Abstract
The lack of complete genome sequence information for Mycobacterium caprae complicates a robust differentiation of M. caprae and Mycobacterium bovis. In this study, the presence or absence of M. caprae-specific single nucleotide polymorphisms in lepA and gyrB genes was assessed. The region of difference 4 (RD4) was analyzed for the identification and characterization of M. caprae. Molecular characteristics were evaluated in 12 recent M. caprae isolates from livestock and wildlife collected over a 3-year period in Bavaria, Germany. Conventional PCR strategies, sequence analysis of PCR fragments, and data from a next-generation sequencing approach together with variable-number tandem-repeat genotyping were utilized. Single nucleotide polymorphisms in the lepA and gyrB genes indicating the presence of M. caprae were detected in all the isolates. At least three different RD4 variants were found for Alpine M. caprae isolates. The results demonstrate that the RD4 region is rather heterogeneous in M. caprae genomes. As assumed by others, the presence of RD4 is critical for PCR-based differentiation of M. caprae from M. bovis, but in addition, the observed variability of RD4 allows the identification of M. caprae genotypes and may be indicative of a geographical-type appearance.
INTRODUCTION
Mycobacterium caprae was first described in samples from goats in Spain (1) and was established in 2003 as a unique species in the Mycobacterium tuberculosis complex (MTC) (2) based on the presence or absence of regions of difference (RDs) and single nucleotide polymorphisms (SNPs) (3). M. bovis and M. caprae are the most relevant pathogens for livestock and wildlife large animal hosts. In some countries bovine tuberculosis (TB) is still a problem, particularly when a wildlife reservoir exists and TB transmission from wildlife to cattle and cattle to wildlife on the pasture is a permanent danger (4). Three wildlife reservoirs of TB caused by M. bovis have been described extensively (4–7). To date a wildlife reservoir harboring M. caprae has not been described. However, there have been several reports about M. caprae infections in red deer (Cervus elaphus) (8–12) and wild boars (Sus scrofa) (8, 9, 13) from continental European countries, mainly from Spain (8, 9) and from countries with Alpine wildlife habitats (10–13).
During the years 2007 and 2012 in the Bavarian Alpine “Allgäu” region, an area with plenty of cattle summer pastures also habituated by red deer, infections in cattle have been found to be mainly caused by M. caprae, whereas M. bovis was more frequent among animals found in the northern part of Germany, according to international unpublished findings of the national animal infectious disease reporting system, TierSeuchenNachrichten 3.0 (TSN 3.0).
The majority of molecular strategies for M. caprae differentiation refer to sporadically examined M. caprae isolates (3, 14, 15). In addition, a complete genome sequence of M. caprae is not available in the NCBI GenBank so far, in contrast to genome sequences of M. tuberculosis and M. bovis. Comparative genomic studies with the complete DNA sequence of M. tuberculosis strain H37Rv have identified several Mycobacterium tuberculosis-specific genomic RDs which have been deleted in most other members of the MTC (14, 15). M. caprae isolates displayed the same RD deletions as M. bovis strains except for RD4, a 12.7-kb-spanning genomic region that is absent in M. bovis but present in M. caprae (14). However, to our knowledge, only very few M. caprae isolates, including most isolates originating from Spain, have been tested with regard to the presence of the complete RD4 sequence; 4 M. caprae isolates originate from Spain (14, 16) and 10 isolates are without detailed origin information (15). Rodriguez et al. (8) confirmed the existence of 545-bp RD4-specific sequences in 62 of 63 M. caprae isolates. Notably, the detection of an RD4 deletion has been used as a major criterion for distinguishing M. bovis from the other members of the MTC, especially from M. caprae (16–21).
One M. caprae-specific T-to-G substitution at position 1311 in the gyrB sequence has been described in several M. caprae isolates from central Europe (12, 22, 23) and Spain (3, 8). However, Kubica et al. (24) reported that 3 of 55 German M. caprae isolates from human patients did not show this M. caprae-specific SNP. To date, the only commercially available DNA strip-based assay for the differentiation of MTC species, the GenoType MTBC line probe assay (Hain Lifescience GmbH, Nehren, Germany), takes advantage of this SNP to differentiate M. caprae from other members of the MTC (25).
Reddigton et al. (26) described a novel M. caprae-specific C-to-T substitution at position 690 in the M. tuberculosis H37Rv lepA gene, which was conserved in all five tested M. caprae isolates originating from Germany and the Netherlands.
However, little is known about the genomic features of M. caprae concerning markers for genotyping. Rapid and robust diagnostic differentiation of M. caprae is complicated by this fact.
We used different PCR protocols to determine whether genetic characteristics can be found to accurately differentiate M. caprae from the other MTC members. These PCR protocols were sequence analysis of PCR fragments and two standard PCR-based genotyping methods, spoligotyping (27) and mycobacterial interspersed repetitive-unit-variable-number tandem-repeat (MIRU-VNTR) typing (28, 29). We assessed the presence or absence of M. caprae-specific SNPs in lepA, gyrB gene, and RD4 sequences in the genomes of 12 recent Alpine M. caprae isolates from livestock and wildlife. In addition, we compared the results of a next-generation sequencing approach used to analyze DNA from seven M. caprae isolates. PCR and whole-genome sequencing revealed that three different RD4 variations exist for M. caprae isolates originating from delimited Alpine regions of Bavaria.
MATERIALS AND METHODS
Bacterial strains and DNA preparation.
