Abstract
Typing of methicillin-resistant Staphylococcus aureus (MRSA) remains necessary in order to assess whether transmission of MRSA occurred and to what extent infection prevention measures need to be taken. Raman spectroscopy (SpectraCellRA [SCRA]; RiverD International, Rotterdam, The Netherlands) is a recently developed tool for bacterial typing. In this study, the performance (typeability, discriminatory power, reproducibility, workflow, and costs) of the SCRA system was evaluated for typing of MRSA strains isolated from patients and patients' household members who were infected with or colonized by MRSA. We analyzed a well-documented collection of 113 MRSA strains collected from 54 households. The epidemiological relationship between the MRSA strains within one household was used as the gold standard. Pulsed-field gel electrophoresis (PFGE) was used for discrepancy analysis. The results of SCRA analysis on the strain level corresponded with epidemiological data for 108 of 113 strains, a concordance of 95.6%. When analyzed at the household level, the results of SCRA were correct for 49 out of 54 households, a concordance of 90.7%. Concordance on the strain level with epidemiological data for PFGE was 93.6% (103/110 isolates typed). Concordance on the household level with epidemiological data for PFGE was 93.5% (49/53 households analyzed). With PFGE regarded as the reference standard, the conclusions reached with Raman spectroscopy were identical to those reached with PFGE in 100 of 105 cases (95.2%). The reproducibility of SCRA was found to be 100%. We conclude that the SpectraCellRA system is a fast, easy-to-use, and highly reproducible typing platform for outbreak analysis that can compete with the currently used typing techniques.
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) continues to be a major problem in health care. Typing of MRSA remains necessary for the assessment of transmission so that targeted infection prevention measures can be applied. An ideal typing system for epidemiological surveillance of MRSA should be user friendly, fast, and reliable and have high throughput capacity (1). However, the two most important requirements for a typing system are high discriminatory power and reproducibility (1).
Currently, different typing methods are in use in diagnostic laboratories, with pulsed-field gel electrophoresis (PFGE) of SmaI macrorestriction analysis of genomic DNA still being considered the gold standard because of its high discriminatory power (2, 3). However, PFGE is technically demanding and has limited portability due to low intercenter reproducibility, with a long time to results (4). Raman spectroscopy (SpectraCellRA [SCRA]; RiverD International, Rotterdam, The Netherlands) has been described as an adequate tool for typing of bacteria (5). This vibrational spectroscopy-based technique does not require any labels or dyes, and its high throughput, ease of use, and short time to result, combined with a high concordance of 95% with PFGE, make SCRA a valuable tool for outbreak analysis in routine diagnostic laboratories.
In Dutch hospitals, patients who are found to be colonized or infected with MRSA (primary case or index patients) are placed in isolation as a precaution to prevent transmission. In general, eradication therapy is not prescribed during hospitalization, because risk factors for therapy failure, such as wounds or indwelling catheters, are usually present in hospitalized patients (6, 7) (see www.swab.nl). Also, the policy in the Erasmus MC is to test all household members of the patient for MRSA before any eradication therapy is started. All positive household members (secondary cases) are then treated simultaneously with the primary case. Since the prevalence of MRSA is still low in The Netherlands (8, 9), there is a very low a priori chance of these household members having acquired MRSA from another source. Furthermore, we recently showed that all isolates from MRSA-positive household members (n = 56) had the same PFGE type as the isolate from their index person (n = 29) (10). Therefore, the presence of multiple MRSA carriers within one household is considered to be a consequence of household transmission or exposure to the same source and can be regarded as a single cluster.
The primary aim of this study was to assess whether SCRA is indeed able to determine clonal relationships between MRSA strains isolated from household members infected or colonized with MRSA and to determine the performance of this typing system. PFGE was used as the typing-verification method in case of discrepancies between SCRA typing and the epidemiological data.
(Part of this work was presented during the European Congress of Clinical Microbiology and Infectious Diseases [ECCMID], London, United Kingdom, 2012.)
MATERIALS AND METHODS
Bacterial strains.
A total of 113 MRSA strains from a well-documented set of 54 households were included in the analysis (10). Strains were collected in 2005, 2006, and 2007 from primary cases (index patients, n = 54) and corresponding secondary cases (household members of index patients, n = 59). Households consisted of 4 positive members (n = 3), 3 positive members (n = 14), 2 positive members (n = 22), or 1 positive member (n = 15).
