Abstract
Rapid and accurate identification of the pathogens involved in bloodstream infections is crucial for the prompt initiation of appropriate therapy, as this can decrease morbidity and mortality rates. A PCR-reverse blot hybridization assay for sepsis, the reverse blot hybridization assay (REBA) Sepsis-ID test, was developed; it uses pan-probes to distinguish Gram-positive and -negative bacteria and fungi. In addition, the assay was designed to identify bacteria and fungi using six genus-specific and 13 species-specific probes; it uses additional probes for antibiotic resistance genes, i.e., the mecA gene of methicillin-resistant Staphylococcus aureus (MRSA) and the vanA and vanB genes of vancomycin-resistant enterococci (VRE). The REBA Sepsis-ID test successfully identified clinical isolates and blood culture samples as containing Gram-positive bacteria, Gram-negative bacteria, or fungi. The results matched those obtained with conventional microbiological methods. For the REBA Sepsis-ID test, of the 115 blood culture samples tested, 47 (40.8%) and 49 (42.6%) samples were identified to the species and genus levels, respectively, and the remaining 19 samples (16.5%), which included five Gram-positive rods, were identified as Gram-positive bacteria, Gram-negative bacteria, or fungi. The antibiotic resistances of the MRSA and VRE strains were identified using both conventional microbiological methods and the REBA Sepsis-ID test. In conclusion, the REBA Sepsis-ID test developed for this study is a fast and reliable test for the identification of Gram-positive bacteria, Gram-negative bacteria, fungi, and antibiotic resistance genes (including mecA for MRSA and the vanA and vanB genes for VRE) in bloodstream infections.
INTRODUCTION
Sepsis is an increasingly common cause of morbidity and mortality, particularly in elderly, immunocompromised, and critically ill patients (1), and it represents one of the greatest challenges in intensive care medicine. The causative agents of sepsis are Gram-positive bacteria, Gram-negative bacteria, anaerobes, and fungi (2–6). Moreover, this situation has worsened with the emergence of methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE) (7, 8).
Conventional identification and susceptibility testing systems have several limitations, including a lack of both speed and sensitivity. The current gold standard for sepsis diagnosis, which is blood culture sampling, usually requires 6 to 12 h of incubation and a further 24 to 48 h for definitive identification of the infectious agent and the determination of its susceptibility to antibiotics (9, 10). Therefore, the development of a rapid, sensitive, and specific assay is important for the identification of bloodstream pathogens, as well as for the implementation of timely, appropriate, and accurate antimicrobial therapy (11).
Currently, molecular methods, such as PCR, real-time PCR, and microarray analyses, are used to detect pathogens, and these methods have shown enhanced sensitivities and specificities for microbial pathogens (12). For the diagnosis of sepsis, a new assay that can detect and identify a panel of the most relevant bacterial and fungal pathogens is needed, as broad-range pathogens are increasingly linked to bloodstream infections. At present, various assays are commercially available, such as SeptiFast (Roche Diagnostics GmbH, Mannheim, Germany), SepsiTest (Molzym GmbH & Co. KG, Bremen, Germany), VYOO (SIRS-Lab GmbH, Jena, Germany), and the microarray-based system Prove-it Sepsis (Mobidiag, Helsinki, Finland) (9). These assays can detect broad-range bloodstream pathogens and their drug resistance profiles. However, a new assay that can distinguish between bacteria and fungi or between Gram-positive and -negative bacteria is required for the diagnosis of bloodstream infections. Moreover, for the appropriate treatment of bloodstream infections, the new assay should (i) include current causative agents as targets, (ii) identify pathogens to the genus and species levels, and (iii) simultaneously determine the antibiotic resistance profiles of the detected pathogens.
The reverse blot hybridization assay (REBA), which is a molecular tool for the assessment of multiple samples using multiple probes, allows for simultaneous detection and identification of pathogens. REBA is increasingly used as a diagnostic tool for several pathogens (13, 14).
The aim of the present study was to develop and evaluate the REBA Sepsis-ID test for the rapid diagnosis of sepsis-causing microorganisms. The REBA Sepsis-ID test was developed to detect Gram-positive bacteria, Gram-negative bacteria, and fungi using pan-probes, to identify sepsis-causing organisms to the genus and species levels, and to detect antibiotic resistance profiles using probes for the mecA gene of MRSA and the vanA and vanB genes of VRE.
MATERIALS AND METHODS
Bacterial strains and clinical specimens.
For the development and optimization of the REBA Sepsis-ID test, 48 reference strains and 120 clinical isolates were used (Tables 1 and 2). In total, 120 bloodstream infection-causing clinical isolates were obtained from December 2011 through January 2012 at a university hospital laboratory in Wonju, South Korea. All strains were grown on sheep blood agar and MacConkey agar (BD Diagnostic Systems, Sparks, MD) at 37°C overnight and were identified by the microplate method (11) and the MicroScan system (Siemens Healthcare Diagnostics, Sacramento, CA) for Gram-positive and -negative bacteria, and the Vitek2 system (bioMérieux, Durham, NC) with yeast identification (YST ID) card for Candida spp. The antimicrobial resistance profiles for the Gram-positive bacteria were determined using the MicroScan Pos Breakpoint Combo Panel Type 28 (PBC28) (Siemens Healthcare Diagnostics), and those of the Gram-negative bacteria and fungi were determined according to the guidelines of the Clinical and Laboratory Standards Institute (CSLI). The analyses were performed according to the standard protocol provided by each manufacturer.
