of bound K+ (74); NH4+ was abundant (25 mM) in the low-K+ media employed in these experiments. However, ionic substitution also could have been more general, since other fungi, especially S. cerevisiae, have been found to grow when intracellular K+ is replaced by a variety of alkali and alkali-metal cations (73, 75, 76).
Signaling.
The liquid growth measurements (Fig. 3), together with simultaneous potassium determinations (Fig. 8A), show that the growth rate in Neurospora is not dependent on intracellular [K+] per se; rather, it is dependent on extracellular [K+]. An important implication of this finding is that potassium's primary effect in metabolic regulation, at least in the presence of abundant ammonium ions, should be on signaling rather than upon simple enhancement of cytoplasmic enzyme activities. The same conclusion has also been reached via theoretical analysis of K+ transport in Saccharomyces (77, 78). That seems like a sensible arrangement; given that K+ is a bulk constituent of cytoplasm (also partially sequestered in vacuoles), its actual concentration should respond only slowly to threats. However, a sudden drop of [K+]o itself would be a direct threat. We have made some effort, thus far unsuccessful, to discover possible signaling vectors. In particular, deletion experiments have failed to show involvement of either Neurospora's silent K+ transporter (Trk2p) or its plasma membrane K+ channel (Tok1p) in modulating growth itself or the particular properties of Trk1p or Hak1p.
Further attempts to investigate the primary signaling mechanisms in potassium homeostasis will attempt to bypass the so-called hugeness feature of most metabolic networks and focus on those circuits that are central to K+ homeostasis while avoiding minor paths and side branches. Potassium accumulation in this particular model organism, N. crassa, is propitious for critical pathway mapping for several reasons: (i) only two effector proteins are involved in K+ uptake; (ii) those proteins behave in identifiably different fashions; (iii) a single energizing protein (Pma1p) drives transport through both effector proteins (via membrane voltage) and must be coregulated; and (iv) only the efflux pathway(s) remains uncertain. Changes in the expression of Pma1p still need to be characterized, but the integrated system regulating K+ content of Neurospora inversely adjusts the amount of Hak1p present, while holding Trk1p nearly constant, under varied conditions of potassium starvation or repletion. The same system clearly enhances expression of Hak1p and suppresses expression of Trk1p under carbon starvation. Modulations of Trk1p, Hak1p, and Pma1p under other conditions, e.g., osmotic stress, ammonium starvation, and pH stress, still need to be characterized, but the main thrust for future experiments is that genes/proteins central to the regulation of K+ transport should, when perturbed, displace all three effector proteins in predictable, coordinated fashion. This idea is a kind of protein triangulation.
There are now two main conduits for such investigation. The most direct is to screen for transcription factors which modulate pHAK, the promoter element for the HAK1 gene. This can be done, starting with {HAK1}, by inserting a selectable reporting marker behind pHAK in the HAK1 locus and then subjecting the strain to insertional mutagenesis. By varying background [K+]o during selection on the reporter agonist, this procedure can be biased to identify mutants that either enhance or suppress the activity of pHAK. Alternatively, Neurospora homologues of genes that have been implicated in K+ regulation elsewhere can be tested in deletion and/or overexpression experiments. Prime candidates at present are two serine/threonine (S/T) protein kinases, Hal4p and Hal5p, described as positive regulators of the TRK proteins in Saccharomyces (79, 80), and two phosphatase-related peptides, Ppz1p and Ppz2p, plus calcineurin, described as negative regulators (81, 82). Homologues of all of these are present in Neurospora and can be evaluated by crossing the existing single-gene deletion strains (25) with {HA-HAK1} or {TRK1-HA} or with other constructs described above. A global strategy for characterizing S/T protein kinase genes in Neurospora has already been described (107 total) (83), and other candidate K+ regulatory genes can be extracted from full-genome microarray studies (see, e.g., Tian and collaborators [84, 85]) under different conditions of K+ limitation.
