Abstract
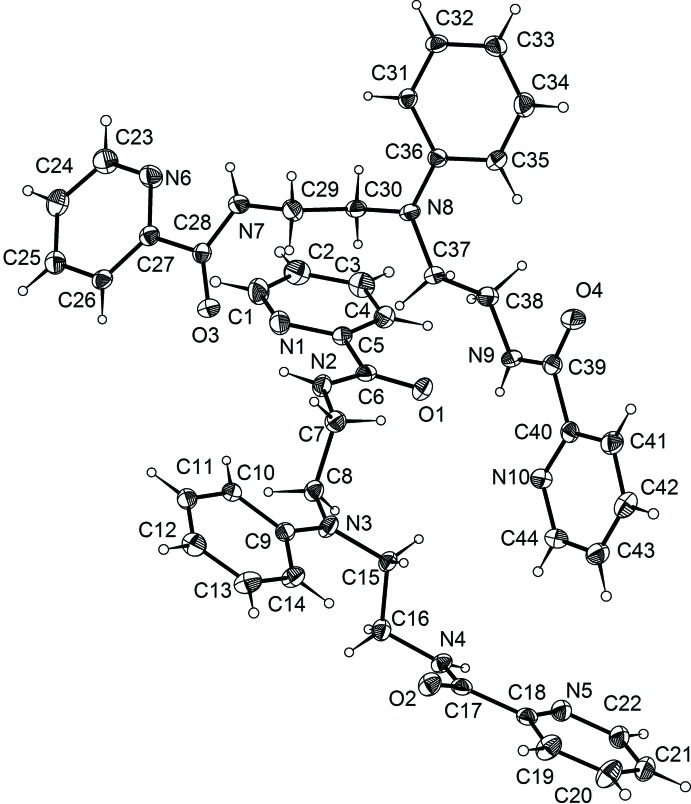
The asymmetric unit of the title compound, C22H23N5O2, contains two independent molecules with similar conformations; the terminal pyridine rings are oriented at dihedral angles of 23.99 (8) and 18.07 (8)° with respect to the central benzene ring in one molecule and 28.99 (8) and 23.22 (8)° in the other. In the crystal, N—H⋯O and weak C—H⋯O hydrogen bonds link the molecules into a three-dimensional supramolecular structure. Weak intermolecular C—H⋯π interactions are also observed in the crystal.
Related literature
For background to bis(pyridinecarboxamide) derivatives, see: Cornman et al. (1999 ▶); Song et al. (2010 ▶); Singh et al. (2008 ▶). For the synthesis, see: Jain et al. (2004 ▶); Lee et al. (2006 ▶); Barnes et al. (1978 ▶). For related structures, see: Adolph et al. (2012 ▶); Munro & Wilson (2010 ▶); Yan et al. (2012 ▶).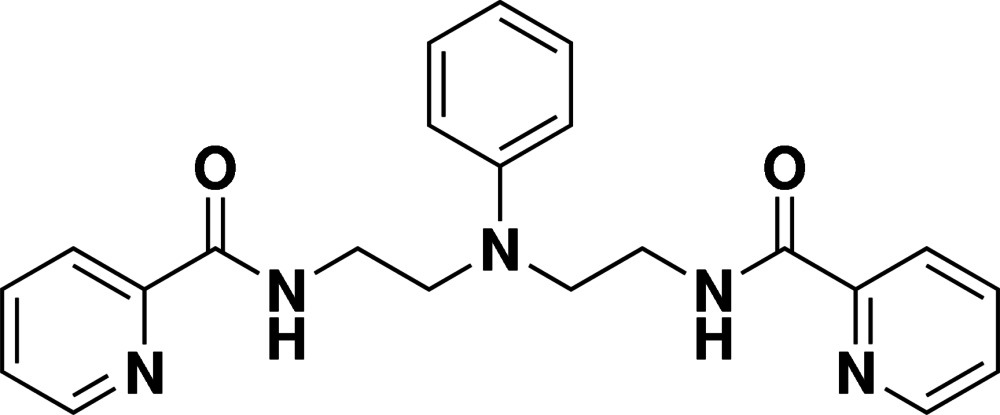
Experimental
Crystal data
C22H23N5O2
M r = 389.45
Monoclinic,
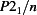
a = 8.64349 (7) Å
b = 24.8210 (3) Å
c = 18.40861 (18) Å
β = 90.5648 (8)°
V = 3949.20 (7) Å3
Z = 8
Cu Kα radiation
μ = 0.70 mm−1
T = 100 K
0.12 × 0.08 × 0.07 mm
Data collection
Agilent Xcalibur Atlas Gemini ultra diffractometer
Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2012 ▶) T min = 0.886, T max = 0.950
28468 measured reflections
7050 independent reflections
6128 reflections with I > 2σ(I)
R int = 0.038
Refinement
R[F 2 > 2σ(F 2)] = 0.044
wR(F 2) = 0.097
S = 1.14
7050 reflections
523 parameters
H-atom parameters constrained
Δρmax = 0.17 e Å−3
Δρmin = −0.26 e Å−3
Data collection: CrysAlis PRO (Agilent, 2012 ▶); cell refinement: CrysAlis PRO; data reduction: CrysAlis PRO; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: DIAMOND (Brandenburg, 1999 ▶); software used to prepare material for publication: OLEX2 (Dolomanov et al., 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813008696/xu5691sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813008696/xu5691Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg5 and Cg6 are the centroids of the C9–C14 benzene and C31–C36 benzene rings, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H2⋯O3 | 0.86 | 2.44 | 3.1099 (18) | 135 |
| N7—H7⋯O1i | 0.86 | 2.42 | 3.0998 (18) | 136 |
| C24—H24⋯O2ii | 0.93 | 2.48 | 3.311 (2) | 149 |
| C25—H25⋯O4iii | 0.93 | 2.54 | 3.213 (2) | 129 |
| C8—H8A⋯Cg6iv | 0.97 | 2.72 | 3.6468 (18) | 161 |
| C30—H30B⋯Cg5v | 0.97 | 2.74 | 3.6584 (18) | 159 |
Symmetry codes: (i) 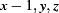 ; (ii)
; (ii) 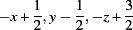 ; (iii)
; (iii) 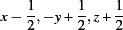 ; (iv)
; (iv) 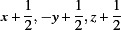 ; (v)
; (v) 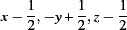 .
.
