Abstract
The title compound, C21H42O5Si2, was synthesized from (3R,4S,5R)-methyl 3,5-bis[(tert-butyldimethylsilyl)oxy]-4-hydroxycyclohex-1-enecarboxylate by an esterification reaction. The cyclohexene ring adopts a half-chair conformation. In the crystal, molecules are linked via C—H⋯O hydrogen bonds, forming helical chains propagating along [010].
Related literature
The title compound is an intermediate in the synthesis of vandetanib {systematic name: N-(4-bromo-2-fluorophenyl)-6-methoxy-7-[(1-methyl-4-piperidinyl)methoxy]-4-quinazolinamine} derivatives. For vandetanib as a tyrosine kinase inhibitor, see: Heymach (2005 ▶); Morabito et al. (2009 ▶); Wells et al. (2010 ▶); Natale et al. (2009 ▶).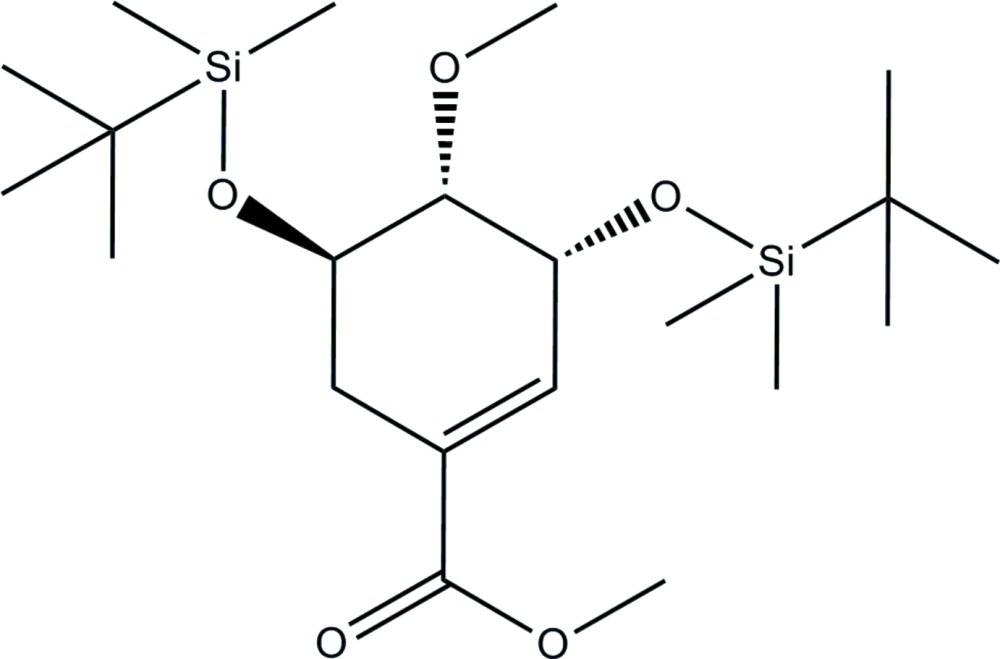
Experimental
Crystal data
C21H42O5Si2
M r = 430.72
Monoclinic,

a = 10.760 (5) Å
b = 8.321 (4) Å
c = 14.601 (7) Å
β = 98.997 (9)°
V = 1291.3 (10) Å3
Z = 2
Mo Kα radiation
μ = 0.16 mm−1
T = 113 K
0.20 × 0.18 × 0.12 mm
Data collection
Rigaku Saturn724 CCD diffractometer
Absorption correction: multi-scan (CrystalClear; Rigaku, 2007 ▶) T min = 0.968, T max = 0.981
13589 measured reflections
6015 independent reflections
4456 reflections with I > 2σ(I)
R int = 0.058
Refinement
R[F 2 > 2σ(F 2)] = 0.048
wR(F 2) = 0.073
S = 0.98
6015 reflections
265 parameters
1 restraint
H-atom parameters constrained
Δρmax = 0.21 e Å−3
Δρmin = −0.29 e Å−3
Absolute structure: Flack (1983 ▶), 2745 Friedel pairs
Flack parameter: −0.04 (9)
Data collection: CrystalClear (Rigaku, 2007 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: CrystalStructure (Rigaku/MSC, 2006 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536813007551/vm2187sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813007551/vm2187Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813007551/vm2187Isup4.cdx
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C7—H7B⋯O3i | 0.98 | 2.55 | 3.410 (3) | 147 |
| C9—H9A⋯O3ii | 0.98 | 2.59 | 3.527 (3) | 161 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Acknowledgments
The synthesis and evaluation of the title compound was undertaken as part of the National Science and Technology Major Project "The synthesis and anticancer activity screening of novel chalcone derivatives". The authors thank the State Key Laboratory of Elemento-organic Chemistry Nankai University, for the X-ray data collection.