Cultures for MRSA were performed at the diagnostic laboratory of the Department of Medical Microbiology and Infectious Diseases (Erasmus MC). Results were confirmed by using AccuProbe (Gen-Probe Incorporated, San Diego, CA) and the MRSA-screen latex agglutination test (Denka Seiken Co., Tokyo, Japan). Furthermore, all MRSA strains were sent to the National Institute for Public Health and the Environment (RIVM) (Bilthoven, The Netherlands) for confirmation and PFGE typing.
For all MRSA isolates, epidemiological data were available, including household information, date of admission, sampling date, and initial PFGE results. We defined secondary cases as MRSA-positive household contacts from index patients, and this epidemiological household relationship was defined as the gold standard. Household contacts were defined according to Mollema et al. (10) as persons living in the same house as the initial MRSA index person or having frequent contact in the same house (more than 2 h per day) with the index person.
Raman spectroscopy. (i) SpectraCellRA analysis.
Clonal relationships among the MRSA isolates were tested by SCRA. Cultures, sample preparation, and SCRA measurements were performed according to the operators manual (version 1.7) (5).
Briefly, isolates were inoculated on blood agar (BD Diagnostics, Erembodegem, Belgium). After incubation for 18 to 20 h, isolates were subcultured for 20 h on Trypticase soy agar (TSA) plates (BD Diagnostics). Biomass was taken from this culture to fill a 1-μl loop. This biomass was suspended in 5 μl of distilled water, and 3 μl of this suspension was transferred onto a microslide sample carrier.
Pearson correlation coefficients (r2 values) were calculated between replicate measurements of the same isolate and between spectra of different isolates. The r2 values between replicates account for any signal variance due to differences in culturing, sample preparation, or actual Raman measurements. For discrimination of 2 isolates, the r2 value between replicate measurements of isolates had to be higher than the r2 values between isolates.
SCRA type analyses of sets of spectra were performed using the pair-wise similarities as a distance matrix in combination with Ward's cluster algorithm. This resulted in a dendrogram in which each node represented the lowest correlation coefficient (or similarity) between all isolates combined in the cluster defined by this node. These SCRA clusters were then compared with the epidemiological household clusters to determine concordance. This concordance was calculated on both the strain level and the household level.
Discriminatory power was evaluated by calculation of the index of discrimination (ID) according to Hunter and Gaston (11, 12). The ID value describes the probability that two unrelated isolates drawn at random from a given population will be placed into different typing groups. An ID value of 1 would indicate that the typing method was able to distinguish each isolate from all others, whereas an ID of 0 would indicate that all isolates were of an identical type. An index of discrimination of 0.5 would mean that if one strain was chosen at random from a strain population, there would be a 50% probability that the next strain chosen at random would be indistinguishable from the first.
(ii) Discrepancy analysis.
Verification of discrepant results between SCRA and the gold standard of household relationships was done by performing a second PFGE of SmaI macrorestriction analysis of genomic DNA, as described previously (3). Criteria according to Belkum et al. were used to determine genetic relatedness (4). Furthermore, we analyzed discrepant household members for possible transmission of MRSA from another known source or via contact with a person with known risk factors as described by the Dutch Workingparty on Infection Prevention (WIP) (www.wip.nl/uk).
(iii) Reproducibility.
For the establishment of reproducibility, 3 reference isolates were processed on 5 different, independent days by 2 different technicians. These reference isolates were obtained from a reference MRSA strain collection that had been used to study interlaboratory reproducibility of PFGE (4). Reference isolates 811 and 814 were genetically related isolates, and reference isolate 806 was chosen as a unique isolate. Furthermore, 26 MRSA isolates from the study were analyzed in duplicate on 2 different days by 2 different technicians. Full biological replicate analysis was performed for the reference isolates and duplicates; isolates were processed from the freezer on different days of the study. Mean correlation coefficients and standard deviations (SD) were calculated with SPSS for the 5 independent measurements of each reference isolate.
(iv) Workflow and costs.
Total hands-on time, turnaround time, and costs (in Euros) were determined for SCRA analysis. These parameters were then compared with those of PFGE.
RESULTS
Raman spectroscopy. (i) SpectraCellRA analysis.