ACKNOWLEDGMENTS
We are indebted to Brett Mason for calibrated samples of HA-tagged Saccharomyces PMA1 protein, for use of the CanoScan digital scanner, and for much helpful advice concerning membrane protein assays; to Tong Wang for use of the Instrumentation Laboratory atomic absorption photometer; and to Efim Golub for the high-fidelity DNA polymerase, as well as for advice on bacterial transformations.
The work was supported in part by NIH research grant GM60696.
Footnotes
Published ahead of print 8 March 2013
REFERENCES
- 1. Slayman CL, Sanders D. 1985. Steady-state kinetic analysis of an electroenzyme. Biochem. Soc. Symp. 50:11–29 [PubMed] [Google Scholar]
- 2. Blatt MR, Slayman CL. 1987. Role of “active” potassium transport in the regulation of cytoplasmic pH by non-animal cells. Proc. Natl. Acad. Sci. U. S. A. 84:2737–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schroeder JI. 1988. K+ transport properties of K+ channels in the plasma membrane of Vicia faba guard cells. J. Gen. Physiol. 92:667–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lebaudy A, Véry Sentenac A-AH. 2007. K+ channel activity in plants: genes, regulations and functions. FEBS Lett. 581:2357–2366 [DOI] [PubMed] [Google Scholar]
- 5. Rodriguez-Navarro A, Blatt MR, Slayman CL. 1986. A potassium-proton symport in Neurospora crassa. J. Gen. Physiol. 87:649–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maathuis FJM, Sanders D. 1994. Mechanism of high-affinity potassium uptake in roots of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 91:9272–9276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rubio F, Gassmann W, Schroeder JI. 1995. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270:1660–1663 [DOI] [PubMed] [Google Scholar]
- 8. Rodríguez-Navarro A, Rubio F. 2006. High-affinity potassium and sodium transport systems in plants. J. Exp. Bot. 57:1149–1160 [DOI] [PubMed] [Google Scholar]
- 9. Ariño J, Ramos J, Sychrová H. 2010. Alkali metal cation transport and homeostasis in yeasts. Microbiol. Mol. Biol. Rev. 74:95–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Souza FSJ, Gomes SL. 1998. A P-type ATPase from the aquatic fungus Blastocladiella emersonii similar to animal Na,K-ATPases. Biochim. Biophys. Acta 1383:183–187 [DOI] [PubMed] [Google Scholar]
- 11. Benito B, Garciadeblás B, Schreier P, Rodríguez-Navarro A. 2004. Novel P-type ATPases mediate high-affinity potassium or sodium uptake in fungi. Eukaryot. Cell 3:359–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jardetzky O. 1966. Simple allosteric model for membrane pumps. Nature 211:969–970 [DOI] [PubMed] [Google Scholar]
- 13. Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. 2003. Structure and mechanism of the lactose permease of Escherichia coli. Science 301:610–615 [DOI] [PubMed] [Google Scholar]
- 14. Law CJ, Maloney PC, Wang D-N. 2008. Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 62:289–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smirnova I, Kasho V, Kaback HR. 2011. Lactose permease and the alternating access mechanism. Biochemistry 50:9684–9693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Durell SR, Guy HR. 1999. Structural models of the KtrB, TrkH, and Trk1,2 symporters, based on the crystal structure of the KcsA K+ channel. Biophys. J. 77:789–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Durell SR, Hao Y, Nakamura T, Bakker EP, Guy HR. 1999. Evolutionary relationship between K+ channels and symporters. Biophys. J. 77:775–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Doyle DA, Cabral JM, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280:69–76 [DOI] [PubMed] [Google Scholar]