supplementary crystallographic information
Comment
Vandetanib is a small molecule tyrosine kinase inhibitor, which can act on the tumor cells epidermal growth factor receptor (EGFR), vascular endothelial growth factor receptor (VEGFR) and the RET tyrosine kinase (Heymach, 2005; Morabito et al., 2009). Vandetanib has a good therapeutic effect for Medullary thyroid cancer and Non-small cell lung cancer (Wells et al., 2010; Natale et al., 2009).
(3R,4S,5R)-Methyl 3,5-bis[(tert-butyldimethylsilyl)oxy]-4-methoxycyclohex-1-enecarboxylate (Fig. 1) is an intermediate to synthetize Vandetanib derivatives. Here, the synthesis and crystallographic characterization of the compound are reported.
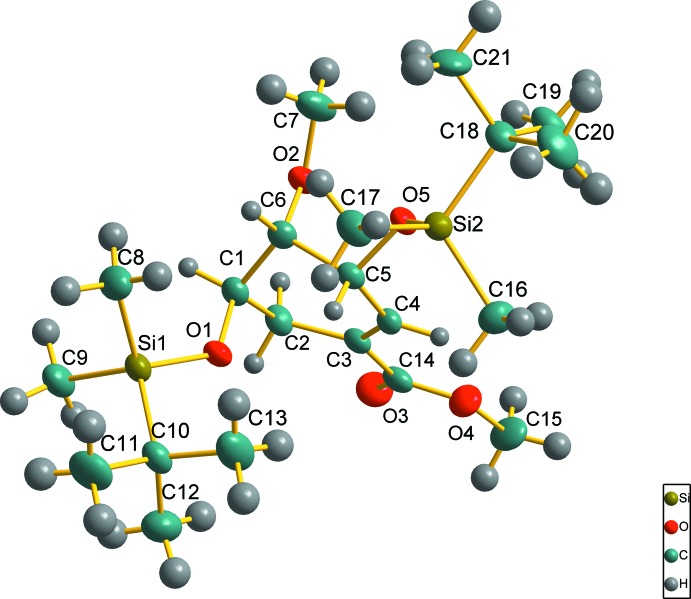
The crystal structure of the compound has monoclinic (P21) symmetry at 113 K. No hydrogen-bonding or π–π interactions are observed in the crystal structure. Despite the relatively large steric size of substituent groups, the cyclohexene still has a nearly ideal half-chair form with carbon atoms C3, C4,C5 and C2 lying in one plane.
Experimental
Solid sodium hydroxide (2.8 g, 0.07 mol) was added to a stirred solution of (3R,4S,5R)-methyl 3,5-bis((tert-butyldimethylsilyl)oxy)-4-hydroxycyclohex-1-εnecarboxylate (5.6 g, 0.134 mol) in acetonitrile (60 ml) at room temperature.The reaction mixture was added dropwise to a solution of dimethyl sulfate (4.2 ml, 0.044 mol) in acetonitrile (60 ml). After the dropwise addition, the temperature was raised to 40 °C. The reaction was completed within 15 hrs at 40 °C stirring. The solvent was removed under reduced pressure, and ethyl acetate (300 ml) and H2O (100 ml) were added three times to extract the solid. The ethyl acetate layer was dried with anhydrous magnesium sulfate and a white solid was obtained after removal of the solvent. The yield was 4.8 g (82.7%). About 0.5g of the product was put in an ampoule bottle and 10 ml absolute methanol was added. The white single crystals suitable for X-ray diffraction analysis were obtained by slow evaporation of the solvent at room temperature after 1 week.
Refinement
H atoms were placed at calculated positions with C-H = 0.98 Å (methyl), 0.99 Å (methylene), 1.00 Å (methine sp3) and 0.95 Å (methine sp2) and refined as riding atoms with Uiso(H) = 1.5Ueq(C) (methyl) or 1.2Ueq(C) (others).
Figures
Fig. 1.
Molecular structure of C21H42O5Si2 with atom-labelling scheme and ellipsoids drawn at the 50% probability level.
