Abstract
The development of an accurate antigen detection assay for the diagnosis of active tuberculosis (TB) would represent a major clinical advance. Here, we demonstrate that the Mycobacterium tuberculosis Rv1681 protein is a biomarker for active TB with potential diagnostic utility. We initially identified, by mass spectroscopy, peptides from the Rv1681 protein in urine specimens from 4 patients with untreated active TB. Rabbit IgG anti-recombinant Rv1681 detected Rv1681 protein in lysates and culture filtrates of M. tuberculosis and immunoprecipitated it from pooled urine specimens from two TB patients. An enzyme-linked immunosorbent assay formatted with these antibodies detected Rv1681 protein in unconcentrated urine specimens from 11/25 (44%) TB patients and 1/21 (4.8%) subjects in whom TB was initially clinically suspected but then ruled out by conventional methods. Rv1681 protein was not detected in urine specimens from 10 subjects with Escherichia coli-positive urine cultures, 26 subjects with confirmed non-TB tropical diseases (11 with schistosomiasis, 5 with Chagas' disease, and 10 with cutaneous leishmaniasis), and 14 healthy subjects. These results provide strong validation of Rv1681 protein as a promising biomarker for TB diagnosis.
INTRODUCTION
Tuberculosis (TB) remains the second most common cause of death from an infectious disease in the world. According to a 2012 World Health Organization (WHO) report, there were an estimated 8.7 million new cases of TB globally in 2011 and approximately 1.4 million TB-related deaths (430,000 of which involved HIV-positive individuals) (1). Failure to control the spread of TB is due in large part to failure to adequately detect (and thus to efficiently treat) infectious cases (2); therefore, a major focus of the WHO's global plan to stop TB (3) is the development of new simple and cost-effective diagnostic methods to improve case detection. Historically, culture of Mycobacterium tuberculosis from a clinical specimen has been the gold standard for TB diagnosis; however, it is expensive, time-consuming, and not available worldwide. Thus, in resource-limited settings, diagnosis of active TB relies primarily on clinical evaluation and, where available, microscopic evaluation of sputum specimens. Sputum smears have widely varying sensitivities in different settings (20 to 80%) and are of limited utility in paucibacillary (e.g., pediatric and HIV-associated) TB (3–5). Due to the lack of accessible and effective diagnostic methods, many patients in resource-limited settings are treated empirically based on clinical information alone, exposing some patients to unneeded and potentially toxic drugs and missing others who have active TB.
The ideal diagnostic method for tuberculosis would be a low-cost, point-of-care (POC), rapid, simple, and accurate test that could distinguish individuals with active TB from those with latent disease or no infection (including Mycobacterium bovis BCG-vaccinated individuals) (2, 6). We previously demonstrated the identification by mass spectroscopy (MS) of four M. tuberculosis antigens in the urine of patients with active pulmonary TB (7, 8). The gene (Rv1681) (Institute for Genomic Research [TIGR] no. MT_1721) coding for one of these antigens, the putative molybdopterin biosynthesis protein MoeX/MoeA, is unique to the M. tuberculosis complex, and no homologues of the Rv1681 protein have been found in the available databases for other representative Mycobacterium species (7). This uniqueness suggests that this molecule could be a particularly useful tool for the diagnosis of TB. Here, we report the biochemical characterization of the Rv1681 protein and the clinical validation of this molecule as a diagnostic biomarker for active pulmonary tuberculosis.
MATERIALS AND METHODS
Clinical specimens. (i) Specimens for antigen identification and biochemical validation.
(a) Brazil (antigen discovery). Urine specimens from 9 subjects with active pulmonary TB were collected as described previously (7, 8).
(b) Peru (Foundation for Innovative New Diagnostics) (antigen discovery). Urine specimens were first-morning specimens from untreated smear-positive TB suspects who were ultimately confirmed to have sputum culture-positive pulmonary TB. Specimens were stored at 4°C and frozen (−80°C) within 24 h.
(c) Lemuel Shattuck Hospital, Boston, MA (affinity chromatography). Overnight urine specimens (∼600 ml) were collected from two patients enrolled as TB suspects based on symptoms, chest X-ray (CXR) findings, and sputum smear results; the two patients were ultimately confirmed to have growth of M. tuberculosis from their sputum samples. There was no clinical suspicion of renal TB for either patient. Urine specimens were collected after the patients had received 8 to 9 days of treatment (both were receiving treatment on admission). The urine was centrifuged at 2,000 rpm, sterile filtered (0.2 μm), and stored at 4°C.
(ii) Specimens for diagnostic assay validation.
(a) Beth Israel Deaconess Medical Center, Boston, MA (Escherichia coli culture-positive control urine specimens). Aliquots of urine specimens processed for routine clinical culture were frozen at −80°C. Culture results were obtained by chart extraction. There was no clinical suspicion of TB for any patient.
(b) Federal University of Minas Gerais, Belo Horizonte, Brazil (non-TB tropical disease control specimens). Urine specimens were obtained from patients with cutaneous leishmaniasis (n = 10), Chagas' disease (n = 5), or schistosomiasis (n = 11). These samples were collected from patients from the area of Montes Claros City, Brazil, with diagnoses confirmed by the Department of Parasitology, Federal University of Minas Gerais, Belo Horizonte, Brazil.
The diagnosis of cutaneous leishmaniasis was confirmed by direct observation of the parasite amastigotes in biopsy specimens from typical skin lesions. The diagnosis of Chagas' disease was confirmed by two or more positive results on three standard serological tests (indirect immunofluorescence, enzyme-linked immunosorbent assay [ELISA], and indirect hemagglutination). The diagnosis of schistosomiasis was confirmed by the finding of schistosome eggs in the patient's stool. There was no clinical suspicion of pulmonary TB for any of these patients, and all of them had normal CXR findings.