Acknowledgments
This work was supported financially by the Natural Science Foundation of Hainan Province, China (grant 212014) and the Scientific Research Foundation of Hainan Normal University.
supplementary crystallographic information
Comment
Bis(2-pyridinecarboxamide) derivatives comprise a large group of organic compounds that are ideal for the mono- or dinuclear chelation of metal ions (Cornman et al., 1999; Song et al., 2010; Singh et al., 2008). In order to explore coordination chemistry of bis(pyridinecarboxamide) ligands, we have synthesized the title compound (I) and report here its crystal structure.
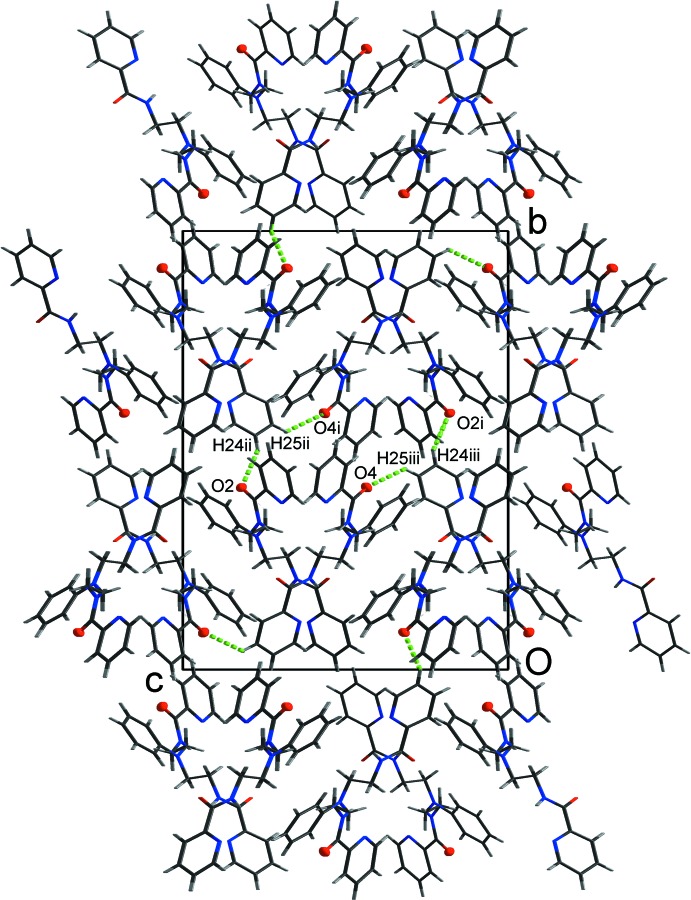
The asymmetric unit of the title compound (I), C22H23N5O2, contains two independent molecules with the similar structure, two terminal pyridine rings are oriented with respect to the central benzene ring at 23.99 (8) and 18.07 (8)° in one molecule and at 28.99 (8) and 23.22 (8)° in the other (Fig. 1). Every independent molecule contains two symmetrical N-ethyl(pyridine-2-carboxiamide) moieties linked by a phenylamino bridge. It is noteworthy that the C═O bonds are oriented trans to the pyridine nitrogen atom, which are in accord with those reported for previous structures (Munro et al., 2010; Yan et al., 2012). In the crystal, intermolecular N–H···O (Fig. 2) and weak C—H···O hydrogen bonds (Fig. 3) link the molecules into the three-dimensional supramolecular structure. The crystal packing exhibits also weak intermolecular C–H···π interaction, proved by short distance C30–H30B···Cg5 [3.6584 (18) Å] and C8–H8A···Cg6 [3.6468 (18) Å], where Cg5 and Cg6 are the centroids of the C9–benzene and C31–benzene rings, respectively [symmetry code: (iv) x+1/2, –y+1/2, z+1/2; (v) x–1/2, –y+1/2, z–1/2] (Table 1).
Experimental
N-(3-Dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (EDCI, 375 mg, 1.95 mmol) and hydroxybenzotriazole (HOBt, 440 mg, 3.26 mmol) were added to a solution of picolinic acid (200 mg, 1.62 mmol) in dry DMF (10 ml) at room temperature. After the mixture was stirred for 30 min, N1-(2-aminoethyl)-N1-phenylethane-1,2-diamine (133 mg, 0.74 mmol) was added (Lee et al., 2006). The reaction mixture was stirred overnight under N2 atmosphere. H2O (30 ml) was added to quench the reaction, and the mixture was extracted with ethyl acetate (3×20 ml). The combined organic phase was washed with brine, dried over Na2SO4, filtrated and concentrated in vacuum. The residue was purified by column chromatography (PE:EA = 2:1~1:2) to give compound (I) (264 mg, yield: 91.8%) as a white solid. Colorless single crystals suitable for X-ray structural analysis were obtained by slow evaporation of a mixture solution of dichloromethane and methanol at room temperature. 1H NMR (400 MHz, CDCl3): 3.48~3.61 (m, 8H), 6.62 (t, J = 8.0 Hz, 1H), 6.85 (d, J = 7.2 Hz, 2H), 7.15 (t, J = 8.0 Hz, 2H), 7.28~7.32 (m, 2H), 7.73 (t, J = 8.0 Hz, 2H), 8.09 (d, J = 7.2 Hz, 2H), 8.30~8.40 (m, 4H).
Refinement
H-atoms were placed in calculated positions (C–H 0.93–0.97 Å, N–H 0.86 Å), and were included in the refinement in the riding model, with Uiso(H) = 1.2Ueq(C,N).
Figures
Fig. 1.
The molecular structure of (I), showing the atom-labelling scheme. Displacement ellipsoids are drawn at the 50% probability level and H atoms are shown as small spheres of arbitrary radii.
Fig. 2.
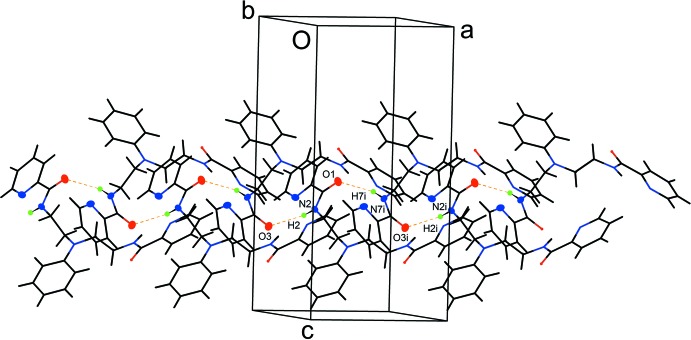
Representation of part of the lattice contents of (I), viewed approximately down the cell diagonal bisecting the b O a angle. Only selected interactions are shown for clarity. Atoms involved in hydrogen bonds are shown as balls of arbitrary radii. All other atoms and covalent bonds are represented as wires or sticks. [symmetry code: (i) 1+ x, y, z].