A total of 12 M. caprae isolates from domestic and wildlife hosts in different parts of Bavaria, Germany, were analyzed (Table 1). For standardization of PCR setups, five isolates of MTC and seven isolates from nontuberculosis mycobacteria (NTM) species were used (Table 1). Clinical isolates were characterized using GenoType MTBC, GenoType mycobacterium AS, and GenoType mycobacterium CS (all from Hain Lifescience GmbH). The DNA of the NTM strains was obtained from the strain collection of the Bavarian Health and Food Safety Authority, Oberschleissheim, Germany.
Table 1.
Origins of Mycobacterium DNA samples
| Species | Isolate identification no. | Host, yr of origina |
|---|---|---|
| M. caprae | 09/RW044 | Red deer isolate, 2009 |
| M. caprae | 10/RW314 | Red deer isolate, 2010 |
| M. caprae | 10/RW079 | Red deer isolate, 2010 |
| M. caprae | 11/D028 | Red deer isolate, 2011b |
| M. caprae | 11/D133 | Red deer isolate, 2011b |
| M. caprae | 11/D102 | Red deer isolate, 2011b |
| M. caprae | 11/Roe010 | Roe deer isolate, 2011b |
| M. caprae | V-09-425 | Cattle isolate, 2009 |
| M. caprae | D6431 | Cattle isolate, 2009 |
| M. caprae | 11/Rd152461 | Cattle isolate, 2011 |
| M. caprae | 12/Rd52799 | Cattle isolate, 2012 |
| M. caprae | V-09-274 | Sheep isolate, 2009 |
| Other MTC members | ||
| M. tuberculosis | ATCC 25177 | NA |
| M. africanum | TT70/03 | Nilgai isolate, 2004 |
| M. bovis | V207 | Cattle isolate, 2009 |
| M. bovis | DSM 43990 | NA |
| M. bovis BCG | ATCC 27289 | NA |
| NTM members | ||
| M. arupense | V-09-1049 | Red deer isolate, 2009 |
| M. bohemicum | V-09-1241 | Red deer isolate, 2009 |
| M. vanbaalenii | RW444 | Red deer isolate, 2010 |
| M. nonchromogenicum | RW712 | Red deer isolate, 2010 |
| M. interjectum | RW871 | Red deer isolate, 2010 |
| M. avium subsp. paratuberculosis | 10016571500101 | Cattle isolate, 2010 |
| M. avium subsp. paratuberculosis | 10016792400101 | Cattle isolate, 2010 |
All the isolates originated in Bavaria, Germany. NA, not applicable.
Isolates were obtained during the Tuberculosis in Alpine Wildlife project, carried out by 17 partners of the four Alpine countries (Austria, Germany, Switzerland, and Italy) and funded by their respective national research funding organizations and supported by the EMIDA ERA-Net (http://tb-alpine-wildlife.org).
MTC isolates were grown in liquid medium (Middlebrook 7H9 broth; Becton Dickson, Heidelberg, Germany) supplemented with 800 μl polymyxin B, amphotericin B, nalidixic acid, trimethoprim, and azlocillin (BBL MGIT PANTA) (Becton Dickinson) and were incubated at 37°C for 4 weeks. After two washing steps and heat inactivation (30 min at 95°C), genomic DNA from MTC isolates for PCR and whole-genome sequencing was extracted from 1.5 ml liquid culture using the cetyltrimethylammonium bromide method and purification with phenol-chloroform. DNA concentrations were determined with a Nanodrop ND-1000 device (Thermo Fisher Scientific, Schwerte, Germany).
Molecular typing.
Two PCR-based genotyping methods were applied for the molecular characterization of M. caprae isolates, spoligotyping (27) and MIRU-VNTR typing (28, 29). The latter determines the copy numbers in 24 or 25 VNTR loci and is regarded as the actual gold standard for comparison of MTC strains (29). DNA for PCR amplification was obtained by the boiled-prep method (11). Spoligotyping was performed as described by Kamerbeek et al. (27). MIRU-VNTR typing was carried out using single PCRs for each locus and DNA fragments were analyzed by gel electrophoresis, as described earlier (11). Fingerprint patterns were digitally compared using BioNumerics software version 6.6 (Applied Maths, Sint-Martens-Latem, Belgium).
Whole-genome sequencing.
Seven isolates of M. caprae were sequenced on a MiSeq-Illumina instrument with paired-end sequencing and a read length of 150 bp in each direction. For the isolates 09/RW044, 10/RW079, 11/D028, and 11/D102, sequencing libraries were prepared from 100 ng of fragmented DNA sonicated with a Bioruptor (NextGen, Diagenode, Liege, Belgium) (for 25 cycles with 30 s on/30 s off) and the Rapid Library preparation kit (NuGen Inc., San Carlos, CA) and were amplified by 6 cycles of PCR with Illumina P5 and P7 primers (P5, AATGATACGGCGACCACCGA; P7, CAAGCAGAAGACGGCATACGA). After the PCR step, libraries were purified with AMPure XP beads (Beckman Coulter, Brea, CA) and quantified with a Bioanalyzer (Agilent, Santa Clara, CA). From the isolates 11/Rd152461, 12/Rd52759, and 11/Roe010, respectively, 1 ng of AMPure XP-purified DNA was used to prepare fragmented libraries with the XT kit (Illumina, San Diego, CA) according to the manufacturer's instructions.