Crystal data
| C21H42O5Si2 | F(000) = 472 |
| Mr = 430.72 | Dx = 1.108 Mg m−3 |
| Monoclinic, P21 | Mo Kα radiation, λ = 0.71073 Å |
| a = 10.760 (5) Å | Cell parameters from 4632 reflections |
| b = 8.321 (4) Å | θ = 1.9–27.9° |
| c = 14.601 (7) Å | µ = 0.16 mm−1 |
| β = 98.997 (9)° | T = 113 K |
| V = 1291.3 (10) Å3 | Prism, colourless |
| Z = 2 | 0.20 × 0.18 × 0.12 mm |
Data collection
| Rigaku Saturn724 CCD diffractometer | 6015 independent reflections |
| Radiation source: rotating anode | 4456 reflections with I > 2σ(I) |
| Multilayer monochromator | Rint = 0.058 |
| Detector resolution: 14.22 pixels mm-1 | θmax = 27.9°, θmin = 1.9° |
| ω and φ scans | h = −14→14 |
| Absorption correction: multi-scan (CrystalClear; Rigaku, 2007) | k = −10→10 |
| Tmin = 0.968, Tmax = 0.981 | l = −19→19 |
| 13589 measured reflections |
Refinement
| Refinement on F2 | Secondary atom site location: difference Fourier map |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.048 | H-atom parameters constrained |
| wR(F2) = 0.073 | w = 1/[σ2(Fo2) + (0.010P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 0.98 | (Δ/σ)max = 0.001 |
| 6015 reflections | Δρmax = 0.21 e Å−3 |
| 265 parameters | Δρmin = −0.29 e Å−3 |
| 1 restraint | Absolute structure: Flack (1983), 2745 Friedel pairs |
| Primary atom site location: structure-invariant direct methods | Flack parameter: −0.04 (9) |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Si1 | 0.43693 (6) | 0.59832 (8) | 0.75544 (5) | 0.02755 (16) | |
| Si2 | 0.93837 (6) | 0.92816 (8) | 0.82849 (4) | 0.02554 (16) | |
| O1 | 0.55081 (13) | 0.59788 (19) | 0.69212 (10) | 0.0277 (4) | |
| O2 | 0.66743 (13) | 0.92780 (19) | 0.57018 (10) | 0.0307 (4) | |
| O3 | 0.75863 (16) | 0.3984 (2) | 0.44370 (12) | 0.0421 (5) | |
| O4 | 0.93371 (15) | 0.4208 (2) | 0.54942 (11) | 0.0367 (4) | |
| O5 | 0.86351 (13) | 0.89726 (17) | 0.72306 (10) | 0.0246 (4) | |
| C1 | 0.5623 (2) | 0.7011 (3) | 0.61583 (16) | 0.0274 (6) | |
| H1 | 0.4777 | 0.7436 | 0.5885 | 0.033* | |
| C2 | 0.61777 (19) | 0.6039 (3) | 0.54295 (15) | 0.0290 (5) | |
| H2A | 0.5717 | 0.5011 | 0.5319 | 0.035* | |
| H2B | 0.6066 | 0.6644 | 0.4838 | 0.035* | |
| C3 | 0.7562 (2) | 0.5697 (3) | 0.57327 (15) | 0.0250 (5) | |
| C4 | 0.8238 (2) | 0.6447 (3) | 0.64413 (15) | 0.0238 (5) | |
| H4 | 0.9101 | 0.6167 | 0.6598 | 0.029* | |
| C5 | 0.7728 (2) | 0.7704 (3) | 0.70088 (15) | 0.0242 (5) | |
| H5 | 0.7564 | 0.7202 | 0.7601 | 0.029* | |
| C6 | 0.6494 (2) | 0.8400 (3) | 0.65060 (15) | 0.0266 (6) | |
| H6 | 0.6093 | 0.9098 | 0.6935 | 0.032* | |
| C7 | 0.6928 (2) | 1.0957 (3) | 0.58602 (16) | 0.0488 (7) | |
