Abstract
Here, we report latent infections with Bartonella quintana and a hemotropic Mycoplasma sp. in a research colony of cynomolgus monkeys (Macaca fascicularis). Sequence alignments, evolutionary analysis, and signature nucleotide sequence motifs of the hemotropic Mycoplasma 16S rRNA and RNase P genes indicate the presence of a novel organism.
INTRODUCTION
Hemotropic Mycoplasma spp. (hemoplasmas) are obligate epierythrocytic bacteria that infect numerous animal species, including Homo sapiens. Infections are often chronic and subclinical; however, some animals and humans develop hemolytic anemia, particularly when stressed or immunosuppressed (1, 2). Phylogenetic analyses of 16S rRNA gene sequences have defined two major subclusters of hemoplasmas, namely, the Mycoplasma haemosuis and Mycoplasma haemofelis groups (3–7).
Historically, diagnosis of hemoplasma infections has relied on cytological examination of stained blood smears. In 1994, Dillberger and colleagues described Haemobartonella-like parasites in five wild-caught anemic cynomolgus monkeys (Macaca fascicularis) that originated from the Philippines; however, the organisms were not characterized phylogenetically (8). For some animal species, the diagnostic sensitivity of a blood smear examination is very poor and unspecific (3, 4, 9). The development of molecular assays, primarily targeting the 16S rRNA and the RNase P genes of these cell wall-deficient uncultivable microbes, has resulted in the recent recognition of several novel animal hemoplasmas (4, 5, 10–13).
Bartonella spp. are facultative intracellular bacteria that also infect erythrocytes in numerous animal species, including Homo sapiens. Previously, Bartonella quintana DNA was amplified, cloned, and sequenced from lysed erythrocytes, and cultured colonies were grown from peripheral blood collected from a captive-bred cynomolgus monkey (Macaca fascicularis) (14). Bartonella quintana was subsequently isolated from 2 of 36 captive rhesus macaques in China, of which 12 of 33 were B. quintana seroreactive (15).
Hemotropic Mycoplasma and Bartonella organisms often cause persistent occult infection in immunocompetent hosts. The extent to which an infection with these bacteria in cynomolgus monkeys involved in a research study might influence assessments or outcomes associated with drug development studies is poorly characterized. This report describes PCR amplification and DNA sequence characterization of a novel hemotropic Mycoplasma sp. found in the blood of 44 of 52 cynomolgus research monkeys (Macaca fascicularis) and the isolation of Bartonella quintana from one monkey. These animals were in a chronic toxicity study, and data from the pretest phase of the study are presented. The animals were tested for Mycoplasma and Bartonella on the basis of the findings in a previous toxicity study that raised the possibility of latent infections. Based on the analysis of the 16S rRNA and RNase P gene sequences, we propose “Candidatus Mycoplasma haemomacaque” as the name for the novel hemotropic Mycoplasma sp. identified in this study.
MATERIALS AND METHODS
Blood from 52 cynomolgus monkeys (Macaca fascicularis) was analyzed prior to the initiation of dosing in a toxicity study for the presence of hemotropic Mycoplasma and Bartonella spp. The monkeys were considered healthy on the basis of multiple pretest physical clinical evaluations, including Coomb's tests and microscopic blood smear evaluations.
Blood samples were collected in EDTA-containing Vacutainers and shipped overnight to Galaxy Diagnostics, Inc., to test for the presence of Mycoplasma spp. and Bartonella spp. Blood samples were analyzed for the presence of Mycoplasma DNA by PCR testing, targeting the 16S rRNA (a 1,200-bp fragment) and RNase P (a 160-bp fragment) genes as reported previously (16). Similarly, blood samples were analyzed for the presence of Bartonella spp. using the Bartonella alphaproteobacterial growth medium (BAPGM) enrichment culture PCR as described previously (16–18). DNAs from naive dog and human blood extracted at the same time and in the same manner were used as negative controls for the PCR testing.
Nucleotide sequence accession number.
The nucleotide sequence of the partial 16S rRNA gene has been deposited in GenBank under accession no. KC512401.