The raw sequencing data were imported to a locally installed instance of the Galaxy platform (30). Raw reads were demultiplexed, adapter and quality trimmed, and mapped to the M. tuberculosis reference genome H37Rv (GenBank database, accession number CP003248) with the program BWA (31). Regions of interest were inspected on the Integrative genomics viewer (IGV), a lightweight visualization tool that enables intuitive real-time exploration of diverse, large-scale genomic data sets on standard desktop computers (32) in order to find large deletions.
Primer and probe design.
The primers and the probe are listed in Table 2. Nucleotide sequence data for the primers and the probe were deduced from various reports (14, 16, 18, 26, 33) or were generated in this study using the NCBI/Primer-BLAST program (http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome) based on the sequences derived from whole-genome sequencing analysis.
Table 2.
Probe and primers for lepA specific real-time PCR and primers for PCR detection of the gyrB gene and RD4 region
| Primer | PCR type | Nucleotide sequence (5′ to 3′) | Reference no. |
|---|---|---|---|
| MTC_Fw | lepA-based species specific | AGA CCG TGC GGA TCT TG | 26 |
| MTC_Rv | lepA-based species specific | CAT GGA GAT CAC CCG TGA | 26 |
| M. caprae-specific probe | lepA-based species specific | FAM TAT CGG GTA CAC AAA GAC GA-BHQ1 | 26 |
| MTUBf | gyrB-based species specific | TCG GAC GCG TAT GCG ATA TC | 33 |
| MTUBr | gyrB-based species specific | ACA TAC AGT TCG GAC TTG CG | 33 |
| RD4F | RD4 internal left part | CCA CGA CTA TGA CTA GGA CAG CAA | 18 |
| RD4R | RD4 internal left part | AAG AAC TAT CAA TCG GGC AAG ATC | 18 |
| Rv1510F | RD4 internal central part | GTG CGC TCC ACC CAA ATA GTT GC | 16 |
| Rv1510R | RD4 internal central part | TGT CGA CCT GGG GCA CAA ATC AGT C | 16 |
| RD4-flankF | RD4 5-kb deletion flanking PCR | CTC GTC GAA GGC CAC TAA AG | 14 |
| RD4-5kbDelR | RD4 5-kb deletion flanking PCR | TTC GTT AGC CGC ACA TCC ATG A | |
| RD4-DelF | RD4 38-kb deletion flanking PCR | GAA GGA AGC GGC TGC GAT TGG TGC | |
| RD4-DelR | RD4 38-kb deletion flanking PCR | TGG TTA CAC GCT CGG CGC AGT CA |
All primers and probes were purchased from Primer-Management (Eurofins MWG Operon, Ebersberg, Germany).
Real-time PCR.
The reactions were run in a Stratagene Mx3005P real-time thermocycler (Agilent Technologies, Waldbronn, Germany). Thermocycling, fluorescent data collection, and data analysis were performed with MxProTM quantitative PCR (qPCR) software (Agilent Technologies).
lepA-based species-specific real-time PCR for M. caprae.
Each reaction consisted of 1 U Taq DNA polymerase recombinant (Invitrogen, Life Technologies, Darmstadt, Germany), 1× PCR buffer, 5 mM MgCl2, 1 mM deoxynucleoside triphosphate (dNTP), 450 nM forward (MTC_Fw) and reverse (MTC_Rv) primers, 125 nM M. caprae-specific probe, 0.5 μl of internal control (IC)-primer-probe mix (2.5 pmol primer/μl plus 1.25 pmol probe/μl), 1 μl ruminant IC DNA sample (200 ng/μl), 5 μl of template (0.1 to 0.3 ng/μl), and RNase-free water to a final volume of 25 μl. The IC for the detection of the beta-actin mRNA was added to each setup in order to assess Taq polymerase inhibition (34). The cycling parameters were: 5 min incubation at 95°C and 45 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 30 s.
Conventional PCR.
PCR amplifications were performed in a T3000 thermocycler (Biometra, Goettingen, Germany). The PCR-amplified DNA fragments were separated by electrophoresis in 0.8 to 2% agarose gels and visualized under UV after ethidium bromide staining.
lepA-based species-specific conventional PCR for the MTC.
The PCR mixture was prepared with 0.05 U of HotStarTaq DNA polymerase (Qiagen, Hilden, Germany), 1× reaction buffer, 1.25 mM MgCl2, 0.2 mM dNTP, 0.5 μM each primer (MTC_Fw/MTC_Rv), 1 μl template, and RNase-free water to 50 μl per reaction to amplify a 155-bp lepA fragment. The cycling parameters were denaturation for 15 min at 95°C, 35 cycles of 95°C for 1 min, 60°C for 30 s, and 72°C for 30 s and a final elongation step of 10 min at 72°C (program 1).
gyrB-based species-specific PCR for the MTC.
Primers MTUBf/MTUBr were used to amplify a 1,020-bp gyrB fragment that comprises the discriminative regions from six M. caprae isolates. The PCR mixture contained 0.05 U of Phusion high-fidelity DNA polymerase (Biozym Scientific GmbH, Hessisch Oldendorf, Germany), 1× reaction buffer, 0.2 mM dNTP, 0.5 μM each primer, 1 μl template, and RNase-free water to 50 μl. The cycling parameters were denaturation for 30 s at 98°C, 35 cycles of 10 s at 98°C and 30 s at 72°C, and a final elongation step of 10 min at 72°C.
RD4-based species-specific PCR for MTC detection.