Table 1.
List of reference strains used in this study
| Gram-positive bacteria |
Gram-negative bacteria |
||
|---|---|---|---|
| Strain | ATCC no. | Strain | ATCC no. |
| Enterococcus cecorum | 43198 | E. coli | 35218 |
| Enterococcus durans | 19432 | Haemophilus influenzae | 49247 |
| Enterococcus faecalis | 29212 | Klebsiella oxytoca | 700324 |
| E. faecalis (vanB) | 51299 | Klebsiella pneumoniae | 13883 |
| Enterococcus faecium | 35667 | Leclercia adecarboxylata | 23216 |
| E. faecium (vanA) | CCUG 36804a | Proteus mirabilis | 7002 |
| Enterococcus flavescens | 49997 | Proteus vulgaris | 49132 |
| Enterococcus gallinarum (vanC-1) | 49573 | Providencia alcalifaciens | 51902 |
| Enterococcus hirae | 9790 | Pseudomonas aeruginosa | 27853 |
| Enterococcus malodoratus | 43197 | Pseudomonas cepacia | 25608 |
| Enterococcus mundtii | 43186 | Salmonella enterica serovar Enteritidis | 13076 |
| Enterococcus raffinosus | 49427 | Salmonella enterica serovar Newport | 6962 |
| Enterococcus saccharolyticus | 43076 | Salmonella enterica serovar Paratyphi | 11511 |
| Enterococcus solitarius | 49428 | Shigella flexneri | 9199 |
| Enterococcus sulfureus | 49903 | ||
| Micrococcus luteus | 49732 | Fungi |
|
| Mycobacterium marinum | 927 | Candida albicans | 10231 |
| Staphylococcus aureus | 25923 | C. albicans | 36802 |
| S. aureus | 29213 | Candida glabrata | 38326 |
| S. aureus (MRSA) | 43300 | Candida guilliermondii | 56822 |
| Staphylococcus xylosus | 29971 | Candida lusitaniae | 34449 |
| Streptococcus agalactiae | 13813 | Candida parapsilosis | 7330 |
| Streptococcus pneumoniae | 33400 | Candida tropicalis | 14506 |
CCUG, Culture Collection, University of Göteborg, Sweden.
Table 2.
Comparison of conventional methods and PCR-REBA: screening and identification results in 120 clinical isolatesa
| Conventional methodsa |
REBA Sepsis-IDa |
||
|---|---|---|---|
| Genus and species | Antibiotic resistance | Genus and species | Antibiotic resistance |
| Gram-positive bacteria (43) | |||
| Staphylococcus aureus (12) | MRSA (9) | S. aureus (12) | MRSA (9) |
| MSSA (3) | MSSA (3) | ||
| Staphylococcus epidermidis (4) | MRCoNS (4) | Staphylococcus spp. (4) | MRCoNS (4) |
| Staphylococcus haemolyticus (3) | MRCoNS (3) | Staphylococcus spp. (3) | MRCoNS (3) |
| Staphylococcus capitis (1) | MSCoNS (1) | Staphylococcus spp. (1) | MSCoNS (1) |
| Streptococcus agalactiae (1) | Streptococcus spp. (1) | ||
| Streptococcus mitis (2) | Streptococcus spp. (2) | ||
| Streptococcus parasanguinis (1) | Streptococcus spp. (1) | ||
| Streptococcus salivarius (1) | Streptococcus spp. (1) | ||
| Streptococcus pyogenes (1) | Streptococcus spp. (1) | ||
| Streptococcus pneumoniae (1) | S. pneumoniae (1) | ||
| Enterococcus faecalis (4) | Enterococcus spp. (4) | ||
| Enterococcus faecium (10) | VRE (7) | Enterococcus spp. (10) | VRE-vanA (7) |
| VSE (3) | VSE (3) | ||
| Enterococcus mundtii (1) | Enterococcus spp. (1) | ||
| Corynebacterium spp. (1) | Gram positive (1) | ||
| Gram-negative bacteria (64) | |||
| Escherichia coli (16) | E. coli (16) | ||
| Enterobacter asburiae (1) | Gram negative (1) | ||
| Enterobacter cloacae (1) | Gram negative (1) | ||
| Klebsiella pneumoniae (14) | K. pneumoniae (14) | ||
| Citrobacter freundii (1) | C. freundii (1) | ||
| Morganella morganii (1) | Gram negative (1) | ||
| Proteus mirabilis (1) | Gram negative (1) | ||
| Serratia marcescens (1) | Gram negative (1) | ||
| Providencia rettgeri (1) | Gram negative (1) | ||
| Acinetobacter baumannii (11) | Acinetobacter baumannii (11) | ||
| Pseudomonas aeruginosa (13) | P. aeruginosa (13) | ||
| Aeromonas spp. (1) | Gram negative (1) | ||
| Haemophilus influenzae (1) | H. influenzae (1) | ||
| Moraxella catarrhalis (1) | Gram negative (1) | ||
| Fungi (13) | |||
| Candida albicans (5) | C. albicans (5) | ||
| Candida parapsilosis (3) | C. parapsilosis (3) | ||
| Candida glabrata (1) | C. glabrata (1) | ||
| Candida tropicalis (2) | C. tropicalis (2) | ||
| Saccharomyces cerevisiae (2) | Fungi (2) | ||
The number in parentheses is the number of isolates of each genus and/or species identified.