| H7A | 0.6265 | 1.1434 | 0.6163 | 0.073* | |
| H7B | 0.6950 | 1.1496 | 0.5266 | 0.073* | |
| H7C | 0.7743 | 1.1086 | 0.6260 | 0.073* | |
| C8 | 0.4174 (2) | 0.8026 (3) | 0.80365 (17) | 0.0441 (7) | |
| H8A | 0.4949 | 0.8335 | 0.8444 | 0.066* | |
| H8B | 0.3473 | 0.8016 | 0.8392 | 0.066* | |
| H8C | 0.3998 | 0.8801 | 0.7527 | 0.066* | |
| C9 | 0.2855 (2) | 0.5375 (3) | 0.68386 (17) | 0.0412 (7) | |
| H9A | 0.2537 | 0.6259 | 0.6423 | 0.062* | |
| H9B | 0.2239 | 0.5122 | 0.7246 | 0.062* | |
| H9C | 0.2992 | 0.4426 | 0.6470 | 0.062* | |
| C10 | 0.4906 (2) | 0.4488 (3) | 0.84924 (16) | 0.0327 (6) | |
| C11 | 0.4060 (3) | 0.4598 (3) | 0.92525 (17) | 0.0537 (8) | |
| H11A | 0.4329 | 0.3794 | 0.9733 | 0.081* | |
| H11B | 0.3184 | 0.4396 | 0.8978 | 0.081* | |
| H11C | 0.4131 | 0.5673 | 0.9529 | 0.081* | |
| C12 | 0.4852 (3) | 0.2784 (3) | 0.80998 (19) | 0.0501 (8) | |
| H12A | 0.5357 | 0.2727 | 0.7598 | 0.075* | |
| H12B | 0.3977 | 0.2505 | 0.7858 | 0.075* | |
| H12C | 0.5186 | 0.2028 | 0.8591 | 0.075* | |
| C13 | 0.6274 (2) | 0.4848 (3) | 0.89552 (17) | 0.0515 (8) | |
| H13A | 0.6519 | 0.4094 | 0.9467 | 0.077* | |
| H13B | 0.6325 | 0.5950 | 0.9194 | 0.077* | |
| H13C | 0.6842 | 0.4727 | 0.8496 | 0.077* | |
| C14 | 0.8129 (2) | 0.4543 (3) | 0.51520 (17) | 0.0299 (6) | |
| C15 | 0.9983 (3) | 0.3166 (3) | 0.49216 (19) | 0.0448 (7) | |
| H15A | 0.9561 | 0.2119 | 0.4853 | 0.067* | |
| H15B | 1.0858 | 0.3022 | 0.5216 | 0.067* | |
| H15C | 0.9964 | 0.3656 | 0.4309 | 0.067* | |
| C16 | 1.0379 (2) | 0.7499 (3) | 0.86556 (17) | 0.0409 (7) | |
| H16A | 0.9843 | 0.6549 | 0.8664 | 0.061* | |
| H16B | 1.0847 | 0.7684 | 0.9278 | 0.061* | |
| H16C | 1.0973 | 0.7324 | 0.8220 | 0.061* | |
| C17 | 0.8227 (2) | 0.9581 (3) | 0.90962 (15) | 0.0384 (7) | |
| H17A | 0.7633 | 1.0430 | 0.8857 | 0.058* | |
| H17B | 0.8674 | 0.9893 | 0.9707 | 0.058* | |
| H17C | 0.7768 | 0.8578 | 0.9151 | 0.058* | |
| C18 | 1.0368 (2) | 1.1119 (3) | 0.81908 (16) | 0.0314 (6) | |
| C19 | 1.1006 (2) | 1.1013 (3) | 0.73279 (15) | 0.0429 (7) | |
| H19A | 1.1597 | 1.1908 | 0.7326 | 0.064* | |
| H19B | 1.0367 | 1.1069 | 0.6771 | 0.064* | |
| H19C | 1.1462 | 0.9993 | 0.7333 | 0.064* | |
| C20 | 1.1383 (2) | 1.1213 (4) | 0.90549 (17) | 0.0510 (8) | |
| H20A | 1.1989 | 1.0339 | 0.9039 | 0.076* | |
| H20B | 1.0988 | 1.1113 | 0.9613 | 0.076* | |
| H20C | 1.1820 | 1.2248 | 0.9064 | 0.076* | |
| C21 | 0.9558 (3) | 1.2647 (3) | 0.8138 (2) | 0.0521 (9) | |
| H21A | 0.9137 | 1.2710 | 0.8685 | 0.078* | |
| H21B | 0.8924 | 1.2613 | 0.7577 | 0.078* | |