Primers chosen to amplify an internal fragment of RD4 (Fig. 1) were used in two PCRs with DNA from all M. caprae isolates. The two fragments that were amplified applied to the left and central parts of RD4 of the M. tuberculosis H37Rv genome in the 5′-to-3′ direction.
Fig 1.
Screenshot of the mapping of the M. caprae whole-genome sequencing data to the M. tuberculosis reference genome. RD4 nucleotide sequence variations for M. caprae isolates from red deer living in the Alpine region of Bavaria, Germany, are shown. The graph was constructed by the Integrative Genomics Viewer (IGV) using Illumina MiSeq paired-end reads of isolate numbers 09/RW044, 10/RW079, and 11/D102. RD4 is represented as a bold black line. The gray histograms indicate the sequence coverage, and the small gray bars indicate individual reads mapped to the corresponding genomic positions. Absence of coverage indicates the respective deletions (marked by solid lines in the histograms) in the RD4 region that were confirmed by PCR with primers (positions shown by boxes and arrows) flanking the deletions and yielding product sizes (shown between the arrows) consistent with the positions and sizes of the respective deletions.
For the RD4 internal left part, an 88-bp fragment of the left part of RD4 (Fig. 1) was amplified in a 50-μl reaction mixture containing 0.05 U of HotStarTaq DNA polymerase (Qiagen), 1× reaction buffer, 1 mM MgCl2, 0.2 mM dNTP, 0.5 μM each primer (RD4F/RD4R), 1 μl template, and RNase-free water (PCR mixture 1). The cycling parameters were those for program 1 but with an annealing of 55°C for 30 s.
For the RD4 internal central part, a 1,031-bp fragment of the central part of RD4 (Fig. 1.) was amplified using PCR mixture 1 but with 1× Q solution and primer Rv1510F/Rv1510R. The cycling parameters were those for program 1 but with an annealing of 60°C for 1 min and an extension step of 72°C for 1 min.
RD4 5-kb deletion-flanking PCR.
Previous results from contig sequences in the next-generation sequencing approach suggested a 5-kb deletion present in the left part of the RD4 sequence in some M. caprae genomes. Hence, flanking primers specific to the deletion were chosen for a PCR setup with DNA from the M. caprae isolates (Fig. 1). To amplify a calculated DNA fragment of approximately 700 bp, we used PCR mixture 1 but with 1× Q solution and primer (RD4-flankF/RD4-5kbDelR). Instead of dGTP, 7-deaza-2′-deoxy-GTP (Roche, Mannheim, Germany) for GC-rich templates was used. The cycling parameters were program 1 but with an annealing step of 55°C for 1 min and an extension step of 72°C for 1 min.
In addition, PCR mixture 1 but with 1× Q solution and primer (RD4-flankF/Rv1510R) was used for amplification of a 2.5-kb deletion-flanking fragment from the M. caprae isolates 11/D028 and 10/RW079. The cycling parameters were program 1 but with an annealing step of 55°C for 1 min and an extension step of 72°C for 2 min.
For amplification of the 6.9-kb DNA fragment that comprised the left and central parts of the RD4 sequence in M. caprae isolates without deletion, the PCR master mix contained 0.05 U of Phusion high-fidelity DNA polymerase (Biozym Scientific, GmbH), 1× GC buffer, 0.2 mM dNTP, 0.5 μM each primer (RD4-flankF/Rv1510R), 1.5 μl dimethyl sulfoxide (DMSO), 1 μl template, and RNase-free water to 50 μl. The cycling parameters were denaturation for 1 min at 98°C followed by 35 cycles of 10 s at 98°C, 30 s at 65°C, and 260 s at 72°C and a final elongation step of 10 min at 72°C.
RD4 38-kb deletion-flanking PCR.
The results of next-generation sequencing suggested that a 38-kb deletion spanning the whole RD4 region might be present in some M. caprae genomes (Fig. 1). Hence, flanking primers adjacent to the suspected deletion were designed for amplification of a small approximately 250-bp fragment representing flanking sequences. PCR mixture 1 was used but with 1× Q solution and primer (PDelF/PDelR). The cycling parameters were program 1 but with an annealing step of 55°C for 1 min and an extension step of 72°C for 1 min.
Sequencing and sequence analysis.
DNA fragments were prepared for sequencing by the QIAquick gel extraction kit (Qiagen) and sequencing was performed by Eurofins MWG Operon. The obtained sequences were compared with each other and with genomic sequences of members of the MTC, available from the genome database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/genome/browse/). The sequence data were aligned by using the Clustal W2 program (http://www.ebi.ac.uk/Tools/msa/clustalw2/) (35).
Nucleotide sequence accession numbers.
Sequence reads obtained from genomic sequencing of the M. caprae isolates RW/044, RW/079, 11/D028, and 11/D102 have been deposited in SRA (http://trace.ncbi.nlm.nih.gov/Traces/sra/). Bam files of these isolates mapped to H37Rv are available under the accession numbers SRS386245, SRS386249, SRS386254, and SRS386255. The obtained PCR sequences of the 5-kb and 38-kb deletion-flanking regions and the fragment of the gyrB gene amplification were deposited at NCBI GenBank under the accession numbers listed in Table 3.
Table 3.
