Abstract
Diagnosis of invasive aspergillosis (IA) remains challenging. With a relatively low incidence of disease, the use of expensive empirical antifungal therapy exposes many patients to unnecessary toxicity. Diagnosis places emphasis on specific but temporal radiological evidence. Circulating biomarker diagnosis has shown potential, but assays show variable performance, take several hours to perform, and require a degree of technical ability. A novel and simple lateral-flow device (LFD) using monoclonal antibody JF5, which targets an extracellular glycoprotein, has been developed and potentially removes any technical requirements, reducing processing time considerably. In this study, we evaluate the performance of this LFD compared to real-time PCR (targeting the 28S rRNA gene) and galactomannan (GM) detection when testing serum from a European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group, National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG)-defined hematological population. In a proven/probable-IA population versus a no-IA population, the LFD performance was comparable to that of both PCR and galactomannan enzyme immunoassay. Specificity (98.0%) was similar to that of PCR (96.6%) and slightly superior to that of GM (91.5%). Sensitivity (81.8%) was inferior to that of PCR (95.5%) but better than that of GM (77.3%). In combination with PCR, it provided both 100% sensitivity and 100% specificity. The LFD permits rapid testing of easily obtainable specimens, to be used as an adjunct test, before confirmation by other investigations. Its simplicity provides centers without specialist diagnostics with a test with clinical performance superior to that of classical microbiological approaches and results that can be used to direct antifungal management. In summary, microbiological diagnosis of IA is difficult and options are limited, with variable performance. An LFD assay targeting a novel specific biomarker has been developed, one which is methodologically simple and provides good clinical performance, particularly if combined with PCR.
INTRODUCTION
Invasive aspergillosis (IA) is a serious infection in immunocompromised patients. Delays in diagnosis contribute to the high morbidity and mortality associated with this disease. The detection of circulating biomarkers (DNA or antigens) indicative of IA is an attractive strategy that allows frequent testing, increasing the opportunity for detecting early infection before overt disease develops. High assay sensitivity and negative predictive value (NPV) can exclude disease, preventing unnecessary treatment in patients who are consistently negative. The number of biomarker test options is limited. The detection of circulating antigens is recognized by the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group, National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG), definitions as a mycological criterion for defining IA disease (1). Diagnostic assays that detect galactomannan (GM) or fungal β-d-glucan (BDG) are commercially available, and meta-analyses have shown similar performances for IA detection (BDG: sensitivity, 77% [95% confidence interval {CI}, 67 to 84]; specificity, 85% [95% CI, 79 to 89]; GM: sensitivity, 78% [95% CI, 61 to 89]; specificity, 81% [95% CI, 72 to 88]) (2, 3). Performance can vary considerably, and GM testing has been shown to be affected by various factors, leading to both false-positive and false-negative results (4). β-d-Glucan is also not specific for Aspergillus spp., detecting other species, including Candida, Pneumocystis, Fusarium, and Scedosporium, but not Cryptococcus or mucoraceous molds. Both tests could benefit from the use of adjunct diagnostic assays for IA.
For many years, PCR has been used as alternative or adjunct test, and diagnostic performance using whole blood (WB) is comparable to that of antigen detection (meta-analysis: sensitivity, 75% [95% CI, 54 to 88]; specificity, 87% [95% CI, 78 to 93]) (5). Combined antigen and PCR testing has been shown to provide improved diagnosis compared to individual assay performance (6). Nevertheless, a lack of standardization and limited commercial interest have prevented incorporation of PCR into disease-defining criteria, restricting its application as a diagnostic procedure. Recently, attempts to standardize Aspergillus PCR testing provided methodological guidance for testing both whole blood and serum (7, 8), and together with the launch of commercial PCR methodology, it is hoped that PCR testing will gain wider acceptance (9, 10). However, PCR testing still requires the availability of expensive and sophisticated equipment and, even with thorough standardization, may be restricted to specialist molecular diagnostic laboratories. The availability of a simple, inexpensive, and quick adjunct test for IA diagnosis would be extremely useful.
A lateral-flow device (LFD) for the serodiagnosis of IA has been described elsewhere (11). The immunochromatographic LFD incorporates a monoclonal antibody (MAb), JF5, that binds to an abundant extracellular glycoprotein antigen secreted during active growth of Aspergillus spp. The MAb is highly specific for Aspergillus species and does not cross-react with a wide range of clinically relevant fungi, including Candida, Fusarium, and Scedosporium species or agents of mucormycosis (11). In a well-established animal model of IA, the LFD provided performance comparable with those of commercial GM and β-d-glucan tests but with the added bonus of earlier positivity (12). With results generated within 10 to 15 min and simplicity of use that negates the need for expensive laboratory equipment, the LFD could potentially be used as a point-of-care (POC) test. Clinical assessment is required to determine its best clinical use (confirming or excluding a diagnosis). In a recent trial in hematological malignancy and solid-organ transplant recipients, the test was shown to have excellent negative predictive value in diagnosing IA by using bronchoalveolar lavage fluids (13). The LFD has, however, not been tested in a large-scale study using noninvasively obtained samples such as serum.