RESULTS
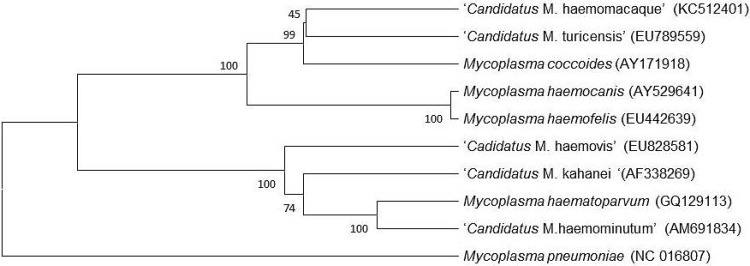
All animals were considered healthy on the basis of the pretest screening. In particular, there was no evidence of anemia, hyperbilirubinemia, or bilirubinuria. By targeting the 16S rRNA and RNase P genes, DNA of a novel hemotropic Mycoplasma sp. was amplified from 44 of 52 (84.6%) cynomolgus monkey blood samples but not from any of the negative controls tested. Sequence analyses of both genes identified a distinct genotype compared with those of sequences for other Mycoplasma spp. deposited in GenBank. When the 1,164-bp nucleotide sequence of the partial 16S rRNA gene was compared with those of M. coccoides (AY171918), “Candidatus Mycoplasma turicensis” (EU789559), M. haemofelis (EU442639), M. haemocanis (AY529641), “Candidatus Mycoplasma haemovis” (EU828581), “Candidatus Mycoplasma haematoparvum” (GQ129113), and “Candidatus Mycoplasma haemominutum” (AM691834), the novel hemoplasma from cynomolgus monkeys shared 90.9% (1,058/1,164 bp) homology with “Candidatus Mycoplasma turicensis,” followed by 90.4% (1,052/1,164 bp) homology with M. coccoides. The nucleotide sequence homology was lower for M. haemocanis (87.3%), M. haemofelis (85.8%), and “Candidatus Mycoplasma kahanei” (79.6%) found in squirrel monkeys (19) (AF338269) and for M. pneumoniae (NC_016807) (76.5%). The bootstrap percentage values are given at the nodes of the phylogenetic tree shown in Fig. 1.
Fig 1.
Phylogenetic tree based on 16S rRNA gene sequences, comparing the positions of “Candidatus Mycoplasma haemomacaque” and other hemotropic Mycoplasma spp. Bootstrap percentages are given at the nodes of the tree.
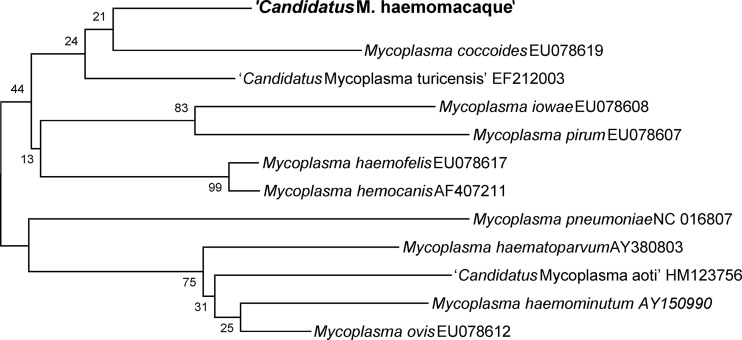
Similarly, when the nucleotide sequence of the partial RNase P gene obtained from Macaca fascicularis was compared with those of other reported Mycoplasma spp., M. coccoides (GenBank accession no. EU078619), “Candidatus Mycoplasma aoti” (HM123756), M. iowae (EU078608), M. pirum (EU078607), “Candidatus Mycoplasma turicensis” (EF212003), M. haemofelis (EU078617), M. haemocanis (AF407211), M. haemovis (EU078612), M. haematoparvum (AY380803), and “Candidatus Mycoplasma haemominutum” (AY150990), there was very low homology. “Candidatus Mycoplasma haemomacaque” shared 78% homology with “Candidatus Mycoplasma aoti” and 74.8% homology with M. haemofelis and M. haemocanis, followed by “Candidatus Mycoplasma turicensis” (72.8%), M. coccoides (68%), M. pirum (66%), M. haematoparvum (62%), and M. iowae (59.2%). Phylogenetic analysis of the partial RNase P gene, including comparisons with sequences available for hemotropic Mycoplasma spp., are shown in Fig. 2.
Fig 2.
Phylogenetic tree based on RNase P gene sequences, comparing the positions of “Candidatus Mycoplasma haemomacaque” and other hemotropic Mycoplasma spp. Bootstrap percentages are given at the nodes of the tree.
