Abstract
Chikungunya virus (CHIKV) is a mosquito-borne alphavirus that causes chikungunya fever in Africa, South Asia, and Southeast Asia. Because the mosquito vector Aedes albopictus is present in habitats across Europe, North America, and East Asia, CHIKV has become a serious worldwide public health concern. Infection with CHIKV typically causes fever, rash, myalgia, and arthralgia. One of the important questions yet to be answered is how the host immune system is involved in the development of this disease. In this study, we prepared a CHIKV-pseudotyped lentiviral vector for use in a safe and convenient neutralization (NT) assay and analyzed its efficacy. The CHIKV-pseudotyped lentiviral vector was prepared by cotransfection with plasmids encoding the CHIKV glycoproteins E3, E2, 6k, and E1, packaging elements, and a luciferase reporter. This alternative to native CHIKV can be safely handled in a biosafety level 2 facility. The NT assay was optimized using sera from CHIKV-immunized mice and then applied to human patient sera. The majority of the serum samples from patients with chikungunya in Thailand showed robust neutralization activities, with titers that were tightly correlated with those determined by a conventional NT assay. Moreover, there was a strong correlation with the CHIKV antibody titers as determined by enzyme-linked immunosorbent assay. Thus, the CHIKV-pseudotyped-lentiviral-vector-based NT assay system is a powerful tool for examining the neutralization activity of patient sera, which will lead to a better understanding of the immune responses involved in CHIKV infection.
INTRODUCTION
Chikungunya virus (CHIKV), a mosquito-borne alphavirus in the family Togaviridae, was first isolated in Tanzania in 1952, and it recently reemerged in Kenya in 2004 (1, 2, 3). The virus has since spread to the Indian Ocean region, South Asia, and Southeast Asia. During the recent outbreak, CHIKV evolved and adapted to an atypical vector species, Aedes albopictus, which is found not only in tropical areas but also across Europe, North America, and East Asia (4, 5, 6, 7, 8). Because CHIKV has been imported to many countries where it is not endemic by infected travelers returning from areas where it is endemic, it might be transmitted locally and then establish endemicity, as seen in Italy in 2007 (9, 10). Chikungunya (CHIK) is now a serious worldwide public health concern. Currently, there are no approved vaccines or specific antiviral drugs available to treat this illness.
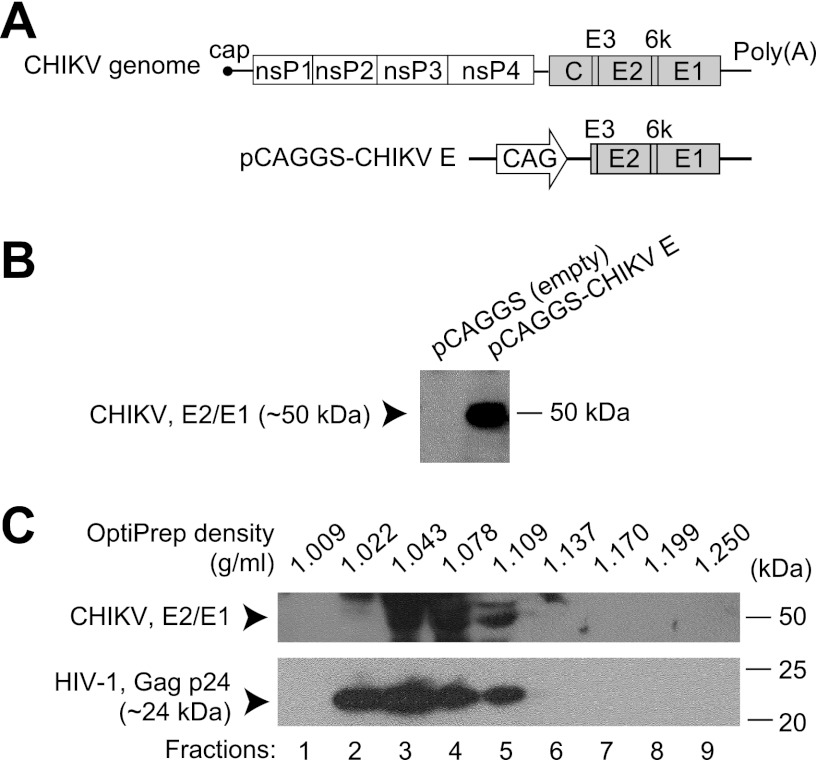
CHIKV is a positive-sense single-stranded RNA virus, and its genome is ∼11.8 kb long with 5′ cap and poly(A) tail structures (11, 12). It contains two open reading frames, one for a nonstructural polyprotein that is processed into four proteins and the other for a structural polyprotein that produces the capsid and the E3, E2, 6k, and E1 proteins (Fig. 1A). E2 and E1 form a heterodimer that projects as an outward-facing spike on the surface of CHIKV particles and, as with other alphaviruses, the E2 protein is located at the tip of the spike so that it is likely to become the target of neutralizing antibodies (13).
Fig 1.
Production of a CHIKV-pseudotyped lentiviral vector. (A) Schematic drawing of the CHIKV genome and an expression vector for CHIKV glycoproteins. The region encoding the polyprotein E3-E2-6k-E1 was inserted into an expression vector to produce the pCAGGS-CHIKV E vector, which was used for pseudotyping. nsP, nonstructural protein; C, capsid; E, envelope; CAG, a promoter consisting of the cytomegalovirus early enhancer element and chicken beta actin promoter. (B) Expression of pCAGGS-CHIKV E vector. Cells were transfected with an empty expression vector or pCAGGS-CHIKV E, and cell lysates were analyzed. CHIKV E2 and E1 proteins were detected by Western blot analysis with mouse anti-CHIKV serum. (C) Buoyant density gradient sedimentation analysis of the CHIKV-pseudotyped lentiviral vector. Supernatant containing CHIKV-pseudotyped lentiviral vector particles was purified by an OptiPrep density gradient. The OptiPrep density of each fraction (1 to 9) is shown at the top. CHIKV E2 and E1 proteins and the lentiviral vector component HIV-1 Gag p24 were detected by Western blot analysis.