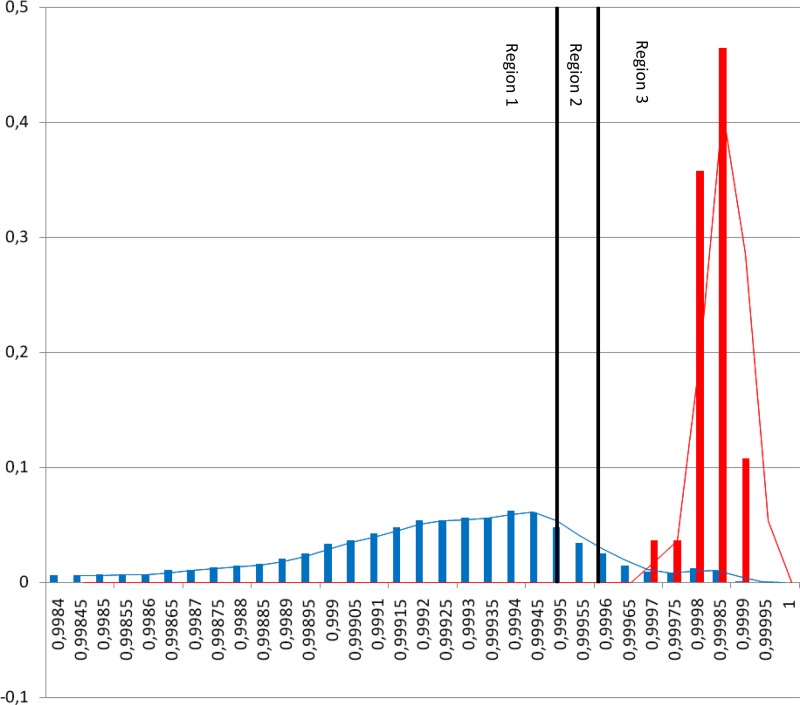
The distribution of the r2 values between replicates of isolates (red bars) and between isolates (blue bars) is given in Fig. 1.
Fig 1.
Histogram of r2 values found for our study. The r2 values between replicates are indicated by red bars; r2 values between isolates are indicated by blue bars. Region 1 indicates nonrelated isolates in the SCRA analysis. When the isolates are in region 3, these isolates are indistinguishable in the SCRA analysis. Isolates in between these regions (in region 2) are possibly related. The cutoff is chosen in region 2, based on the presumption that ≥95% of all replicates must have an r2 value above this cutoff. This similarity cutoff is set to 0.9996.
Of the 113 isolates, 103 cases (91.2%) were identified for which the PFGE cluster was identical to that of the primary case. Among the 113 MRSA isolates, 59 SCRA types could be distinguished (Table 1). Based on the epidemiological household data, results of SCRA analysis were correct for 108 of 113 strains. This resulted in a concordance of 95.6% (Table 2).
Table 1.
Results of SpectraCellRA analysis
| Household cluster no. | No. of household members | SCRA type(s) (n) | Initial PFGE type(s) (n) |
|---|---|---|---|
| 1 | 3 | 2 (3) | 155 (3) |
| 2 | 2 | 57 (2) | 55 (2) |
| 3 | 4 | 11 (4) | 18 (4) |
| 4 | 3 | 10 (3) | 18 (3) |
| 5 | 3 | 21 (3) | 27 (3) |
| 6 | 2 | 69 (2) | 37 (2) |
| 7a | 2 | 48 (1), 67 (1) | 68 (2) |
| 8 | 2 | 41 (2) | 55 (2) |
| 9 | 3 | 28 (3) | 109a |
| 10 | 2 | 43 (2) | 587 |
| 11 | 3 | 9 (3) | 23 (2), 28 (1) |
| 12 | 1 | 37 (1) | 28 (1) |
| 13 | 2 | 62 (2) | 293 (2) |
| 14 | 3 | 68 (3) | NTb ST398 (3) |
| 15 | 1 | 34 (1) | 263 (1) |
| 16 | 1 | 3 (1) | 15 (1) |
| 17 | 3 | 16 (3) | 23 (3) |
| 18 | 4 | 39 (4) | 22 (1), 68 (2), 665 (1) |
| 19a | 2 | 47 (1), 58 (1) | 55 (2) |
| 20a | 2 | 56 (1), 59 (1) | 55 (2) |
| 21 | 2 | 52 (2) | 50a (1), 675 (1) |
| 22 | 1 | 19 (1) | 22 (1) |
| 23 | 1 | 60 (1) | 23 (1) |
| 24 | 2 | 22 (2) | 24 (2) |
| 25 | 1 | 51 (1) | 25 (1) |
| 26 | 3 | 50 (3) | 26 (3) |
| 27 | 2 | 65 (2) | 27 (2) |
| 29 | 2 | 35 (2) | 50a (2) |
| 30 | 1 | 73 (1) | 30 (1) |
| 31 | 3 | 25 (3) | 31 (3) |
| 32 | 3 | 6 (3) | 32 (3) |
| 33 | 1 | 54 (1) | 381 (1) |
| 34 | 1 | 76 (1) | 34 (1) |
| 35 | 2 | 30 (2) | 35 (2) |
| 36 | 2 | 29 (2) | 199 (2) |
| 37 | 2 | 5 (2) | 37 (2) |
| 38 | 2 | 71 (2) | 38 (2) |
| 39 | 1 | 31 (1) | 39 (1) |
| 40 | 3 | 75 (3) | 40 (3) |
| 41 | 1 | 32 (1) | 41 (1) |
| 42 | 2 | 77 (2) | 42 (2) |
| 43 | 3 | 82 (3) | 43 (3) |
| 44 | 1 | 27 (1) | 44 (1) |
| 45 | 4 | 14 (4) | 27 (3), 28 (1) |
| 46 | 2 | 12 (2) | 46 (2) |
| 47a | 2 | 4 (1), 18 (1) | 47 (2) |
| 49 | 2 | 36 (2) | 49 (2) |
| 50 | 1 | 17 (1) | 50 (1) |
| 52a | 3 | 40 (2), 45 (1) | 52 (3) |
| 54 | 2 | 61 (2) | 54 (2) |