Fig. 3.
Representation of part of the lattice contents of (I), along the a axis, showing the hydrogen-bonded chains. Only selected interactions are shown for clarity. Atoms involved in hydrogen bonds are shown as balls of arbitrary radii. All other atoms and covalent bonds are represented as wires or sticks. [symmetry code: (i) 1–x, 1–y, 1–z; (ii) 1/2–x, 1/2+y, 3/2–z; (iii) 1/2+x, 1/2–y, –1/2+z].
Crystal data
| C22H23N5O2 | F(000) = 1648 |
| Mr = 389.45 | Dx = 1.310 Mg m−3 |
| Monoclinic, P21/n | Cu Kα radiation, λ = 1.5418 Å |
| Hall symbol: -P 2yn | Cell parameters from 13020 reflections |
| a = 8.64349 (7) Å | θ = 3.0–67.1° |
| b = 24.8210 (3) Å | µ = 0.70 mm−1 |
| c = 18.40861 (18) Å | T = 100 K |
| β = 90.5648 (8)° | Block, colorless |
| V = 3949.20 (7) Å3 | 0.12 × 0.08 × 0.07 mm |
| Z = 8 |
Data collection
| Agilent Xcalibur Atlas Gemini ultra diffractometer | 7050 independent reflections |
| Radiation source: Enhance Ultra (Cu) X-ray Source | 6128 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.038 |
| Detector resolution: 10.5095 pixels mm-1 | θmax = 67.2°, θmin = 3.0° |
| ω scans | h = −8→10 |
| Absorption correction: multi-scan (CrysAlis PRO; Agilent, 2012) | k = −29→29 |
| Tmin = 0.886, Tmax = 0.950 | l = −21→21 |
| 28468 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.044 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.097 | H-atom parameters constrained |
| S = 1.14 | w = 1/[σ2(Fo2) + (0.0328P)2 + 1.7734P] where P = (Fo2 + 2Fc2)/3 |
| 7050 reflections | (Δ/σ)max < 0.001 |
| 523 parameters | Δρmax = 0.17 e Å−3 |
| 0 restraints | Δρmin = −0.26 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O3 | 0.01667 (13) | 0.20717 (5) | 0.70802 (6) | 0.0213 (3) | |
| O4 | 0.42663 (14) | 0.41541 (5) | 0.43618 (7) | 0.0262 (3) | |
| N10 | 0.72634 (16) | 0.38111 (6) | 0.56121 (7) | 0.0211 (3) | |
| N8 | 0.05731 (16) | 0.30231 (6) | 0.48259 (8) | 0.0210 (3) | |
| N6 | −0.24832 (18) | 0.10919 (6) | 0.64032 (8) | 0.0253 (3) | |
| N9 | 0.47932 (15) | 0.33680 (6) | 0.49331 (7) | 0.0194 (3) | |
| H9 | 0.5429 | 0.3194 | 0.5208 | 0.023* | |
| C40 | 0.64956 (18) | 0.41244 (7) | 0.51379 (8) | 0.0189 (3) | |
| N7 | −0.14356 (16) | 0.20756 (6) | 0.60938 (7) | 0.0211 (3) | |
| H7 | −0.2159 | 0.1904 | 0.5868 | 0.025* | |
| C39 | 0.50887 (18) | 0.38841 (7) | 0.47694 (9) | 0.0195 (3) | |
| C28 | −0.08427 (18) | 0.18567 (7) | 0.66985 (8) | 0.0172 (3) | |
| C27 | −0.15311 (18) | 0.13184 (7) | 0.68976 (8) | 0.0178 (3) | |
| C36 | −0.02993 (18) | 0.32728 (7) | 0.42846 (8) | 0.0177 (3) | |
| C41 | 0.6897 (2) | 0.46544 (8) | 0.49946 (10) | 0.0251 (4) | |
| H41 | 0.6331 | 0.4858 | 0.4661 | 0.030* | |
| C32 | −0.26386 (18) | 0.33195 (7) | 0.35478 (9) | 0.0195 (3) | |
| H32 | −0.3619 | 0.3187 | 0.3437 | 0.023* | |
| C37 | 0.19986 (18) | 0.32603 (7) | 0.50953 (9) | 0.0194 (3) | |
| H37A | 0.2156 | 0.3153 | 0.5597 | 0.023* | |
| H37B | 0.1901 | 0.3650 | 0.5085 | 0.023* | |
| C23 | −0.3065 (2) | 0.06086 (8) | 0.65665 (10) | 0.0282 (4) | |
| H23 | −0.3722 | 0.0445 | 0.6230 | 0.034* | |
| C26 | −0.11910 (18) | 0.10871 (7) | 0.75624 (9) | 0.0187 (3) | |
| H26 | −0.0554 | 0.1263 | 0.7896 | 0.022* | |
| C31 | −0.17940 (18) | 0.30923 (7) | 0.41132 (8) | 0.0175 (3) | |
| H31 | −0.2227 | 0.2815 | 0.4384 | 0.021* | |
| C24 | −0.2747 (2) | 0.03374 (7) | 0.72098 (10) | 0.0256 (4) | |
| H24 | −0.3149 | −0.0004 | 0.7293 | 0.031* | |
| C30 | 0.01142 (18) | 0.25116 (7) | 0.51395 (9) | 0.0186 (3) | |
| H30A | 0.1032 | 0.2313 | 0.5285 | 0.022* | |
| H30B | −0.0431 | 0.2301 | 0.4775 | 0.022* | |
| C33 | −0.20453 (19) | 0.37409 (8) | 0.31463 (9) | 0.0238 (4) | |