RD4 mapping and genotypes of 12 representative M. caprae isolates from southern Bavaria and accession numbers of amplified gyrB and RD4 deletion-adjacent sequences
| Isolate no. | PCR result |
Accession no. |
Genotyping |
Type | |||
|---|---|---|---|---|---|---|---|
| RD4 internal left portion | RD4 internal central portion | Amplified gyrB sequencea | RD4 deletion-adjacent sequence | MIRU-VNTR codeb | Spoligotypec | ||
| 11/D028 | − | + | d | 2364212535222534133363322 | SB2174 | Karwendel | |
| 10/RW079 | − | + | JX420304 | KC007398d,e | 2364212535222534133363322 | SB2174 | Karwendel |
| V-09-425 | − | + | d | 2364212535222534133363322 | SB2174 | Karwendel | |
| 11/D133 | − | + | d | 2364212535222534133363322 | SB2174 | Karwendel | |
| V-09-274 | − | − | JX420311 | 2264242433224534132553323 | SB0418 | Lechtal | |
| 11/D102 | − | − | JX308291 | JX420308 | 2264242433224534132553323 | SB0418 | Lechtal |
| 11/Roe010 | − | − | JX420306 | JX420309 | 2264242433224534132553323 | SB0418 | Lechtal |
| 12/Rd52799 | − | − | JX420307 | JX420310 | 2264242433224534132553323 | SB0418 | Lechtal |
| 09/RW044 | + | + | JX420305 | 2354242534224534122243322 | SB0418 | Allgäu | |
| 10/RW314 | + | + | JX420303 | 2354242534224534122243322 | SB0418 | Allgäu | |
| 11/Rd152461 | + | + | 2354242534224534122243322 | SB0418 | Allgäu | ||
| D6431 | + | + | 2354242534224534122243322 | SB0418 | Allgäu | ||
In addition to the GenoType MTBC assay, sequences provide information about the SNPs in the gyrB gene.
Copy numbers in loci MIRU2-4-10-16-20-23-24-26-27-31-39-40 and VNTR424-577-1982-2401-3690-4156-1955-2163b-2165-2347-2461-3171-4052. Copy numbers differing from the Lechtal genotype are in bold type.
According to www.mbovis.org.
A 700-bp fragment was obtained, but the nucleotide sequence could not be determined because of high guanine-cytosine content.
Results from contig sequences in the next-generation sequencing of the 2.5-kb PCR approach produced a 2,224-bp fragment spanning the 5-kb deletion.
RESULTS
SNP analysis.
The M. caprae-specific gyrB gene SNPs were detected using GenoType MTBC (Hain Lifescience GmbH) in all 12 M. caprae isolates. Additionally, these SNPs of the gyrB gene (22) were confirmed by the determined nucleotide sequences of six M. caprae isolates (Table 3).
Using real-time PCR with an M. caprae-specific probe (26), we identified the M. caprae characteristic C-to-T substitution at position 690 of the lepA gene in all 12 isolates. Sequences indicative of other members of the MTC and NTM, respectively, were not detected. A 155-bp PCR fragment of the lepA gene was obtained from all MTBC members in a conventional PCR setup, but not from NTM members. Hence the data confirm that the M. caprae-specific probe binding within the lepA gene allows the identification of M. caprae isolates.
RD4 and genotyping analysis.
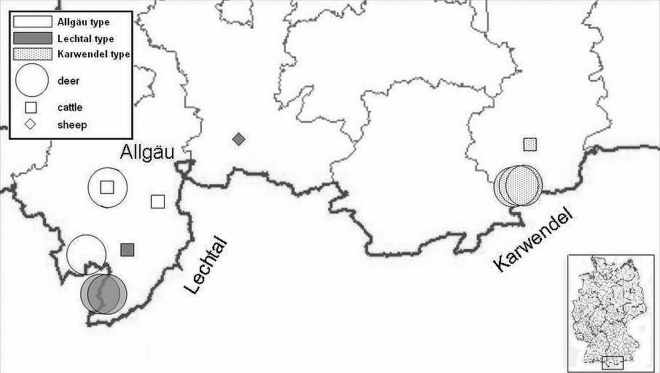
A MIRU-VNTR and spoligotyping code was obtained for M. caprae samples (Table 3), and two RD4-based PCR setups were chosen to amplify an internal fragment of the left and central parts in the RD4 sequence. The results of the cluster analysis by MIRU-VNTR genotyping and the RD4-based PCR setups allowed a discrimination of three groups of M. caprae isolates (Table 3). The geographic origin of the M. caprae isolates supported the diversity of genotyping and RD4 pattern results (Fig. 2). According to the RD4 differences and the MIRU-VNTR code, we chose the origin-related designations Allgäu, Lechtal, and Karwendel types for the three groups of our Alpine M. caprae isolates (Table 3).
Fig 2.
Geographic distribution of the three types of Bavarian M. caprae isolates included in this study. Each symbol indicates the host species. The M. caprae types are represented by shading, and the number of symbols reflects the number of tuberculosis cases. (Primary artwork reproduced with permission of TSN 3.0.)
The complete sequence of RD4 proved to be conserved in the Alpine M. caprae isolates of the Allgäu type (Fig. 1). The results of analysis of the next-generation sequencing data sets of eight M. caprae isolates performed using the program IGV suggested the presence of two different deletions of 5 kb in the genome of the Karwendel type and of 38 kb in the genome of the Lechtal type (Fig. 1). To confirm this finding, we designed flanking primers specific for deletion-adjacent sequences.
The sequence analysis of amplified deletion-flanking fragments showed that the 38-kb deletion includes the nucleotide sequences from position 1.694595 to 1.732924 in the M. tuberculosis H37Rv genome encompassing the whole RD4.