CHIKV infection causes fever, rash, myalgia, and arthralgia. Polyarthralgia is the most significant symptom because it causes severe pain in most patients (1, 2). Accurate diagnosis is sometimes difficult because similar symptoms are also caused by several other tropical diseases, and, in terms of symptoms and areas of endemicity, chikungunya is very similar to dengue (DEN) fever (1, 2). Although patients usually fully recover within a couple of weeks after the disease onset, some suffer from recurrent arthralgia that can last for months to years. In the recent chikungunya outbreak, severe cases of neurological diseases and death were reported for which CHIKV was suggested to be partly responsible (14). Both the innate and adaptive immune systems are important in CHIKV infection, but how these immune responses are involved in the disease pathogenesis and development has yet to be explained (15, 16, 17).
In this study, we applied lentiviral vector technology to develop a safe and convenient neutralization (NT) assay system for CHIKV, as described previously (18, 19). Lentiviral vector technology has proven to be a safe, efficient, and powerful gene delivery system and is being applied in many fields of research (20, 21, 22). These vectors have the advantages that they can infect both dividing and nondividing cells, that the target genes are delivered and maintained in cells as a result of stable integration into the genome, and that viral proteins are not expressed after vector transduction. If a vector containing a reporter gene, such as luciferase, is cotransfected, the infection level can be easily measured. Moreover, because viral glycoproteins generally mediate cell entry, lentiviral vectors show tropism based on the expressed glycoprotein. A commonly used glycoprotein is the vesicular stomatitis virus (VSV)-G protein, which functions with a wide range of cell types. A lentiviral vector pseudotyped with CHIKV should show the same cell tropism as native CHIKV and will allow the study of the host immune response without the dangers of working with a highly pathogenic native virus.
In this study, we evaluated the efficacy of the CHIKV-pseudotyped-lentiviral-vector-based NT assay. The assay was optimized using sera from CHIKV-immunized mice and was applied to samples from patients with chikungunya that had been collected in Thailand. Most of the patient samples had high neutralization activities, and these results were consistent with the neutralization activity that was measured by a conventional NT assay using native CHIKV. This research offers important information on a safe and convenient alternative NT assay system. This assay has the potential for various applications, such as for a large-scale study of patient sera, which would contribute to a better understanding of the immune responses involved in CHIKV infection.
MATERIALS AND METHODS
Cell culture.
The four cell lines used in this study were incubated at 37°C under a 5% CO2 atmosphere. Vero (African green monkey kidney epithelial) and HeLa (human cervical adenocarcinoma) cells were cultured in Eagle's minimum essential medium (Sigma-Aldrich, St. Louis, MO), whereas BHK-21 (baby hamster kidney) and human embryonic kidney (HEK) 293T cells were cultured in Dulbecco's modified Eagle medium (Life Technologies/Gibco, Foster City, CA). Media were supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA) and 100 U/ml penicillin–100 μg/ml streptomycin (Life Technologies/Gibco).
Mouse and human serum samples.
Four female 6-week-old BALB/c mice were immunized three times at 2-week intervals with formalin-inactivated CHIKV, which was prepared as described previously (23), and were sacrificed for whole-blood collection a week after the last immunization. Whole blood was also collected from a healthy unexposed adult mouse and used as a control (“normal” serum). A total of 23 human serum samples were collected in Thailand from chikungunya (CHIK)-positive individuals by the National Institute of Health (NIH), Thailand. Nine samples were collected in 2009 (NIH Thailand codes: 13-52-20882, 13-52-20883, 13-52-20968, 13-52-21029, 13-52-21080, 13-52-22440, 13-52-23199, 13-52-23346, 13-52-24798), nine in 2010 (13-53-32095, 13-53-32097, 13-53-32221, 13-53-04736, 13-53-04864, 13-53-03450, 13-53-02639, 13-53-05460, 13-53-23661), one in 2011 (13-54-02473), and four in 2012 (13-55-06862, 13-55-06863, 13-55-06864, 13-55-06865). Serum samples collected in Thailand from four dengue (DEN)-positive individuals (one of each of the four serotypes R11-410c1, R12-031c1, R12-079c1, and R12-083c1) and two Japanese encephalitis (JE)-positive individuals (13-55-07351, 13-55-07967) were also obtained. NIH Thailand confirmed the reactivity of all serum samples by enzyme-linked immunosorbent assay (ELISA) or by a hemagglutination inhibition test. In addition, we used commercial pooled healthy human sera (Cosmo Bio, Koto-ku, Japan), as well as human serum that we obtained from a healthy individual, as negative controls (“normal” sera). This research was approved and performed in accordance with the ethical guidelines of NIH Thailand.
Plasmid vector preparation.
The genome from CHIKV strain 37997 (GenBank accession no. EU224270) was used for plasmid construction. DNA sequences encoding the structural polyprotein E3-E2-6k-E1 were optimized for expression in human-derived cells (GenScript, Piscataway, NJ) and amplified by PCR using a sense primer containing a start codon and a SacI site (5′-AAAGAGCTCATGAGTCTGGCCCTGCCAGT-3′) and an antisense primer containing a stop codon and an NheI site (5′-TTTGCTAGCTCAATGCCTGGAAAATGA-3′). The PCR product was inserted into a pCAGGS.MCSII eukaryotic expression vector (24) to construct pCAGGS-CHIKV E. The expression was confirmed by transfecting 293T cells using Lipofectamine 2000 reagent (Life Technologies/Gibco) according to the manufacturer's protocols and detecting CHIKV E2 and E1 proteins by Western blot analysis. Plasmid pMD2.G, which contains the VSV-G gene, pLenti CMV Puro LUC (w168-1), which contains the firefly luciferase reporter gene, and the HIV-based lentivirus packaging vector psPAX2, were purchased (Addgene, Cambridge, MA). An expression vector for the GP70 protein of murine leukemia virus (MLV), pFBASALF, was provided by T. Miyazawa, Kyoto University.
Pseudotyped lentiviral vector production.