| H33 | −0.2608 | 0.3889 | 0.2762 | 0.029* | |
| C34 | −0.0595 (2) | 0.39369 (8) | 0.33289 (10) | 0.0262 (4) | |
| H34 | −0.0198 | 0.4228 | 0.3073 | 0.031* | |
| C43 | 0.8975 (2) | 0.45549 (8) | 0.58395 (10) | 0.0259 (4) | |
| H43 | 0.9832 | 0.4691 | 0.6088 | 0.031* | |
| C44 | 0.8496 (2) | 0.40288 (7) | 0.59447 (9) | 0.0241 (4) | |
| H44 | 0.9063 | 0.3814 | 0.6265 | 0.029* | |
| C29 | −0.0926 (2) | 0.25855 (7) | 0.57982 (9) | 0.0210 (4) | |
| H29A | −0.0365 | 0.2783 | 0.6171 | 0.025* | |
| H29B | −0.1823 | 0.2797 | 0.5658 | 0.025* | |
| C42 | 0.8154 (2) | 0.48745 (8) | 0.53567 (10) | 0.0272 (4) | |
| H42 | 0.8442 | 0.5230 | 0.5277 | 0.033* | |
| C38 | 0.34208 (19) | 0.30960 (7) | 0.46552 (9) | 0.0211 (4) | |
| H38A | 0.3565 | 0.2709 | 0.4686 | 0.025* | |
| H38B | 0.3260 | 0.3190 | 0.4149 | 0.025* | |
| C35 | 0.02825 (19) | 0.37092 (7) | 0.38850 (9) | 0.0218 (4) | |
| H35 | 0.1259 | 0.3846 | 0.3994 | 0.026* | |
| C25 | −0.18174 (19) | 0.05870 (7) | 0.77240 (9) | 0.0218 (4) | |
| H25 | −0.1616 | 0.0423 | 0.8169 | 0.026* | |
| O1 | 0.51720 (13) | 0.20741 (5) | 0.55318 (6) | 0.0224 (3) | |
| O2 | 0.92905 (13) | 0.41536 (5) | 0.82007 (6) | 0.0233 (3) | |
| N5 | 1.22283 (16) | 0.37938 (6) | 0.69577 (7) | 0.0204 (3) | |
| N3 | 0.55536 (15) | 0.30360 (6) | 0.77265 (7) | 0.0206 (3) | |
| N2 | 0.35218 (16) | 0.20593 (6) | 0.64857 (7) | 0.0210 (3) | |
| H2 | 0.2804 | 0.1881 | 0.6699 | 0.025* | |
| N4 | 0.97851 (15) | 0.33578 (6) | 0.76493 (7) | 0.0183 (3) | |
| H4 | 1.0416 | 0.3178 | 0.7385 | 0.022* | |
| N1 | 0.24906 (17) | 0.10774 (6) | 0.61151 (8) | 0.0251 (3) | |
| C5 | 0.35355 (18) | 0.13041 (7) | 0.56756 (9) | 0.0183 (3) | |
| C6 | 0.41614 (18) | 0.18486 (7) | 0.58916 (8) | 0.0175 (3) | |
| C17 | 1.00987 (18) | 0.38738 (7) | 0.78022 (8) | 0.0179 (3) | |
| C18 | 1.15194 (18) | 0.41036 (7) | 0.74477 (9) | 0.0179 (3) | |
| C9 | 0.47424 (18) | 0.32890 (7) | 0.82781 (8) | 0.0174 (3) | |
| C11 | 0.24738 (19) | 0.33473 (7) | 0.90377 (9) | 0.0201 (3) | |
| H11 | 0.1500 | 0.3219 | 0.9160 | 0.024* | |
| C22 | 1.3487 (2) | 0.39964 (7) | 0.66449 (9) | 0.0225 (4) | |
| H22 | 1.4002 | 0.3784 | 0.6308 | 0.027* | |
| C21 | 1.4069 (2) | 0.45028 (8) | 0.67933 (10) | 0.0259 (4) | |
| H21 | 1.4951 | 0.4628 | 0.6562 | 0.031* | |
| C14 | 0.53710 (19) | 0.37270 (7) | 0.86626 (9) | 0.0223 (4) | |
| H14 | 0.6336 | 0.3863 | 0.8538 | 0.027* | |
| C15 | 0.69885 (18) | 0.32599 (7) | 0.74607 (9) | 0.0190 (3) | |
| H15A | 0.6910 | 0.3650 | 0.7458 | 0.023* | |
| H15B | 0.7138 | 0.3142 | 0.6964 | 0.023* | |
| C8 | 0.50815 (18) | 0.25151 (7) | 0.74369 (9) | 0.0188 (3) | |
| H8A | 0.4570 | 0.2311 | 0.7814 | 0.023* | |
| H8B | 0.5992 | 0.2316 | 0.7291 | 0.023* | |
| C13 | 0.4555 (2) | 0.39580 (8) | 0.92285 (10) | 0.0258 (4) | |
| H13 | 0.4988 | 0.4247 | 0.9480 | 0.031* | |
| C10 | 0.32600 (18) | 0.31133 (7) | 0.84680 (8) | 0.0176 (3) | |
| H10 | 0.2797 | 0.2835 | 0.8207 | 0.021* | |
| C12 | 0.3113 (2) | 0.37699 (7) | 0.94294 (9) | 0.0241 (4) | |
| H12 | 0.2588 | 0.3923 | 0.9817 | 0.029* | |
| C7 | 0.39857 (19) | 0.25749 (7) | 0.67850 (9) | 0.0208 (3) | |
| H7A | 0.3071 | 0.2771 | 0.6933 | 0.025* | |
| H7B | 0.4493 | 0.2784 | 0.6411 | 0.025* | |
| C16 | 0.84045 (19) | 0.30946 (7) | 0.79192 (9) | 0.0198 (3) | |
| H16A | 0.8245 | 0.3195 | 0.8422 | 0.024* | |
| H16B | 0.8536 | 0.2707 | 0.7898 | 0.024* | |
| C1 | 0.1974 (2) | 0.05894 (8) | 0.59321 (10) | 0.0290 (4) | |
| H1 | 0.1244 | 0.0427 | 0.6229 | 0.035* | |
| C4 | 0.40508 (19) | 0.10617 (7) | 0.50462 (9) | 0.0219 (4) | |
| H4A | 0.4750 | 0.1237 | 0.4747 | 0.026* | |
| C3 | 0.3509 (2) | 0.05546 (8) | 0.48688 (10) | 0.0269 (4) | |