(c) Forsyth Institute, Cambridge, MA (healthy control specimens). Spot urine specimens were collected from 14 healthy donors and frozen at −80°C. Ten of the 14 donors were from countries in which TB is endemic, had been BCG vaccinated, and were known to be purified protein derivative of tuberculin (PPD) positive; the other 4 donors were BCG and PPD negative.
(d) Texas/Mexico (University of Texas Health Science Center Houston, Brownsville Regional Campus, Brownsville, TX) (confirmed-TB or TB-ruled-out specimens). Participants were enrolled at TB clinics in southern Texas (Hidalgo County Health Department) or northeastern Mexico (Secretaria de Salud de Tamaulipas, Matamoros, Mexico). Eligible subjects were either (i) patients with suspected pulmonary TB based on clinical findings and positive sputum smears or abnormal CXR results or (ii) individuals who initially presented with symptoms potentially consistent with TB but who subsequently had three serial negative sputum smears and for whom TB treatment was not planned (“ruled-out” controls). Jail inmates, individuals <18 years of age, and patients receiving TB treatment were excluded. Participants were interviewed to document sociodemographic data and medical TB risk factors (diabetes and HIV), as described previously (9). Available preenrollment CXR and smear findings were extracted from medical records.
Random, non-“clean-catch” urine specimens (≥30 ml) were collected in standard sterile cups from all subjects, put at 4°C shortly thereafter, and aliquoted/frozen (−80°C) within 24 h. Noninduced “spot” sputum specimens were collected in parallel with urine specimens for both presumed TB cases and ruled-out controls. A second (first-morning) sputum specimen was subsequently collected prior to or within the first week of anti-TB treatment (if applicable) and was used only if the first sputum specimen had insufficient volume for analysis. Sputum was transported to Brownsville, TX, concentrated (10, 11), and inoculated into a mycobacterial growth indicator tube (MGIT) (BD Diagnostics, Sparks, MD) and a Lowenstein-Jensen-Gruft slant (LJ) (Remel, Lenexa, KS). The remainder was frozen (−70°C) for subsequent DNA extraction and PCR. Cultures were incubated for up to 42 days (MGIT 960) or 8 weeks (LJ). Positive cultures were evaluated by smear microscopy (TB acid-fast stain; BD Diagnostics, Sparks, MD), and the mycobacterial species was confirmed by M. tuberculosis AccuProbe testing (Gen-Probe, Inc., San Diego, CA) (Brownsville or Texas Department of State Health Services) and/or high-performance liquid chromatography (HPLC) (Texas Department of State Health Services) (12). DNA was extracted from concentrated sputum, and M. tuberculosis was detected using an in-house TaqMan assay for IS6110, RD1, and IS1081, as described previously (10, 13).
For subjects enrolled as controls who had not yet had CXRs (not routinely obtained for TB suspects in Mexico), CXRs (posterior-anterior) were obtained (typically within 1 to 2 weeks of enrollment) and reviewed by TB clinic physicians. For any subjects who received CXRs >2 weeks after enrollment, a follow-up telephone interview was performed 2 to 4 months after enrollment to confirm that the prior symptoms had fully resolved without any TB therapy and that no new symptoms potentially consistent with TB had developed.
Participants were classified as “confirmed TB” when M. tuberculosis was isolated from sputum culture and as “TB ruled out” when all microbiological findings (sputum smears [preenrollment], cultures, and PCR for M. tuberculosis) were negative and (i) CXR results were normal or not suggestive of TB, (ii) a follow-up interview revealed no concerns regarding untreated TB, or (iii) both. One subject's culture grew non-TB mycobacteria, and this subject was included in the TB-ruled-out group. Participants with inconclusive or clinical TB classifications (e.g., negative smear/culture but abnormal CXR results consistent with TB) were excluded from further analysis.
Investigational review board approval.
The urine collection protocols were approved by the investigational review boards/ethics committees of the Medical School of Triângulo Mineiro, Instituto de Medicina Tropical Alexander von Humboldt (Universidad Peruana Cayetano Heredia, Lima, Peru), Federal University of Minas Gerais (Belo Horizonte, Brazil), Lemuel Shattuck Hospital (LSH), Beth Israel Deaconess Medical Center (BIDMC), Forsyth Institute, University of Texas Health Science Center Houston, and Secretaria de Salud de Tamaulipas. For all except the discarded-specimen studies, the participants provided written informed consent.
Generation of specific polyclonal antibodies.
Female New Zealand rabbits were immunized with purified recombinant Rv1681 (rRv1681) using the same methods as those described previously (7, 8). The IgG fraction was purified by standard protein G affinity chromatography. Two milligrams of the rabbit IgG pool was biotinylated with an EZ-Link Sulfo-NHS-LC biotinylation kit (Thermo Fisher Scientific, PA) according to the manufacturer's protocol. A range of 2 to 4 biotin molecules per IgG molecule was obtained. This determination was obtained using the kit manufacturer's instructions, which are based on published work (14). Western blot analyses using these antibodies were performed as described previously (7, 8).
Biochemical characterization of native Rv1681 protein in the urine of patients with active TB.