Recombinant lentiviral vectors that express glycoproteins from CHIKV, VSV, or MLV and carry a luciferase reporter gene were produced as described previously (21), with some modifications. Briefly, 293T cells were seeded in a 60-mm culture dish, and 24 h later, the subconfluent cells were cotransfected with 1.3 μg of glycoprotein expression vector, 4.5 μg of luciferase reporter vector, and 4.5 μg of packaging vector by the calcium phosphate-mediated method using N,N-bis-(2-hydroxyethyl)-2-aminoethanesulfonic acid-buffered saline (Sigma-Aldrich) (25). After overnight incubation at 37°C under a 5% CO2 atmosphere, the medium was replaced with fresh medium and incubated for two more days. The supernatant containing the pseudotyped lentiviral vectors was harvested, filtered through a 0.45-μm syringe filter, and stored in aliquots at −85°C. The lentiviral vectors were normalized based on the amount of HIV-1 Gag p24 protein present as measured by an HIV-1 p24 antigen capture assay (Advanced BioScience Laboratories, Rockville, MD).
Buoyant density gradient sedimentation analysis of lentiviral vectors.
The supernatant containing the lentiviral vectors was layered onto 7.5% (wt/vol) OptiPrep (iodixanol) medium (Invitrogen) and concentrated by centrifugation at 50,000 × g for 1.5 h in an SW 32 Ti rotor (Beckman Coulter, Indianapolis, IN). The pellet was resuspended in buffer containing 50 mM Tris-HCl, 100 mM NaCl, and 0.5 mM EDTA (pH 7.4). A gradient was prepared by layering equal amounts of 45, 40, 35, 30, 25, 20, and 15% (wt/vol) OptiPrep solution and incubating at room temperature for 4 h. The suspension was placed on the top of the gradient and centrifuged at 152,000 × g for 20 h in an SW 55 Ti rotor (Beckman Coulter). Fractions were collected from the top using a pipette and were mixed with sample buffer containing 2-mercaptoethanol to prepare the samples for Western blot analysis. CHIKV E2 and E1 proteins and the lentiviral vector component, Gag p24, were detected by Western blot analysis using mouse anti-CHIKV (mouse no. 3, prepared by ourselves by immunization) and rabbit anti-p24 (BioAcademia, Ibaraki, Japan) sera, respectively.
Transduction of cells with pseudotyped lentiviral vectors.
The lentiviral vectors were titrated by 4-fold serial dilutions starting with 200 ng/ml of p24. One day prior to infection, 3 × 104 cells were seeded per well in a 24-well plate. In each well, 250 μl of serially diluted pseudotyped lentiviral vector was added. After overnight incubation, the supernatant was replaced with fresh medium (with 2% FBS), and the cells were incubated for two more days before determining the transduction efficiency by measuring luciferase activity with Glo Lysis buffer and the Steady-Glo luciferase assay system (both from Promega, Madison, WI). Luciferase activity was measured with a Centro LB 960 microplate luminometer (Berthold Technologies, Bad Wildbad, Germany). The experiments were independently repeated three times, each with a single sample per dilution. Based on data from three experiments, nonlinear regression analysis was carried out with Prism 5 software (GraphPad, La Jolla, CA).
CHIKV-pseudotyped-lentiviral-vector-based NT assay.
The assay was performed on HeLa cells in the same way as transduction, with the following modifications. The lentiviral vectors were diluted to 24 ng/ml of p24 and mixed with a 4-fold serial dilution of serum samples from mice and humans beginning at a dilution of 1:200. The mixture was incubated at room temperature for 1 to 2 h and then added to the cells in each well. The percent inhibition rate was calculated as [1 − (luciferase activity in cells treated with a mixture of pseudotyped lentiviral vector and diluted serum sample)/(luciferase activity in cells treated with a mixture of pseudotyped lentiviral vector and control healthy serum diluted at the same ratio)] × 100. The experiments were independently repeated three times, each with a single sample per dilution. Based on the data from three experiments, nonlinear regression analysis was carried out, and the serum dilution at 50% inhibition was calculated using Prism 5 software and referred to as the Luc NT titer in the comparison analyses. For samples that did not show neutralization activity, such as the control healthy sera, the Luc NT titer was set to the lowest value obtained.
pH-dependent cell entry analysis.
One day prior to infection, 3 × 104 HeLa cells were seeded per well in a 24-well plate. Ammonium chloride or chloroquine (both from Sigma-Aldrich) was 2-fold serially diluted with medium (FBS reduced to 2%), and cells were treated with either chemical for 1 h before being infected with the pseudotyped lentiviral vector solution (200 ng/ml of p24), which also contained either chemical. After 6 h, the supernatant was replaced with fresh medium (with 2% FBS). The luciferase activity was measured as described above. The experiments were independently repeated three times, each in triplicate per dilution. The average of three replicates was calculated for each experiment and used for analysis. Based on the data from three experiments, nonlinear regression analysis was carried out with Prism 5 software. Cell viability was assessed using a WST-1 assay kit (Roche, Basel, Switzerland). Briefly, HeLa cells were treated with ammonium chloride or chloroquine as above except that no pseudotyped lentiviral vectors were added to the cells. At the time of the luciferase assay, these cells were used for the cell viability test. The optical density (OD) was measured with a Multiskan FC microplate photometer (Thermo Fisher Scientific). Infectivity and percent cell viability were both calculated relative to the untreated cells (i.e., without ammonium chloride or chloroquine) as follows: (OD of treated cells/OD of untreated cells) × 100. The experiments were independently repeated three times, each with a single sample per dilution.
Stock CHIKV preparation.
A CHIKV isolate (13-52-16856) was derived from a human serum sample collected in 2009 in Thailand by NIH Thailand. The strain was plaque cloned three times on Vero cells, and the stock virus was prepared by a single passage in BHK-21 cells. The virus titer was determined by a plaque assay and microplate 50% cell culture infectious dose (CCID50) method on Vero cells. The virus was concentrated by centrifugation at 110,000 × g for 2 h in an SW 32 Ti rotor and then purified by cesium chloride equilibrium density gradient centrifugation at 100,000 × g for 16 h in an SW 55 Ti rotor. Purified virus was quantified with the Quick Start Bradford 1× dye reagent (Bio-Rad, Hercules, CA) and stored at −85°C.
Conventional NT assay.