Using flanking primers specific for the 5-kb deletion, we obtained an approximately 700-bp PCR fragment for all M. caprae isolates of the Karwendel type. The nucleotide sequence of the PCR fragment could not be determined because of a high guanine-cytosine content. However, results from contig sequences in the Illumina sequencing of the 2.5-kb PCR approach of isolate 10/RW079 resulted in a 2,224-bp fragment spanning the 5-kb deletion. In this fragment, the 5-kb deletion contains the sequence stretch from position 1.695834 to 1.700422 spanning the left part of RD4. Of particular interest is the finding that the deletion is located 188 bp upstream of the previously assumed RD4 deletion in the genome of M. bovis subsp. bovis strain AF2122/97 (14, 21, 36). Notably, these genomic characteristics of the Allgäu, Lechtal, and Karwendel types isolated from red deer were also present in isolates from cattle from the respective geographic regions (Table 2; Fig. 2).
DISCUSSION
Twelve M. caprae isolates originating from wildlife and livestock in the Bavarian Alpine region of Germany were evaluated for genomic traits. The presence or absence of M. caprae-specific SNPs in the lepA and gyrB genes and of RD4-specific sequence peculiarities were assessed for identification and characterization of the M. caprae isolates.
The M. caprae-specific T-to-G substitution at position 1331 of the gyrB sequence and the C-to-T substitution at position 690 of the lepA sequence were confirmed to be conserved in all tested M. caprae isolates. The presence of these single-nucleotide changes was shown to be a stable marker in our Alpine M. caprae isolates as well as for M. caprae infections of different host species. However, it remains to be tested whether the sensitivity of the PCR is sufficient when DNA is prepared directly from native tissue samples.
Interestingly, three different RD4 variations were found in Bavarian Alpine M. caprae isolates which corresponded with their geographic origins and genotypes (Fig. 1 and Fig. 2). The absence of RD4 has previously been described as a marker for M. bovis (14–21). However, only a few M. caprae isolates have been tested so far with refined sequence analysis of RD4, and most isolates originated from Spain (14–16). Rodríguez et al. (8) verified the existence of RD4-specific sequences in 62 Spanish M. caprae isolates without giving detailed information about the primers used. Hence it is unknown which part of RD4 was detected and it has not been confirmed that the whole RD4 sequence is conserved. The results of our study demonstrate that RD4 is not uniformly present in Alpine M. caprae genomes as assumed to date. Due to this heterogeneity, RD4 cannot be recommended for use in effective differentiation of M. caprae from M. bovis. It cannot be excluded that further genomic deletions may be found in other M. caprae isolates.
The 5-kb deletion in the Karwendel type includes the genes Rv1505c to Rv1509, whereas the 38-kb deletion in the Lechtal type has led to the loss of the genes Rv1504c to Rv1531. This finding is of particular interest because RD4 is predicted to encode proteins involved in membrane degradation (Rv1508c) and exopolysaccharide synthesis (Rv1516c) (37). In addition, Al-Khodari et al. (38) identified in TB-infected persons immunodominant peptides reactive with antibodies recognizing amino acid (aa) epitopes 241 to 265 of Rv1508c and aa 325 to 336 of Rv1516c. Thus, some peptide sequences eliciting an antibody-mediated immune response are not well conserved in Alpine M. caprae isolates, and their loss may be a result of long-term host interactions.
During the past 10 years, a low but consistent incidence of M. caprae was registered in the Alpine part of Bavaria, Germany, according to TSN 3.0 reports. Efficient TB control should include epidemiological surveillance systems based on mycobacterial species identification to gain a better knowledge about the regional occurrence and host range of MTC members and other mycobacterial species. DNA fingerprinting methods have proven to be powerful epidemiologic tools for the differentiation of the MTC, the current gold standard being MIRU-VNTR typing (29). The presence of three types of the Bavarian Alpine M. caprae isolates, all three found in both cattle and red deer, clearly demonstrates tuberculosis transmission between livestock and wildlife (Fig. 2). However, the primary host of the strains cannot be predicted. Notably, the three types have shown a remarkable genetic stability for at least 3 years (Table 1).
To date, the Allgäu type has not been described. The region-specific types, genotype Lechtal and genotype Karwendel, have been found in bordering regions in Austria and were characterized by spoligotyping and MIRU-VNTR for 12 loci (10–12). The genotype Lechtal has been identified in a “hot spot” area of TB in red deer (prevalence 23.1%) in the Lechtal, Austria (12). Because of the local high incidence in the red deer population, a high red deer transmission rate of the Lechtal type can be assumed. A spillover of the Lechtal type to the German Allgäu region seems likely. On the other hand, the Allgäu type exists in the same area of Bavaria and contains the complete RD4. This raises the question of whether the RD4 deletion in the Lechtal type might have evolved from the Allgäu type by animal passages. It became evident that a detection of the three variants of RD4 sequences was not possible using spoligotyping or MIRU-VNTR based on repetitive DNA elements. However, informative SNPs are valuable markers for classifying MTC members into phylogenetic lineages because of very low degrees of homoplasy (39, 40). Whole-genome sequencing provides complete information for the identification of a sufficient number of reliable SNPs and the genomic characteristics of M. caprae (41).
ACKNOWLEDGMENTS
We thank the EMIDA ERA-Net coordination board for helpful discussions and the consortium for agreement with publication of the manuscript. We are grateful to Eva Maria Gerstmair, Ludmilla Naumann, and the Tuberculosis Culture Collection Laboratory of the Bavarian Health and Food Safety Authority for providing isolates.