For evaluation of the REBA Sepsis-ID test, blood samples were collected from 115 hospitalized patients and were cultured using both the Bactec FX system (BD Diagnostic Systems) and the BacT/Alert 3D system (bioMérieux). Cultures found to contain growing bacteria in the bottles were regarded as positive cultures and were further processed for identification using the MicroScan system (Siemens Healthcare Diagnostics). The panels used for positive cultures were selected according to the results of Gram staining.
Preparation of oligonucleotide probes for REBA Sepsis-ID.
To maximize the sensitivities and specificities of the species- or genus-specific primers and probes, sequences from the most variable regions within the bacterial 16S ribosomal DNA (16S rDNA) for Gram-positive and -negative bacteria, 18S to 5.8S internal transcribed sequence (ITS) for fungi, the nuc gene for S. aureus (with the mecA gene for MRSA), and the vanA and vanB genes for VRE, which were the target DNAs, were selected by multiple alignment using the ClustalW program (http://www.cmbi.kun.nl/bioinf/tools/clustalw.shtml). The specific primers and probes used for the genus- and species-level identifications of bacteria and fungi and for antibiotic resistance detection are listed in Table 3. The sequences of the primers and probes were compared by nucleotide-nucleotide NCBI BLAST (BLASTn) searches to determine their sequence homologies. Oligonucleotide probes were selected while taking into consideration our desire to avoid self-complementarity of more than three nucleotides per probe. After evaluation of the probes, 17 bacterial probes, six fungal probes, and three antibiotic resistance probes were used for the REBA Sepsis-ID test.
Table 3.
Primers and probes used in this study
| Target and primer/probe name | Nucleotide sequence (5′–3′) |
|---|---|
| 16S rRNA | |
| F-16S | TAAYACATGCAAGTCGARCG |
| R-16S | TGGCACGDAGTTRGCCGKKGCTT |
| Staphylococcus aureus | |
| F-Saur | AGCGATTGATGGTGATACGGT |
| R-Saur | ATGCACTTGCTTCAGGACCA |
| mecA | |
| F-mecA | GGTGTTGGTGAAGATATACCAAGTG |
| R-mecA | GAAAGGATCTGTACTGGGTTAATCAT |
| vanA | |
| F-VanA | TCAATAGCGCGGACGAATTG |
| R-VanA | GCGGGAACGGTTATAACTGCGTTT |
| vanB | |
| F-VanB | TACCTACCCTGTCTTTGTGAAGCC |
| R-VanB | GCTGCTTCTATCGCAGCGTTTAGT |
| Fungus | |
| F-Fung | AACGCANMTTGCRCYCHHTG |
| R-Fung | CAGCGGGTADYCCYACCTGA |
| Bacteria: p-panBact | AGYGGCGGACGGGTGAGTAA |
| Gram(+) bacteria: p-GrPo | TGAGTAACACGTGGGYAACC |
| Gram(−) bacteria: p-GrNg | ATGTCTGGGAAACTGCCTGATG |
| Fungus: p-Fung | TGCGTTCAARRAYTCGATGA |
| Enterococcus spp.: p-Ent | CCATCAGARGGGGATAACACTT |
| Mycobacterium spp.: p-Mycob | TGGTGSAAAGCTTTTGCGGT |
| Salmonella spp.: p-Salm | CGGAAGCCTCCGCTAATTTGAT |
| Shigella spp.: p-Shig | AGTTCAGTAAGATGGTTGTGCGCA |
| Staphylococcus spp.: p-Stap | CACGTRGCTAACCTACCTATAAGACTG |
| Streptococcus spp.: p-Str | CGCGTAGGTAACCTGCCTGGTA |
| Acinetobacter baumannii: p-Abaum | AGCTTGCTACCGGACCTAGC |
| Candida albicans: p-Calb | AATAGTGGTAAGGCGGGATC |
| Candida glabrata: p-Cgrab | AGCGCAAGCTTCTCTATTAATCTG |
| Candida krusei: p-Ckrus | AGCGGAGCGGACGACGTGTA |
| Candida parapsilosis: p-Cpara | AGGCGGAGTATAAACTAATGGATAGGT |
| Candida tropicalis: p-Ctrp | ACGTGGAAACTTATTTTAAGCGA |
| Citrobacter freundii: P-Cfreun | TAGCACAGAGGAGCTTGCTCCTTG |
| Escherichia coli: p-Ecoli | AAAGGGAGTAAAGTTAATACCTTTGCTCA |
| Haemophilus influenzae: p-Hinfl | CGTATTATCGGAAGATGAAAGTGC |
| Klebsiella pneumoniae: p-KPneum | AAAAAAAGGTTAATAACCTCATCGATTGAC |
| Pseudomonas aeruginosa: p-Paer | ATACGTCCTGAGGGAGAAAGTG |
| Streptococcus pneumoniae: p-Stpneum | TCAGTGTCGCTGTTTTAGCAGAT |
| S. aureus: p-Saur | TTGGTTGATACACCTGAAACAAAG |
| mecA: p-MecA | AGCTGATTCAGGTTACGGACAAGGT |
| vanA: p-VanA | TCGTATTCATCAGGAAGTCGAGCC |
| vanB: p-VanB | TCGTCCTTTGGCGTAACCAA |
DNA preparation.