One day prior to infection, Vero cells were seeded in a 96-well plate. Serum samples were 2-fold serially diluted with medium (FBS reduced to 2%), starting at a 1:20 dilution to a 1:2,560 dilution (eight dilutions). For mouse sera and human control sera, the starting dilution was set at 1:2. Equal amounts of CHIKV (100 CCID50/well) and serum solutions were mixed and incubated at 37°C for 2 h and then added to semiconfluent Vero cells. For each sample, two wells were used per dilution. After incubation for 1 week, the cells were examined under the microscope to look for cytopathic effects. This experiment was performed once. The highest dilution of serum at which infectivity was inhibited in 50% of the wells was calculated and referred to as the NT titer in the comparison analyses.
ELISA for anti-CHIKV.
Purified CHIKV was diluted with phosphate-buffered saline (PBS) and immobilized on an F96 MaxiSorp Nunc-Immuno plate (Thermo Fisher Scientific) by overnight incubation at 4°C (5 ng CHIKV per well). The plate was blocked with 5% skim milk in PBS containing 0.05% Tween 20 (PBST) at 37°C for 1 h. Serum samples were 2-fold serially diluted with 1% skim milk in PBST starting at a 1:200 dilution to a 1:409,600 dilution (12 dilutions). A single sample was prepared for each dilution. After the wells were washed four times with PBST, the serum samples were added and the plate was incubated at 37°C for 1 h. After four washes with PBST, horseradish peroxidase (HRP)-conjugated goat anti-human IgG (H+L; KPL, Gaithersburg, MD) diluted to 1:32,000 with 1% skim milk in PBST was added and the plate was incubated at 37°C for 1 h. After four additional washes with PBST, 100 μl of SureBlue Reserve TMB microwell peroxidase substrate 1 (KPL) was added to each well, and the plate was incubated at room temperature for 30 min in the dark. Stop solution (0.6 N H2SO4) was then added (100 μl per well). The OD at 450 nm, as well as at 620 nm for background subtraction, was immediately measured with a Multiskan FC microplate photometer. The experiment was performed once. Nonlinear regression analysis was done, and the antibody (Ab) dilution at a cutoff value (OD = 0.100) was calculated using Prism 5 software and referred to as the ELISA Ab titer in the comparison analyses.
RESULTS
Preparation of CHIKV-pseudotyped lentiviral vector.
The region encoding the CHIKV glycoproteins E3, E2, 6k, and E1 was optimized for expression in human-derived cells and inserted into a eukaryotic expression vector to construct pCAGGS-CHIKV E (Fig. 1A). When 293T cells were transfected with pCAGGS-CHIKV E, the expression of E2 and E1 proteins was confirmed by Western blot analysis with mouse anti-CHIKV serum (Fig. 1B). Subconfluent 293T cells were cotransfected with three components: pCAGGS-CHIKV E, a lentiviral packaging vector, and a luciferase reporter plasmid. After incubation for several days, the supernatant containing lentiviral vector particles was collected, and buoyant density gradient sedimentation analysis was performed to analyze the incorporation of CHIKV glycoproteins into the lentiviral vector. Western blot analysis revealed that the CHIKV proteins E2 and E1 cofractionated with the HIV-1 capsid protein Gag p24 (Fig. 1C), supporting the hypothesis that the lentiviral vectors were pseudotyped to CHIKV.
In addition, VSV- and MLV-pseudotyped lentiviral vectors were produced by transfection with VSV-G- and MLV-glycoprotein-expressing vectors, respectively, instead of pCAGGS-CHIKV E. A negative-control lentiviral vector was produced by transfecting empty pCAGGS.MCSII vector instead. The quantities of the lentiviral vectors were standardized based on the amount of HIV-1 Gag p24 present.
Characterization of the CHIKV-pseudotyped lentiviral vector.
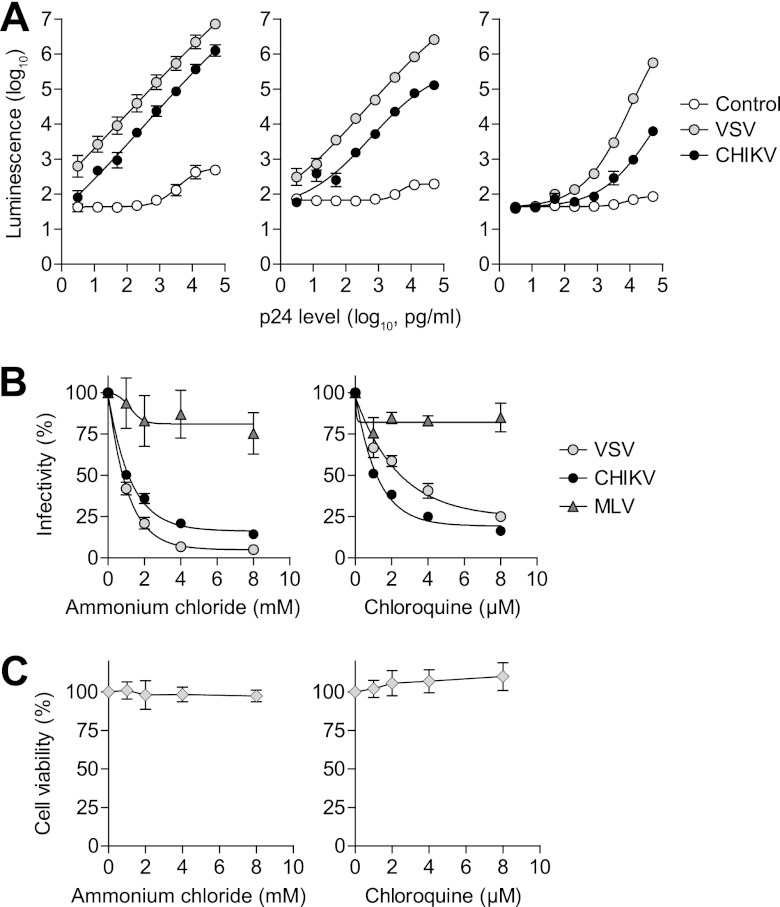
To determine whether the CHIKV-pseudotyped lentiviral vector could infect cells in the same manner as native CHIKV, BHK-21, HeLa, and Vero cells were transduced with pseudotyped lentiviral vectors, and a luciferase reporter assay was performed. These three cell lines are permissive for CHIKV infection (26). Cells transduced with the VSV-pseudotyped lentiviral vector showed high, dose-dependent luciferase activity, whereas the empty lentiviral vector control did not produce significant luciferase activity in any of the cell lines tested (Fig. 2A). The CHIKV-pseudotyped lentiviral vector produced dose-dependent luciferase activity in the three cell lines, although this activity was orders of magnitude lower than what was seen with the VSV-pseudotyped lentiviral vector (Fig. 2A).