In this case-control study, retrospective serum samples were tested to determine the performance of the LFD compared to those of GM enzyme-linked immunosorbent assay (ELISA) and real-time PCR used in a prospective diagnostic capacity. To our knowledge, this is the first large-scale clinical evaluation of the LFD as a diagnostic aid for IA detection using serum.
MATERIALS AND METHODS
Study design and patient population.
The case-control study was performed as a retrospective anonymous evaluation of LFD performance in IA diagnosis. All samples were sent for prospective Aspergillus PCR and antigen testing as part of the routine diagnostic strategy within the local neutropenic fever care pathway (6). Diagnosis of IA was determined according to the revised EORTC/MSG criteria, with a single GM ELISA sample considered significant (1). LFD testing was performed retrospectively and had no impact on diagnosis or patient management. The study was conducted as a retrospective performance assessment not requiring ethical approval.
The study comprised 103 adult hematology patients at high risk of developing IA, including eight proven cases of IA, 14 probable cases, 22 possible cases, and 59 controls not achieving an EORTC/MSG diagnosis. Cases were selected on the basis of diagnosis and sample availability, whereas controls were randomly selected but were from a period of testing similar to that of the IA cases. Patient demographics and underlying hematological malignancy are summarized in Table 1.
Table 1.
Patient demographics with EORTC/MSG diagnosis of aspergillosisa
| Demographic characteristic | Proven IA (n = 8) | Probable IA (n = 14) | Possible IA (n = 22) | No IFD (n = 59) |
|---|---|---|---|---|
| No. male/no. female | 5/3 | 10/4 | 15/7 | 43/16 |
| Mean age (yr) | 48.3 | 45.1 | 53.6 | 55.9 |
| Hematological malignancy (type, no.) | AML/MDS, 6; lymphoma, 1; CLL, 1 | AML/MDS, 6; ALL, 3; CML, 2; lymphoma, 1; CLL, 1; SAA, 1 | AML/MDS, 15; lymphoma, 4; ALL, 2; CML, 1 | AML/MDS, 21; lymphoma, 16; myeloma, 12; SAA, 3; ALL, 2; CML, 2; CLL, 1; other, 2 |
| HSCT (type, no.) | Allo, 2 | Allo, 9; Auto, 1 | Allo, 5 | Allo, 13; Auto, 17 |
| Disease manifestation (type, no.) | IPA, 3; IPA/sinusitis, 3; cerebral, 1; IPA/Dissem, 1 | IPA, 10; IPA/sinusitis, 1; cerebral/sinusitis, 1; IPA/Dissem, 1; IPA/cerebral/sinusitis, 1 | IPA, 16; sinusitis, 5; cerebral/sinusitis, 1 | None |
Abbreviations: AML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia; CML, chronic myeloid leukemia; CLL, chronic lymphoblastic leukemia; MDS, myelodysplastic syndrome; SAA, severe aplastic anemia; lymphoma, Hodgkin's lymphoma, non-Hodgkin's lymphoma, lymphoproliferative disorder, and diffuse large B cell lymphoma; other, natural killer cell leukemia and chronic pancytopenia; HSCT, hematopoietic stem cell transplantation; Allo, allogeneic; Auto, autologous; IPA, invasive pulmonary aspergillosis; Dissem, disseminated disease.
Samples.
EDTA blood and serum were routinely sent for prospective diagnostic PCR and GM ELISA testing, respectively. As part of routine practice, serum is stored at −80°C for quality assurance purposes and performance assessment. A total of 529 samples were tested by GM and LFD (203 from proven/probable cases, 151 from possible cases, and 175 samples from controls).
In addition, 490 samples were also tested by PCR (183 from proven/probable cases, 140 from possible cases, and 167 from controls).
PCR testing.
Molecular testing was performed as described previously (14), in line with published recommendations (7, 8, 15). The Aspergillus real-time PCR targeted the 28S rRNA gene, and the limit of detection was 3 input copies/reaction. Positive (101 conidia) and negative simulated specimens were used as extraction controls, and PCR performance was monitored by the inclusion of cloned PCR products (300, 30, and 3 input copies) and no-template molecular-grade water. PCR positivity was determined using a threshold of 45 cycles. PCR efficiency was ca. 90% when testing DNA extracts from WB. A separate internal control PCR was performed to monitor for inhibition limiting the reporting of false-negative results (10).
GM ELISA.
Platelia kits (Bio-Rad, United Kingdom) were used for the detection of galactomannan (GM). GM results were determined using a threshold index (optical density at 450 nm/620 nm [OD450/620] of sample/OD450/620 of threshold control). According to the manufacturer's instructions, any value above 0.5 was considered positive.
Aspergillus lateral-flow device.