| H3 | 0.3842 | 0.0381 | 0.4451 | 0.032* | |
| C19 | 1.2014 (2) | 0.46160 (8) | 0.76317 (10) | 0.0272 (4) | |
| H19 | 1.1488 | 0.4819 | 0.7975 | 0.033* | |
| C20 | 1.3310 (2) | 0.48193 (8) | 0.72935 (11) | 0.0310 (4) | |
| H20 | 1.3666 | 0.5164 | 0.7401 | 0.037* | |
| C2 | 0.2463 (2) | 0.03102 (7) | 0.53257 (10) | 0.0285 (4) | |
| H2A | 0.2095 | −0.0035 | 0.5228 | 0.034* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O3 | 0.0171 (6) | 0.0243 (6) | 0.0225 (6) | −0.0026 (5) | −0.0031 (5) | −0.0003 (5) |
| O4 | 0.0194 (6) | 0.0311 (7) | 0.0280 (6) | 0.0025 (5) | −0.0047 (5) | 0.0053 (5) |
| N10 | 0.0197 (7) | 0.0244 (8) | 0.0191 (7) | −0.0006 (6) | −0.0016 (5) | −0.0002 (6) |
| N8 | 0.0154 (7) | 0.0250 (8) | 0.0226 (7) | −0.0057 (6) | −0.0056 (5) | 0.0064 (6) |
| N6 | 0.0304 (8) | 0.0253 (8) | 0.0200 (7) | −0.0056 (6) | −0.0018 (6) | −0.0029 (6) |
| N9 | 0.0112 (7) | 0.0268 (8) | 0.0203 (7) | 0.0014 (5) | −0.0017 (5) | 0.0021 (6) |
| C40 | 0.0159 (8) | 0.0244 (9) | 0.0164 (8) | 0.0026 (7) | 0.0040 (6) | 0.0003 (7) |
| N7 | 0.0205 (7) | 0.0231 (8) | 0.0196 (7) | −0.0061 (6) | −0.0039 (5) | 0.0019 (6) |
| C39 | 0.0156 (8) | 0.0264 (9) | 0.0166 (8) | 0.0031 (7) | 0.0022 (6) | −0.0003 (7) |
| C28 | 0.0147 (8) | 0.0208 (8) | 0.0163 (8) | 0.0016 (6) | 0.0031 (6) | −0.0018 (6) |
| C27 | 0.0156 (8) | 0.0195 (9) | 0.0184 (8) | 0.0007 (6) | 0.0020 (6) | −0.0029 (6) |
| C36 | 0.0148 (8) | 0.0224 (9) | 0.0157 (8) | 0.0014 (6) | 0.0001 (6) | −0.0013 (6) |
| C41 | 0.0189 (9) | 0.0280 (10) | 0.0284 (9) | 0.0027 (7) | 0.0011 (7) | 0.0054 (7) |
| C32 | 0.0130 (8) | 0.0256 (9) | 0.0198 (8) | 0.0029 (6) | −0.0012 (6) | −0.0037 (7) |
| C37 | 0.0155 (8) | 0.0240 (9) | 0.0187 (8) | −0.0035 (6) | −0.0024 (6) | 0.0016 (7) |
| C23 | 0.0348 (10) | 0.0245 (10) | 0.0253 (9) | −0.0089 (8) | −0.0011 (8) | −0.0053 (7) |
| C26 | 0.0134 (8) | 0.0222 (9) | 0.0205 (8) | 0.0020 (6) | 0.0013 (6) | −0.0013 (7) |
| C31 | 0.0155 (8) | 0.0202 (8) | 0.0170 (8) | 0.0003 (6) | 0.0023 (6) | −0.0020 (6) |
| C24 | 0.0270 (9) | 0.0190 (9) | 0.0310 (10) | −0.0018 (7) | 0.0059 (7) | −0.0005 (7) |
| C30 | 0.0149 (8) | 0.0206 (9) | 0.0201 (8) | −0.0001 (6) | −0.0036 (6) | 0.0021 (7) |
| C33 | 0.0189 (9) | 0.0315 (10) | 0.0209 (8) | 0.0065 (7) | −0.0015 (7) | 0.0043 (7) |
| C34 | 0.0233 (9) | 0.0285 (10) | 0.0269 (9) | 0.0007 (7) | 0.0022 (7) | 0.0093 (8) |
| C43 | 0.0217 (9) | 0.0307 (10) | 0.0254 (9) | −0.0055 (7) | −0.0003 (7) | −0.0031 (8) |
| C44 | 0.0219 (9) | 0.0292 (10) | 0.0209 (8) | −0.0017 (7) | −0.0047 (7) | 0.0017 (7) |
| C29 | 0.0244 (9) | 0.0206 (9) | 0.0181 (8) | −0.0013 (7) | −0.0006 (7) | 0.0012 (7) |
| C42 | 0.0222 (9) | 0.0243 (10) | 0.0353 (10) | −0.0037 (7) | 0.0042 (7) | 0.0014 (8) |
| C38 | 0.0188 (8) | 0.0254 (9) | 0.0191 (8) | −0.0002 (7) | −0.0018 (6) | −0.0010 (7) |
| C35 | 0.0145 (8) | 0.0266 (9) | 0.0243 (9) | −0.0023 (7) | 0.0000 (6) | 0.0028 (7) |
| C25 | 0.0198 (8) | 0.0220 (9) | 0.0238 (9) | 0.0040 (7) | 0.0029 (7) | 0.0037 (7) |
| O1 | 0.0169 (6) | 0.0250 (6) | 0.0253 (6) | −0.0022 (5) | 0.0045 (5) | −0.0001 (5) |
| O2 | 0.0182 (6) | 0.0255 (7) | 0.0262 (6) | 0.0028 (5) | 0.0042 (5) | −0.0038 (5) |
| N5 | 0.0199 (7) | 0.0221 (8) | 0.0192 (7) | −0.0011 (6) | 0.0012 (5) | −0.0008 (6) |
| N3 | 0.0152 (7) | 0.0252 (8) | 0.0214 (7) | −0.0044 (6) | 0.0057 (5) | −0.0062 (6) |
| N2 | 0.0204 (7) | 0.0239 (8) | 0.0187 (7) | −0.0054 (6) | 0.0038 (5) | −0.0026 (6) |
| N4 | 0.0126 (6) | 0.0212 (7) | 0.0211 (7) | 0.0018 (5) | 0.0015 (5) | −0.0030 (6) |
| N1 | 0.0278 (8) | 0.0275 (8) | 0.0199 (7) | −0.0065 (6) | 0.0022 (6) | 0.0007 (6) |