Fig 2.
Characterization of the CHIKV-pseudotyped lentiviral vector. (A) Cell tropism of the CHIKV-pseudotyped lentiviral vector in BHK-21 (left), HeLa (middle), and Vero (right) cell lines. Pseudotyped lentiviral vectors were serially diluted and added to cells, and the transduction rate was determined by measuring luciferase reporter gene activity (luminescence). A VSV-pseudotyped lentiviral vector served as a positive control. The negative-control lentiviral vector produced by transfection with an empty expression vector did not infect cells. (B) pH dependence of cell entry for the CHIKV-pseudotyped lentiviral vector. Cells were pretreated with serially diluted ammonium chloride or chloroquine and then transduced by pseudotyped lentiviral vectors. Infectivity was calculated relative to the control cells without chemical treatment. (C) Cell viability after treatment with ammonium chloride or chloroquine. Cells were treated in the same way as the cell entry analysis except that no pseudotyped lentiviral vectors were added to the cells. Percent cell viability was tested by WST-1 reagent and was calculated relative to control cells without chemical treatment. Error bars show the standard errors of the mean (SEM) of results from three independent experiments.
To test the pH dependence of cell entry for the CHIKV-pseudotyped lentiviral vector, we preincubated cells with ammonium chloride or chloroquine, both of which prevent the acidification of the endosome and thus interfere with viral infection (19, 26, 27). The transduction rates of both the CHIKV- and VSV-pseudotyped lentiviral vectors decreased in a dose-dependent manner when treated with either chemical (Fig. 2B). In contrast, the transduction of the MLV-pseudotyped lentiviral vector was not affected by the presence of either chemical (Fig. 2B). Cell viability was not significantly affected by either ammonium chloride or chloroquine (Fig. 2C). These results suggest that CHIKV-pseudotyped lentiviral vector cell entry is pH dependent, as has been demonstrated with native CHIKV (26).
Development of an NT assay using the CHIKV-pseudotyped lentiviral vector.
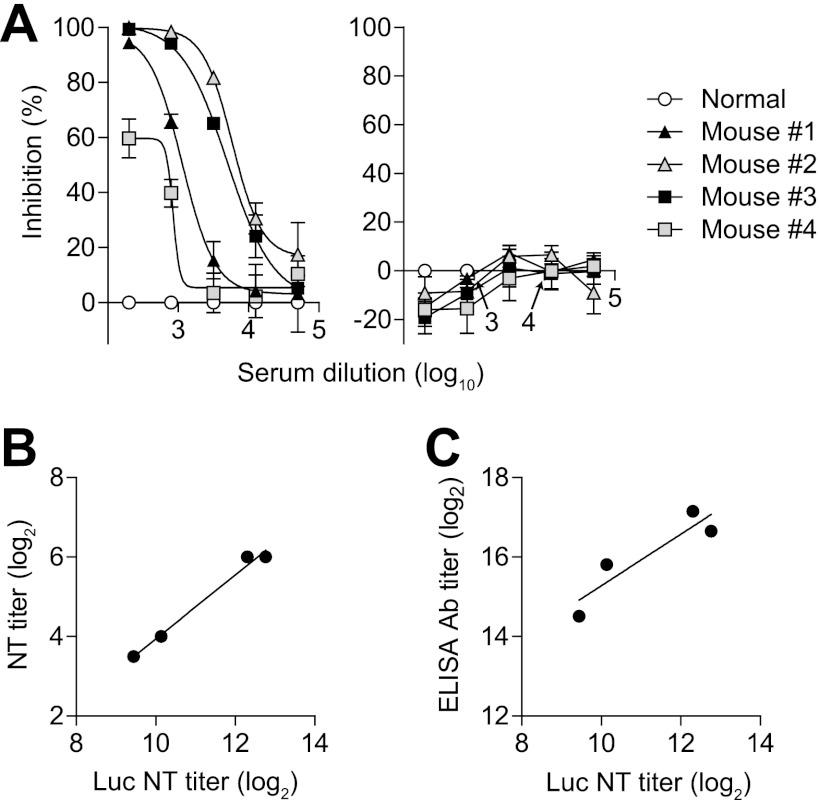
The CHIKV-pseudotyped lentiviral vector was incubated with serially diluted serum samples collected from four mice immunized with formalin-inactivated CHIKV. The mixture was added to cells, and luciferase activity was measured after several days to analyze the effects on the transduction rate. Luciferase activity was reduced to basal levels when the CHIKV-pseudotyped lentiviral vector was preabsorbed with the lowest dilution (1:200) of mouse serum (Fig. 3A, left). There was little or no effect on luciferase activity when the vector was incubated with the most-diluted mouse serum (1:51,200) (Fig. 3A, left). Thus, neutralization was dose dependent. Luciferase activity in the cells transduced with the VSV-pseudotyped lentiviral vector was not affected by incubation with the anti-CHIKV mouse sera, showing that the neutralization is CHIKV specific (Fig. 3A, right).
Fig 3.
Neutralization of the CHIKV-pseudotyped lentiviral vector by CHIKV-immunized mouse sera. (A) Dose-dependent neutralization of CHIKV-pseudotyped lentiviral vector by CHIKV-immunized mouse serum samples (left) and control experiment with a VSV-pseudotyped lentiviral vector (right). Pseudotyped lentiviral vectors were normalized to a p24 level of 24 ng/ml and incubated with serially diluted serum. The mixture was added to cells and luciferase activity was measured. The percent inhibition rate by CHIKV-immunized mouse sera (mice no. 1–4) was calculated relative to healthy mouse serum. Error bars show the SEM of results from three independent experiments. (B) Comparison between NT titers determined by NT assay using CHIKV-pseudotyped lentiviral vector (Luc NT titer) and by a conventional assay using native CHIKV (NT titer). (C) Comparison between Luc NT titer and CHIKV antibody titer determined by ELISA (ELISA Ab titer).