| 55 | 2 | 55 (2) | 55 (2) |
| 57 | 3 | 49 (3) | 15 (3) |
| 58 | 1 | 79 (1) | 58 (1) |
| 59 | 1 | 80 (1) | 59 (1) |
| Reference 806 | 5 | 29 | |
| Reference 811 | 5 | 51 | |
| Reference 814 | 5 | 50 |
Indicates discrepant results between SCRA and the epidemiological data.
NT, nontypeable with PFGE; ST, sequence type.
Table 2.
Discriminatory power expressed as ID and concordance of SCRA and PFGE compared to epidemiological data
| Typing method | All isolates (n = 113) |
Concordance (%) with epidemiological data on the: |
||
|---|---|---|---|---|
| No. of types | IDc | Strain level (n = 113) | Household level (n = 54) | |
| PFGEa | 48 | 0.98 | 103 (91.2) | 49 (90.7) |
| SCRAb | 59 | 0.99 | 108 (95.6) | 49 (90.7) |
PFGE of SmaI macrorestriction analysis of genomic DNA.
SCRA, SpectraCellRA.
ID, index of discrimination according to Hunter and Gaston (12).
When SCRA types were analyzed at the level of household clusters, results of the SCRA analysis were concordant for 49 out of 54 households (90.7%).
Of the 110 PFGE typeable isolates, 105 cases (95.4%) were identified for which the PFGE cluster was identical to that of the primary case. In 108/113 cases (95.5%), the same applied for the Raman type. When PFGE was regarded as the reference standard, the conclusions based on the results of Raman spectroscopy were identical to those based on PFGE in 100 of 105 cases (95.2%).
ID values were calculated from the distributions of types and were 0.98 and 0.99 for PFGE and SCRA, respectively (Table 2).
All MRSA isolates within household cluster 14 (n = 3) were nontypeable using PFGE of SmaI macrorestriction analysis (sequence type 398, which is typeable using PFGE when using another restriction enzyme), whereas SCRA resulted in 1 SCRA type (type 68).
Concordance on the strain level with epidemiological data for PFGE was 93.6% (103/110 isolates typed). Concordance on the household level with epidemiological data for PFGE was 93.5% (49/53 households analyzed).
(ii) Discrepancy analysis.
Five discrepant results were observed between SCRA analysis and epidemiological household data. PFGE was performed on all of the isolates from the discrepant households 7, 19, 20, 47, and 52. Household clusters 7, 19, 20, and 47 consisted of 1 primary case and 1 secondary case each. PFGE showed 2 identical isolates, whereas SCRA analysis resulted in 2 unique isolates for each of these clusters. Household cluster 52 consisted of 3 members. For this household, PFGE showed 3 identical isolates, whereas SCRA resulted in 2 identical isolates and 1 unique isolate. Analysis for the risk of acquisition of MRSA outside the household did not result in any plausible risk of transmission from another known source or via contact with a person with known risk factors.