| C5 | 0.0146 (8) | 0.0220 (9) | 0.0183 (8) | 0.0008 (6) | −0.0011 (6) | 0.0026 (7) |
| C6 | 0.0131 (8) | 0.0218 (9) | 0.0177 (8) | 0.0015 (6) | −0.0027 (6) | 0.0004 (6) |
| C17 | 0.0146 (8) | 0.0226 (9) | 0.0165 (8) | 0.0032 (6) | −0.0032 (6) | 0.0004 (7) |
| C18 | 0.0159 (8) | 0.0210 (9) | 0.0168 (8) | 0.0026 (6) | −0.0033 (6) | 0.0010 (6) |
| C9 | 0.0145 (8) | 0.0207 (9) | 0.0172 (8) | 0.0026 (6) | 0.0003 (6) | 0.0015 (6) |
| C11 | 0.0156 (8) | 0.0251 (9) | 0.0197 (8) | 0.0050 (7) | 0.0016 (6) | 0.0042 (7) |
| C22 | 0.0214 (9) | 0.0273 (9) | 0.0188 (8) | −0.0027 (7) | 0.0035 (6) | −0.0019 (7) |
| C21 | 0.0233 (9) | 0.0286 (10) | 0.0260 (9) | −0.0062 (7) | 0.0043 (7) | 0.0012 (7) |
| C14 | 0.0160 (8) | 0.0250 (9) | 0.0257 (9) | −0.0005 (7) | 0.0009 (7) | −0.0039 (7) |
| C15 | 0.0146 (8) | 0.0240 (9) | 0.0185 (8) | −0.0029 (6) | 0.0033 (6) | −0.0022 (7) |
| C8 | 0.0161 (8) | 0.0215 (9) | 0.0190 (8) | 0.0001 (6) | 0.0037 (6) | −0.0029 (7) |
| C13 | 0.0199 (9) | 0.0261 (10) | 0.0312 (10) | 0.0028 (7) | −0.0015 (7) | −0.0093 (8) |
| C10 | 0.0164 (8) | 0.0198 (8) | 0.0166 (8) | 0.0007 (6) | −0.0013 (6) | 0.0021 (6) |
| C12 | 0.0231 (9) | 0.0275 (9) | 0.0216 (8) | 0.0081 (7) | 0.0021 (7) | −0.0040 (7) |
| C7 | 0.0203 (8) | 0.0216 (9) | 0.0205 (8) | −0.0005 (7) | 0.0011 (6) | −0.0017 (7) |
| C16 | 0.0182 (8) | 0.0214 (9) | 0.0198 (8) | 0.0001 (7) | 0.0033 (6) | 0.0009 (7) |
| C1 | 0.0351 (10) | 0.0271 (10) | 0.0250 (9) | −0.0109 (8) | 0.0020 (8) | 0.0043 (8) |
| C4 | 0.0189 (8) | 0.0243 (9) | 0.0225 (8) | 0.0006 (7) | 0.0020 (6) | −0.0002 (7) |
| C3 | 0.0299 (10) | 0.0250 (10) | 0.0260 (9) | 0.0030 (7) | 0.0010 (7) | −0.0057 (7) |
| C19 | 0.0217 (9) | 0.0260 (10) | 0.0339 (10) | −0.0010 (7) | 0.0048 (7) | −0.0086 (8) |
| C20 | 0.0259 (10) | 0.0246 (10) | 0.0427 (11) | −0.0076 (8) | 0.0060 (8) | −0.0068 (8) |
| C2 | 0.0345 (10) | 0.0199 (9) | 0.0310 (10) | −0.0042 (8) | −0.0046 (8) | 0.0006 (8) |
Geometric parameters (Å, º)
| O3—C28 | 1.236 (2) | O1—C6 | 1.235 (2) |
| O4—C39 | 1.227 (2) | O2—C17 | 1.232 (2) |
| N10—C40 | 1.340 (2) | N5—C18 | 1.338 (2) |
| N10—C44 | 1.337 (2) | N5—C22 | 1.335 (2) |
| N8—C36 | 1.390 (2) | N3—C9 | 1.390 (2) |
| N8—C37 | 1.449 (2) | N3—C15 | 1.449 (2) |
| N8—C30 | 1.452 (2) | N3—C8 | 1.455 (2) |
| N6—C27 | 1.344 (2) | N2—H2 | 0.8600 |
| N6—C23 | 1.336 (2) | N2—C6 | 1.337 (2) |
| N9—H9 | 0.8600 | N2—C7 | 1.448 (2) |
| N9—C39 | 1.341 (2) | N4—H4 | 0.8600 |
| N9—C38 | 1.454 (2) | N4—C17 | 1.338 (2) |
| C40—C39 | 1.509 (2) | N4—C16 | 1.452 (2) |
| C40—C41 | 1.387 (3) | N1—C5 | 1.342 (2) |
| N7—H7 | 0.8600 | N1—C1 | 1.333 (2) |
| N7—C28 | 1.336 (2) | C5—C6 | 1.508 (2) |
| N7—C29 | 1.448 (2) | C5—C4 | 1.383 (2) |
| C28—C27 | 1.509 (2) | C17—C18 | 1.508 (2) |
| C27—C26 | 1.381 (2) | C18—C19 | 1.383 (3) |
| C36—C31 | 1.400 (2) | C9—C14 | 1.404 (2) |
| C36—C35 | 1.405 (2) | C9—C10 | 1.401 (2) |
| C41—H41 | 0.9300 | C11—H11 | 0.9300 |
| C41—C42 | 1.382 (3) | C11—C10 | 1.383 (2) |
| C32—H32 | 0.9300 | C11—C12 | 1.385 (3) |
| C32—C31 | 1.385 (2) | C22—H22 | 0.9300 |
| C32—C33 | 1.382 (3) | C22—C21 | 1.380 (3) |
| C37—H37A | 0.9700 | C21—H21 | 0.9300 |
| C37—H37B | 0.9700 | C21—C20 | 1.381 (3) |
| C37—C38 | 1.534 (2) | C14—H14 | 0.9300 |
| C23—H23 | 0.9300 | C14—C13 | 1.388 (2) |
| C23—C24 | 1.387 (3) | C15—H15A | 0.9700 |
| C26—H26 | 0.9300 | C15—H15B | 0.9700 |
| C26—C25 | 1.388 (2) | C15—C16 | 1.536 (2) |
| C31—H31 | 0.9300 | C8—H8A | 0.9700 |
| C24—H24 | 0.9300 | C8—H8B | 0.9700 |
| C24—C25 | 1.382 (3) | C8—C7 | 1.529 (2) |
| C30—H30A | 0.9700 | C13—H13 | 0.9300 |
| C30—H30B | 0.9700 | C13—C12 | 1.384 (3) |
| C30—C29 | 1.528 (2) | C10—H10 | 0.9300 |