For the pseudotyped-lentiviral-vector-based NT assay, we calculated the extent to which luciferase activity was decreased in the cells incubated with a mixture of lentiviral vector and serum samples compared to the luciferase activity in those incubated with a mixture of lentiviral vector and control healthy serum (percent inhibition rate). We then performed statistical analyses to determine the serum dilution at 50% inhibition and set it as the NT titer. We also performed a conventional neutralization assay in which we set the NT titer as the highest dilution of serum at which infectivity was inhibited in 50% of the wells. When the luciferase assay NT titers were compared with those determined by a conventional NT assay, there was a strong correlation among the four serum samples (y = 0.81x − 4.13, r2 = 0.98), showing that the NT assay using the CHIKV-pseudotyped lentiviral vector is likely to be as reliable as the conventional assay (Fig. 3B). There was also a relatively strong correlation among the four serum samples (y = 0.65x + 8.79, r2 = 0.83) between the luciferase assay NT titers and anti-CHIKV antibody titers determined by ELISA (Fig. 3C), suggesting that the ratio of neutralizing antibodies to all CHIKV antibodies was fairly constant among serum samples from different mice. Therefore, it appears that the NT assay using CHIKV-pseudotyped lentiviral vector might serve as a valuable alternative to a conventional NT assay.
NT assay with sera from patients with chikungunya.
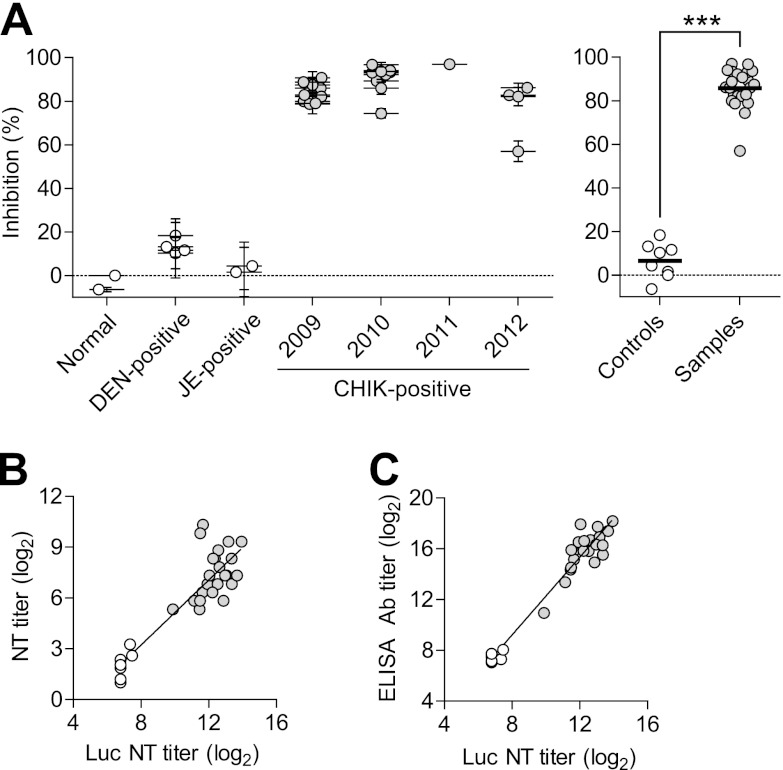
A total of 23 human serum samples, collected in Thailand between 2009 and 2012, were analyzed with the NT assay that we have discussed here. Four DEN-positive and two JE-positive samples collected in Thailand were used as controls. In addition, commercial pooled healthy human serum samples and a healthy human serum sample collected for this study were used as negative controls. Most of the CHIK-positive human serum samples showed strong neutralization activity, whereas the DEN-positive, JE-positive, and healthy human serum samples showed little or no neutralization activity (Fig. 4A, and see Fig. S1A in the supplemental material). As seen with mouse serum samples, luciferase activity was reduced to basal levels for most of the CHIK-positive human serum samples when the pseudotyped lentiviral vector was absorbed with serum at the lowest dilution (1:200), whereas there was little or no effect on luciferase activity when the pseudotyped lentiviral vector was incubated with the highest dilution of CHIK-positive serum (1:51,200) (see Fig. S1B to D in the supplemental material). Thus, neutralization activity was dose dependent. When the luciferase assay NT titers were compared with those determined by a conventional assay, there was a good correlation (y = 0.96x − 4.47, r2 = 0.78) (Fig. 4B). When the luciferase assay NT titers were compared with antibody titers, they were highly correlated (y = 1.53x − 3.03, r2 = 0.94) (Fig. 4C). As seen with the anti-CHIKV mouse sera, the ratio of neutralizing antibodies to all CHIKV antibodies seems to be fairly constant among different patient serum samples. Thus, the NT assay using the CHIKV-pseudotyped lentiviral vector will be very useful in the study of patient serum samples.
Fig 4.
Neutralization activity analysis of CHIK-positive patient sera. (A) Neutralization activity of serum samples from 23 patients with chikungunya that were collected in Thailand from 2009 to 2012 was analyzed using the CHIKV-pseudotyped lentiviral vector. Four DEN-positive and two JE-positive serum samples collected in Thailand, as well as two healthy human serum samples, were used as controls. Pseudotyped lentiviral vectors were incubated with serially diluted serum and added to cells, and luciferase activity was measured. The percent inhibition rate was calculated relative to pooled healthy human serum plotted in the normal sample group. CHIK-positive sera, gray circles; control sera, white circles. Left, neutralization activity of serum samples diluted to 1:800. Error bars show the SEM of results from three independent experiments. Right, the data shown on the left were rearranged into control and sample groups; thick black lines show the grand mean. ***, P < 0.0001 (Student's t test). (B) Comparison between NT titers determined by NT assay using the CHIKV-pseudotyped lentiviral vector (Luc NT titer) and by a conventional assay using native CHIKV (NT titer). (C) Comparison between Luc NT titer and CHIKV antibody titer determined by ELISA (ELISA Ab titer).