For the initial identification of Rv1681 protein in the urine of patients with active TB, urine specimens were processed and mass spectroscopy was performed as described previously (7, 8). For purification of native Rv1681 protein in the patients' urine specimens, standard immunoaffinity was performed using a 1-ml column made with purified rabbit IgG anti-rRv1681 antibody immobilized on Sepharose 4B resin (BrCN method), over which was run a pool of two urine specimens (∼800 ml) collected from two TB patients from LSH. The column was exhaustively washed with phosphate-buffered saline (PBS) (∼50 ml), followed by elution with 0.1 M glycine buffer (pH 2.5). After elution, the pH was adjusted to 7.5 with 10× PBS. Eluted material was then concentrated to a final volume of ∼150 μl with a 5-kDa-cutoff Centricon filter, subjected to SDS-PAGE, and Coomassie blue stained. Protein bands were excised from the gel and submitted to MS for determination of the protein sequences.
Antigen detection assay.
A capture enzyme-linked immunosorbent assay (ELISA) was developed using purified anti-rRv1681 IgG obtained from antisera produced in two different rabbits. Briefly, the wells of 96-well ELISA plates (high-binding enzyme immunoassay/radioimmunoassay plates; Corning International, Corning, NY) were coated overnight at 4°C with 0.2 g purified IgG (obtained from one of the immunized rabbits) diluted in bicarbonate buffer (pH 9.0). The wells were washed with PBS and 0.1% Tween 20 (PBST buffer) and then blocked with 1% bovine serum albumin (BSA) in PBST buffer for 2 h at room temperature. Urine specimens for testing were thawed, centrifuged at 2,000 rpm to remove debris, and sterile-filtered with 0.2-μm filters. Unconcentrated urine samples (200 μl/well) were then added to the plates and incubated overnight at 4°C. Plates were washed and then incubated for 2 h with biotin-labeled IgG (obtained from the second immunized rabbit) at 2 μg/ml, a dilution previously determined by conventional sandwich ELISA. Following three rinses with PBST buffer, peroxidase-labeled streptavidin at a 1:2,000 dilution (BD Bioscience, San Jose, CA) was added for 30 min. The plates were then washed with PBST buffer, the reactions were developed with 3,3′,5,5′-tetramethylbenzidine substrate (Sureblue TMB substrate; KPL, Gaithersburg, MD), and the absorbance was read at 450 nm. Each plate included a recombinant protein standard curve and a blank sample. Each experimental condition was assayed in triplicate, and the means and standard deviations (SDs) were calculated.
Statistical analysis.
ELISA results were analyzed with Excel (2010 Microsoft Office package). The cutoff for a positive result was calculated as the average of the results for the combined control samples from TB-ruled-out subjects, healthy donors, patients with E. coli culture-positive urine specimens, and patients with confirmed non-TB tropical diseases, plus 3 SDs (see Fig. 4).
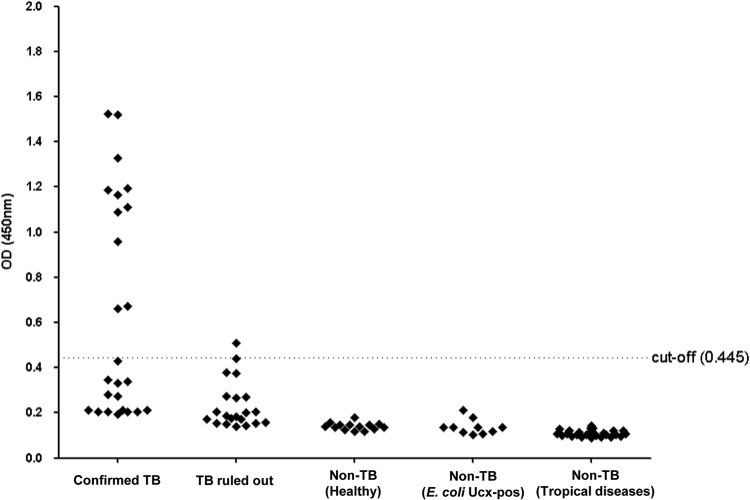
Fig 4.
Validation of Rv1681 protein as a diagnostic biomarker for active pulmonary tuberculosis. Five separate sets of urine specimens were tested for the presence of Rv1681 protein by the capture ELISA: confirmed TB, 25 patients with culture-confirmed active pulmonary TB (all pretreatment); TB ruled out, 21 patients in whom TB was initially clinically suspected but ultimately ruled out by conventional methods (20/21 had a history of BCG vaccination); non-TB (healthy), 14 healthy subjects (10/14 were from countries in which TB is endemic, had a history of BCG vaccination, and were known to be PPD positive); non-TB (E. coli Ucx-pos), 10 non-TB patients with E. coli-positive urine cultures; and non-TB (tropical diseases), 26 non-TB patients with confirmed tropical diseases, i.e., Chagas' disease (n = 5), cutaneous leishmaniasis (n = 10), or schistosomiasis (n = 11). The dotted line represents the cutoff for positive results, i.e., 0.445 (calculated by taking the average of the results for the four control groups plus 3 standard deviations).
RESULTS
Discovery of Rv1681 protein in the urine of TB patients by mass spectroscopy.
We previously described the discovery, by MS analysis, of Rv1681 protein in urine specimens from untreated Brazilian patients with culture-confirmed active pulmonary TB (7, 8). Peptides matching the amino acid sequence of Rv1681 protein were found in the urine specimens from three of the nine Brazilian subjects studied. The Rv1681 locus codes for a putative MoeX/MoaA-related protein (TIGR no. MT_1721, Rv no. Rv1681, NCBI reference sequence ZP_07480440.2). Subsequently, 12 additional urine specimens from Peruvian patients (pretreatment) with culture-confirmed pulmonary TB (see Materials and Methods) were similarly analyzed by MS. Urine from one of those subjects also yielded peptides matching the amino acid sequence of Rv1681 protein. A summary of the MS and sequence data obtained for the peptides previously identified in the urine specimens from each of the three subjects from Brazil (subjects 1 to 3) and more recently from the one subject from Peru (subject 4) is shown in Table 1. These peptide sequences provided highly significant MS correlation score (XCorr) values (2.1400 to 3.1587), and the peptide sequences are unique to the members of the M. tuberculosis complex (E value of ≤1e−05).