| C33—H33 | 0.9300 | C12—H12 | 0.9300 |
| C33—C34 | 1.383 (3) | C7—H7A | 0.9700 |
| C34—H34 | 0.9300 | C7—H7B | 0.9700 |
| C34—C35 | 1.388 (3) | C16—H16A | 0.9700 |
| C43—H43 | 0.9300 | C16—H16B | 0.9700 |
| C43—C44 | 1.384 (3) | C1—H1 | 0.9300 |
| C43—C42 | 1.382 (3) | C1—C2 | 1.384 (3) |
| C44—H44 | 0.9300 | C4—H4A | 0.9300 |
| C29—H29A | 0.9700 | C4—C3 | 1.381 (3) |
| C29—H29B | 0.9700 | C3—H3 | 0.9300 |
| C42—H42 | 0.9300 | C3—C2 | 1.381 (3) |
| C38—H38A | 0.9700 | C19—H19 | 0.9300 |
| C38—H38B | 0.9700 | C19—C20 | 1.382 (3) |
| C35—H35 | 0.9300 | C20—H20 | 0.9300 |
| C25—H25 | 0.9300 | C2—H2A | 0.9300 |
| C44—N10—C40 | 116.80 (15) | C22—N5—C18 | 117.06 (15) |
| C36—N8—C37 | 121.20 (14) | C9—N3—C15 | 120.88 (14) |
| C36—N8—C30 | 121.79 (13) | C9—N3—C8 | 121.83 (13) |
| C37—N8—C30 | 117.01 (13) | C15—N3—C8 | 117.07 (13) |
| C23—N6—C27 | 116.90 (15) | C6—N2—H2 | 118.6 |
| C39—N9—H9 | 119.3 | C6—N2—C7 | 122.80 (14) |
| C39—N9—C38 | 121.41 (14) | C7—N2—H2 | 118.6 |
| C38—N9—H9 | 119.3 | C17—N4—H4 | 119.2 |
| N10—C40—C39 | 117.07 (15) | C17—N4—C16 | 121.59 (13) |
| N10—C40—C41 | 123.47 (16) | C16—N4—H4 | 119.2 |
| C41—C40—C39 | 119.42 (15) | C1—N1—C5 | 117.02 (15) |
| C28—N7—H7 | 118.2 | N1—C5—C6 | 117.30 (14) |
| C28—N7—C29 | 123.57 (14) | N1—C5—C4 | 123.14 (16) |
| C29—N7—H7 | 118.2 | C4—C5—C6 | 119.55 (14) |
| O4—C39—N9 | 123.26 (16) | O1—C6—N2 | 124.18 (16) |
| O4—C39—C40 | 121.20 (16) | O1—C6—C5 | 121.22 (14) |
| N9—C39—C40 | 115.52 (14) | N2—C6—C5 | 114.60 (14) |
| O3—C28—N7 | 124.11 (15) | O2—C17—N4 | 123.34 (15) |
| O3—C28—C27 | 121.46 (14) | O2—C17—C18 | 120.88 (15) |
| N7—C28—C27 | 114.42 (14) | N4—C17—C18 | 115.78 (14) |
| N6—C27—C28 | 116.48 (14) | N5—C18—C17 | 117.03 (15) |
| N6—C27—C26 | 123.36 (15) | N5—C18—C19 | 123.43 (15) |
| C26—C27—C28 | 120.16 (15) | C19—C18—C17 | 119.54 (15) |
| N8—C36—C31 | 120.78 (15) | N3—C9—C14 | 121.52 (14) |
| N8—C36—C35 | 121.65 (15) | N3—C9—C10 | 120.73 (15) |
| C31—C36—C35 | 117.57 (15) | C10—C9—C14 | 117.75 (15) |
| C40—C41—H41 | 120.7 | C10—C11—H11 | 119.5 |
| C42—C41—C40 | 118.70 (17) | C10—C11—C12 | 121.09 (16) |
| C42—C41—H41 | 120.7 | C12—C11—H11 | 119.5 |
| C31—C32—H32 | 119.5 | N5—C22—H22 | 118.1 |
| C33—C32—H32 | 119.5 | N5—C22—C21 | 123.70 (16) |
| C33—C32—C31 | 120.95 (15) | C21—C22—H22 | 118.1 |
| N8—C37—H37A | 108.9 | C22—C21—H21 | 120.8 |
| N8—C37—H37B | 108.9 | C22—C21—C20 | 118.38 (16) |
| N8—C37—C38 | 113.23 (14) | C20—C21—H21 | 120.8 |
| H37A—C37—H37B | 107.7 | C9—C14—H14 | 120.0 |
| C38—C37—H37A | 108.9 | C13—C14—C9 | 120.08 (16) |
| C38—C37—H37B | 108.9 | C13—C14—H14 | 120.0 |
| N6—C23—H23 | 118.1 | N3—C15—H15A | 109.0 |
| N6—C23—C24 | 123.76 (17) | N3—C15—H15B | 109.0 |
| C24—C23—H23 | 118.1 | N3—C15—C16 | 113.09 (14) |
| C27—C26—H26 | 120.6 | H15A—C15—H15B | 107.8 |
| C27—C26—C25 | 118.81 (16) | C16—C15—H15A | 109.0 |
| C25—C26—H26 | 120.6 | C16—C15—H15B | 109.0 |
| C36—C31—H31 | 119.4 | N3—C8—H8A | 109.3 |
| C32—C31—C36 | 121.14 (15) | N3—C8—H8B | 109.3 |
| C32—C31—H31 | 119.4 | N3—C8—C7 | 111.76 (14) |
| C23—C24—H24 | 120.8 | H8A—C8—H8B | 107.9 |
| C25—C24—C23 | 118.43 (17) | C7—C8—H8A | 109.3 |
| C25—C24—H24 | 120.8 | C7—C8—H8B | 109.3 |
| N8—C30—H30A | 109.2 | C14—C13—H13 | 119.1 |
| N8—C30—H30B | 109.2 | C12—C13—C14 | 121.79 (17) |
| N8—C30—C29 | 112.07 (14) | C12—C13—H13 | 119.1 |
| H30A—C30—H30B | 107.9 | C9—C10—H10 | 119.5 |
| C29—C30—H30A | 109.2 | C11—C10—C9 | 121.06 (16) |
| C29—C30—H30B | 109.2 | C11—C10—H10 | 119.5 |
| C32—C33—H33 | 120.8 | C11—C12—H12 | 120.9 |
| C32—C33—C34 | 118.44 (16) | C13—C12—C11 | 118.16 (15) |