DISCUSSION
In this study, we developed a safe and convenient assay system using a CHIKV-pseudotyped lentiviral vector with a luciferase reporter to analyze the neutralization activities of serum samples from patients with chikungunya. The CHIKV-pseudotyped lentiviral vector system allows for the investigation of CHIKV-host interactions without the danger of exposure to the highly pathogenic native CHIKV, and it allows experiments to be performed in a biosafety level 2 facility.
The West African CHIKV strain 37997 was selected to prepare the glycoprotein-expressing vector because it worked successfully in a previous study (19). Sequence comparison of CHIKV strains showed that the genome is highly conserved in coding regions, even among strains of the three divergent genotypes (Asian, East/Central/South African, and West African) (28, 29). The strain 37997-based CHIKV-pseudotyped lentiviral vectors that we developed reacted efficiently with serum samples from the mice that were immunized and the patients who were infected with the current endemic strains in Thailand.
The CHIKV-pseudotyped lentiviral vector transduced three cell lines that are permissive for CHIKV infection, but the transduction was less efficient than that with a VSV-pseudotyped lentiviral vector (Fig. 2A), as was shown in a previous study (19). Viral glycoprotein-mediated cell entry is generally either pH dependent or independent. For example, some viruses, such as CHIKV and VSV, infect cells in a pH-dependent manner, whereas other viruses, such as amphotropic MLV, infect cells in a pH-independent manner (19, 26, 27). Chemicals that prevent the acidification of endosomes, such as ammonium chloride and chloroquine, can interfere with pH-dependent viral cell entry. Transduction of the CHIKV-pseudotyped lentiviral vector into the cells was found to be pH dependent (Fig. 2B), as is the case with the native CHIKV (26), suggesting a similar mode of infection as that of native CHIKV. When considered together, the results from the buoyant density gradient sedimentation analysis and from cell tropism and pH-dependence characterization (Fig. 1C and 2A and B) demonstrate that the lentiviral vector was successfully pseudotyped with CHIKV.
Serum samples obtained from CHIKV-immunized mice showed different degrees of neutralization activities (Fig. 3A). Luciferase assay NT titers with mouse sera were highly similar to those obtained with a conventional NT assay, as well as being highly correlated with those obtained by CHIKV antibody ELISA (Fig. 3B and C). Similarly, luciferase assay NT titers obtained with serum samples from infected human patients correlated well with those obtained by conventional NT assay or CHIKV antibody ELISA (Fig. 4B and C). Although more samples should be analyzed for a reliable comparison analysis using mouse sera, this high degree of correlation supports the reliability of the NT assay system that was developed for this study. While the NT titers of the pseudotyped-lentiviral-vector-based and conventional assays resulted in fairly different scales (Fig. 3B and 4B) due to the ways NT titers were set in the two methods, they both showed the neutralizing activities of sera and provided consistent results. Furthermore, this system required only a small amount of serum, less than one-fifth of that required by the conventional NT assay, making it more useful in cases where sample quantities are limited.
When the NT assay using the CHIKV-pseudotyped lentiviral vector is compared against a conventional NT assay, there are some disadvantages. For example, the production of pseudotyped lentiviral vectors by the calcium phosphate-mediated method can be technically difficult and tedious (21, 25), and the assay reagents and equipment for luciferase reporter assays are relatively expensive. The pseudotyped lentiviral vector is, however, much safer to work with than the native virus, and the reporter assay is much easier and faster than conventional methods. In fact, many options are now available to readily produce high titers of lentiviral vectors, and various reporter genes are available, depending on the intended purpose, budget, and availability of laboratory equipment (22).
The outbreak of chikungunya fever in 2005 to 2006 on Réunion Island had a significant public health impact, in part because almost a third of the population on the island was infected (1, 6). CHIKV infection is thus a real threat to populations with low herd immunity. Whereas chronic arthralgia is a hallmark of CHIKV infection and causes significant burden to patients, current endemic CHIKV strains can also cause severe symptoms, such as neurological complications or even death (14). A better understanding of the pathogenesis, disease development, and host immune responses during CHIKV infection is crucial to countering this reemerging disease. Antibodies against CHIKV clear the virus, and cytokine activation acts against the viruses and possibly induces cellular immune responses, although the mechanism is poorly understood (15, 16, 17). Important questions remain as to whether adaptive immunity, such as that from autoimmune antibodies, is involved in the development of chronic arthralgia, as there seem to be similarities between chikungunya chronic arthralgia and autoimmune rheumatoid arthritis (15, 16). In addition, it is not clear why IgM was detected for an extended period of time after recovery in some patients. Detailed analysis of chikungunya-infected patients will help reveal the role of immune responses in disease development, and the NT assay system discussed here will be a valuable tool in these investigations.
Supplementary Material
ACKNOWLEDGMENTS
We thank Shigeyuki Hamada (RCC-ERI) for valuable managerial support for this study.
This work was supported by the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID) sponsored by the Ministry of Education, Culture, Sports, Science, and Technology of Japan. This study was also supported by the Department of Medical Sciences, Ministry of Public Health, Thailand. The sponsors had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 13 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03109-12.