This work was funded by the EMIDA ERA-Net Project Tuberculosis in Alpine Wildlife (BMBF-FK0315857A) and the Bavarian State Ministry of Environment and Health.
Footnotes
Published ahead of print 13 February 2013
REFERENCES
- 1. Aranaz A, Liébana E, Gómez-Mampaso E, Galán JC, Cousins D, Ortega A, Blázquez J, Baquero F, Mateos A, Súarez G, Domínguez L. 1999. Mycobacterium tuberculosis subsp. caprae subsp. nov.: a taxonomic study of a new member of the Mycobacterium tuberculosis complex isolated from goats in Spain. Int. J. Syst. Bacteriol. 49:1263–1273 [DOI] [PubMed] [Google Scholar]
- 2. Wayne LG, Kubica GP. 1986. The mycobacteria, p 1435–1457 In Sneath PHA, Holt JG. (ed), Bergey's manual of systematic bacteriology, vol 2 The Williams & Wilkins Co., Baltimore, MD [Google Scholar]
- 3. Aranaz A, Cousins D, Mateos A, Domínguez L. 2003. Elevation of Mycobacterium tuberculosis subsp. caprae Aranaz et al. 1999 to species rank as Mycobacterium caprae comb. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 53:1785–1789 [DOI] [PubMed] [Google Scholar]
- 4. Jenkins HE, Woodroffe R, Donnelly CA. 2008. The effects of annual widespread badger culls on cattle tuberculosis following the cessation of culling. Int. J. Infect. Dis. 12:457–465 [DOI] [PubMed] [Google Scholar]
- 5. O'Brien DJ, Schmitt SM, Rudolph BA, Nugent G. 2011. Recent advances in the management of bovine tuberculosis in free-ranging wildlife. Vet. Microbiol. 151:23–33 [DOI] [PubMed] [Google Scholar]
- 6. Ryan TJ, Livingstone PG, Ramsey DS, de Lisle GW, Nugent G, Collins DM, Buddle BM. 2006. Advances in understanding disease epidemiology and implications for control and eradication of tuberculosis in livestock: the experience from New Zealand. Vet. Microbiol. 112:211–219 [DOI] [PubMed] [Google Scholar]
- 7. Conner MM, Ebinger MR, Blanchong JA, Cross PC. 2008. Infectious disease in cervids of North America: data, models, and management challenges. Ann. N. Y. Acad. Sci. 1134:146–172 [DOI] [PubMed] [Google Scholar]
- 8. Rodríguez S, Bezos J, Romero B, de Juan L, Álvarez J, Castellanos E, Moya N, Lozano F, Javed MT, Sáez-Llorente JL, Liébana E, Mateos A, Domínguez L, Aranaz A; Spanish Network on Surveillance and Monitoring of Animal Tuberculosis 2011. Mycobacterium caprae infection in livestock and wildlife, Spain. Emerg. Infect. Dis. 17:532–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vicente J, Höfle U, Garrido JM, Fernández-De-Mera IG, Juste R, Barral M, Gortazar C. 2006. Wild boar and red deer display high prevalences of tuberculosis-like lesions in Spain. Vet. Res. 37:107–119 [DOI] [PubMed] [Google Scholar]
- 10. Prodinger WM, Eigentler A, Allerberger F, Schönbauer M, Glawischnig W. 2002. Infection of red deer, cattle, and humans with Mycobacterium bovis subsp. caprae in western Austria. J. Clin. Microbiol. 40:2270–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Prodinger WM, Brandstätter A, Naumann L, Pacciarini M, Kubica T, Boschiroli ML, Aranaz A, Nagy G, Cvetnic Z, Ocepek M, Skrypnyk A, Erler W, Niemann S, Pavlik I, Moser I. 2005. Characterization of Mycobacterium caprae isolates from Europe by mycobacterial interspersed repetitive unit genotyping. J. Clin. Microbiol. 43:4984–4992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schoepf K, Prodinger WM, Glawischnig W, Hofer E, Revilla-Fernandez S, Hofrichter J, Fritz J, Köfer J, Schmoll F. 2012. A two-years' survey on the prevalence of tuberculosis caused by Mycobacterium caprae in red deer (Cervus elaphus) in the Tyrol, Austria. ISRN Vet. Sci. doi:10.5402/2012/245138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Erler W, Martin G, Sachse K, Naumann L, Kahlau D, Beer J, Bartos M, Nagy G, Cvetnic Z, Zolnir-Dovc M, Pavlik I. 2004. Molecular fingerprinting of Mycobacterium bovis subsp. caprae isolates from central Europe. J. Clin. Microbiol. 42:2234–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, Garnier T, Gutierrez C, Hewinson G, Kremer K, Parsons LM, Pym AS, Samper S, van Soolingen D, Cole ST. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. U. S. A. 99:3684–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mostowy S, Cousins D, Brinkman J, Aranaz A, Behr MA. 2002. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J. Infect. Dis. 186:74–80 [DOI] [PubMed] [Google Scholar]
- 16. Huard RC, Lazzarini LC, Butler WR, van Soolingen D, Ho JL. 2003. PCR-based method to differentiate the subspecies of the Mycobacterium tuberculosis complex on the basis of genomic deletions. J. Clin. Microbiol. 41:1637–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gordon SV, Brosch R, Billault A, Garnier T, Eiglmeier K, Cole ST. 1999. Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol. Microbiol. 32:643–655 [DOI] [PubMed] [Google Scholar]