(iii) Reproducibility.
For the three reference samples 806, 811, and 814, the mean correlation coefficients (±SD) were 0.9998 (±0.0), 0.9998 (±0.0), and 0.9999 (±0.0). Furthermore, for all duplicate measurements of the 26 isolates, the results were identical. Therefore, the reproducibility was 100%.
(iv) Workflow and costs.
Total hands-on time for 24 samples was around 3 h for SCRA and 7 h for PFGE. Total turnaround time for 24 samples was 36 to 48 h for SCRA and 96 h for PFGE, with a maximum of 72 samples per day for SCRA and 50 samples for PFGE.
SCRA analysis for 24 samples required a subculture on TSA on day 0 (15 min for inoculation, 15 min for documentation, and 18 to 20 h of incubation). On day 1, secondary subcultures on TSA were performed (15 min for inoculation, 20 h of incubation). Then, isolates were processed and measured on day 2, followed by analysis of the results (30 min for preparation of the slides, 60 min for measurements, and 30 min. for analysis).
Costs of SCRA and PFGE are comparable and are approximately €50 per sample, including personnel expenses and consumables. The cost for the SCRA apparatus will be about €125,000 (price levels for The Netherlands).
DISCUSSION
An ideal typing system for the epidemiological surveillance of MRSA should be fast and come with high levels of discriminatory power and reproducibility (1).
In this study, the discriminatory power of our ID values were 0.98 for PFGE and 0.99 for SCRA. The acceptable level of discrimination will depend on a number of factors, such as the epidemiological question, but an ID value of >0.900 might be desirable if the typing results are to be interpreted with confidence (12).
Our results for SCRA analysis were 95.6% concordant with the gold standard of household epidemiology. When our results were analyzed at the household cluster level, results for both PFGE and SCRA were 90.7% concordant. This indicates that the discriminatory power of SCRA might be too high for adequate outbreak analysis of small clusters.
However, initial PFGE results were 93.6% concordant with the gold standard of epidemiology on the strain level and 93.5% concordant on the household level. This implies that outbreak analysis by PFGE may lead to missing of small outbreaks to the same extent as analysis by SCRA.
Multiple independent measurements of 3 reference isolates and the duplicate measurements of 26 MRSA strains resulted in a reproducibility of 100%. This result indicates that SCRA is stable over a longer period of time, which has been reported before (5). Furthermore, with PFGE some MRSA isolates (e.g., ST398) are nontypeable (13), whereas with SCRA these isolates can be typed using the same protocol. Typeability (as shown by the ST398 isolates in household cluster 14) was 100% for SCRA and 97.3% for PFGE.
Our findings are comparable to those of a study performed by Wulf et al. (14). In this study, the observed concordance between SCRA and the gold standard of PFGE was 97%, whereas in our study the observed concordance between SCRA and PFGE was 95%. Our study differs from the study of Wulf et al. in setting and strains. Whereas Wulf et al. concentrated on large numbers of major MRSA clones (including ST 398) with multiple isolates, we focused on outbreak analysis, with large numbers of multiple clones with few isolates. This may explain the small difference in concordance between PFGE and SCRA in the two studies. Reproducibility in our study was better (100% versus 95%). In both studies isolates were tested as full biological replicates at different points in the study.
PFGE is still considered the gold standard for typing of MRSA. However, due to limited portability, with a long time to result, alternative typing techniques, such as Staphylococcus aureus protein A typing (spa typing), multilocus sequence typing (MLST), and multilocus variable number of tandem repeats analysis (MLVA) are used for outbreak analysis. Although spa typing and MLST have good portability due to standard nomenclature, the discriminatory power of these methods is too limited for adequate outbreak analysis (15, 16). The discriminatory power of MLVA is comparable with that of PFGE; however, the MLVA method cannot be easily used in routine diagnostic laboratories because of the required technical expertise (17). A commercially available automated repetitive-sequence-based PCR system, the DiversiLab system (bioMérieux, Marcy l'Etiole, France), offers advances in ease of use and reproducibility of the procedure over manual typing systems (18). However, although investigators in four independent studies concluded that the DiversiLab system is a rapid and reproducible technique, it clearly lacks resolution for typing of Gram-positive bacteria such as MRSA (19, 20, 21, 22).