In addition to the novel Mycoplasma sp., the Bartonella quintana 16S rRNA-23S rRNA intergenic spacer region DNA was sequenced from the extracted blood, from 7- and 14-day BAPGM enrichment cultures, and from a subculture isolate (20–23) from one monkey. Sequence analysis of the Bartonella internal transcribed spacer (ITS) region revealed 100% homology (420/420 bp) with Bartonella quintana (GenBank accession no. L35100).
DISCUSSION
Infection with a novel hemotropic Mycoplasma sp. was documented in 44/52 (84.6%) monkeys, and B. quintana was isolated from 1/52 (1.9%) cynomolgus monkeys in a research colony. Analysis of the hemoplasma 16S rRNA gene sequences derived from Macaca fascicularis in this study identified a 92.3% similarity to M. coccoides and a 90.6% similarity to “Candidatus Mycoplasma turicensis.” The RNase P sequence, used to discriminate among hemotropic Mycoplasma organisms, as described by Birkenheuer et al. in 2002 (5) and Tasker et al. in 2003 (3), revealed low similarities with other hemotropic Mycoplasma spp., including M. haemocanis, M. haemofelis, “Candidatus Mycoplasma turicensis,” and M. coccoides. Based on the phylogenetic analysis of DNA sequences found in these cynomolgus monkeys compared with those of other hemoplasma and nonhemotropic Mycoplasma spp., the low percentage of similarities of this bacteria supports its designation as a novel hemoplasma. Based on differences in the 16S rRNA and partial RNase P gene homologies and according to the guidelines for naming uncultivated prokaryotes (24, 25), we propose a “Candidatus” designation for this newly recognized macaque hemoplasma and recommend that it be named “Candidatus Mycoplasma haemomacaque.”
This report represents the third time that B. quintana has been isolated from nonhuman primates raised in research facilities (14, 15). Infections resulting in chronic bacteremia have also been established experimentally in rhesus macaque monkeys (Macaca mulatta) inoculated with B. quintana isolates derived from infected humans (26, 27), which supports the fact that nonhuman primates might be able to acquire B. quintana from humans or from other monkeys.
Hemotropic Mycoplasma spp. (hemoplasmas, formerly classified as Haemobartonella and Eperythrozoon spp.) (4, 12, 28, 29) appear to have coevolved with animals, including dogs, cats, humans, alpacas, capybaras, and sea lions (1, 10, 13, 30–40). The development of molecular assays, which target primarily the 16S rRNA gene of these microbes, has resulted in the more recent recognition of several novel animal hemoplasmas (5, 12, 37, 41). Hemoplasmas are obligate epierythrocytic organisms that attach to erythrocytes, appear to be relatively nonpathogenic, and are visualized on blood smears more often during periods of stress, hard work, or concurrent infection (1, 2, 7, 10, 42, 43). In some animals, hemoplasma infection is associated with hemolytic anemia of variable severity, ranging from nonclinical hemolysis to severe anemia (7, 40, 44). There were no pretest hematological or serum biochemical abnormalities associated with the novel hemotropic Mycoplasma sp. or B. quintana in the cynomolgus monkeys in this study.
ACKNOWLEDGMENTS
E.B.B., in conjunction with Sushama Sontakke and North Carolina State University, holds U.S. patent 7,115,385, Media and Methods for Cultivation of Microorganisms, which was issued 3 October 2006.
E.B.B. is the chief scientific officer for Galaxy Diagnostics, Inc., a newly formed company that provides diagnostic testing for the detection of Bartonella species infection in animals and human patients. R.G.M. has led research efforts to optimize the BAPGM platform and is the scientific technical advisor for Galaxy Diagnostics. P.E.M. was a part-time research scientist at Galaxy Diagnostics. N.B., C.M.R., C.M.K., L.R., and M.W.L. have no potential conflicts of interest.
The use of animal subjects was approved by the Pfizer institutional review board (IACUC approval HLS 10-3497, Pfizer 10MA066).