Table 1.
Summary of mass spectroscopy data for Rv1681 peptides found in urine specimens from four TB patients from Brazil and Peru
| Patient origin | Patient identification no. | No. of Rv1681 peptides found in the urine | Peptide sequence | Peptide rank charge | No. of ions seen/no. of ions predicted | ΔCna | XCorr | Positions in the Rv1681 protein |
|---|---|---|---|---|---|---|---|---|
| Brazil | 1 | 1 | MVIIELMR | 2 | 13/14 | 0.150 | 2.2400 | 1–8 |
| 2 | 1 | MVIIELMR | 2 | 13/14 | 0.100 | 2.3700 | 1–8 | |
| 3 | 1 | MVIIELMR | 2 | 13/14 | 0.080 | 2.1400 | 1–8 | |
| Peru | 4 | 2 | TVEPLTPALDMVSR | 2 | 16/26 | 0.410 | 3.1587 | 299–312 |
| LFAEQWTR | 2 | 13/14 | 0.095 | 2.7127 | 313–320 |
ΔCn, difference between the XCorr values for the best match and the second-best match.
Biochemical characterization of native Rv1681 protein in M. tuberculosis.
rRv1681 was expressed, purified, and used to produce rabbit antisera (see Materials and Methods). Validation of Rv1681 protein as a genuine secreted M. tuberculosis molecule was then performed by Western blotting using rabbit anti-rRv1681 IgG antibodies, a M. tuberculosis whole-cell lysate, and a culture filtrate (supplied by John Belisle, Colorado State University). This analysis demonstrated that the anti-rRv1681 IgG recognized the corresponding native Rv1681 protein in both the mycobacterial lysate and culture filtrate and, as expected, the recombinant protein (Fig. 1). The molecular mass of the band recognized in the mycobacterial lysate and the culture filtrate matched the predicted molecular mass of the native Rv1681 protein. These results confirm that Rv1681 protein is produced by M. tuberculosis and is present among the secreted proteins in the M. tuberculosis culture filtrate.
Fig 1.
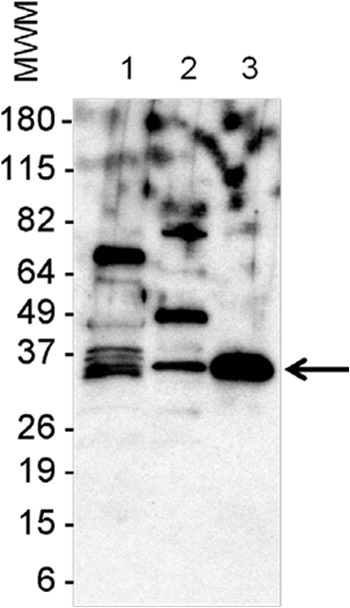
Rv1681 protein expression and secretion by M. tuberculosis. The validation of Rv1681 protein as a genuine secreted M. tuberculosis molecule was performed by Western blotting using an M. tuberculosis whole-cell lysate and a culture filtrate. Lane 1, crude whole-cell lysate; lane 2, culture filtrate; lane 3, purified recombinant antigen (rRv1681). Proteins were electrophoresed under reducing conditions in a 4-to-20% gradient gel and were transferred to a nitrocellulose membrane, followed by probing with rabbit anti-rRv1681. Reactivity was detected with peroxidase-labeled goat anti-rabbit immunoglobulin, and signals were developed using an enhanced chemiluminescence reagent. Numbers on the left are the molecular weights of the markers (MWM). The arrow points to rRv1681 (lane 3) and corresponding protein bands in the crude bacterial cell lysate (lane 1) and culture filtrate (lane 2).
Biochemical identification and characterization of Rv1681 protein in the urine of TB patients.