The feasibility of the SCRA system with respect to hands-on time (∼3 h) and time to result (36 to 48 h) was better than that of PFGE (∼7 h hands-on time and ∼96 h time to result). Time to result may be improved even further by applying 1 subculture instead of 2 subcultures. In this way, results will be available the next day, which is of great value for infection prevention. Furthermore, SCRA has the advantage that all isolates (including, e.g., ST398) can be typed using the same protocol.
In many studies on typing of MRSA, the focus is on a small set of MRSA isolates and the relevant epidemiological data of the isolates are often (partially) unknown. We had the unique opportunity to use the epidemiological relationships within households as the gold standard, together with PFGE.
SCRA analysis can be initiated the next day after detection of 2 (or more) MRSA isolates. For 90 to 95% of cases a considerable amount of time for implementing prevention measures is gained with SCRA compared with PFGE (2 to 3 days instead of 4 to 5 days). This is especially important in a country with low MRSA prevalence; before closing a ward you will be sure that this action really is necessary, e.g., as transmission is shown (or not shown) by SCRA. For the remaining 5 to 10% of cases, further analysis will be necessary.
We conclude that the SpectraCellRA system is a highly reproducible, easy-to-use, and fast typing platform that can compete with the currently used typing techniques.
ACKNOWLEDGMENTS
Kees Maquelin is acknowledged for technical support. Both the Infection Prevention and the Diagnostics Units of the Department of Medical Microbiology and Infectious Diseases are acknowledged for collection of epidemiological data and isolates.
This research was funded by the European Community Seventh Framework Programme FP7/2007-2013 under grant agreement no. 395 241742 (TEMPOtest-QC).
Footnotes
Published ahead of print 20 February 2013
REFERENCES
- 1. van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, Fussing V, Green J, Feil E, Gerner-Smidt P, Brisse S, Struelens M, European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group on Epidemiological Markers (ESGEM) 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13(Suppl 3):1–46 [DOI] [PubMed] [Google Scholar]
- 2. Goering RV, Duensing TD. 1990. Rapid field inversion gel electrophoresis in combination with an rRNA gene probe in the epidemiological evaluation of staphylococci. J. Clin. Microbiol. 28:426–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ichiyama S, Ohta M, Shimokata K, Kato N, Takeuchi J. 1991. Genomic DNA fingerprinting by pulsed-field gel electrophoresis as an epidemiological marker for study of nosocomial infections caused by methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 29:2690–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van Belkum A, van Leeuwen W, Kaufmann ME, Cookson B, Forey F, Etienne J, Goering R, Tenover F, Steward C, O'Brien F, Grubb W, Tassios P, Legakis N, Morvan A, El Solh N, de Ryck R, Struelens M, Salmenlinna S, Vuopio-Varkila J, Kooistra M, Talens A, Witte W, Verbrugh H. 1998. Assessment of resolution and intercenter reproducibility of results of genotyping Staphylococcus aureus by pulsed-field gel electrophoresis of SmaI macrorestriction fragments: a multicenter study. J. Clin. Microbiol. 36:1653–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Willemse-Erix DF, Scholtes-Timmerman MJ, Jachtenberg JW, van Leeuwen WB, Horst-Kreft D, Bakker Schut TC, Deurenberg RH, Puppels GJ, van Belkum A, Vos MC, Maquelin K. 2009. Optical fingerprinting in bacterial epidemiology: Raman spectroscopy as a real-time typing method. J. Clin. Microbiol. 47:652–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mollema FP, Severin JA, Nouwen JL, Ott A, Verbrugh HA, Vos MC. 2010. Successful treatment for carriage of methicillin-resistant Staphylococcus aureus and importance of follow-up. Antimicrob. Agents Chemother. 54:4020–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wertheim HF, Ammerlaan HS, Bonten MJ, van den Broek PJ, Troelstra A, Vandenbroucke-Grauls CM, Vos MC, Voss A, Nouwen JL, Kluytmans JA. 2008. Optimisation of the antibiotic policy in the Netherlands. XII. The SWAB guideline for antimicrobial eradication of MRSA in carriers. Ned. Tijdschr. Geneeskd. 152:2667–2671 (In Dutch.) [PubMed] [Google Scholar]