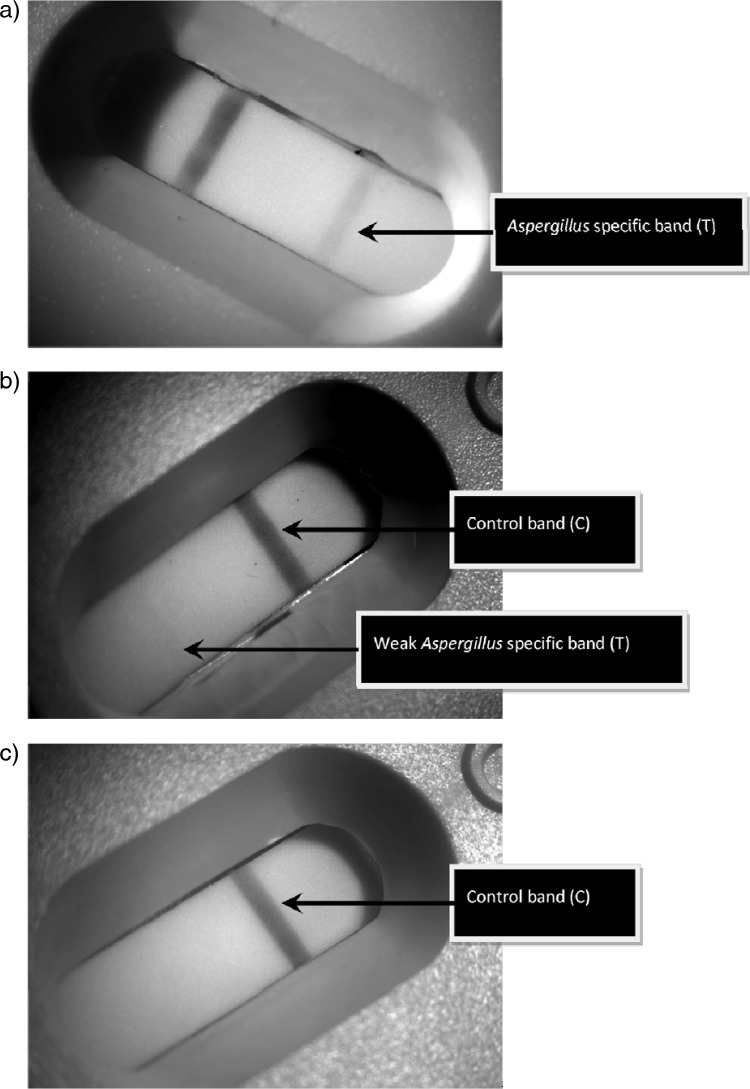
To dissociate immune complexes and precipitate proteins, 200 μl of serum was mixed with 75 μl of EDTA acid solution (Bio-Rad Platelia kit) and boiled for 3 min. The heat-treated mixture was centrifuged at 10,000 × g for 10 min, and 100 μl of the supernatant was removed with a pipette and added to the release port on the LFD device. The device was incubated at room temperature for 10 min, during which the supernatant had migrated along the strip as indicated by the development of the control line (C) in the result window. The development of the Aspergillus specific test line (T) was determined in a period 10 to 20 min after the initial addition. Results were determined as positive or negative as indicated in Fig. 1. For an LFD to be considered positive, a clearly defined band across the entire result window was required. Each LFD device was independently checked by two users. Concurrent results were recorded, whereas incongruent results were ratified by a third user.
Fig 1.
Typical LFD results: strongly positive (a), weakly positive (b), and negative (c).
Statistical analysis.
Performance parameters for each assay were determined by construction of 2 × 2 tables with true positives being cases, with degree of certainty as defined by the EORTC/MSG criteria, and true negatives being controls, with no EORTC/MSG evidence of invasive fungal disease (IFD) (Table 1). For each proportion-based parameter, 95% confidence intervals were calculated (16). The significance of differences between parameters was determined by the generation of confidence intervals (16) and by using Fisher's exact test to generate two-sided P values, with a P value of <0.05 being considered significant. In addition to observed agreement, a kappa statistic was determined and interpreted as described previously (17). Values greater than 0.8 represented excellent agreement between tests, values of 0.61 to 0.8 represented substantial agreement, values of 0.41 to 0.6 represented moderate agreement, values of 0.21 to 0.4 represented fair agreement, and values below 0.2 represented poor agreement (17).
RESULTS
Sample positivity.
Assay positivity rates according to disease classification are shown in Table 2. For an assay to be diagnostically useful, it is essential that positivity in cases is significantly greater than false positivity in controls. When comparing proven/probable cases with no-IFD controls, positivity was significantly greater in cases than in controls for all three assays (GM ELISA difference: 21.3%, 95% CI of 13.4 to 28.9, P = 0.0001; PCR difference: 17.1%, 95% CI of 8.5 to 24.8, P = 0.0001; LFD difference: 20.9%, 95% CI of 13.7 to 27.9, P = 0.0001). There was a trend for greater GM positivity in proven/probable cases, but this was not significant. False positivity was greatest for PCR testing; again, the difference from the other tests was not significant (Table 2). PCR positivity was significantly greater than both GM and LFD positivity for combined proven/probable/possible IA cases and significantly greater than GM positivity for proven cases (Table 2).
Table 2.