- 19. Tholema N, Bakker EP, Suzuki A, Nakamura T. 1999. Change to alanine of one out of four selectivity filter glycines in KtrB causes a two orders of magnitude decrease in the affinities for both K+ and Na+ of the Na+-dependent K+-uptake system KtrAB from Vibrio alginolyticus. FEBS Lett. 450:217–220 [DOI] [PubMed] [Google Scholar]
- 20. Kato Y, Sakaguchi M, Mori Y, Saito K, Nakamura T, Bakker EP, Sato Y, Goshima S, Uozumi N. 2001. Evidence in support of a four transmembrane-pore-transmembrane topology model for the Arabidopsis thaliana Na+/K+ translocating AtHKT1 protein, a member of the superfamily of K+ transporters. Proc. Natl. Acad. Sci. U. S. A. 98:6488–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mäser P, Hosoo Y, Goshima S, Horie T, Eckelman B, Yamada K, Yoshida K, Bakker EP, Shinmyo A, Oiki S, Schroeder JI, Uozumi N. 2002. Glycine residues in potassium channel-like selectivity filters determine potassium selectivity in four-loop-per subunit HKT transporters from plants. Proc. Natl. Acad. Sci. U. S. A. 99:6428–6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zeng G-F, Pypaert M, Slayman CL. 2004. Epitope tagging of the yeast K+-carrier, TRK2, demonstrates folding which is consistent with a channel-like structure. J. Biol. Chem. 279:3003–3013 [DOI] [PubMed] [Google Scholar]
- 23. Cao Y, Jin X-S, Huang H, Derebe MG, Levin EJ, Kabaleeswaran V, Pan Y-P, Punta M, Love J, Weng J, Quick M, Kloss B, Bruni R, Martinez-Hackert E, Hendrickson WA, Rost B, Javitch JA, Rajashankar KR, Jiang Y, Zhou M. 2011. Crystal structure of a potassium ion transporter TrkH. Nature 471:336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haro R, Sainz L, Rubio F, Rodríguez-Navarro A. 1999. Cloning of two genes encoding potassium transporters in Neurospora crassa and expression of the corresponding cDNAs in Saccharomyces cerevisiae. Mol. Microbiol. 31:511–520 [DOI] [PubMed] [Google Scholar]
- 25. Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, Weiss RL, Borkovich KA, Dunlap JC. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. U. S. A. 103:10353–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ninomiya Y, Suzuki K, Ishii C, Inoue H. 2004. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. U. S. A. 101:12248–12253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Davis RH, de Serres FJ. 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 17A:79–143 [Google Scholar]
- 28. Vogel HJ. 1956. A convenient growth medium for Neurospora (Medium N). Microb. Gen. Bull. 13:42–46 [Google Scholar]
- 29. Slayman CW, Tatum EL. 1964. Potassium transport in Neurospora. I. Intracellular sodium and potassium concentrations, and cation requirements for growth. Biochim. Biophys. Acta 88:578–592 [PubMed] [Google Scholar]
- 30. Rose MD, Winston F, Hieter P. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31. Bowman EJ, Bowman BJ, Slayman CW. 1981. Isolation and characterization of plasma membranes from wild type Neurospora crassa. J. Biol. Chem. 256:12336–12342 [PubMed] [Google Scholar]
- 32. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265–275 [PubMed] [Google Scholar]
- 33. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 34. Slayman CL, Long WS, Lu CY-H. 1973. The relationship between ATP and an electrogenic pump in the plasma membrane of Neurospora crassa. J. Membr. Biol. 14:305–338 [DOI] [PubMed] [Google Scholar]
- 35. Bowman BJ, Slayman CW. 1977. Characterization of plasma membrane adenosine triphosphatase of Neurospora crassa. J. Biol. Chem. 252:3357–3363 [PubMed] [Google Scholar]