- 8. Bode LG, Wertheim HF, Kluytmans JA, Bogaers-Hofman D, Vandenbroucke-Grauls CM, Roosendaal R, Troelstra A, Box AT, Voss A, van Belkum A, Verbrugh HA, Vos MC. 2011. Sustained low prevalence of meticillin-resistant Staphylococcus aureus upon admission to hospital in The Netherlands. J. Hosp. Infect. 79:198–201 [DOI] [PubMed] [Google Scholar]
- 9. Wertheim HF, Vos MC, Boelens HA, Voss A, Vandenbroucke-Grauls CM, Meester MH, Kluytmans JA, van Keulen PH, Verbrugh HA. 2004. Low prevalence of methicillin-resistant Staphylococcus aureus (MRSA) at hospital admission in the Netherlands: the value of search and destroy and restrictive antibiotic use. J. Hosp. Infect. 56:321–325 [DOI] [PubMed] [Google Scholar]
- 10. Mollema FP, Richardus JH, Behrendt M, Vaessen N, Lodder W, Hendriks W, Verbrugh HA, Vos MC. 2010. Transmission of methicillin-resistant Staphylococcus aureus to household contacts. J. Clin. Microbiol. 48:202–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hunter PR. 1990. Reproducibility and indices of discriminatory power of microbial typing methods. J. Clin. Microbiol. 28:1903–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rasschaert G, Vanderhaeghen W, Dewaele I, Janez N, Huijsdens X, Butaye P, Heyndrickx M. 2009. Comparison of fingerprinting methods for typing methicillin-resistant Staphylococcus aureus sequence type 398. J. Clin. Microbiol. 47:3313–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wulf MW, Willemse-Erix D, Verduin CM, Puppels G, van Belkum A, Maquelin K. 2012. The use of Raman spectroscopy in the epidemiology of methicillin-resistant Staphylococcus aureus of human- and animal-related clonal lineages. Clin. Microbiol. Infect. 18:147–152 [DOI] [PubMed] [Google Scholar]
- 15. Malachowa N, Sabat A, Gniadkowski M, Krzyszton-Russjan J, Empel J, Miedzobrodzki J, Kosowska-Shick K, Appelbaum PC, Hryniewicz W. 2005. Comparison of multiple-locus variable-number tandem-repeat analysis with pulsed-field gel electrophoresis, spa typing, and multilocus sequence typing for clonal characterization of Staphylococcus aureus isolates. J. Clin. Microbiol. 43:3095–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Struelens MJ, Hawkey PM, French GL, Witte W, Tacconelli E. 2009. Laboratory tools and strategies for methicillin-resistant Staphylococcus aureus screening, surveillance and typing: state of the art and unmet needs. Clin. Microbiol. Infect. 15:112–119 [DOI] [PubMed] [Google Scholar]
- 17. Schouls LM, Spalburg EC, van Luit M, Huijsdens XW, Pluister GN, van Santen-Verheuvel MG, van der Heide HG, Grundmann H, Heck ME, de Neeling AJ. 2009. Multiple-locus variable number tandem repeat analysis of Staphylococcus aureus: comparison with pulsed-field gel electrophoresis and spa-typing. PLoS One 4:e5082 doi:10.1371/journal.pone.0005082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Healy M, Huong J, Bittner T, Lising M, Frye S, Raza S, Schrock R, Manry J, Renwick A, Nieto R, Woods C, Versalovic J, Lupski JR. 2005. Microbial DNA typing by automated repetitive-sequence-based PCR. J. Clin. Microbiol. 43:199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Babouee B, Frei R, Schultheiss E, Widmer AF, Goldenberger D. 2011. Comparison of the DiversiLab repetitive element PCR system with spa typing and pulsed-field gel electrophoresis for clonal characterization of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol 49:1549–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fluit AC, Terlingen AM, Andriessen L, Ikawaty R, van Mansfeld R, Top J, Cohen Stuart JW, Leverstein-van Hall MA, Boel CH. 2010. Evaluation of the DiversiLab system for detection of hospital outbreaks of infections by different bacterial species. J. Clin. Microbiol. 48:3979–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Te Witt R, Kanhai V, van Leeuwen WB. 2009. Comparison of the DiversiLab system, pulsed-field gel electrophoresis and multi-locus sequence typing for the characterization of epidemic reference MRSA strains. J. Microbiol. Methods 77:130–133 [DOI] [PubMed] [Google Scholar]
- 22. Tenover FC, Gay EA, Frye S, Eells SJ, Healy M, McGowan JE., Jr 2009. Comparison of typing results obtained for methicillin-resistant Staphylococcus aureus isolates with the DiversiLab system and pulsed-field gel electrophoresis. J. Clin. Microbiol. 47:2452–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]