Footnotes
Published ahead of print 13 February 2013
REFERENCES
- 1. dos Santos AP, dos Santos RP, Biondo AW, Dora JM, Goldani LZ, de Oliveira ST, de Sa Guimaraes AM, Timenetsky J, de Morais HA, Gonzalez FH, González FH, Messick JB. 2008. Hemoplasma infection in HIV-positive patient, Brazil. Emerg. Infect. Dis. 14:1922–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Messick JB. 2004. Hemotrophic mycoplasmas (hemoplasmas): a review and new insights into pathogenic potential. Vet. Clin. Pathol. 33:2–13 [DOI] [PubMed] [Google Scholar]
- 3. Tasker S, Helps CR, Day MJ, Harbour DA, Shaw SE, Harrus S, Baneth G, Lobetti RG, Malik R, Beaufils JP, Belford CR, Gruffydd-Jones TJ. 2003. Phylogenetic analysis of hemoplasma species: an international study. J. Clin. Microbiol. 41:3877–3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Willi B, Boretti FS, Tasker S, Meli ML, Wengi N, Reusch CE, Lutz H, Hofmann-Lehmann R. 2007. From Haemobartonella to hemoplasma: molecular methods provide new insights. Vet. Microbiol. 125:197–209 [DOI] [PubMed] [Google Scholar]
- 5. Birkenheuer AJ, Breitschwerdt EB, Alleman AR, Pitulle C. 2002. Differentiation of Haemobartonella canis and Mycoplasma haemofelis on the basis of comparative analysis of gene sequences. Am. J. Vet. Res. 63:1385–1388 [DOI] [PubMed] [Google Scholar]
- 6. Neimark H. 1984. Deletions, duplications and rearrangements in mycoplasma ribosomal RNA gene sequences. Isr. J. Med. Sci. 20:765–767 [PubMed] [Google Scholar]
- 7. Willi B, Boretti FS, Baumgartner C, Cattori V, Meli ML, Doherr MG, Reusch CE, Hofmann-Lehmann R. 2006. Feline hemoplasmas in Switzerland: identification of a novel species, diagnosis, prevalence, and clinical importance. Schweiz. Arch. Tierheilkd. 148:139–140, 142, 144 passim. (In German.) [DOI] [PubMed] [Google Scholar]
- 8. Dillberger JE, Loudy DE, Adler RR, Gass JH. 1994. Hemobartonella-like parasites in cynomolgus monkeys (Macaca fascicularis). Vet. Pathol. 31:301–307 [DOI] [PubMed] [Google Scholar]
- 9. Tasker S, Lappin MR. 2002. Haemobartonella felis: recent developments in diagnosis and treatment. J. Feline Med. Surg. 4:3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sykes JE, Lindsay LL, Maggi RG, Breitschwerdt EB. 2010. Human coinfection with Bartonella henselae and two hemotropic mycoplasma variants resembling Mycoplasma ovis. J. Clin. Microbiol. 48:3782–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Neimark H, Peters W, Robinson BL, Stewart LB. 2005. Phylogenetic analysis and description of Eperythrozoon coccoides, proposal to transfer to the genus Mycoplasma as Mycoplasma coccoides comb. nov. and request for an opinion. Int. J. Syst. Evol. Microbiol. 55:1385–1391 [DOI] [PubMed] [Google Scholar]
- 12. Peters IR, Helps CR, McAuliffe L, Neimark H, Lappin MR, Gruffydd-Jones TJ, Day MJ, Hoelzle LE, Willi B, Meli M, Hofmann-Lehmann R, Tasker S. 2008. RNase P RNA gene (rnpB) phylogeny of hemoplasmas and other Mycoplasma species. J. Clin. Microbiol. 46:1873–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vieira RF, Molento MB, dos Santos LC, Moraes W, Cubas ZS, Santos AP, Guimaraes AM, Mohamed A, Barros Filho IR, Biondo AW, Messick JB. 2009. Detection of a novel hemoplasma based on 16S rRNA gene DNA in captive and free-ranging capybaras (Hydrochaeris hydrochaeris). Vet. Microbiol. 139:410–413 [DOI] [PubMed] [Google Scholar]
- 14. O'Rourke LG, Pitulle C, Hegarty BC, Kraycirik S, Killary KA, Grosenstein P, Brown JW, Breitschwerdt EB. 2005. Bartonella quintana in cynomolgus monkey (Macaca fascicularis). Emerg. Infect. Dis. 11:1931–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang R, Liu Q, Li G, Li D, Song X, Birtles RJ, Zhao F. 2011. Bartonella quintana infections in captive monkeys, China. Emerg. Infect. Dis. 17:1707–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Compton SM, Maggi RG, Breitschwerdt EB. 2012. Candidatus Mycoplasma haematoparvum and Mycoplasma haemocanis infections in dogs from the United States. Comp. Immunol. Microbiol. Infect. Dis. 35:557–562 [DOI] [PubMed] [Google Scholar]