- 36. Hager KM, Mandala SM, Davenport JW, Speicher DW, Benz EJ, Jr, Slayman CW. 1986. Amino acid sequence of the plasma membrane ATPase of Neurospora crassa: deduction from genomic and cDNA sequences. Proc. Natl. Acad. Sci. U. S. A. 83:7693–7697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Capieaux E, Vignais ML, Sentenac A, Goffeau A. 1989. The yeast H+-ATPase gene is controlled by the promoter binding factor TUF. J. Biol. Chem. 264:7437–7446 [PubMed] [Google Scholar]
- 38. Figler RA, Omote H, Nakamoto RK, Al-Shawi MK. 2000. Use of chemical chaperones in the yeast Saccharomyces cerevisiae to enhance heterologous membrane protein expression: high-yield expression and purification of human P-glycoprotein. Arch. Biochem. Biophys. 376:34–46 [DOI] [PubMed] [Google Scholar]
- 39. Freitag M, Hickey PC, Raju NB, Selker EU, Read ND. 2004. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet. Biol. 41:897–910 [DOI] [PubMed] [Google Scholar]
- 40. Roberts SK. 2003. TOK homologue in Neurospora crassa: First cloning and functional characterization of an ion channel in a filamentous fungus. Eukaryot. Cell 2:181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen CB, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-Pedersen D, Nelson MA, Werner-Washburne M, Selitrennikoff CP, Kinsey JA, Braun EL, Zelter A, Schulte U, Kothe GO, Jedd G, Mewes W, Staben C, Marcotte E, Greenberg D, Roy A, Foley K, Naylor J, Stange-Thomann N, Barrett R, Gnerre S, Kamal M, Kamvysselis M, Mauceli E, Bielke C, Rudd S, Frishman D, Krystofova S, Rasmussen C, Metzenberg RL, Perkins DD, Kroken S, Cogoni C, Macino G, Catcheside D, Li W, Pratt RJ, Osmani SA, DeSouza CP, Glass L, Orbach MJ, Berglund JA, Voelker R, Yarden O, Plamann M, Seiler S, Dunlap J, Radford A, Aramayo R, Natvig DO, Alex LA, Mannhaupt G, Ebbole DJ, Freitag M, Paulsen I, Sachs MS, Lander ES, Nusbaum C, Birren B. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859–868 [DOI] [PubMed] [Google Scholar]
- 42. Kiranmayi P, Mohan PM. 2006. Metal transportome of Neurospora crassa. In Silico Biol. 6:169–180 [PubMed] [Google Scholar]
- 43. Ren Q, Chen K, Paulsen IT. 2007. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acid Res. 35:D275–D279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bihler H, Slayman CL, Bertl A. 1998. NSC1: a novel high-current inward rectifier for cations in the plasma membrane of Saccharomyces cerevisiae. FEBS Lett. 432:59–64 [DOI] [PubMed] [Google Scholar]
- 45. Bihler H, Slayman CL, Bertl A. 2002. Low-affinity potassium uptake by Saccharomyces cerevisiae is mediated by NSC1, a calcium-blocked non-specific cation channel. Biochim. Biophys. Acta 1558:109–118 [DOI] [PubMed] [Google Scholar]
- 46. Bertl A, Slayman CL, Gradmann D. 1993. Gating and conductance in an outward-rectifying K+ channel from the plasma membrane of Saccharomyces cerevisiae. J. Membr. Biol. 132:183–199 [DOI] [PubMed] [Google Scholar]
- 47. Fairman C, Zhou X, Kung C. 1999. Potassium uptake through the TOK1 K+ channel in the budding yeast. J. Membr. Biol. 168:149–157 [DOI] [PubMed] [Google Scholar]
- 48. Roller A, Natura G, Bihler H, Slayman CL, Bertl A. 2008. Functional consequences of leucine and tyrosine mutations in the dual pore motifs of the yeast K+ channel, Tok1p. Pflügers Arch. Eur. J. Physiol. 456:883–896 [DOI] [PubMed] [Google Scholar]
- 49. Slayman CL. 1977. Energetics and control of transport in Neurospora, p 69–86 In Jungreis AM, Hodges TK, Kleinzeller A, Schultz SG. (ed), Water relations in membrane transport in plants and animals. Academic Press, New York, NY [Google Scholar]
- 50. Slayman CL. 1980. Transport control phenomena in Neurospora, p 179–190 In Spanswick RM, Lucas WJ, Dainty J. (ed), Plant membrane transport: current conceptual issues. Elsevier, Amsterdam, the Netherlands [Google Scholar]
