Abstract
Among 30 urinary isolates of Staphylococcus saprophyticus identified by sequencing methods, the rate of accurate identification was 100% for Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), 86.7% for the Phoenix PID and Vitek 2 GP systems, 93.3% for the MicroScan GP33 system, and 46.7% for the BBL CHROMagar Orientation system.
TEXT
Staphylococcus saprophyticus, a coagulase-negative staphylococcus (CoNS), is a common pathogen associated with community-acquired uncomplicated urinary tract infections (UTIs) in humans, particularly women younger than 50 years of age (1). Of the urinary CoNS isolates, S. saprophyticus is identified presumptively by simplified conventional methods and commercially automated identification methods (2–9). However, misidentification of S. saprophyticus by these simplified conventional and automated phenotypic methods often occurs. Although matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has been used for many years in the fields of proteomics and toxicology, the technology has only recently been applied to the field of microbiology for species identification of bacteria (2, 10–16). Direct detection of urinary tract pathogens from urine samples with high accuracy by MALDI-TOF MS has been reported, especially for Gram-negative bacteria with high bacterial counts (17).
In this study, we evaluated a total of 34 presumptive isolates of S. saprophyticus obtained from the urine of patients with UTIs at the National Taiwan University Hospital during the period January 2004 to December 2011. The 34 isolates were presumptively identified as S. saprophyticus based on colonial and microscopic morphotypes characteristic of the organism, the absence of hemolytic activity or coagulase production, resistance to novobiocin (5-μg disk; no inhibition zone), and susceptibility to polymyxin (300-IU disk; inhibition zone of ≥10 mm). Identification of the 34 isolates to the species level was performed using the BBL CHROMagar Orientation system (Becton Dickinson, Diagnostic Systems, Sparks, MD) and three commercially automated identification systems, namely, the Phoenix PMIC/ID-30 automated system (Becton Dickinson), the Vitek 2 GP system (bioMérieux Inc., La Balme les Grottes, France), and the MicroScan Walkaway automated system (Dade Behring, Inc., West Sacramento, CA). S. saprophyticus ATCC 15305 was used as a quality control strain for the BBL CHROMagar Orientation system.
For performance of the Bruker Biotyper MALDI-TOF MS system (Bruker Daltonics), a single colony from a blood agar plate was subjected to an ethanol-formic acid extraction procedure for microorganism profiling (4). Spectra were analyzed using MALDI Biotyper automation control and the Bruker Biotyper 3.1 software and library (version 3.1.66, with 4,613 entries; Bruker Daltonics). Identification scores of ≥2.000 indicated species-level identification, scores of 1.700 to 1.999 indicated genus-level identification, and scores of <1.700 indicated no identification (4). Cluster analysis of the isolates was performed using ClinProtools 3.0 (Bruker Daltonics).
PCR amplification of the nearly complete 16S rRNA gene (1,463 bp) from the 34 isolates was performed with two primers (8FPL and 1492) as previously described (18). The sequences obtained were compared with published sequences in the GenBank database, using the BLASTN algorithm (http://www.ncbi.nlm.nih.gov/blast). PCR amplification and sequencing of the partial rpoB gene of the 34 isolates were performed using primers 2491F and 3241R (16, 19, 20).
Of the 34 isolates, 30 were identified as S. saprophyticus, 2 were identified as S. cohnii, 1 was identified as S. xylosus, and 1 was identified as S. sciuri by 16S rRNA and rpoB gene sequencing. The 30 isolates of S. saprophyticus were recovered from 30 voided urine samples from 24 patients, including 23 women and 1 man, with a mean age of 36 years (range, 19 to 63 years). Six female patients had two different urine samples that were positive for S. saprophyticus. Of the 30 isolates that had been identified by sequencing as being S. saprophyticus, 22 (73.3%) were correctly identified as S. saprophyticus by the Phoenix PMIC/ID-30 system (confidence values, 90 to 99%), the Vitek 2 GP system (probability of identification, 99%), and the MicroScan system (99.4 to 99.9% identification). The Phoenix PID system misidentified 13.3% (4/30 isolates) of the isolates, the Vitek 2 GP system resulted in misidentification of an equal number of isolates (13.3% [4/30 isolates]), and the MicroScan system misidentified 6.7% (2/30 isolates) of the isolates (Table 1). Figure 1 shows the colony characteristics of two S. saprophyticus isolates and of one isolate each of S. cohnii, S. xylosus, and S. sciuri on BBL CHROMagar Orientation agar. Among the 30 isolates of S. saprophyticus, 14 (46.7%; class I) yielded pink/opaque colonies that are characteristic of S. saprophyticus on BBL CHROMagar Orientation agar, and 16 (53.3%; class II) showed white colonies.