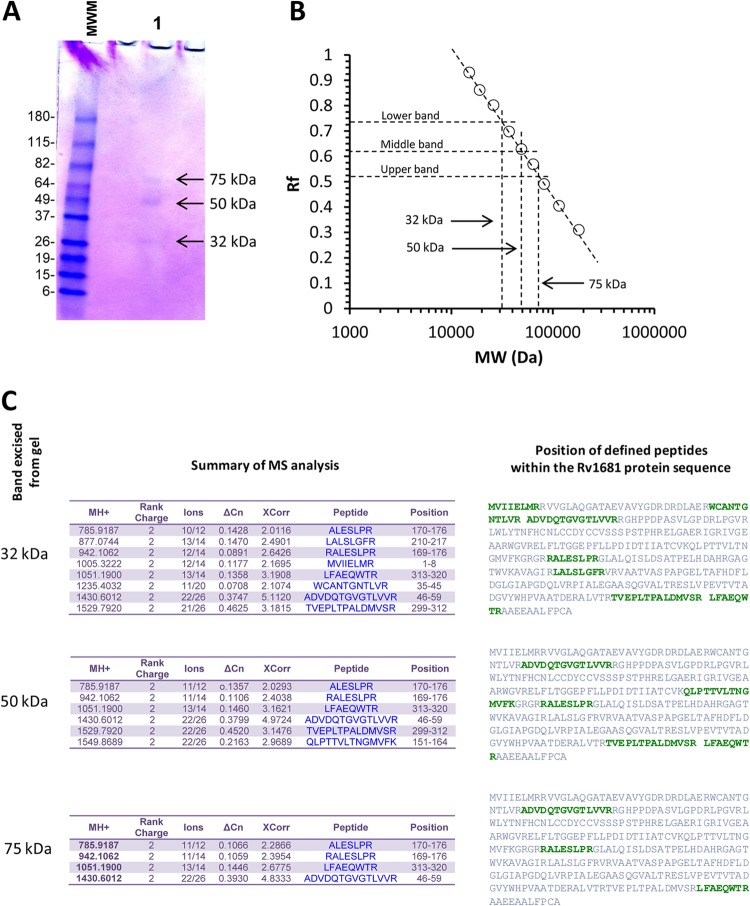
As described above, the initial discovery of Rv1681 protein in the urine of TB patients was by mass spectroscopy, a method that deduces peptide sequences but does not actually detect protein molecules biochemically in the sample. To definitively demonstrate that Rv1681 protein could be detected immunologically in the urine of TB patients, we performed immunoaffinity purification. Large-volume urine specimens (∼800 ml) were collected and pooled from two patients with active pulmonary TB who were enrolled at LSH (see Materials and Methods). The two subjects had been on standard TB treatment regimens for 8 and 7 days at the time of urine collection (both were already receiving treatment on admission to LSH). The pooled urine was run over an affinity column containing purified rabbit IgG anti-rRv1681 antibody immobilized on Sepharose 4B resin, and the proteins which bound to the column were subsequently acid eluted and run on SDS-PAGE gels (Fig. 2A). The eluate contained three discrete protein bands of 75, 50, and 32 kDa (Fig. 2B). The band of 32 kDa approximately matches the deduced molecular mass of the full-length native Rv1681 protein (35.3 kDa). The 75-kDa, 50-kDa, and 32-kDa bands were cut from the gel and sent for protein sequencing by MS (see Materials and Methods). The results identified the three affinity-purified molecules as Rv1681 protein (Fig. 2C). Although the two bands of 50 kDa and 75 kDa have higher molecular masses than the deduced molecular mass of the native Rv1681 protein, they might represent polymerization (Rv1681 contains 8 cysteine residues), aggregates of the native molecule with itself (or with fragments of itself), or complexes of the native molecule with other proteins. Understanding why Rv1681 peptide sequences can be found in the urine of TB patients in molecular forms that have higher molecular masses than the predicted molecular mass of the native Rv1681 molecule is beyond the scope of this article. Nonetheless, this result demonstrates that M. tuberculosis proteins can be immunochemically detected in the urine of TB patients, therefore establishing the proof of concept for our approach. Because the peptides detected by MS from the 32-kDa band aligned with amino acid sequences across multiple regions of the Rv1681 protein (Fig. 2C), these results also suggest that at least some of the native proteins might be present in urine as full-length molecules.
Fig 2.
Detection of Rv1681 protein, by immunoaffinity chromatography, in urine specimens from patients with active TB. (A) SDS-PAGE gel of immunoaffinity-purified protein from pooled urine specimens from two TB patients. MWM, molecular weight markers; lane 1, eluted affinity-purified fraction. The arrows point to bands with molecular masses of 32, 50, and 75 kDa, calculated using their respective relative mobilities in the gel (B). (C) Summary data for the MS analysis performed using each one of the three bands individually excised from the gel. The position of each identified peptide within the amino acid sequence of Rv1681 protein is highlighted in green letters.
Analytic performance of conventional capture ELISA for the detection of rRv1681.
The analytical performance of the capture ELISA (see Materials and Methods) was evaluated by spiking rRv1681 at decreasing concentrations (100, 10, 1, 0.1, and 0.01 ng/ml) into either 20 mM Tris buffer or urine from healthy donors (Fig. 3). The limit of detection for this assay (arbitrarily defined as an optical density [OD] 50% above the OD for the background reading) for protein spiked into either urine or buffer ranged from ∼20 to 50 pg/ml. The background (no added rRv1681) OD readings for the assays performed with buffer versus those performed with urine did not differ.
Fig 3.
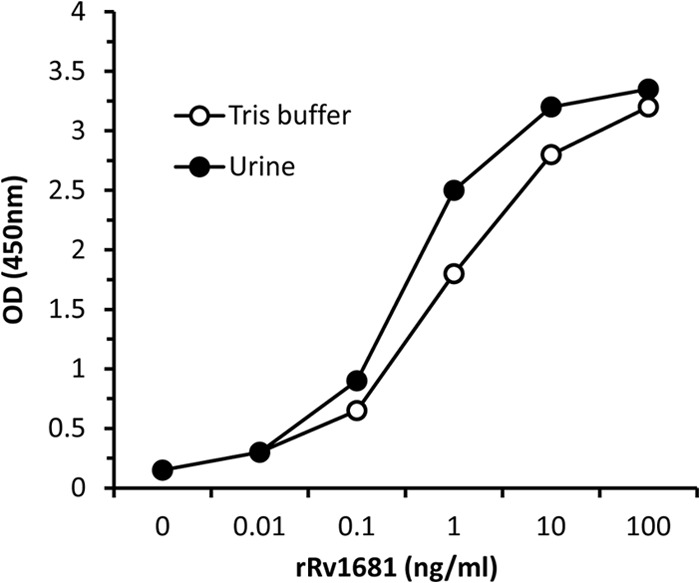
Dose-response curves for the rRv1681 capture ELISA in buffer and urine. rRv1681 was spiked at decreasing concentrations (e.g., 100 ng/ml and 10 ng/ml) into either 20 mM Tris buffer or urine from healthy donors. The capture ELISA was performed as described (see Materials and Methods), and the results were read at 450 nm.
Assessment of the utility of Rv1681 protein as a diagnostic biomarker for active pulmonary TB.