- 51. Slayman CW, Tatum EL. 1965. Potassium transport in Neurospora. II. Measurement of steady-state potassium fluxes. Biochim. Biophys. Acta 102:149–160 [PubMed] [Google Scholar]
- 52. Slayman CL, Slayman CW. 1968. Net uptake of potassium in Neurospora: exchange for sodium and hydrogen ions. J. Gen. Physiol. 52:424–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ramos J, Rodríguez-Navarro A. 1985. Rubidium transport in Neurospora crassa. Biochim. Biophys. Acta 815:97–101 [Google Scholar]
- 54. Ramos J, Haro R, Rodríguez-Navarro A. 1990. Regulation of potassium fluxes in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1029:211–217 [DOI] [PubMed] [Google Scholar]
- 55. Slayman CL, Slayman CW. 1974. Depolarization of the plasma membrane of Neurospora during active transport of glucose: evidence for a proton-dependent co-transport system. Proc. Natl. Acad. Sci. U. S. A. 71:1935–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schneider RP, Wiley WR. 1971. Regulation of sugar transport in Neurospora crassa. J. Bacteriol. 106:487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Alberghina FAM. 1973. Growth regulation in Neurospora crassa. Effects of nutrients and of temperature. Arch. Mikrobiol. 89:83–94 [DOI] [PubMed] [Google Scholar]
- 58. Madi L, McBride SA, Bailey LA, Ebbole DJ. 1997. rco-3, a gene involved in glucose transport and conidiation in Neurospora crassa. Genetics 146:499–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Slayman CL, Moussatos VV, Webb WW. 1994. Endosomal accumulation of pH indicator dyes delivered as acetoxymethyl esters. J. Exp. Biol. 196:419–438 [DOI] [PubMed] [Google Scholar]
- 60. Kane PM. 1995. Disassembly and reassembly of the yeast vacuolar H+-ATPase in vivo. J. Biol. Chem. 270:17025–17032 [PubMed] [Google Scholar]
- 61. Rodríguez-Navarro A. 2000. Potassium transport in fungi and plants. Biochim. Biophys. Acta 1469:1–30 [DOI] [PubMed] [Google Scholar]
- 62. Senn ME, Rubio F, Bañuelos MA, Rodríguez-Navarro A. 2001. Comparative functional features of plant potassium HvHAK1 and HvHAK2 transporters. J. Biol. Chem. 276:44563–44569 [DOI] [PubMed] [Google Scholar]
- 63. Britto DV, Kronzucker HJ. 2008. Cellular mechanisms of potassium transport in plants. Physiol. Plant 133:637–650 [DOI] [PubMed] [Google Scholar]
- 64. Gomez-Parras JL, Riaño-Pachón DM, Benito B, Haro R, Sklodowski K, Rodríguez-Navarro A, Dreyer I. 2012. Phylogenetic analysis of K+ transporters in bryophytes, lycophytes, and flowering plants indicates a specialization of vascular plants. Front. Plant Sci. 167:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Benito B, Garciadeblás B, Fraile-Escanciano A, Rodríguez-Navarro A. 2011. Potassium and sodium uptake systems in fungi. The transporter diversity of Magnaporthe oryzae. Fungal Genet. Biol. 48:812–822 [DOI] [PubMed] [Google Scholar]
- 66. Slayman CW, Slayman CL. 1970. Potassium transport in Neurospora: evidence for a multi-site carrier at high pH. J. Gen. Physiol. 55:758–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cabrera E, Alvarez MC, Martín Y, Siverio J, Ramos J. 2012. K+ uptake systems in the yeast Hansenula polymorpha. Transcriptional and post-translational mechanisms involved in high-affinity K+ transporter regulation. Fungal Genet. Biol. 49:755–763 [DOI] [PubMed] [Google Scholar]
- 68. Ramos J, Rodríguez-Navarro A. 1986. Regulation and interconversion of the potassium transport systems of Saccharomyces cerevisiae as revealed by rubidium transport. Eur. J. Biochem. 154:307–311 [DOI] [PubMed] [Google Scholar]