- 18. Halse TA, Escuyer VE, Musser KA. 2011. Evaluation of a single-tube multiplex real-time PCR for differentiation of members of the Mycobacterium tuberculosis complex in clinical specimens. J. Clin. Microbiol. 49:2562–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pinsky BA, Banaei N. 2008. Multiplex real-time PCR assay for rapid identification of Mycobacterium tuberculosis complex members to the species level. J. Clin. Microbiol. 46:2241–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Taylor GM, Murphy E, Hopkins R, Rutland P, Chistov Y. 2007. First report of Mycobacterium bovis DNA in human remains from the Iron Age. Microbiology 153:1243–1249 [DOI] [PubMed] [Google Scholar]
- 21. Warren RM, Gey van Pittius NC, Barnard M, Hesseling A, Engelke E, de Kock M, Gutierrez MC, Chege GK, Victor TC, Hoal EG, van Helden PD. 2006. Differentiation of Mycobacterium tuberculosis complex by PCR amplification of genomic regions of difference. Int. J. Tuberc. Lung Dis. 10:818–822 [PubMed] [Google Scholar]
- 22. Niemann S, Harmsen D, Rüsch-Gerdes S, Richter E. 2000. Differentiation of clinical Mycobacterium tuberculosis complex isolates by gyrB DNA sequence polymorphism analysis. J. Clin. Microbiol. 38:3231–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Boniotti MB, Goria M, Loda D, Garrone A, Benedetto A, Mondo A, Tisato E, Zanoni M, Zoppi S, Dondo A, Tagliabue S, Bonora S, Zanardi G, Pacciarini ML. 2009. Molecular typing of Mycobacterium bovis strains isolated in Italy from 2000 to 2006 and evaluation of variable-number tandem repeats for geographically optimized genotyping. J. Clin. Microbiol. 47:636–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kubica T, Rüsch-Gerdes S, Niemann S. 2003. Mycobacterium bovis subsp. caprae caused one-third of human M. bovis-associated tuberculosis cases reported in Germany between 1999 and 2001. J. Clin. Microbiol. 41:3070–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Richter E, Weizenegger M, Fahr AM, Rüsch-Gerdes S. 2004. Usefulness of the GenoType MTBC assay for differentiating species of the Mycobacterium tuberculosis complex in cultures obtained from clinical specimens. J. Clin. Microbiol. 42:4303–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Reddington K, O'Grady J, Dorai-Raj S, Niemann S, van Soolingen D, Barry T. 2011. A novel multiplex real-time PCR for the identification of mycobacteria associated with zoonotic tuberculosis. PLoS One 6:e23481 doi:10.1371/journal.pone.0023481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mazars E, Lesjean S, Banuls AL, Gilbert M, Vincent V, Gicquel B, Tibayrenc M, Locht C, Supply P. 2001. High-resolution minisatellitebased typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. U. S. A. 98:1901–1906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rusch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, Bifani P, Kurepina N, Kreiswirth B, Sola C, Rastogi N, Vatin V, Gutierrez MC, Fauville M, Niemann S, Skuce R, Kremer K, Locht C, van Soolingen D. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 44:4498–4510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goecks J, Nekrutenko A, Taylor J, The Galaxy Team 2010. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11:R86 doi:10.1186/gb-2010-11-8-r86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kasai H, Ezaki T, Harayama S. 2000. Differentiation of phylogenetically related slowly growing mycobacteria by their gyrB sequences. J. Clin. Microbiol. 38:301–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Toussaint JF, Sailleau C, Breard E, Zientara S, De Clercq K. 2007. Bluetongue virus detection by two real-time RT-qPCRs targeting two different genomic segments. J. Virol. Methods 140:115–123 [DOI] [PubMed] [Google Scholar]
- 35. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Garnier T, Eiglmeier K, Camus JC, Medina N, Mansoor H, Pryor M, Duthoy S, Grondin S, Lacroix C, Monsempe C, Simon S, Harris B, Atkin R, Doggett J, Mayes R, Keating L, Wheeler PR, Parkhill J, Barrell BG, Cole ST, Gordon SV, Hewinson RG. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. U. S. A. 100:7877–7882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pym AS, Brodin P, Brosch R, Huerre M, Cole ST. 2002. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol. Microbiol. 46:709–717 [DOI] [PubMed] [Google Scholar]
- 38. Al-Khodari NY, Al-Attiyah R, Mustafa AS. 2011. Identification, diagnostic potential, and natural expression of immunodominant seroreactive peptides encoded by five Mycobacterium tuberculosis-specific genomic regions. Clin. Vaccine Immunol. 18:477–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Comas I, Homolka S, Niemann S, Gagneux S. 2009. Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS One 4:e7815 doi:10.1371/journal.pone.0007815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schürch AC, van Soolingen D. 2012. DNA fingerprinting of Mycobacterium tuberculosis: from phage typing to whole-genome sequencing. Infect. Genet. Evol. 12:602–609 [DOI] [PubMed] [Google Scholar]
- 41. Stucki D, Malla B, Hostettler S, Huna T, Feldmann J, Yeboah-Manu D, Borrell S, Fenner L, Comas I, Coscollà M, Gagneux S. 2012. Two new rapid SNP-typing methods for classifying Mycobacterium tuberculosis complex into the main phylogenetic lineages. PLoS One 7:e41253 doi:10.1371/journal.pone.0041253 [DOI] [PMC free article] [PubMed] [Google Scholar]