To prepare DNA templates, one colony per type strain and clinical isolate was suspended in 100 μl of DNA extraction solution (M&D, Wonju, South Korea). Suspended bacterial solutions were boiled for 10 min. After centrifugation at 13,000 × g for 10 min, the supernatant was used as the DNA template. To prepare DNA template from blood culture samples, 0.5 ml of culture sample was taken directly from the Bactec FX system (BD Diagnostic System) or from the BacT/Alert 3D system (bioMérieux), added to 1 ml of phosphate-buffered saline (PBS) buffer (pH 8.0), and centrifuged at 13,000 × g for 1 min. The supernatant was removed and the pellet resuspended in 1 ml of sterile water for the lysis of red blood cells. It was then centrifuged at 13,000 × g for 1 min. This washing step was repeated twice, and the pellet was resuspended in DNA extraction solution as described above for the clinical isolates.
PCR amplification.
PCR was performed according to the manufacturer's instructions using a 50-μl reaction mixture (Bioneer, Daejeon, South Korea) that contained 2× Master mix, 1× primer mix, and 5 μl of sample DNA. Sterile distilled water was added to give a final volume of 50 μl. For the REBA Sepsis-ID test, the first 10 PCR cycles comprised initial denaturation at 95°C for 30 s, followed by annealing and extension at 65°C for 30 s. These 10 cycles were followed by 40 cycles of denaturation at 95°C for 30 s, followed by annealing and extension at 60°C for 30 s. After the final cycle, samples were maintained at 72°C for 7 min to complete the synthesis of all strands. In all PCR assays, sterile distilled water was added to the PCR mixture in place of bacterial DNA as a negative control. Following amplification, 5 μl of each PCR product was electrophoresed on a 2.0% Tris–borate–EDTA (TBE) agarose gel to confirm successful amplification. The gel was stained with 0.05 mg/ml ethidium bromide (EtBr) solution, visualized using the AgaroPower system (Bioneer), and photographed using the Gel Doc EQ system (Bio-Rad, Hercules, CA).
PCR-reverse blot hybridization assay.
The REBA Sepsis-ID test was performed according to the standard protocols provided by the manufacturer (M&D). For PCR-REBA, hybridization of the PCR products with probes immobilized on a membrane was carried out according to the manufacturer's instructions. Briefly, denatured PCR products in hybridization solution (HS) were incubated with REBA Sepsis-ID membrane strips at 55°C and 90 rpm for 30 min on a mini incubation tray (Bio-Rad). Following washing with washing solution (WS) at 55°C and 90 rpm for 10 min, streptavidin-alkaline phosphatase conjugate diluted 1:2,000 in conjugate diluent solution (CDS) was added and the membrane was incubated at 25°C and 90 rpm for 30 min. Finally, the membrane was washed twice with CDS at 25°C and 90 rpm for 10 min. For detection, the membrane was incubated with nitroblue tetrazolium–5-bromo-4-chloro-3-indolyphosphate (NBT-BCIP) staining solution (SS) for 10 min. Data interpretation was conducted using the provided REBA Sepsis-ID data sheet.
RESULTS
Development of the REBA Sepsis-ID test using type strains.
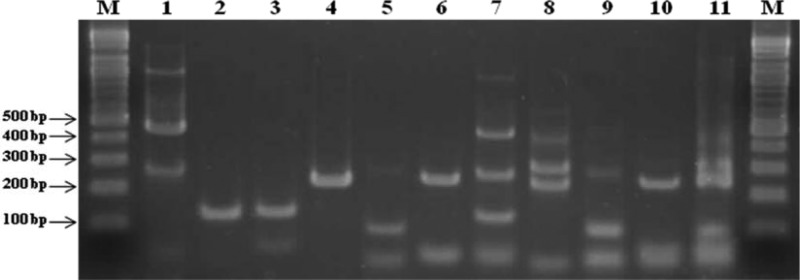
The PCR amplification results for the REBA Sepsis-ID test are shown in Fig. 1. The PCR products for the 16S rRNA genes of Gram-positive and -negative bacteria (470 and 250 bp, respectively), the 18S to 5.8S internal transcribed sequence (ITS) of fungi (230 bp), the nuc gene of S. aureus (120 bp), the mecA gene of MRSA (120 bp), and the vanA and vanB genes of VRE (250 and 100 bp, respectively) were of the expected sizes.