- 69. Rodríguez-Navarro A, Ramos J. 1986. Two systems mediate rubidium uptake in Neurospora crassa: one exhibits the dual-uptake isotherm. Biochim. Biophys. Acta 857:229–237 [Google Scholar]
- 70. Chapman JB, Johnson EA, Kootsey JM. 1983. Electrical and biochemical properties of an enzyme model of the sodium pump. J. Membr. Biol. 74:139–153 [DOI] [PubMed] [Google Scholar]
- 71. Sanders D, Hansen U-P, Gradmann D, Slayman CL. 1984. Generalized kinetic analysis of ion cotransport systems: a unified interpretation of selective ionic effects on Michaelis parameters. J. Membr. Biol. 77:123–152 [DOI] [PubMed] [Google Scholar]
- 72. Madrid R, Gómez MJ, Ramos J, Rodríguez-Navarro A. 1998. Ectopic potassium uptake in trk1 trk2 mutants of Saccharomyces cerevisiae correlates with a highly hyperpolarized membrane potential. J. Biol. Chem. 273:14838–14844 [DOI] [PubMed] [Google Scholar]
- 73. Conway EJ, Gaffney HM. 1966. The further preparation of inorganic cationic yeasts and some of their chief properties. Biochem. J. 101:385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Page MJ, Di Cera E. 2006. Role of Na+ and K+ in enzyme function. Physiol. Rev. 86:1049–1092 [DOI] [PubMed] [Google Scholar]
- 75. Conway EJ, Breen J. 1945. An “ammonia”-yeast and some of its properties. Biochem. J. 39:368–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Conway EJ, Moore PT. 1954. A sodium-yeast and some of its properties. Biochem. J. 57:523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kschischo M, Kahm M, Navarrete C, Ramos J. 2011. Actuators of yeast potassium homeostasis, p 10 Abstr. 8th Eur. Conf. Math. Theor. Biol. [Google Scholar]
- 78. Kahm M, Navarrete C, Llopis-Torregrosa V, Herrera R, Barreto L, Yenush L, Ariño J, Ramos J, Kschischo M. 2012. Potassium starvation in yeast: mechanisms of homeostasis revealed by mathematical modeling. PLoS Biol. 8:e1002548 doi:10.1371/journal.pcbi.1002548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Yenush L, Mulet J, Ariño J, Serrano R. 2002. The Ppz protein phosphatases are key regulators of K+ and pH homeostasis: implications for salt tolerance, cell wall integrity, and cell cycle progression. EMBO J. 21:920–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Yenush L, Merchan S, Holmes J, Serrano R. 2005. pH-responsive, posttranslational regulation of the Trk1 potassium transporter by the type 1-related Ppz1 phosphatase. Mol. Cell. Biol. 25:8683–8692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mulet JM, Leube MP, Kron SJ, Rios G, Fink GR, Serrano R. 1999. A novel mechanism of ion homeostasis and salt tolerance in yeast: the Hal4 and Hal5 protein kinases modulate the Trk1-Trk2 potassium transporter. Mol. Cell. Biol. 19:3328–3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Goossens A, de la Fuente N, Formet J, Serrano R, Portillo F. 2000. Regulation of yeast H+-ATPase by protein kinases belonging to a family dedicated to activation of plasma membrane transporters. Mol. Cell. Biol. 20:7654–7661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Park G, Servin JA, Turner GE, Altamirano L, Colot HV, Collopy P, Litvinkova L, Li L, Jones CA, Diala F-G, Dunlap JC, Borkovich KA. 2011. Global analysis of serine-threonine protein kinase genes in Neurospora crassa. Eukaryot. Cell 10:1553–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tian C, Beeson WT, Iavarone AT, Sun J, Marletta MA, Cate JHD, Glass NL. 2009. Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc. Natl. Acad. Sci. U. S. A. 106:22157–22162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tian C, Li J, Glass NL. 2011. Exploring the bZIP transcription factor regulatory network in Neurospora crassa. Microbiology 157:747–759 [DOI] [PMC free article] [PubMed] [Google Scholar]