| C34—C33—H33 | 120.8 | C13—C12—H12 | 120.9 |
| C33—C34—H34 | 119.2 | N2—C7—C8 | 112.33 (14) |
| C33—C34—C35 | 121.54 (16) | N2—C7—H7A | 109.1 |
| C35—C34—H34 | 119.2 | N2—C7—H7B | 109.1 |
| C44—C43—H43 | 120.7 | C8—C7—H7A | 109.1 |
| C42—C43—H43 | 120.7 | C8—C7—H7B | 109.1 |
| C42—C43—C44 | 118.59 (17) | H7A—C7—H7B | 107.9 |
| N10—C44—C43 | 123.74 (17) | N4—C16—C15 | 110.18 (13) |
| N10—C44—H44 | 118.1 | N4—C16—H16A | 109.6 |
| C43—C44—H44 | 118.1 | N4—C16—H16B | 109.6 |
| N7—C29—C30 | 112.14 (14) | C15—C16—H16A | 109.6 |
| N7—C29—H29A | 109.2 | C15—C16—H16B | 109.6 |
| N7—C29—H29B | 109.2 | H16A—C16—H16B | 108.1 |
| C30—C29—H29A | 109.2 | N1—C1—H1 | 118.2 |
| C30—C29—H29B | 109.2 | N1—C1—C2 | 123.69 (17) |
| H29A—C29—H29B | 107.9 | C2—C1—H1 | 118.2 |
| C41—C42—C43 | 118.68 (17) | C5—C4—H4A | 120.5 |
| C41—C42—H42 | 120.7 | C3—C4—C5 | 118.91 (16) |
| C43—C42—H42 | 120.7 | C3—C4—H4A | 120.5 |
| N9—C38—C37 | 110.24 (13) | C4—C3—H3 | 120.7 |
| N9—C38—H38A | 109.6 | C4—C3—C2 | 118.60 (17) |
| N9—C38—H38B | 109.6 | C2—C3—H3 | 120.7 |
| C37—C38—H38A | 109.6 | C18—C19—H19 | 120.8 |
| C37—C38—H38B | 109.6 | C20—C19—C18 | 118.40 (17) |
| H38A—C38—H38B | 108.1 | C20—C19—H19 | 120.8 |
| C36—C35—H35 | 119.9 | C21—C20—C19 | 119.02 (18) |
| C34—C35—C36 | 120.28 (16) | C21—C20—H20 | 120.5 |
| C34—C35—H35 | 119.9 | C19—C20—H20 | 120.5 |
| C26—C25—H25 | 120.7 | C1—C2—H2A | 120.7 |
| C24—C25—C26 | 118.65 (16) | C3—C2—C1 | 118.58 (17) |
| C24—C25—H25 | 120.7 | C3—C2—H2A | 120.7 |
Hydrogen-bond geometry (Å, º)
Cg5 and Cg6 are the centroids of the C9–C14 benzene and C31–C36 benzene rings, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| N2—H2···O3 | 0.86 | 2.44 | 3.1099 (18) | 135 |
| N7—H7···O1i | 0.86 | 2.42 | 3.0998 (18) | 136 |
| C24—H24···O2ii | 0.93 | 2.48 | 3.311 (2) | 149 |
| C25—H25···O4iii | 0.93 | 2.54 | 3.213 (2) | 129 |
| C8—H8A···Cg6iv | 0.97 | 2.72 | 3.6468 (18) | 161 |
| C30—H30B···Cg5v | 0.97 | 2.74 | 3.6584 (18) | 159 |
Symmetry codes: (i) x−1, y, z; (ii) −x+1/2, y−1/2, −z+3/2; (iii) x−1/2, −y+1/2, z+1/2; (iv) x+1/2, −y+1/2, z+1/2; (v) x−1/2, −y+1/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: XU5691).
References
- Adolph, M., Zevaco, T. A., Walter, O., Dinjus, E. & Döring, M. (2012). Polyhedron, 48, 92–98.
- Agilent (2012). CrysAlis PRO Agilent Technologies Inc., Santa Clara, CA, USA.
- Barnes, D. J., Chapman, R. L., Vagg, R. S. & Watton, E. C. (1978). J. Chem. Eng. Data, 23, 349–350.
- Brandenburg, K. (1999). DIAMOND Crystal Impact GbR, Bonn, Germany.
- Cornman, C. R., Zovinka, E. P., Boyajian, Y. D., Olmstead, M. M. & Noll, B. C. (1999). Inorg. Chim. Acta, 285, 134–137.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Jain, S. L., Bhattacharyya, P., Milton, H. L., Slawin, A. M. A., Crayston, J. A. & Woollins, J. D. (2004). Dalton Trans. pp. 862–871. [DOI] [PubMed]
- Lee, S. J., Lee, S. S., Lee, J. Y. & Jung, J. H. (2006). Chem. Mater. 18, 4713–4715.
- Munro, O. Q. & Wilson, C. (2010). Acta Cryst. C66, o535–o539. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Singh, A. K., Jacob, W., Boudalis, A. K., Tuchagues, J.-P. & Mukherjee, R. (2008). Eur. J. Inorg. Chem. pp. 2820–2829.
- Song, Y. J., Lee, J. H., Koo, H. G., Lee, T. G., Myoung, S.-H., Kim, C., Kim, S.-J. & Kim, Y. (2010). Inorg. Chem. Commun. 13, 753–756.
- Yan, Y., Zhao, W., Bhagavathy, G. V., Faurie, A., Mosey, N. J. & Petitjean, A. (2012). Chem. Commun. 48, 7829–7831. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536813008696/xu5691sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813008696/xu5691Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report