Given that the ultimate goal of this work was to develop a diagnostic test for active TB, we proceeded to a preliminary clinical validation of Rv1681 protein as a diagnostic biomarker by performing conventional capture ELISA for Rv1681 protein in urine specimens from patients with or without active TB (see Materials and Methods). Patients with suspected pulmonary TB and control subjects for whom active TB had been preliminarily ruled out by three negative sputum smears were enrolled from TB clinics in Texas/Mexico and subsequently classified as “confirmed TB” (sputum culture positive for M. tuberculosis, n = 25) or “TB ruled out” (n = 21) by a combination of sputum culture/PCR, CXR, and clinical follow-up. Urine specimens from TB suspects were collected prior to initiating TB therapy. As additional controls, 10 discarded clinical urine specimens submitted for routine culture were collected; none of the donors had suspected TB, all donors had suspected urinary tract infection (UTI), 8/10 donors had urinalysis results consistent with UTI, and all 10 urine cultures yielded pure E. coli (≥10,000 to 100,000 organisms/ml). An additional 26 control urine specimens were collected from patients with confirmed non-TB tropical diseases (11 with schistosomiasis, 5 with Chagas' disease, and 10 with cutaneous leishmaniasis). Finally, an additional 14 control urine specimens were collected from healthy subjects; 10/14 subjects were from countries in which TB is endemic, had been BCG vaccinated, and were known to be PPD positive, and the other 4/14 subjects had not received BCG vaccine and were PPD negative. All urine specimens were frozen (−80°C) prior to testing and were handled in the same manner upon thawing. The Rv1681 protein capture ELISA was performed as described using 200 μl of unconcentrated urine per well.
Using a standard cutoff value calculated as the average of the results for the four groups of control samples plus 3 SDs, 11/25 (44%) confirmed TB patients tested positive by capture ELISA (Fig. 4); all 11 were sputum smear positive. Of the 14 confirmed TB patients whose urine specimens tested negative by ELISA, 10/14 (71%) were sputum smear positive and 4/14 (29%) were sputum smear negative. We did not attempt to correlate urine results with M. tuberculosis burden by sputum smear (e.g., 1+, 2+, etc.) because those smears were prepared by nonstudy personnel prior to urine collection and smear results represented cumulative data from up to 3 smears.
All 10 subjects with E. coli-positive urine cultures, all 26 subjects with confirmed non-TB tropical diseases, and all 14 healthy subjects tested negative by ELISA (Fig. 4). Of the 21 patients for whom TB was ruled out by conventional methods (20/21 of whom had a history of BCG vaccination), 1 (4.8%) tested positive by ELISA (Fig. 4). The clinical history and laboratory data for this individual were examined in detail. The subject had had 3 negative sputum smears prior to enrollment, per the inclusion criteria, and negative culture and PCR results for sputum collected at enrollment. However, only one (rather than two) study sputum sample was collected for the subject, and low volume enabled MGIT but not LJ culture (see Materials and Methods). This subject had experienced 3 days of productive cough, chest pain, and difficulty breathing at the time of enrollment but was one of the few subjects who did not have a CXR; these symptoms resolved without TB treatment. The subject had diabetes but was HIV negative. It is thus difficult to completely exclude the possibility that this subject had active TB at the time of urine collection, but this diagnosis was not made by conventional methods. Finally, one subject grew a non-TB mycobacterium from acid-fast bacillus smear-positive sputum and was thus included in the TB-ruled-out group; this patient's urine tested negative in the ELISA.
DISCUSSION
Our study has validated Rv1681 protein as a biomarker of active TB. The detection of full-length Rv1681 protein in the urine of patients with active pulmonary TB by immunoaffinity precipitation supports our initial suggestion that this molecule has promise as a urinary diagnostic biomarker; our detection of the protein in the urine specimens of 11/25 (44%) culture-confirmed TB patients further strengthens this argument. Our study included four types of key clinical negative controls, i.e., healthy individuals, patients with high urinary burdens of E. coli (a frequent colonizer and cause of UTI), patients with non-TB tropical diseases, and patients in whom TB was initially suspected but then ruled out by conventional methods. The clinical specificity of the assay is supported by the complete absence of urine reactivity in the healthy control individuals (most of whom were PPD positive, were from countries in which TB is endemic, and might have had latent TB), patients with E. coli culture-positive urine specimens (particularly given that our polyclonal antibodies were generated against rRv1681 produced in E. coli), and patients with confirmed non-TB tropical diseases. We made the decision to include the TB-ruled-out controls (Fig. 4) in the determination of the cutoff because we felt that this was more reflective of real-world application than a lower cutoff calculated using only healthy, E. coli culture-positive, and confirmed non-TB tropical disease controls (Fig. 4). TB-ruled-out controls are a critical population for the evaluation of assay specificity, but gold standard diagnosis in this group is inherently complicated by the limitations of existing diagnostic methods. Our assay detected Rv1681 protein in the urine of 1/21 (4.8%) individuals in this group; further rigorous evaluation of our assay in this type of control population will be key to confirming the specificity of our assay.