Fig 1.
Typical PCR results for REBA Sepsis-ID test. Lanes M, 100-bp DNA ladder (Bioneer, Daejeon, South Korea); lane 1, 16S PCR (470 bp, 250 bp); lane 2, S. aureus (120 bp); lane 3, mecA (120 bp); lane 4, vanA (250 bp); lane 5, vanB (100 bp); lane 6, fungus (230 bp); lane 7, 16S-S. aureus-mecA mix; lane 8, fungus-vanA; lane 9, fungus-vanB; lane 10, fungus; lane 11, fungus-vanA-vanB.
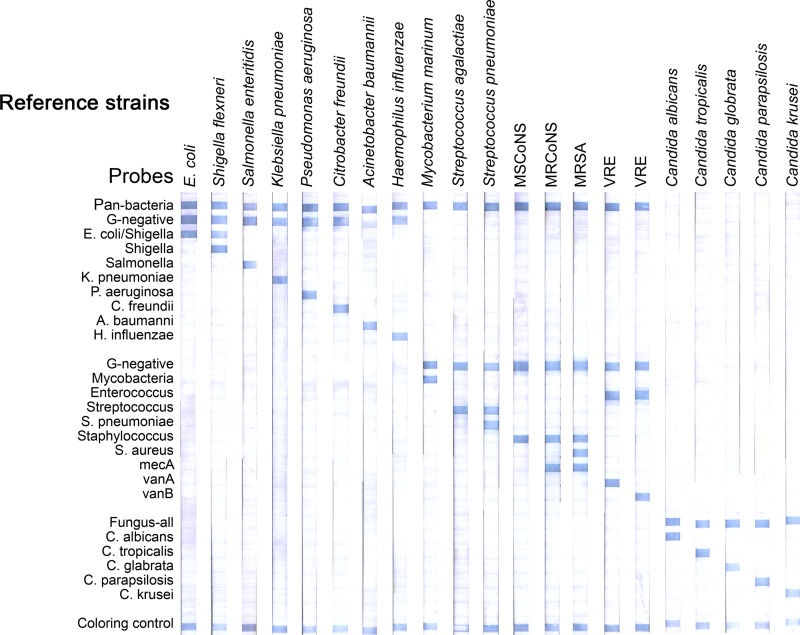
To validate the usefulness of the selected probes, target DNA samples amplified from 48 reference strains were applied to REBA Sepsis-ID membrane strips spotted with the selected probes. The hybridization results are shown in Fig. 2. The amplified PCR products reacted with universal probes (including the pan-bacterial, Gram-positive bacterial, Gram-negative bacterial, and fungal probes) and genus- and species-specific probes and had unique hybridization patterns, as expected. The target DNA samples hybridized strongly with the probes derived from their targets and showed no cross-reactivities. All of the 48 type strains showed strong specific hybridization signals at the positions of the corresponding probes derived from their respective sequences, suggesting that sepsis-causing microbial pathogens can be specifically detected and identified using this test.
Fig 2.
Typical results of the REBA Sepsis-ID test using reference strains. All Gram-negative bacteria, Gram-positive bacteria, and fungi show strong specific hybridization signals at the positions of the corresponding probes. For the development of PCR-REBA, methicillin-resistant coagulase-negative staphylococci (MRCoNS) were used as the clinical isolates in the tests shown.
Validation of the REBA Sepsis-ID test.
In a verification test, 120 clinical isolates that had been identified by culture-based methods were analyzed by the REBA Sepsis-ID test. The results of a confirmation test for clinical isolates indicated high concordance between the routine culture-based methods and the REBA Sepsis-ID test. Among the 120 clinical isolates, all 43 isolates of Gram-positive bacteria, all 64 isolates of Gram-negative bacteria, and all 13 isolates of fungi hybridized at the position of the corresponding universal probe (pan-bacteria, Gram-negative, Gram-positive, and fungus). In addition, the REBA Sepsis-ID test identified the following clinical isolates to the genus and/or species level: Staphylococcus spp., Streptococcus spp., Enterococcus spp., Candida spp., Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii, Haemophilus influenzae, Citrobacter freundii, Streptococcus pneumoniae, S. aureus, Candida albicans, Candida tropicalis, and Candida glabrata (Table 2).
Antibiotic resistance gene-carrying clinical isolates, such as MRSA and VRE, were detected by the REBA Sepsis-ID test. In an antibiotic susceptibility test, nine of 12 S. aureus strains were MRSA, eight of 12 coagulase-negative staphylococci (CoNS) strains were resistant to oxacillin (≥4 μg/ml), and seven of 15 Enterococcus spp. were resistant to vancomycin (≥32 μg/ml). All antibiotic-resistant clinical isolates, including MRSA, methicillin-resistant coagulase-negative staphylococci (MRCoNS), and VRE, hybridized with resistance gene probes, such as those for mecA and vanA (data not shown).