| H21C | 1.0095 | 1.3593 | 0.8116 | 0.078* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Si1 | 0.0247 (4) | 0.0286 (4) | 0.0307 (4) | −0.0038 (3) | 0.0084 (3) | 0.0005 (3) |
| Si2 | 0.0258 (4) | 0.0250 (3) | 0.0258 (4) | −0.0017 (3) | 0.0038 (3) | −0.0023 (3) |
| O1 | 0.0305 (9) | 0.0244 (8) | 0.0305 (9) | −0.0039 (8) | 0.0118 (7) | 0.0027 (8) |
| O2 | 0.0330 (10) | 0.0296 (9) | 0.0293 (9) | −0.0063 (9) | 0.0039 (8) | 0.0070 (8) |
| O3 | 0.0526 (12) | 0.0404 (11) | 0.0344 (10) | −0.0022 (10) | 0.0103 (9) | −0.0126 (9) |
| O4 | 0.0402 (11) | 0.0333 (9) | 0.0389 (10) | 0.0074 (10) | 0.0137 (9) | −0.0083 (9) |
| O5 | 0.0239 (9) | 0.0259 (9) | 0.0238 (8) | −0.0070 (7) | 0.0027 (7) | 0.0000 (7) |
| C1 | 0.0271 (15) | 0.0303 (13) | 0.0261 (13) | −0.0032 (11) | 0.0076 (12) | 0.0023 (11) |
| C2 | 0.0293 (14) | 0.0325 (13) | 0.0261 (13) | −0.0070 (13) | 0.0071 (11) | −0.0070 (13) |
| C3 | 0.0284 (13) | 0.0223 (13) | 0.0269 (13) | −0.0053 (10) | 0.0121 (11) | −0.0011 (10) |
| C4 | 0.0228 (13) | 0.0242 (13) | 0.0258 (13) | −0.0020 (10) | 0.0081 (11) | 0.0010 (10) |
| C5 | 0.0263 (14) | 0.0243 (12) | 0.0228 (13) | −0.0053 (11) | 0.0060 (11) | 0.0002 (10) |
| C6 | 0.0297 (15) | 0.0285 (13) | 0.0223 (13) | −0.0015 (11) | 0.0065 (11) | 0.0026 (11) |
| C7 | 0.0525 (17) | 0.0375 (15) | 0.0521 (18) | −0.0128 (16) | −0.0055 (14) | 0.0143 (16) |
| C8 | 0.0436 (18) | 0.0375 (15) | 0.057 (2) | 0.0024 (14) | 0.0250 (16) | −0.0008 (14) |
| C9 | 0.0345 (16) | 0.0421 (16) | 0.0460 (17) | −0.0101 (12) | 0.0030 (13) | 0.0066 (14) |
| C10 | 0.0306 (15) | 0.0356 (15) | 0.0330 (14) | −0.0086 (12) | 0.0089 (12) | 0.0021 (12) |
| C11 | 0.065 (2) | 0.057 (2) | 0.0444 (18) | −0.0087 (16) | 0.0247 (16) | 0.0125 (16) |
| C12 | 0.055 (2) | 0.0378 (17) | 0.058 (2) | −0.0007 (15) | 0.0082 (17) | 0.0078 (15) |
| C13 | 0.0412 (18) | 0.062 (2) | 0.0471 (18) | −0.0091 (15) | −0.0064 (14) | 0.0198 (16) |
| C14 | 0.0361 (16) | 0.0248 (14) | 0.0320 (14) | −0.0073 (12) | 0.0146 (12) | −0.0002 (11) |
| C15 | 0.0537 (18) | 0.0387 (16) | 0.0472 (17) | 0.0135 (15) | 0.0241 (15) | −0.0056 (14) |
| C16 | 0.0421 (17) | 0.0359 (16) | 0.0429 (17) | 0.0046 (13) | 0.0008 (14) | 0.0045 (14) |
| C17 | 0.0428 (17) | 0.0393 (17) | 0.0353 (15) | −0.0083 (13) | 0.0136 (13) | −0.0057 (13) |
| C18 | 0.0321 (14) | 0.0280 (13) | 0.0344 (14) | −0.0053 (13) | 0.0065 (12) | −0.0077 (13) |
| C19 | 0.0434 (16) | 0.0476 (15) | 0.0392 (16) | −0.0200 (16) | 0.0110 (13) | −0.0045 (15) |
| C20 | 0.0497 (18) | 0.0577 (19) | 0.0436 (18) | −0.0243 (17) | 0.0011 (15) | −0.0130 (16) |
| C21 | 0.059 (2) | 0.0201 (14) | 0.078 (2) | −0.0042 (14) | 0.0140 (19) | −0.0030 (15) |