Comparison of GM ELISA, Aspergillus real-time PCR, and Aspergillus LFD sample positivity in the different patient populationsa
| Patient population |
% assay sample positivitya (% difference; 95% CI; P) |
||
|---|---|---|---|
| GM vs PCR | GM vs LFD | PCR vs LFD | |
| Proven IA | 14.9 vs 28.2 (13.3; 0.43–26.1; 0.0501) | 14.9 vs 17.2 (2.3; −8.8–13.3; 0.8389) | 28.2 vs 17.2 (11.0; 24.0–−2.1; 0.1240) |
| Proven/probable IA | 31.0 vs 27.9 (3.1; 12.1–−6.0; 0.5050) | 31.0 vs 26.6 (4.4; 13.1–−4.4; 0.3807) | 27.9 vs 26.6 (1.3; 10.2–−7.6; 0.8192) |
| Proven/probable/possible IA | 17.8 vs 29.1 (11.3; 4.9–17.5; 0.0007) | 17.8 vs 20.1 (2.3; −3.5–8.0; 0.5019) | 29.1 vs 20.1 (9.0; 2.5–15.4; 0.0072) |
| No IFD | 9.7 vs 10.8 (1.1; −5.3–8.1; 0.7236) | 9.7 vs 5.7 (4.0; 9.9–−1.7; 0.2289) | 10.8 vs 5.7 (5.1; 11.6–−5.1; 0.1128) |
Please note that positivity in cases (proven/probable/possible IA) represents “true positivity” and positivity in controls (No IFD) represents “false positivity.” Significant differences are represented in bold text.
Sample concordance.
Observed sample concordance between antigen tests (LFD and GM ELISA) was moderate/fair, generating an observed agreement of 84.9% (416/490) and a kappa statistic of 0.436. Agreement between PCR and both antigen tests was poor, with observed agreement and kappa statistics of 69.4% (340/490)/0.026 and 72.7% (356/490)/0.118 for concordance with GM ELISA and LFD, respectively.
Patient concordance.
Considering a single positive result for each assay as significant, 72.7% of proven/probable cases were positive by all three tests and 81.8% were positive by at least two assays, leaving just four cases that were positive by only one test (Table 3). For the possible cases, GM negative by definition, 36.4% were positive by both PCR and LFD, 36.4% were positive by PCR alone, 18.2% were positive by LFD only, and 9.1% were negative by all tests. Of the 59 control patients, 6.8% were positive by both LFD and GM ELISA, 27.1% were positive by PCR alone, 11.9% were positive by GM ELISA alone, and 8.5% were positive only by LFD.
Table 3.
Assay concordance on a patient basis
| Test combination | No. of population positive by test or test combination |
|||||
|---|---|---|---|---|---|---|
| Proven/probable (n = 22) |
Possible (n = 22) |
No IFD (n = 59) |
||||
| Single positivea | Multiple positiveb | Single positive | Multiple positive | Single positive | Multiple positive | |
| GM, PCR, and LFD positive | 16 | 7 | 0 | 0 | 0 | 0 |
| GM and PCR positive | 1 | 3 | 0 | 0 | 0 | 0 |
| GM and LFD positive | 0 | 4 | 0 | 0 | 4 | 1 |
| PCR and LFD positive | 1 | 2 | 8 | 2 | 0 | 0 |
| GM positive | 0 | 1 | 0 | 0 | 7 | 4 |
| PCR positive | 3 | 3 | 8 | 11 | 16 | 2 |
| LFD positive | 1 | 0 | 4 | 1 | 5 | 0 |
| GM, PCR, and LFD negative | 0 | 2 | 2 | 8 | 27 | 52 |
“Single positive” represents a threshold where the minimum requirement for patient positivity is a single positive sample per assay.
“Multiple positive” represents a threshold where two or more positive samples are required per patient for an assay to be considered positive, and where more than one assay is combined, all assays must have multiple positive results.
If a multiple (≥2) positive threshold was required, 31.8% of cases were positive by all three tests, 72.7% were positive by at least two assays, and 18.2% were positive by just one assay (Table 3). For the possible cases, 63.6% were positive by PCR and/or LFD. Only 9.1% of proven/probable cases were negative by all three tests, compared to 88.1% of the control group.
Assay performance.
The performance parameters for each assay are shown in Table 4. Data for GM ELISA are affected by ascertainment bias since GM ELISA is an EORTC/MSG factor used to define disease, and this should be noted when comparing performance with PCR or the LFD assay. Despite this, overall performance as demonstrated by the diagnostic odds ratio (DOR) was inferior to that of both PCR and LFD (Table 4). PCR testing provided the highest sensitivity/negative predictive value (NPV), while the LFD provided the best specificity/positive predictive value (PPV), although neither was statistically significant. As shown by a negative likelihood ratio of 0.06, PCR can be used to exclude disease if consistently negative. For all assays, a requirement of two or more positive results improved the positive likelihood ratio and accuracy of ruling in a diagnosis of IA, generating >90% specificity.
Table 4.