Application of PCR-REBA to clinical isolates.
To evaluate the REBA Sepsis-ID test using clinical samples, 115 positive blood culture samples from patients with presumed sepsis were analyzed and the results were compared with those obtained using the conventional identification system. The patients were ≤1 year of age and 12 had septic shock, five had pneumonia, and one had continuous bacteremia. The PCR-REBA results revealed strong concordance between the microbiological identification method and the REBA Sepsis-ID test for all the blood samples.
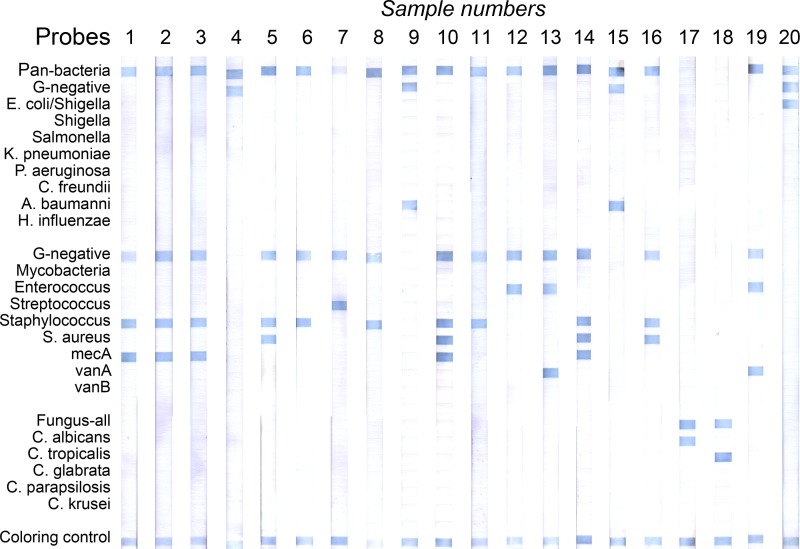
The REBA Sepsis-ID test identified the following clinical isolates at the genus and/or species level: S. aureus (n = 13; 10 MRSA, three methicillin-susceptible S. aureus [MSSA]), Staphylococcus spp. (n = 39; 32 MRCoNS, six methicillin-susceptible coagulase-negative staphylococci [MSCoNS]), Enterococcus spp. (n = 7; two VRE, four vancomycin-susceptible enterococci [VSE]), E. coli (n = 14), K. pneumoniae (n = 7), Haemophilus influenzae (n = 1), P. aeruginosa (n = 1), C. albicans (n = 8), Candida parapsilosis (n = 2), and C. glabrata (n = 1). In addition, eight Gram-positive bacteria, eight Gram-negative bacteria, and two fungi were detected in the blood samples from patients with suspected sepsis (Fig. 3, Table 4).
Fig 3.
Typical results of the REBA Sepsis-ID test using blood culture samples. Lanes 1 to 3, MRCoNS; lane 4, Gram-negative bacteria; lanes 5 and 16, MSSA; lanes 6, 8, and 11, Staphylococcus sp. (MSCoNS); lane 7, Streptococcus sp.; lane 9, P. aeruginosa; lanes 10 and 14, MRSA; lane 12, Enterococcus spp. (VSE); lane 13, VRE; lane 15, A. baumannii; lane 17, C. albicans; lane 18, C. tropicalis; lane 19, E. coli; lane 20, negative control.
Table 4.