It has been proposed that a rapid and widely available diagnostic test for TB with ≥85% sensitivity for smear-positive and smear-negative cases and 97% specificity could save ∼400,000 lives annually (15). While the newly developed Xpert MTB/RIF assay (Cepheid, Inc.) (endorsed by the WHO in 2010 [16]) offers a large step forward by making sensitive nucleic acid amplification-based diagnosis widely available, it does not satisfy the need for a simple inexpensive POC test (6, 17); the technology remains prohibitively expensive in some parts of the world, is optimized for sputum specimens, and is not intended for use as a POC device. Urine has the potential to be both a convenient and a high-yield clinical sample for TB diagnosis (18), given the ease of collection from both adults and children, the fact that collection confers minimal TB transmission risk, and the potential utility for diagnosis of smear-negative and extrapulmonary TB. With these potential advantages in mind, our research has focused on the discovery and evaluation of novel M. tuberculosis antigens that are produced in vivo in humans (7, 8) and mice (19, 20) during active TB disease, enter the blood circulation, and are ultimately eliminated in the urine. The hypothesis underlying these studies has been that microbial antigens found in vivo in the bodily fluids of the host would be potentially effective candidate molecules for the development of an antigen detection assay for the diagnosis of active TB.
Precedent for the use of mycobacterial antigens in diagnostic test kits is provided by assays developed to detect a mycobacterial cell wall lipopolysaccharide antigen, lipoarabinomannan (LAM), in urine (21). Although this precedent supports our hypothesis, LAM tests have shown widely varying sensitivities and specificities (resulting in current utility potentially only for diagnosis of HIV-associated TB in patients with advanced immunodeficiency) (21), and specificity is potentially hindered by the widespread distribution of LAM molecules across mycobacterial species and potential cross-reaction of anti-LAM antibodies with nonmycobacterial species (21–23). In contrast, the specificity of Rv1681 protein for the M. tuberculosis complex suggests that we can expect high clinical specificity of an assay based on this molecule. In this study, only one of our specimens was from a patient who was confirmed to have a pulmonary infection with non-TB mycobacteria, and it tested negative by ELISA. Further evaluation of our assay among subjects with non-TB mycobacterial disease will be important for confirming assay specificity.
We recognize that a sensitivity of 44% would be insufficient for this ELISA to be clinically useful in its current form. We have anticipated that the concentrations of M. tuberculosis proteins in urine might be below the achievable limit of detection of conventional ELISA, which is typically in the pg/ml range (24, 25), and that successful clinical test development will likely require the application of more-sensitive novel protein detection technology; collaborative experiments toward this goal are under way. An additional strategy for increasing clinical sensitivity and specificity would be to combine detection of multiple biomarkers in one assay; in support of this approach, we note that we previously demonstrated the ability to detect the M. tuberculosis biomarker MT_1694 protein in 37.5% (6/16) of patients with active pulmonary TB using a similar conventional ELISA (8). Larger clinical studies and a robust fully developed diagnostic assay will be required to validate this biomarker in the field and ultimately to determine its optimal place in TB diagnostic algorithms.
In sum, the M. tuberculosis Rv1681 protein and the other three proteins that we described previously (7, 8) are the only M. tuberculosis protein biomarkers reported to date that have been identified directly from urine specimens from TB patients from regions where TB is endemic and thus are, we feel, strong candidates for development of an antigen detection test for the diagnosis of active TB. Such a test should be able to distinguish between patients with active versus latent disease and would thus provide an important new strategy for control of this global disease. In particular, an antigen detection test that uses urine could potentially allow POC TB diagnosis—not only in adults with pulmonary TB, but also in difficult-to-diagnose populations such as children and adults with extrapulmonary TB. Such a test might also address the critical need for an assay that can monitor response to TB treatment. Our work with Rv1681 protein provides promising evidence that this biomarker, in particular, represents an attractive candidate for the development of a highly sensitive and specific urine antigen detection test.
ACKNOWLEDGMENTS
This work was supported by grants from the Foundation for Innovative New Diagnostics (FIND) (to A.C.-N.), the Bill and Melinda Gates Foundation (grant OPP1039610 to A.C.-N. and N.R.P.), the National Institutes of Health (grant R01 AI076425 to A.C.-N. and grant K23 AI074638 to N.R.P.), Harvard Catalyst (pilot grant to N.R.P.), and the Pittsfield Anti-Tuberculosis Association (to N.R.P.).
We thank Eloise Valli and Nora Champouillon for their assistance with managing FIND sample logistics and Matt MacKechnie in the BIDMC clinical microbiology laboratory for assistance with collecting discarded clinical urine specimens. We also thank members of the LSH staff, particularly Marie Turner, for assistance with collecting urine specimens from LSH patients.