Geometric parameters (Å, º)
| Si1—O1 | 1.6464 (15) | C9—H9B | 0.9800 |
| Si1—C9 | 1.863 (2) | C9—H9C | 0.9800 |
| Si1—C8 | 1.864 (3) | C10—C12 | 1.527 (3) |
| Si1—C10 | 1.874 (3) | C10—C11 | 1.544 (3) |
| Si2—O5 | 1.6421 (16) | C10—C13 | 1.549 (3) |
| Si2—C16 | 1.860 (2) | C11—H11A | 0.9800 |
| Si2—C17 | 1.864 (2) | C11—H11B | 0.9800 |
| Si2—C18 | 1.877 (3) | C11—H11C | 0.9800 |
| O1—C1 | 1.427 (2) | C12—H12A | 0.9800 |
| O2—C6 | 1.422 (2) | C12—H12B | 0.9800 |
| O2—C7 | 1.435 (3) | C12—H12C | 0.9800 |
| O3—C14 | 1.207 (3) | C13—H13A | 0.9800 |
| O4—C14 | 1.347 (3) | C13—H13B | 0.9800 |
| O4—C15 | 1.454 (3) | C13—H13C | 0.9800 |
| O5—C5 | 1.440 (2) | C15—H15A | 0.9800 |
| C1—C6 | 1.523 (3) | C15—H15B | 0.9800 |
| C1—C2 | 1.530 (3) | C15—H15C | 0.9800 |
| C1—H1 | 1.0000 | C16—H16A | 0.9800 |
| C2—C3 | 1.513 (3) | C16—H16B | 0.9800 |
| C2—H2A | 0.9900 | C16—H16C | 0.9800 |
| C2—H2B | 0.9900 | C17—H17A | 0.9800 |
| C3—C4 | 1.324 (3) | C17—H17B | 0.9800 |
| C3—C14 | 1.475 (3) | C17—H17C | 0.9800 |
| C4—C5 | 1.492 (3) | C18—C19 | 1.529 (3) |
| C4—H4 | 0.9500 | C18—C20 | 1.536 (3) |
| C5—C6 | 1.527 (3) | C18—C21 | 1.537 (3) |
| C5—H5 | 1.0000 | C19—H19A | 0.9800 |
| C6—H6 | 1.0000 | C19—H19B | 0.9800 |
| C7—H7A | 0.9800 | C19—H19C | 0.9800 |
| C7—H7B | 0.9800 | C20—H20A | 0.9800 |
| C7—H7C | 0.9800 | C20—H20B | 0.9800 |
| C8—H8A | 0.9800 | C20—H20C | 0.9800 |
| C8—H8B | 0.9800 | C21—H21A | 0.9800 |
| C8—H8C | 0.9800 | C21—H21B | 0.9800 |
| C9—H9A | 0.9800 | C21—H21C | 0.9800 |
| O1—Si1—C9 | 110.29 (10) | C12—C10—Si1 | 110.67 (18) |
| O1—Si1—C8 | 110.58 (10) | C11—C10—Si1 | 109.69 (17) |
| C9—Si1—C8 | 108.63 (12) | C13—C10—Si1 | 110.70 (16) |
| O1—Si1—C10 | 103.74 (10) | C10—C11—H11A | 109.5 |
| C9—Si1—C10 | 111.85 (11) | C10—C11—H11B | 109.5 |
| C8—Si1—C10 | 111.69 (12) | H11A—C11—H11B | 109.5 |
| O5—Si2—C16 | 108.88 (10) | C10—C11—H11C | 109.5 |
| O5—Si2—C17 | 109.77 (10) | H11A—C11—H11C | 109.5 |
| C16—Si2—C17 | 109.45 (12) | H11B—C11—H11C | 109.5 |
| O5—Si2—C18 | 104.99 (10) | C10—C12—H12A | 109.5 |
| C16—Si2—C18 | 111.41 (12) | C10—C12—H12B | 109.5 |
| C17—Si2—C18 | 112.21 (11) | H12A—C12—H12B | 109.5 |
| C1—O1—Si1 | 126.87 (15) | C10—C12—H12C | 109.5 |
| C6—O2—C7 | 114.49 (18) | H12A—C12—H12C | 109.5 |
| C14—O4—C15 | 115.5 (2) | H12B—C12—H12C | 109.5 |
| C5—O5—Si2 | 122.79 (13) | C10—C13—H13A | 109.5 |
| O1—C1—C6 | 108.68 (18) | C10—C13—H13B | 109.5 |
| O1—C1—C2 | 108.40 (18) | H13A—C13—H13B | 109.5 |
| C6—C1—C2 | 110.27 (18) | C10—C13—H13C | 109.5 |
| O1—C1—H1 | 109.8 | H13A—C13—H13C | 109.5 |
| C6—C1—H1 | 109.8 | H13B—C13—H13C | 109.5 |