Table 1.
Differences among various microbial identification methods in identifying 12 urinary CoNS isolates that were presumptively identified as S. saprophyticus
| Isolate no. | Identification result |
Colony color on BBL CHROMagar Orientation agar/class | ||||
|---|---|---|---|---|---|---|
| Sequencing of 16S rRNA and rpoB genes | MALDI-TOF Biotyper (score/cluster) | Vitek II GP (probability of identification [%]) | Phoenix PID (confidence value [%]) | MicroScan GP33 (% identification) | ||
| 1 | S. saprophyticus | S. saprophyticus (2.11/B) | S. saprophyticus (99) | S. saprophyticus (90) | S. hominis (72.7) | White/II |
| 2 | S. saprophyticus | S. saprophyticus (2.044/B) | S. saprophyticus (99) | S. epidermidis (90) | S. saprophyticus (99.7) | White/II |
| 3 | S. saprophyticus | S. saprophyticus (2.007/B) | S. saprophyticus (99) | S. epidermidis (90) | S. saprophyticus (99.9) | White/II |
| 4 | S. saprophyticus | S. saprophyticus (2.120/B) | S. saprophyticus (99) | S. warneri (90) | S. saprophyticus (86.0) | White/II |
| 5 | S. saprophyticus | S. saprophyticus (2.121/B) | S. hominis (96) | S. saprophyticus (94) | S. schleiferi (71) | White/II |
| 6 | S. saprophyticus | S. saprophyticus (2.154/B) | S. warneri (94) | S. saprophyticus (99) | S. saprophyticus (99.7) | White/II |
| 7 | S. saprophyticus | S. saprophyticus (1.993/B) | S. warneri or S. hominis | S. saprophyticus (99) | S. saprophyticus (99.7) | White/II |
| 8 | S. saprophyticus | S. saprophyticus (2.103/A) | S. hominis (low) | S. epidermidis (92) | S. saprophyticus (99.9) | Pink/I |
| 9 | S. cohnii | S. cohnii (1.495) | S. xylosus (95) | S. equorum (99) | S. xylosus (99.2) | Blue |
| 10 | S. cohnii | S. succinus (1.394) | S. xylosus (95) | S. equorum (99) | S. xylosus (97.3) | Blue |
| 11 | S. xylosus | S. xylosus (2.073) | S. xylosus (98) | S. gallinarum (97) | S. xylosus (90.1) | White |
| 12 | S. sciuri | S. sciuri (2.036) | S. sciuri (99) | S. sciuri (99) | S. sciuri (99.9) | Blue |
Fig 1.
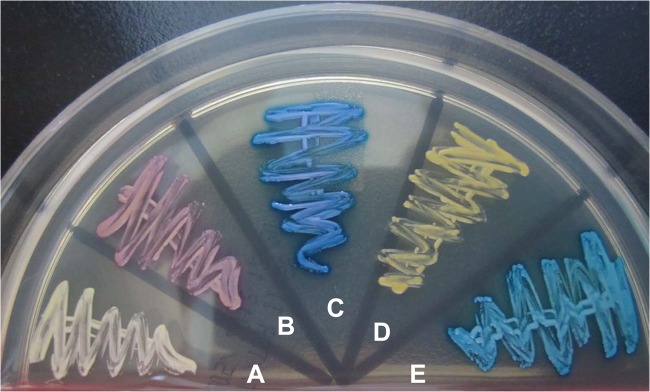
Colonial characteristics on CHROMagar Orientation agar of four different Staphylococcus species that were identified by 16S rRNA gene sequencing and a Biotyper MALDI-TOF MS system. Isolates: A and B, S. saprophyticus; C, S. cohnii; D, S. xylosus; E, S. sciuri.