REFERENCES
- 1. Burt FJ, Rolph MS, Rulli NE, Mahalingam S, Heise MT. 2012. Chikungunya: a re-emerging virus. Lancet 379:662–671 [DOI] [PubMed] [Google Scholar]
- 2. Schwartz O, Albert ML. 2010. Biology and pathogenesis of chikungunya virus. Nat. Rev. Microbiol. 8:491–500 [DOI] [PubMed] [Google Scholar]
- 3. Powers AM, Logue CH. 2007. Changing patterns of chikungunya virus: re-emergence of a zoonotic arbovirus. J. Gen. Virol. 88:2363–2377 [DOI] [PubMed] [Google Scholar]
- 4. Tsetsarkin KA, Chen R, Leal G, Forrester N, Higgs S, Huang J, Weaver SC. 2011. Chikungunya virus emergence is constrained in Asia by lineage-specific adaptive landscapes. Proc. Natl. Acad. Sci. U. S. A. 108:7872–7877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. 2007. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 3:e201 doi:10.1371/journal.ppat.0030201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schuffenecker I, Iteman I, Michault A, Murri S, Frangeul L, Vaney MC, Lavenir R, Pardigon N, Reynes JM, Pettinelli F, Biscornet L, Diancourt L, Michel S, Duquerroy S, Guigon G, Frenkiel MP, Bréhin AC, Cubito N, Desprès P, Kunst F, Rey FA, Zeller H, Brisse S. 2006. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 3:e263 doi:10.1371/journal.pmed.0030263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Butler D. 2012. Europe on alert for flying invaders. Nature 489:187–188 [DOI] [PubMed] [Google Scholar]
- 8. Enserink M. 2008. Entomology. A mosquito goes global. Science 320:864–866 [DOI] [PubMed] [Google Scholar]
- 9. Rezza G, Nicoletti L, Angelini R, Romi R, Finarelli AC, Panning M, Cordioli P, Fortuna C, Boros S, Magurano F, Silvi G, Angelini P, Dottori M, Ciufolini MG, Majori GC, Cassone A. 2007. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet 370:1840–1846 [DOI] [PubMed] [Google Scholar]
- 10. Bonilauri P, Bellini R, Calzolari M, Angelini R, Venturi L, Fallacara F, Cordioli P, Angelini P, Venturelli C, Merialdi G, Dottori M. 2008. Chikungunya virus in Aedes albopictus, Italy. Emerg. Infect. Dis. 14:852–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Solignat M, Gay B, Higgs S, Briant L, Devaux C. 2009. Replication cycle of chikungunya: a re-emerging arbovirus. Virology 393:183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Khan AH, Morita K, Parquet MDC, Hasebe F, Mathenge EG, Igarashi A. 2002. Complete nucleotide sequence of chikungunya virus and evidence for an internal polyadenylation site. J. Gen. Virol. 83:3075–3084 [DOI] [PubMed] [Google Scholar]
- 13. Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA. 2010. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468:709–712 [DOI] [PubMed] [Google Scholar]
- 14. Lemant J, Boisson V, Winer A, Thibault L, André H, Tixier F, Lemercier M, Antok E, Cresta MP, Grivard P, Besnard M, Rollot O, Favier F, Huerre M, Campinos JL, Michault A. 2008. Serious acute chikungunya virus infection requiring intensive care during the Reunion Island outbreak in 2005–2006. Crit. Care Med. 36:2536–2541 [DOI] [PubMed] [Google Scholar]
- 15. Kam YW, Ong EK, Renia L, Tong JC, Ng LF. 2009. Immuno-biology of Chikungunya and implications for disease intervention. Microbes Infect. 11:1186–1196 [DOI] [PubMed] [Google Scholar]
- 16. Jaffar-Bandjee MC, Das T, Hoarau JJ, Krejbich Trotot P, Denizot M, Ribera A, Roques P, Gasque P. 2009. Chikungunya virus takes centre stage in virally induced arthritis: possible cellular and molecular mechanisms to pathogenesis. Microbes Infect. 11:1206–1218 [DOI] [PubMed] [Google Scholar]
- 17. Her Z, Malleret B, Chan M, Ong EK, Wong SC, Kwek DJ, Tolou H, Lin RT, Tambyah PA, Rénia L, Ng LF. 2010. Active infection of human blood monocytes by Chikungunya virus triggers an innate immune response. J. Immunol. 184:5903–5913 [DOI] [PubMed] [Google Scholar]
- 18. Salvador B, Zhou Y, Michault A, Muench MO, Simmons G. 2009. Characterization of Chikungunya pseudotyped viruses: identification of refractory cell lines and demonstration of cellular tropism differences mediated by mutations in E1 glycoprotein. Virology 393:33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Akahata W, Yang ZY, Andersen H, Sun S, Holdaway HA, Kong WP, Lewis MG, Higgs S, Rossmann MG, Rao S, Nabel GJ. 2010. A virus-like particle vaccine for epidemic Chikungunya virus protects nonhuman primates against infection. Nat. Med. 16:334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Naldini L, Blomer U, Gage FH, Trono D, Verma IM. 1996. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc. Natl. Acad. Sci. U. S. A. 93:11382–11388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang ZY, Wei CJ, Kong WP, Wu L, Xu L, Smith DF, Nabel GJ. 2007. Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science 317:825–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sakuma T, Barry MA, Ikeda Y. 2012. Lentiviral vectors: basic to translational. Biochem. J. 443:603–618 [DOI] [PubMed] [Google Scholar]
- 23. Harrison VR, Eckels KH, Bartelloni PJ, Hampton C. 1971. Production and evaluation of a formalin-killed Chikungunya vaccine. J. Immunol. 107:643–647 [PubMed] [Google Scholar]
- 24. Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193–199 [DOI] [PubMed] [Google Scholar]
- 25. Chen C, Okayama H. 1987. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2745–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sourisseau M, Schilte C, Casartelli N, Trouillet C, Guivel-Benhassine F, Rudnicka D, Sol-Foulon N, Le Roux K, Prevost MC, Fsihi H, Frenkiel MP, Blanchet F, Afonso PV, Ceccaldi PE, Ozden S, Gessain A, Schuffenecker I, Verhasselt B, Zamborlini A, Saïb A, Rey FA, Arenzana-Seisdedos F, Desprès P, Michault A, Albert ML, Schwartz O. 2007. Characterization of reemerging chikungunya virus. PLoS Pathog. 3:e89 doi:10.1371/journal.ppat.0030089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yang ZY, Huang Y, Ganesh L, Leung K, Kong WP, Schwartz O, Subbarao K, Nabel GJ. 2004. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 78:5642–5650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arankalle VA, Shrivastava S, Cherian S, Gunjikar RS, Walimbe AM, Jadhav SM, Sudeep AB, Mishra AC. 2007. Genetic divergence of Chikungunya viruses in India (1963–2006) with special reference to the 2005–2006 explosive epidemic. J. Gen. Virol. 88:1967–1976 [DOI] [PubMed] [Google Scholar]
- 29. Volk SM, Chen R, Tsetsarkin KA, Adams AP, Garcia TI, Sall AA, Nasar F, Schuh AJ, Holmes EC, Higgs S, Maharaj PD, Brault AC, Weaver SC. 2010. Genome-scale phylogenetic analyses of chikungunya virus reveal independent emergences of recent epidemics and various evolutionary rates. J. Virol. 84:6497–6504 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.