Performance parameters for GM ELISA, Aspergillus real-time PCR, and Aspergillus LFD when testing cases of proven/probable IA (n = 22) versus no IFD (n = 59)
| Parameter | Positivity thresholda | Assayb |
||
|---|---|---|---|---|
| GM | PCR | LFD | ||
| % sensitivity (95% CI) | Single | 77.27 (56.6–89.9) | 95.45 (78.2–99.2) | 81.82 (61.5–92.7) |
| Multiple | 68.18 (47.3–83.6) | 68.18 (47.3–83.6) | 59.10 (38.7–76.7) | |
| % specificity (95% CI) | Single | 81.36 (69.6–89.3) | 72.88 (60.4–82.6) | 84.75 (73.5–91.8) |
| Multiple | 91.53 (81.7–96.3) | 96.61 (88.5–99.1) | 98.00 (91.0–99.7) | |
| % PPV (95% CI) | Single | 60.71 (42.4–76.4) | 56.76 (40.9–71.3) | 66.67 (47.8–81.4) |
| Multiple | 75.00 (53.1–88.8) | 88.24 (65.7–96.7) | 93.00 (68.5–98.7) | |
| % NPV (95% CI) | Single | 90.60 (79.8–95.9) | 97.73 (88.2–99.6) | 92.59 (82.5–97.1) |
| Multiple | 88.52 (78.2–94.3) | 89.06 (79.1–94.6) | 86.57 (76.4–92.8) | |
| Positive likelihood ratio | Single | 4.15 | 3.52 | 5.37 |
| Multiple | 8.05 | 20.11 | 29.55 | |
| Negative likelihood ratio | Single | 0.28 | 0.06 | 0.21 |
| Multiple | 0.35 | 0.33 | 0.42 | |
| DOR | Single | 14.82 | 58.67 | 25.57 |
| Multiple | 23.00 | 60.94 | 70.36 | |
“Single” represents a threshold where the minimum requirement for patient positivity is a single positive sample per assay; “multiple” represents a threshold where two or more positive samples per patient are required for an assay to be considered positive.
Optimal values for each parameter are shown in bold.
To compare GM ELISA directly with PCR and LFD, it was necessary to determine performance using a case population where GM ELISA was not used as a diagnostic factor, and this could be achieved in two ways: first, by comparing performance in histologically proven cases only (n = 8), and second, by using a population where a diagnosis is made based solely on specific clinical evidence and comparing assay performance in proven/probable/possible cases (n = 44), where current EORTC possible cases are given the same significance as that of previous probable cases. For proven cases, the sensitivity of PCR, LFD, and GM ELISA using a single positive threshold was 100% (95% CI, 67.6 to 100), 87.5% (95% CI, 52.9 to 97.8), and 75.0% (95% CI, 40.9 to 92.9), respectively, and for a multiple (≥2) positive threshold, the sensitivity was 75.0% (95% CI, 40.9 to 92.9) for PCR and 50.0% (95% CI, 21.5 to 78.5) for both LFD and GM ELISA. For a proven/probable/possible population, the sensitivity for PCR, LFD, and GM using a single positive threshold was 84.1% (95% CI, 70.6 to 92.1), 68.2% (95% CI, 53.4 to 80.0), and 38.6% (95% CI, 25.7 to 53.4), respectively. Using a multiple (≥2) positive threshold, the sensitivity was 63.6% (95% CI, 48.9 to 76.2), 36.4% (95% CI, 23.8 to 51.1), and 34.1% (95% CI, 21.9 to 48.9).
Combined assay performance.
Optimal performance was achieved by combining PCR with the LFD assay (Table 5). In doing so, a 100% sensitivity/NPV was attainable, allowing IA to be confidently excluded if both assays were consistently negative. Conversely, if both PCR and LFD assays were positive, then specificity/PPV was 100%, and disease could be accurately diagnosed. Combining PCR with GM ELISA also provided 100% specificity/PPV, but sensitivity/NPV was <100%. Combining the two antigen detection systems was the least successful approach, with performance comparable to that using the antigen assays individually and DOR values inferior to those generated by combining PCR with either antigen test (Tables 4 and 5). Combining all three tests did not improve performance further (results not shown).
Table 5.
Combined assay performance for GM ELISA, Aspergillus real-time PCR, and Aspergillus LFD when testing cases of proven/probable IA (n = 22) versus no IFD (n = 59)
| Parameter | Positivity threshold | Assay combinationa |
|||||
|---|---|---|---|---|---|---|---|
| PCR or LFD | PCR and LFD | PCR or GM | PCR and GM | GM or LFD | GM and LFD | ||
| % sensitivity (95% CI) | Single | 100 (85.1–100) | 77.3 (56.6–89.9) | 95.5 (78.2–99.2) | 77.3 (56.6–89.9) | 86.4 (66.7–95.3) | 72.7 (51.9–86.9) |
| Multiple | 86.4 (66.7–95.3) | 40.9 (23.3–61.3) | 90.9 (72.2–97.5) | 45.5 (26.9–65.3) | 77.3 (56.6–89.9) | 50.0 (30.7–69.3) | |
| % specificity (95% CI) | Single | 57.6 (44.9–69.4) | 100 (93.9–100) | 54.2 (41.7–66.3) | 100 (93.9–100) | 72.9 (60.4–82.6) | 93.2 (83.8–97.3) |
| Multiple | 94.9 (86.1–98.3) | 100 (93.9–100) | 88.1 (77.5–94.1) | 100 (93.9–100) | 91.5 (81.7–96.3) | 98.3 (91.0–99.7) | |
| % PPV (95% CI) | Single | 46.8 (33.3–60.8) | 100 (81.6–100) | 43.8 (30.7–57.7) | 100 (81.6–100) | 54.3 (38.2–69.5) | 80.0 (58.4–91.9) |
| Multiple | 86.4 (66.7–95.3) | 100 (70.1–100) | 74.1 (55.3–86.8) | 100 (72.3–100) | 77.3 (56.6–89.9) | 91.7 (64.6–98.5) | |
| % NPV (95% CI) | Single | 100 (89.9–100) | 92.2 (83.0–96.6) | 97.0 (84.7–99.5) | 92.2 (83.0–96.6) | 94.4 (82.5–97.8) | 90.2 (80.2–95.4) |
| Multiple | 94.9 (86.1–98.3) | 81.9 (71.5–89.1) | 96.3 (87.5–99.0) | 83.1 (72.7–90.1) | 91.5 (81.7–96.3) | 84.1 (73.7–90.9) | |
| Positive likelihood ratio | Single | 2.4 | >772.7 | 2.1 | >772.7 | 3.2 | 10.7 |
| Multiple | 17.0 | >409.1 | 7.7 | >454.5 | 9.1 | 29.6 | |
| Negative likelihood ratio | Single | <0.0017 | 0.23 | 0.08 | 0.23 | 0.19 | 0.29 |