| C2—C1—H1 | 109.8 | O3—C14—O4 | 123.3 (2) |
| C3—C2—C1 | 111.65 (19) | O3—C14—C3 | 124.1 (2) |
| C3—C2—H2A | 109.3 | O4—C14—C3 | 112.5 (2) |
| C1—C2—H2A | 109.3 | O4—C15—H15A | 109.5 |
| C3—C2—H2B | 109.3 | O4—C15—H15B | 109.5 |
| C1—C2—H2B | 109.3 | H15A—C15—H15B | 109.5 |
| H2A—C2—H2B | 108.0 | O4—C15—H15C | 109.5 |
| C4—C3—C14 | 122.0 (2) | H15A—C15—H15C | 109.5 |
| C4—C3—C2 | 122.4 (2) | H15B—C15—H15C | 109.5 |
| C14—C3—C2 | 115.5 (2) | Si2—C16—H16A | 109.5 |
| C3—C4—C5 | 124.0 (2) | Si2—C16—H16B | 109.5 |
| C3—C4—H4 | 118.0 | H16A—C16—H16B | 109.5 |
| C5—C4—H4 | 118.0 | Si2—C16—H16C | 109.5 |
| O5—C5—C4 | 110.08 (18) | H16A—C16—H16C | 109.5 |
| O5—C5—C6 | 109.77 (18) | H16B—C16—H16C | 109.5 |
| C4—C5—C6 | 111.52 (19) | Si2—C17—H17A | 109.5 |
| O5—C5—H5 | 108.5 | Si2—C17—H17B | 109.5 |
| C4—C5—H5 | 108.5 | H17A—C17—H17B | 109.5 |
| C6—C5—H5 | 108.5 | Si2—C17—H17C | 109.5 |
| O2—C6—C1 | 105.72 (18) | H17A—C17—H17C | 109.5 |
| O2—C6—C5 | 111.78 (18) | H17B—C17—H17C | 109.5 |
| C1—C6—C5 | 108.40 (19) | C19—C18—C20 | 109.1 (2) |
| O2—C6—H6 | 110.3 | C19—C18—C21 | 109.3 (2) |
| C1—C6—H6 | 110.3 | C20—C18—C21 | 108.9 (2) |
| C5—C6—H6 | 110.3 | C19—C18—Si2 | 110.14 (17) |
| O2—C7—H7A | 109.5 | C20—C18—Si2 | 108.56 (18) |
| O2—C7—H7B | 109.5 | C21—C18—Si2 | 110.82 (16) |
| H7A—C7—H7B | 109.5 | C18—C19—H19A | 109.5 |
| O2—C7—H7C | 109.5 | C18—C19—H19B | 109.5 |
| H7A—C7—H7C | 109.5 | H19A—C19—H19B | 109.5 |
| H7B—C7—H7C | 109.5 | C18—C19—H19C | 109.5 |
| Si1—C8—H8A | 109.5 | H19A—C19—H19C | 109.5 |
| Si1—C8—H8B | 109.5 | H19B—C19—H19C | 109.5 |
| H8A—C8—H8B | 109.5 | C18—C20—H20A | 109.5 |
| Si1—C8—H8C | 109.5 | C18—C20—H20B | 109.5 |
| H8A—C8—H8C | 109.5 | H20A—C20—H20B | 109.5 |
| H8B—C8—H8C | 109.5 | C18—C20—H20C | 109.5 |
| Si1—C9—H9A | 109.5 | H20A—C20—H20C | 109.5 |
| Si1—C9—H9B | 109.5 | H20B—C20—H20C | 109.5 |
| H9A—C9—H9B | 109.5 | C18—C21—H21A | 109.5 |
| Si1—C9—H9C | 109.5 | C18—C21—H21B | 109.5 |
| H9A—C9—H9C | 109.5 | H21A—C21—H21B | 109.5 |
| H9B—C9—H9C | 109.5 | C18—C21—H21C | 109.5 |
| C12—C10—C11 | 109.4 (2) | H21A—C21—H21C | 109.5 |
| C12—C10—C13 | 108.7 (2) | H21B—C21—H21C | 109.5 |
| C11—C10—C13 | 107.5 (2) | ||
| C9—Si1—O1—C1 | 65.82 (19) | O5—C5—C6—C1 | 171.87 (16) |
| C8—Si1—O1—C1 | −54.4 (2) | C4—C5—C6—C1 | 49.6 (2) |
| C10—Si1—O1—C1 | −174.24 (17) | O1—Si1—C10—C12 | −70.14 (19) |
| C16—Si2—O5—C5 | 63.48 (18) | C9—Si1—C10—C12 | 48.7 (2) |
| C17—Si2—O5—C5 | −56.31 (18) | C8—Si1—C10—C12 | 170.72 (18) |
| C18—Si2—O5—C5 | −177.12 (16) | O1—Si1—C10—C11 | 169.01 (16) |
| Si1—O1—C1—C6 | 97.0 (2) | C9—Si1—C10—C11 | −72.1 (2) |