Comparison of conventional methods and PCR-REBA: screening and identification results in 115 blood samples
| Conventional methodsa |
REBA Sepsis-IDa |
||
|---|---|---|---|
| Genus and species (n) | Antibiotic resistance | Genus and species (n) | Antibiotic resistance |
| Gram-positive bacteria (70) | |||
| Staphylococcus aureus (13) | MRSA (10) | S. aureus (13) | MRSA (10) |
| MSSA (3) | MSSA (3) | ||
| Staphylococcus epidermidis (23) | MRCoNS (20) | Staphylococcus spp. (23) | MRCoNS (20) |
| MSCoNS (3) | MSCoNS (3) | ||
| Staphylococcus capitis (5) | MRCoNS (4) | Staphylococcus spp. (5) | MRCoNS (4) |
| MSCoNS (1) | MSCoNS (1) | ||
| Staphylococcus haemolyticus (5) | MRCoNS (5) | Staphylococcus spp. (5) | MRCoNS (5) |
| Staphylococcus hominis (5) | MRCoNS (3) | Staphylococcus spp. (5) | MRCoNS (3) |
| MSCoNS (2) | MSCoNS (2) | ||
| Staphylococcus warneri (1) | Staphylococcus spp. (1) | ||
| Streptococcus parasanguinis (1) | Streptococcus spp. (1) | ||
| Streptococcus salivarius (1) | Streptococcus spp. (1) | ||
| Enterococcus faecium (6) | VRE (2) | Enterococcus spp. (6) | VRE-vanA (2) |
| VSE (4) | VSE (4) | ||
| Enterococcus faecalis (1) | Enterococcus spp. (1) | ||
| Corynebacterium spp. (1) | Gram positive (1) | ||
| Micrococcus spp. (2) | Gram positive (1) | ||
| Propionibacterium acnes (1) | Gram positive (1) | ||
| Gram-positive rods (5) | Gram positives (5) | ||
| Gram-negative bacteria (32) | |||
| Escherichia coli (14) | E. coli (14) | ||
| Klebsiella pneumoniae (7) | K. pneumoniae (7) | ||
| Klebsiella oxytoca (2) | Gram negatives (2) | ||
| Salmonella group D (1) | Salmonella spp. (1) | ||
| Acinetobacter lwoffii (1) | Gram negative (1) | ||
| Aeromonas spp. (1) | Gram negative (1) | ||
| Citrobacter koseri (1) | Gram negative (1) | ||
| Haemophilus influenzae (1) | H. influenzae (1) | ||
| Neisseria sicca (1) | Gram negative (1) | ||
| Proteus mirabilis (1) | Gram negative (1) | ||
| Pseudomonas aeruginosa (1) | P. aeruginosa (1) | ||
| Sphingomonas paucimobilis (1) | Gram negative (1) | ||
| Fungi (13) | |||
| Candida albicans (8) | C. albicans (8) | ||
| Candida parapsilosis (2) | C. parapsilosis (2) | ||
| Candida glabrata (1) | C. glabrata (1) | ||
| Cryptococcus neoformans (1) | Fungi (1) | ||
| Saccharomyces cerevisiae (1) | Fungi (1) | ||
The number in parentheses is the number of isolates of each genus and/or species identified.
DISCUSSION
We developed a multiplex PCR-REBA for the diagnosis of pathogens in cases of sepsis. The REBA Sepsis-ID test targets the bacteria and fungi that cause bloodstream infections. We evaluated the REBA Sepsis-ID test using clinical bacterial and fungal isolates, as well as using blood culture samples from sepsis patients.
The REBA Sepsis-ID test distinguished Gram-positive bacteria, Gram-negative bacteria, and fungi using pan-probes, and it identified bacteria and fungi using six genus-specific probes and 13 species-specific probes. Moreover, this assay simultaneously identified MRSA and VRE strains.
Conventional methods identified, of 115 blood specimens, 106 specimens (92.2%) at the species level and 4 (3.5%) at the genus level, as well as five specimens (4.3%) as Gram-positive rods. Using the REBA Sepsis-ID test, the majority of the blood specimens, i.e., 47 specimens (40.8%) at the species level and 49 specimens (42.6%) at the genus level, were identified, and 19 specimens (16.5%) were identified as Gram-positive bacteria, Gram-negative bacteria, or fungi. Interestingly, the REBA Sepsis-ID test detected all the antibiotic-resistant isolates, similar to the performance of conventional tests.
In the present study, Gram-positive bacteria were present in 61% and Gram-negative bacteria in 27% of the blood culture samples. The major causative agents of bloodstream infections were (percent and number identified out of 115 blood specimens): Staphylococcus epidermidis (20.0%, 23), S. aureus (11.3%, 13), E. coli (12.2%, 14), K. pneumoniae (6.1%, 7), and C. albicans (7.0%, 8). These results are consistent with those of Koh et al. (4), which showed that Gram-positive and -negative bacteria accounted for 53% and 35.2% of bloodstream infections, respectively. Stoll et al. (6, 15) reported Gram-positive bacteria as the major cause of bloodstream infections, although they found that the incidence of Gram-negative bacteria was increasing. Koh et al. reported that the major causative agents of bloodstream infection were S. aureus (13.8%), E. coli (24.5%), and K. pneumoniae (8.9%). Although S. pneumoniae and group A and B streptococci are regarded as important agents of bloodstream infections, they were not detected in the present study. Overall, our results are consistent with those of previous studies (4, 11, 16), while the nondetection of S. pneumoniae and group B streptococci may be due to the minimal number of specimens we examined.
The REBA Sepsis-ID test has certain advantages over the conventional methods in terms of the identification of Gram-positive and -negative genera and/or species that cause sepsis, and the fact that antibiotic resistance can be determined within a few hours of receipt of the blood culture sample. Moreover, the REBA Sepsis-ID has advantages over other commercial diagnostic assays for sepsis. The SeptiFast Test (Roche) does not include probes for antibiotic resistance, and while the VYOO test (SIRS-Lab) does contain probes for resistance to vancomycin and for beta-lactamase, it does not test for resistance to methicillin, which is a major antibiotic resistance of S. aureus (17).
Since the causative agents of bloodstream infections differ according to region and country, the target panel of the REBA Sepsis-ID should be optimized in additional evaluations to include the most important and virulent pathogens associated with bloodstream infections as targets at the species and/or genus level. This would ensure that patients receive appropriate medical treatment earlier and would prevent deterioration of the patient's condition, perhaps even saving his or her life.