| Multiple | 0.14 | 0.59 | 0.10 | 0.55 | 0.25 | 0.51 | |
| DOR | Single | >1,388.2 | >3,359.6 | 26.1 | >3,359.6 | 16.7 | 37.0 |
| Multiple | 121 | >693.4 | 76.7 | >826.4 | 36.5 | 58.0 | |
For the assay combinations, “or” indicates that at least one of the assays is required to be positive, whereas “and” indicates that both assays are required to be positive before a patient is considered positive. Single and multiple positivity thresholds are as defined in Table 4.
DISCUSSION
Diagnosis of IA remains challenging partially due to a clinical presentation that is frequently nonspecific but also due to a limited range of accepted diagnostic assays with variable performance. The difficulty of PCR is further compounded by limited methodological standardization and commercial interest. The development of a novel methodologically simple assay is beneficial, provided that clinical validity is satisfactory. The LFD investigated here provides a high degree of accuracy as the MAb, JF5, used in the LFD is highly specific for Aspergillus species and does not cross-react with other clinically relevant fungi, including agents of candidiasis, fusariosis, scedosporiosis, or mucormycosis (11). Furthermore, the LFD is easy to perform with minimal training and is quick, taking 10 to 15 min to complete.
Evaluation of the LFD conducted here showed that it had a clinical performance that was comparable to that of other biomarker assays and, when combined with PCR, could be used to exclude or confirm disease. The simplicity of LFDs provides a potential POC approach, to be performed outside the laboratory, by health care workers directly caring for the patient. However, in this study it was necessary to heat treat serum samples to optimize performance, and that would prevent a POC approach. The same is true for the GM ELISA, where it is necessary to dissociate immune complexes and precipitate circulating proteins to improve detection. In the development of this LFD, the analytical sensitivity was shown to be inferior in the presence of serum proteins (1.25 ng/ml versus 35 ng/ml) (11). While this pretreatment may prevent its use as a POC test, the rapid processing time means routine diagnostic testing runs are unnecessary and urgent testing can be undertaken. The LFD could be used as an adjunct test to provide an interim rapid result.
Diagnosis of IA remains multifaceted. Interpretation of the LFD result is qualitative and subjective. In this study, interpretation through consensus was applied, but this may not be possible in diagnostic settings. To be considered positive, a defined line across the entire result window was required; subjectivity may be difficult to apply without prior experience, and variability in interpretation could lead to incorrect results. Changing the positivity threshold to any coloration within the Aspergillus-specific test line provided sensitivity and specificity values of 81.8% (18/22) and 86.4% (51/59), respectively, for proven/probable cases with use of a 10-min incubation time. Using this “all-or-nothing” approach simplifies interpretation, and when retrospectively applied to this study, it did not affect sensitivity/specificity, although overall sample positivity was reduced. Subjectivity can be eliminated by using hand-held densitometers that measure the color intensity of the test line and allow the establishment of assay detection thresholds (18). To provide optimal performance, combining the LFD with another test is recommended. Results in Table 5 indicate that combining the LFD with PCR provides the best strategy, and this mirrors previous recommendations to combine PCR and GM ELISA (6). In this study, a combined PCR-ELISA strategy also provided excellent clinical performance.
Interestingly, sample concordance between PCR and both antigen tests is poor, whereas that between antigen tests is fair. This individual sample discordance provides overall superior performance per patient. In vivo, molecular and antigen biomarkers have different timings and mechanics of release associated with disease progression. Testing for both increases the opportunity for temporal disease detection.
A previous study has shown that GM is released during active growth of Aspergillus fumigatus, whereas DNA is released through hyphal damage autolysis or host defenses and will not be released concomitantly with GM (19). The extracellular glycoprotein targeted by the LFD is also released during active hyphal growth, and greater concordance with GM positivity could be predicted (11). It is important to consider when performing PCR on WB that the protocol used in this study does not target free DNA (DNAemia), and yet it still generates positive results that coincide with IA. Positivity is hypothesized to be linked with the presence of the organism or fragments of organism rather than biomarkers released during growth. It is unlikely that WB PCR is detecting a true “fungemia,” as the organism is rarely isolated from the bloodstream (20). As disease progresses via angioinvasion and dissemination, nonviable and/or phagocytosed fungal cells may be detectable by PCR. In this scenario, positivity would be relatively random and transient but dependent on the degree of angioinvasion.