- 17. Varanat M, Maggi RG, Linder KE, Breitschwerdt EB. 2011. Molecular prevalence of Bartonella, Babesia, and hemotropic Mycoplasma sp. in dogs with splenic disease. J. Vet. Intern. Med. 25:1284–1291 [DOI] [PubMed] [Google Scholar]
- 18. Pérez C, Maggi RG, Diniz PP, Breitschwerdt EB. 2011. Molecular and serological diagnosis of Bartonella infection in 61 dogs from the United States. J. Vet. Intern. Med. 25:805–810 [DOI] [PubMed] [Google Scholar]
- 19. Neimark H, Barnaud A, Gounon P, Michel JC, Contamin H. 2002. The putative haemobartonella that influences Plasmodium falciparum parasitaemia in squirrel monkeys is a haemotrophic mycoplasma. Microbes Infect. 4:693–698 [DOI] [PubMed] [Google Scholar]
- 20. Maggi RG, Mozayeni BR, Pultorak EL, Hegarty BC, Bradley JM, Correa M, Breitschwerdt EB. 2012. Bartonella spp. bacteremia and rheumatic symptoms in patients from Lyme disease-endemic region. Emerg. Infect. Dis. 18:783–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maggi RG, Mascarelli PE, Pultorak EL, Hegarty BC, Bradley JM, Mozayeni BR, Breitschwerdt EB. 2011. Bartonella spp. bacteremia in high-risk immunocompetent patients. Diagn. Microbiol. Infect. Dis. 71:430–437 [DOI] [PubMed] [Google Scholar]
- 22. Maggi RG, Duncan AW, Breitschwerdt EB. 2005. Novel chemically modified liquid medium that will support the growth of seven Bartonella species. J. Clin. Microbiol. 43:2651–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Duncan AW, Maggi RG, Breitschwerdt EB. 2007. A combined approach for the enhanced detection and isolation of Bartonella species in dog blood samples: pre-enrichment liquid culture followed by PCR and subculture onto agar plates. J. Microbiol. Methods 69:273–281 [DOI] [PubMed] [Google Scholar]
- 24. Murray RG, Stackebrandt E. 1995. Taxonomic note: implementation of the provisional status Candidatus for incompletely described procaryotes. Int. J. Syst. Bacteriol. 45:186–187 [DOI] [PubMed] [Google Scholar]
- 25. Woese CR, Stackebrandt E, Ludwig W. 1984. What are mycoplasmas: the relationship of tempo and mode in bacterial evolution. J. Mol. Evol. 21:305–316 [DOI] [PubMed] [Google Scholar]
- 26. MacKichan JK, Gerns HL, Chen YT, Zhang P, Koehler JE. 2008. A SacB mutagenesis strategy reveals that the Bartonella quintana variably expressed outer membrane proteins are required for bloodstream infection of the host. Infect. Immun. 76:788–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang P, Chomel BB, Schau MK, Goo JS, Droz S, Kelminson KL, George SS, Lerche NW, Koehler JE. 2004. A family of variably expressed outer-membrane proteins (Vomp) mediates adhesion and autoaggregation in Bartonella quintana. Proc. Natl. Acad. Sci. U. S. A. 101:13630–13635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neimark H, Johansson KE, Rikihisa Y, Tully JG. 2001. Proposal to transfer some members of the genera Haemobartonella and Eperythrozoon to the genus Mycoplasma with descriptions of “Candidatus Mycoplasma haemofelis”, “Candidatus Mycoplasma haemomuris”, “Candidatus Mycoplasma haemosuis” and “Candidatus Mycoplasma wenyonii”. Int. J. Syst. Evol. Microbiol. 51:891–899 [DOI] [PubMed] [Google Scholar]
- 29. Neimark H, Johansson KE, Rikihisa Y, Tully JG. 2002. Revision of haemotrophic Mycoplasma species names. Int. J. Syst. Evol. Microbiol. 52:683. [DOI] [PubMed] [Google Scholar]