In conclusion, the REBA Sepsis-ID test appears to be a promising assay for the early identification of pathogenic bacteria and fungi, including antibiotic-resistant strains, such as MRSA and VRE, in blood samples, which might not only allow medical staff to make a precise diagnosis but also allow them to administer quick and appropriate treatment to hospitalized patients.
ACKNOWLEDGMENT
This study was supported by a grant of the Korea Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (A121030, H.L.).
Footnotes
Published ahead of print 27 February 2013
REFERENCES
- 1. Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, Schein RM, Sibbald WJ. 1992. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest 101:1644–1655 [DOI] [PubMed] [Google Scholar]
- 2. Cohen J. 2002. The immunopathogenesis of sepsis. Nature 420:885–891 [DOI] [PubMed] [Google Scholar]
- 3. Goldani LZ, Mário PS. 2003. Candida tropicalis fungemia in a tertiary care hospital. J. Infect. 46:155–160 [DOI] [PubMed] [Google Scholar]
- 4. Koh EM, Lee SG, Kim CK, Kim M, Yong D, Lee K, Kim JM, Kim DS, Chong Y. 2007. Microorganisms isolated from blood cultures and their antimicrobial susceptibility patterns at a university hospital during 1994–2003. Korean J. Lab Med. 27:265–275 (In Korean.) [DOI] [PubMed] [Google Scholar]
- 5. Reimer LG, Wilson ML, Weinstein MP. 1997. Update on detection of bacteremia and fungemia. Clin. Microbiol. Rev. 10:444–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stoll BJ, Hansen N, Fararoff AA, Wright LL, Carlo WA, Ehrenkranz RA, Lemons JA, Donovan EF, Stark AR, Tyson JE, Oh W, Bauer CR, Korones SB, Shankaran S, Laptook AR, Stevenson DK, Papile LA, Poole WK. 2002. Changes in pathogens causing early-onset sepsis in very-low-birth-weight infants. N. Engl. J. Med. 347:240–247 [DOI] [PubMed] [Google Scholar]
- 7. DiazGranados CA, Zimmer SM, Klein M, Jernigan JA. 2005. Comparison of mortality associated with vancomycin-resistant and vancomycin-susceptible enterococcal bloodstream infections: a meta-analysis. Clin. Infect. Dis. 41:327–333 [DOI] [PubMed] [Google Scholar]
- 8. Zhang K, McClure JA, Elsayed S, Louie T, Colny JM. 2005. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5026–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hansen WL, Beuving J, Bruggeman CA, Wolffs PF. 2010. Molecular probes for diagnosis of clinically relevant bacterial infections in blood cultures. J. Clin. Microbiol. 48:4432–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu C, Bayer A, Cosgrove SE, Daum RS, Fridkin SK, Gorwitz RJ, Kaplan SL, Karchmer AW, Levine DP, Murray BE, Rybak M, Talan DA, Chambers HF. 2011. Clinical practice guidelines by the Infectious Diseases Society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children: executive summary. Clin. Infect. Dis. 52:285–292 [DOI] [PubMed] [Google Scholar]
- 11. Uh Y, Son JS, Hwang GY, Jang IH, Yoon KJ, Seo DM. 1999. Microplate identification system of Enterobacteriaceae. Korean J. Clin. Microbiol. 2:135–143 (In Korean.) [Google Scholar]
- 12. Lodes U, Bohmeier B, Lippert H, König B, Meyer F. 2012. PCR-based rapid sepsis diagnosis effectively guides clinical treatment in patients with new onset of SIRS. Langenbecks Arch. Surg. 397:447–455 [DOI] [PubMed] [Google Scholar]
- 13. Choi Y, Lee G, Bang H, Kim JB, Lee H. 2010. Detection of waterborne pathogens by PCR-reverse blot hybridization. J. Exp. Biomed. Sci. 16:10–18 (In Korean.) [Google Scholar]
- 14. Kim H, Jin H, Kim S, Wang HY, Choi Y, Bang H, Park JS, Lee JH, Won YH, Ahn KJ, Kim YK, Lee H. 2011. PCR-reverse blot hybridization assay for species identification of dermatophytes. Korean J. Med. Mycol. 16:86–98 (In Korean.) [Google Scholar]
- 15. Martin GS, Mannino DM, Eaton S, Moss M. 2003. The epidemiology of sepsis in the United States from 1979 through 2000. N. Engl. J. Med. 348:1546–1554 [DOI] [PubMed] [Google Scholar]
- 16. Shin JH, Song SA, Kim MN, Lee NY, Kim EC, Kim S, Koo SH, Ryoo NH, Kim JS, Cho JH. 2011. Comprehensive analysis of blood culture performed at nine university hospitals in Korea. Korean J. Lab Med. 31:101–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoo SM, Choi JY, Yun JK, Choi JK, Shin SY, Lee K, Kim JM, Lee SY. 2010. DNA microarray-based identification of bacterial and fungal pathogens in bloodstream infections. Mol. Cell. Probes 24:44–52 [DOI] [PubMed] [Google Scholar]