Diagnosis of IA using the revised EORTC/MSG criteria places emphasis on specific radiological signs (nodules, halos, and cavities), and mycology is used as supportive evidence (1). This leaves a category of patients (possible IA) for whom diagnosis is unclear, and where radiological signs could represent other conditions potentially of noninfective origin (21). This highlights the importance of specific mycological tests, particularly as the radiological signs may be transient (22). Of the 22 possible cases in this study, all, by definition, were negative by GM ELISA. However, 16 were positive by PCR and 12 were positive by LFD, eight being positive by both tests. Only two possible cases were negative by all three tests and could represent other etiologies. It could be argued that 20 possible cases are GM ELISA false-negative results. If LFD and PCR results are given the same status as that of GM ELISA, then the sensitivity of PCR and LFD is significantly greater than that of the GM ELISA.
The LFD evaluated here is currently undergoing design optimization, scale-up, and United Kingdom validation of the FDA-approved manufacturing process, as necessary prerequisites for European CE marking of the test as a medical in vitro diagnostic (IVD) test in April 2013. Sales of the CE-marked test for use in the European Union (EU) will commence in May 2013. The CE-marked device will then be submitted to the U.S. National Institutes of Health Aspergillus Technology Consortium (AsTec) for evaluation of the LFD as a diagnostic test for IA in the United States. The AsTec application process was initiated in December 2012, and the Application Review Committee has undertaken its preliminary review of the application and supporting materials. Should AsTec consent to evaluate the CE-marked device be granted, we envisage the U.S. evaluation process being completed by November 2013. FDA approval for use of the device as a diagnostic test for IA in the United States will then be sought. During the AsTec evaluation and FDA approval process, the CE-marked LFD will be available to U.S. investigators for exploratory studies, from Christopher Thornton, Chief Executive Officer of ISCA Diagnostics Limited, a University of Exeter spinoff company established for commercialization of the Aspergillus LFD.
In conclusion, the development of an additional assay targeting a surrogate biomarker specific to IA is beneficial. Even in proven/probable cases, sample positivity rates may be low (23), but frequent testing for multiple different biomarkers may overcome this. With provision of a methodologically simple assay, the prohibitive limitations of molecular techniques that require the use of specialist equipment and expertise may be avoided. It is likely that the most appropriate use of the LFD would be to combine it with molecular detection, thereby minimizing its main limitation, the subjective interpretation of test results.
ACKNOWLEDGMENTS
Regarding conflicts of interest, P.L.W. is a founding member of the EAPCRI, received project funding from Myconostica and Luminex, and was sponsored to attend international meetings by Myconostica and Gilead Sciences. C.P. has no conflicts of interest. C.T. developed the lateral-flow device. R.A.B. is a founding member of the EAPCRI; received an educational grant and scientific fellowship award from Gilead Sciences and Pfizer; is a member of the advisory board and speakers bureau for Gilead Sciences, MSD, Astellas, and Pfizer; and was sponsored to attend international meetings by Gilead Sciences and Pfizer.
Footnotes
Published ahead of print 13 March 2013
REFERENCES
- 1. De Pauw B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, Pappas PG, Maertens J, Lortholary O, Kauffman CA, Denning DW, Patterson TF, Maschmeyer G, Bille J, Dismukes WE, Herbrecht R, Hope WW, Kibbler CC, Kullberg BJ, Marr KA, Muñoz P, Odds FC, Perfect JR, Restrepo A, Ruhnke M, Segal BH, Sobel JD, Sorrell TC, Viscoli C, Wingard JR, Zaoutis T, Bennett JE, European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group, National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin. Infect. Dis. 46:1813–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Karageorgopoulos DE, Vouloumanou EK, Ntziora F, Michalopoulos A, Rafailidis PI, Falagas ME. 2011. β-d-Glucan assay for the diagnosis of invasive fungal infections: a meta-analysis. Clin. Infect. Dis. 52:750–770 [DOI] [PubMed] [Google Scholar]
- 3. Leeflang MM, Debets-Ossenkopp YJ, Visser CE, Scholten RJ, Hooft L, Bijlmer HA, Reitsma JB, Bossuyt PM, Vandenbroucke-Grauls CM. 2008. Galactomannan detection for invasive aspergillosis in immunocompromised patients. Cochrane Database Syst. Rev. 8(4):CD007394 doi:10.1002/14651858.CD007394 [DOI] [PubMed] [Google Scholar]
- 4. Maertens J, Theunissen K, Lagrou K. 2010. Galactomannan testing, p 105–124 In Pasqualotto AC. (ed), Aspergillosis: from diagnosis to prevention, 1st ed Springer Publishers, London, United Kingdom [Google Scholar]