- 30. Almy FS, Ladd SM, Sponenberg DP, Crisman MV, Messick JB. 2006. Mycoplasma haemolamae infection in a 4-day-old cria: support for in utero transmission by use of a polymerase chain reaction assay. Can. Vet. J. 47:229–233 [PMC free article] [PubMed] [Google Scholar]
- 31. Hackett TB, Jensen WA, Lehman TL, Hohenhaus AE, Crawford PC, Giger U, Lappin MR. 2006. Prevalence of DNA of Mycoplasma haemofelis, “Candidatus Mycoplasma haemominutum,” Anaplasma phagocytophilum, and species of Bartonella, Neorickettsia, and Ehrlichia in cats used as blood donors in the United States. J. Am. Vet. Med. Assoc. 229:700–705 [DOI] [PubMed] [Google Scholar]
- 32. Hoelzle K, Hofmann-Lehmann R, Hoelzle LE. 2010. “Candidatus Mycoplasma haemobos,” a new bovine haemotrophic Mycoplasma species? Vet. Microbiol. 144:525–526 [DOI] [PubMed] [Google Scholar]
- 33. Hoelzle LE. 2008. Haemotrophic mycoplasmas: recent advances in Mycoplasma suis. Vet. Microbiol. 130:215–226 [DOI] [PubMed] [Google Scholar]
- 34. Hoelzle LE, Adelt D, Hoelzle K, Heinritzi K, Wittenbrink MM. 2003. Development of a diagnostic PCR assay based on novel DNA sequences for the detection of Mycoplasma suis (Eperythrozoon suis) in porcine blood. Vet. Microbiol. 93:185–196 [DOI] [PubMed] [Google Scholar]
- 35. Kenny MJ, Shaw SE, Beugnet F, Tasker S. 2004. Demonstration of two distinct hemotropic mycoplasmas in French dogs. J. Clin. Microbiol. 42:5397–5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roura X, Peters IR, Altet L, Tabar MD, Barker EN, Planellas M, Helps CR, Francino O, Shaw SE, Tasker S. 2010. Prevalence of hemotropic mycoplasmas in healthy and unhealthy cats and dogs in Spain. J. Vet. Diagn. Invest. 22:270–274 [DOI] [PubMed] [Google Scholar]
- 37. Sykes JE, Ball LM, Bailiff NL, Fry MM. 2005. “Candidatus Mycoplasma haematoparvum”, a novel small haemotropic mycoplasma from a dog. Int. J. Syst. Evol. Microbiol. 55:27–30 [DOI] [PubMed] [Google Scholar]
- 38. Sykes JE, Terry JC, Lindsay LL, Owens SD. 2008. Prevalences of various hemoplasma species among cats in the United States with possible hemoplasmosis. J. Am. Vet. Med. Assoc. 232:372–379 [DOI] [PubMed] [Google Scholar]
- 39. Volokhov DV, Norris T, Rios C, Davidson MK, Messick JB, Gulland FM, Chizhikov VE. 2011. Novel hemotrophic mycoplasma identified in naturally infected California sea lions (Zalophus californianus). Vet. Microbiol. 149:262–268 [DOI] [PubMed] [Google Scholar]
- 40. Willi B, Filoni C, Catao-Dias JL, Cattori V, Meli ML, Vargas A, Martinez F, Roelke ME, Ryser-Degiorgis MP, Leutenegger CM, Lutz H, Hofmann-Lehmann R. 2007. Worldwide occurrence of feline hemoplasma infections in wild felid species. J. Clin. Microbiol. 45:1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tasker S, Caney SM, Day MJ, Dean RS, Helps CR, Knowles TG, Lait PJ, Pinches MD, Gruffydd-Jones TJ. 2006. Effect of chronic FIV infection, and efficacy of marbofloxacin treatment, on Mycoplasma haemofelis infection. Vet. Microbiol. 117:169–179 [DOI] [PubMed] [Google Scholar]
- 42. Tanahara M, Miyamoto S, Nishio T, Yoshii Y, Sakuma M, Sakata Y, Nishigaki K, Tsujimoto H, Setoguchi A, Endo Y. 2010. An epidemiological survey of feline hemoplasma infection in Japan. J. Vet. Med. Sci. 72:1575–1581 [DOI] [PubMed] [Google Scholar]
- 43. Novacco M, Meli ML, Gentilini F, Marsilio F, Ceci C, Pennisi MG, Lombardo G, Lloret A, Santos L, Carrapico T, Willi B, Wolf W, Hofmann-Lehmann R. 2010. Prevalence and geographical distribution of canine hemotropic mycoplasma infections in Mediterranean countries and analysis of risk factors for infection. Vet. Microbiol. 142:276–284 [DOI] [PubMed] [Google Scholar]
- 44. Groebel K, Hoelzle K, Wittenbrink MM, Ziegler U, Hoelzle LE. 2009. Mycoplasma suis invades porcine erythrocytes. Infect. Immun. 77:576–584 [DOI] [PMC free article] [PubMed] [Google Scholar]