Footnotes
Published ahead of print 6 February 2013
REFERENCES
- 1. World Health Organization 2012. Global tuberculosis report 2012. World Health Organization, Geneva, Switzerland [Google Scholar]
- 2. McNerney R, Maeurer M, Abubakar I, Marais B, McHugh TD, Ford N, Weyer K, Lawn S, Grobusch MP, Memish Z, Squire SB, Pantaleo G, Chakaya J, Casenghi M, Migliori GB, Mwaba P, Zijenah L, Hoelscher M, Cox H, Swaminathan S, Kim PS, Schito M, Harari A, Bates M, Schwank S, O'Grady J, Pletschette M, Ditui L, Atun R, Zumla A. 2012. Tuberculosis diagnostics and biomarkers: needs, challenges, recent advances, and opportunities. J. Infect. Dis. 205(Suppl 2):S147–S158 [DOI] [PubMed] [Google Scholar]
- 3. World Health Organization 2010. The global plan to stop TB 2011-2015. Publication no. WHO/HTM/STB/2010.2. World Health Organization, Geneva, Switzerland [Google Scholar]
- 4. Steingart KR, Ng V, Henry M, Hopewell PC, Ramsay A, Cunningham J, Urbanczik R, Perkins MD, Aziz MA, Pai M. 2006. Sputum processing methods to improve the sensitivity of smear microscopy for tuberculosis: a systematic review. Lancet Infect. Dis. 6:664–674 [DOI] [PubMed] [Google Scholar]
- 5. Steingart KR, Ramsay A, Pai M. 2007. Optimizing sputum smear microscopy for the diagnosis of pulmonary tuberculosis. Expert Rev. Anti Infect. Ther. 5:327–331 [DOI] [PubMed] [Google Scholar]
- 6. Wallis RS, Pai M, Menzies D, Doherty TM, Walzl G, Perkins MD, Zumla A. 2010. Biomarkers and diagnostics for tuberculosis: progress, needs, and translation into practice. Lancet 375:1920–1937 [DOI] [PubMed] [Google Scholar]
- 7. Kashino SS, Pollock N, Napolitano DR, Rodrigues V, Jr, Campos-Neto A. 2008. Identification and characterization of Mycobacterium tuberculosis antigens in urine of patients with active pulmonary tuberculosis: an innovative and alternative approach of antigen discovery of useful microbial molecules. Clin. Exp. Immunol. 153:56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Napolitano DR, Pollock N, Kashino SS, Rodrigues V, Jr, Campos-Neto A. 2008. Identification of Mycobacterium tuberculosis ornithine carboamyltransferase in urine as a possible molecular marker of active pulmonary tuberculosis. Clin. Vaccine Immunol. 15:638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Restrepo BI, Camerlin AJ, Rahbar MH, Wang W, Restrepo MA, Zarate I, Mora-Guzman F, Crespo-Solis JG, Briggs J, McCormick JB, Fisher-Hoch SP. 2011. Cross-sectional assessment reveals high diabetes prevalence among newly-diagnosed tuberculosis cases. Bull. World Health Organ. 89:352–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gomez DI, Mullin CS, Mora-Guzman F, Crespo-Solis JG, Fisher-Hoch SP, McCormick JB, Restrepo BI. 2011. Rapid DNA extraction for specific detection and quantitation of Mycobacterium tuberculosis DNA in sputum specimens using TaqMan assays. Tuberculosis (Edinb.) 91(Suppl 1):S43–S48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kent PT, Kubica GP. 1985. Public health mycobacteriology: a guide for the level III laboratory. Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 12. Butler WR, Jost KC, Jr, Kilburn JO. 1991. Identification of mycobacteria by high-performance liquid chromatography. J. Clin. Microbiol. 29:2468–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Restrepo BI, Gomez DI, Shipley GL, McCormick JB, Fisher-Hoch SP. 2006. Selective enrichment and detection of mycobacterial DNA in paucibacillary specimens. J. Microbiol. Methods 67:220–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Green NM, Konieczny L, Toms EJ, Valentine RC. 1971. The use of bifunctional biotinyl compounds to determine the arrangement of subunits in avidin. Biochem. J. 125:781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Keeler E, Perkins MD, Small P, Hanson C, Reed S, Cunningham J, Aledort JE, Hillborne L, Rafael ME, Girosi F, Dye C. 2006. Reducing the global burden of tuberculosis: the contribution of improved diagnostics. Nature 444(Suppl 1):49–57 [DOI] [PubMed] [Google Scholar]
- 16. World Health Organization 2011. Global tuberculosis control 2011. World Health Organization, Geneva, Switzerland [Google Scholar]
- 17. Weyer K, Carai S, Nunn P. 2011. Viewpoint TB diagnostics: what does the world really need? J. Infect. Dis. 204(Suppl 4):S1196–S1202 [DOI] [PubMed] [Google Scholar]
- 18. Peter J, Green C, Hoelscher M, Mwaba P, Zumla A, Dheda K. 2010. Urine for the diagnosis of tuberculosis: current approaches, clinical applicability, and new developments. Curr. Opin. Pulm. Med. 16:262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mukherjee S, Daifalla N, Zhang Y, Douglass J, Brooks L, Vedvick T, Houghton R, Reed SG, Campos-Neto A. 2004. Potential serological use of a recombinant protein that is a replica of a Mycobacterium tuberculosis protein found in the urine of infected mice. Clin. Diagn. Lab. Immunol. 11:280–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mukherjee S, Kashino SS, Zhang Y, Daifalla N, Rodrigues-Junior V, Reed SG, Campos-Neto A. 2005. Cloning of the gene encoding a protective Mycobacterium tuberculosis secreted protein detected in vivo during the initial phases of the infectious process. J. Immunol. 175:5298–5305 [DOI] [PubMed] [Google Scholar]
- 21. Lawn SD. 2012. Point-of-care detection of lipoarabinomannan (LAM) in urine for diagnosis of HIV-associated tuberculosis: a state of the art review. BMC Infect. Dis. 12:103 doi:10.1186/1471-2334-12-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dheda K, Davids V, Lenders L, Roberts T, Meldau R, Ling D, Brunet L, van Zyl SR, Peter J, Green C, Badri M, Sechi L, Sharma S, Hoelscher M, Dawson R, Whitelaw A, Blackburn J, Pai M, Zumla A. 2010. Clinical utility of a commercial LAM-ELISA assay for TB diagnosis in HIV-infected patients using urine and sputum samples. PLoS One 5:e9848 doi:10.1371/journal.pone.0009848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mishra AK, Driessen NN, Appelmelk BJ, Besra GS. 2011. Lipoarabinomannan and related glycoconjugates: structure, biogenesis and role in Mycobacterium tuberculosis physiology and host-pathogen interaction. FEMS Microbiol. Rev. 35:1126–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Avrameas S. 1992. Amplification systems in immunoenzymatic techniques. J. Immunol. Methods 150:23–32 [DOI] [PubMed] [Google Scholar]
- 25. Giljohann DA, Mirkin CA. 2009. Drivers of biodiagnostic development. Nature 462:461–464 [DOI] [PMC free article] [PubMed] [Google Scholar]