- 5. Mengoli C, Cruciani M, Barnes RA, Loeffler J, Donnelly JP. 2009. Use of PCR for diagnosis of invasive aspergillosis: systematic review and meta-analysis. Lancet Infect. Dis. 9:89–96 [DOI] [PubMed] [Google Scholar]
- 6. Barnes RA, White PL, Bygrave C, Evans N, Healy B, Kell J. 2009. Clinical impact of enhanced diagnosis of invasive fungal disease in high-risk haematology and stem cell transplant patients. J. Clin. Pathol. 62:64–69 [DOI] [PubMed] [Google Scholar]
- 7. White PL, Bretagne S, Klingspor L, Melchers WJ, McCulloch E, Schulz B, Finnstrom N, Mengoli C, Barnes RA, Donnelly JP, Loeffler J, European Aspergillus PCR Initiative 2010. Aspergillus PCR: one step closer to standardization. J. Clin. Microbiol. 48:1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. White PL, Mengoli C, Bretagne S, Cuenca-Estrella M, Finnstrom N, Klingspor L, Melchers WJ, McCulloch E, Barnes RA, Donnelly JP, Loeffler J, European Aspergillus PCR Initiative (EAPCRI) 2011. Evaluation of Aspergillus PCR protocols for testing serum specimens. J. Clin. Microbiol. 49:3842–3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Torelli R, Sanguinetti M, Moody A, Pagano L, Caira M, De Carolis E, Fuso L, De Pascale G, Bello G, Antonelli M, Fadda G, Posteraro B. 2011. Diagnosis of invasive aspergillosis by a commercial real-time PCR assay for Aspergillus DNA in bronchoalveolar lavage fluid samples from high-risk patients compared to a galactomannan enzyme immunoassay. J. Clin. Microbiol. 49:4273–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. White PL, Perry MD, Moody A, Follett SA, Morgan G, Barnes RA. 2011. Evaluation of analytical and preliminary clinical performance of Myconostica MycAssay Aspergillus when testing serum specimens for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 49:2169–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thornton CR. 2008. Development of an immunochromatographic lateral-flow device for rapid serodiagnosis of invasive aspergillosis. Clin. Vaccine Immunol. 15:1095–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wiederhold NP, Thornton CR, Najvar LK, Kirkpatrick WR, Bocanegra R, Patterson TF. 2009. Comparison of lateral flow technology and galactomannan and (1→3)-beta-d-glucan assays for detection of invasive pulmonary aspergillosis. Clin. Vaccine Immunol. 16:1844–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hoenigl M, Koidl C, Duettmann W, Seeber K, Wagner J, Buzina W, Wölfler A, Raggam RB, Thornton CR, Krause R. 2012. Bronchoalveolar lavage lateral-flow device test for invasive pulmonary aspergillosis diagnosis in haematological malignancy and solid organ transplant patients. J. Infect. 65:588–591 [DOI] [PubMed] [Google Scholar]
- 14. White PL, Linton CJ, Perry MD, Johnson EM, Barnes RA. 2006. The evolution and evaluation of a whole blood polymerase chain reaction assay for the detection of invasive aspergillosis in hematology patients in a routine clinical setting. Clin. Infect. Dis. 42:479–486 [DOI] [PubMed] [Google Scholar]
- 15. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55:611–622 [DOI] [PubMed] [Google Scholar]
- 16. Newcombe RG. 1998. Improved confidence intervals for the difference between binomial proportions based on paired data. Stat. Med. 17:2635–2650 [PubMed] [Google Scholar]
- 17. Viera AJ, Garrett JM. 2005. Understanding interobserver agreement: the kappa statistic. Fam. Med. 37:360–363 [PubMed] [Google Scholar]
- 18. Faulstich K, Gruler R, Eberhard M, Lentzsch D, Haberstroh K. 2009. Handheld and portable reader devices for lateral flow immunoassays, p 157–183 In Wong RC, Tse HY. (ed), Lateral flow immunoassay. Humana Press, New York, NY [Google Scholar]
- 19. Mennink-Kersten MA, Ruegebrink D, Wasei N, Melchers WJ, Verweij PE. 2006. In vitro release by Aspergillus fumigatus of galactofuranose antigens, 1,3-beta-d-glucan, and DNA, surrogate markers used for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 44:1711–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morton CO, Loeffler J, De Luca A, Frost S, Kenny C, Duval S, Romani L, Rogers TR. 2010. Dynamics of extracellular release of Aspergillus fumigatus DNA and galactomannan during growth in blood and serum. J. Med. Microbiol. 59:408–413 [DOI] [PubMed] [Google Scholar]
- 21. Green R, Shibuya K, Ando T. 2009. Histology and radiology, p 373–390 In Latge J-P, Steinbach WJ. (ed), Aspergillus fumigatus and aspergillosis. ASM Press, Washington, DC [Google Scholar]
- 22. Caillot D, Couaillier JF, Bernard A, Casasnovas O, Denning DW, Mannone L, Lopez J, Couillault G, Piard F, Vagner O, Guy H. 2001. Increasing volume and changing characteristics of invasive pulmonary aspergillosis on sequential thoracic computed tomography scans in patients with neutropenia. J. Clin. Oncol. 19:253–259 [DOI] [PubMed] [Google Scholar]
- 23. Verweij PE. 2005. Advances in diagnostic testing. Med. Mycol. 43:S121–S124 [DOI] [PubMed] [Google Scholar]