| Si1—O1—C1—C2 | −143.19 (15) | C8—Si1—C10—C11 | 49.9 (2) |
| O1—C1—C2—C3 | −73.1 (2) | O1—Si1—C10—C13 | 50.48 (19) |
| C6—C1—C2—C3 | 45.8 (3) | C9—Si1—C10—C13 | 169.35 (17) |
| C1—C2—C3—C4 | −13.4 (3) | C8—Si1—C10—C13 | −68.6 (2) |
| C1—C2—C3—C14 | 170.15 (18) | C15—O4—C14—O3 | 2.6 (3) |
| C14—C3—C4—C5 | 175.70 (19) | C15—O4—C14—C3 | −175.76 (18) |
| C2—C3—C4—C5 | −0.5 (3) | C4—C3—C14—O3 | −170.6 (2) |
| Si2—O5—C5—C4 | −110.40 (18) | C2—C3—C14—O3 | 5.9 (3) |
| Si2—O5—C5—C6 | 126.48 (16) | C4—C3—C14—O4 | 7.8 (3) |
| C3—C4—C5—O5 | −140.3 (2) | C2—C3—C14—O4 | −175.7 (2) |
| C3—C4—C5—C6 | −18.3 (3) | O5—Si2—C18—C19 | −43.92 (18) |
| C7—O2—C6—C1 | 151.68 (18) | C16—Si2—C18—C19 | 73.8 (2) |
| C7—O2—C6—C5 | −90.6 (2) | C17—Si2—C18—C19 | −163.11 (17) |
| O1—C1—C6—O2 | 173.92 (17) | O5—Si2—C18—C20 | −163.30 (16) |
| C2—C1—C6—O2 | 55.2 (2) | C16—Si2—C18—C20 | −45.6 (2) |
| O1—C1—C6—C5 | 53.9 (2) | C17—Si2—C18—C20 | 77.52 (19) |
| C2—C1—C6—C5 | −64.8 (2) | O5—Si2—C18—C21 | 77.13 (18) |
| O5—C5—C6—O2 | 55.7 (2) | C16—Si2—C18—C21 | −165.17 (17) |
| C4—C5—C6—O2 | −66.5 (2) | C17—Si2—C18—C21 | −42.1 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C7—H7B···O3i | 0.98 | 2.55 | 3.410 (3) | 147 |
| C9—H9A···O3ii | 0.98 | 2.59 | 3.527 (3) | 161 |
Symmetry codes: (i) x, y+1, z; (ii) −x+1, y+1/2, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: VM2187).
References
- Flack, H. D. (1983). Acta Cryst. A39, 876–881.
- Heymach, J. V. (2005). Br. J. Cancer, 92(Suppl 1), 14–20. [DOI] [PMC free article] [PubMed]
- Morabito, A., Piccirillo, M. C., Falasconi, F., De Feo, G., Del Giudice, A., Bryce, J., Di Maio, M., De Maio, E., Normanno, N. & Perrone, F. (2009). Oncologist, 14, 378–390. [DOI] [PubMed]
- Natale, R. B., Thongprasert, S., Greco, F. A., Thomas, M., Tsai, C. M., Sunpaweravong, P., Ferry, D., Langmuir, P., Rowbottom, J. A. & Goss, G. D. (2009). J. Clin. Oncol. 27(15S), abstr. 8009.
- Rigaku (2007). CrystalClear Rigaku Corporation, Tokyo, Japan.
- Rigaku/MSC (2006). CrystalStructure Rigaku/MSC, The Woodlands, Texas, USA.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Wells, S. A., Gosnell, J. E., Gagel, R. F., Moley, J., Pfister, D., Sosa, J. A., Skinner, M., Krebs, A., Vasselli, J. & Schlumberger, M. (2010). J. Clin. Oncol. 28, 767–772. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S1600536813007551/vm2187sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813007551/vm2187Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813007551/vm2187Isup4.cdx
Additional supplementary materials: crystallographic information; 3D view; checkCIF report