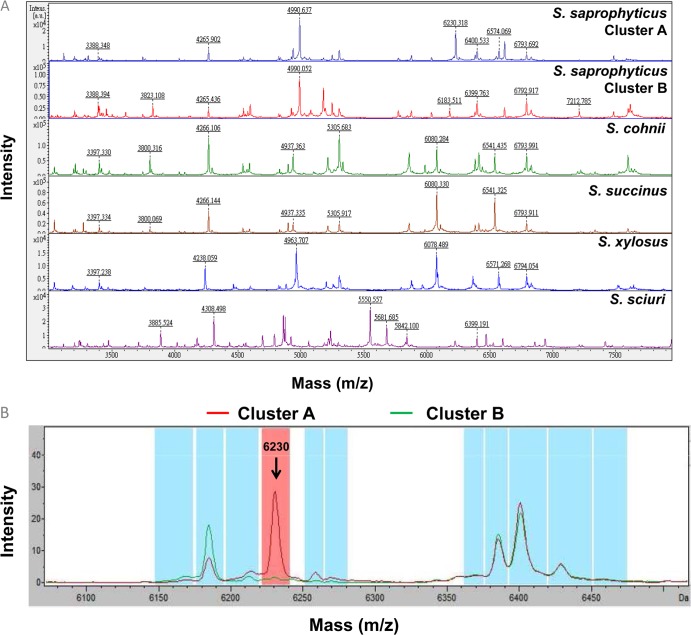
MALDI-TOF MS profiles of the five different CoNS species are illustrated in Fig. 2A. About 10 to 20 prominent ion peaks were noted in the m/z 2,000 to 12,000 range. Of the two isolates of S. cohnii that had been identified by genetic methods, the Biotyper MALDI-TOF system identified one as S. cohnii and one as S. succinus (Table 1); however, the identification scores were low. Of the 30 isolates of S. saprophyticus, 23 (76.7%) had identification scores of >2.0 (ranging from 2.005 to 2.154), and 7 (23.3%) had identification scores ranging from 1.893 to 1.997.
Fig 2.
(A) MALDI-TOF MS identification of Staphylococcus saprophyticus class I (pink colony growth on CHROMagar Orientation agar), S. saprophyticus class II (white colony growth on CHROMagar Orientation agar), S. cohnii, S. succinus, S. xylosus, and S. sciuri. The absolute intensities of the ions are shown on the y axis, and the masses (m/z) of the ions are shown on the x axis. The m/z value represents the mass-to-charge ratio. (B) Two clusters of S. saprophyticus spectra, i.e., cluster A and cluster B, analyzed by clustering analysis of MALDI-TOF results. The cluster A average spectrum is demonstrated in red, and the cluster B average spectrum is demonstrated in green. The arrow shows the peak 6,230 m/z signal. The 6,230 m/z signal could be observed in the cluster A spectrum but not the cluster B spectrum.
Cluster analysis of the 30 isolates was performed based on the colonial characteristics on CHROMagar Orientation agar (pink/opaque colonies for class I and white colonies for class II). Ten spectra for each class were selected randomly for model generation by ClinProtools 3.0, using the QuickClassifier algorithm (Bruker Daltonics). This model included eight peaks (3,115, 5,068, 6,213, 6,230, 6,258, 6,928, 8,223, and 9,926 m/z) for classification. All 30 S. saprophyticus isolates were classified using this model, and the results revealed that a cluster A spectrum was present for 13 (92.8%) of 14 class I isolates, with a cluster B spectrum for 16 (100%) of 16 class II isolates (Fig. 2B). A characteristic 6,230 m/z signal was observed in the cluster A spectrum but not the cluster B spectrum (Fig. 2B). Isolates from the same patients belonged to the same clusters. There was no clinical association of patients (underlying medical conditions or types of UTIs) with isolates belonging to the two clusters. This clustering of S. saprophyticus isolates was not observed previously using MALDI-TOF or other commercial methods.
Several factors are associated with S. saprophyticus UTIs, including outdoor swimming, sexual intercourse, and work in meat and cheese production (1). In contrast, UTIs caused by other CoNS, particularly S. epidermidis, are commonly related to indwelling catheters or surgery and cause complicated hospital-acquired UTIs (1). As a result, accurate identification of CoNS to the species level, or at least rapid differentiation between S. saprophyticus and other CoNS isolates, is important.
In the present study, three important findings were illustrated. First, the accuracy of identification of S. saprophyticus by use of the recommended presumptive identification scheme for S. saprophyticus among urinary CoNS isolates was better than that using the BBL CHROMagar Orientation system (88.2% versus 46.7%). Second, among the 30 isolates genetically identified as S. saprophyticus, misidentification was found more commonly using the Phoenix PID and Vitek 2 GP systems than using the MicroScan system. Third, all 30 of the S. saprophyticus isolates were accurately identified by the Biotyper MALDI-TOF system. A previous study demonstrated that MALDI-TOF MS could accurately identify all CoNS species and subspecies tested (16). However, one isolate of S. cohnii in this study could not be identified accurately to the species level by MALDI-TOF MS.
Few studies have compared the accuracies of identifying S. saprophyticus in urine of the Vitek 2, Phoenix, and MicroScan systems (2, 5, 8). The Vitek 2 system has been shown to be better than the Phoenix system at identifying CoNS isolates to the species level (91.9% versus 88.4%) when molecular reference methods are used (2). In addition, the MicroScan system has been reported to have an accuracy rate of 90% for correctly identifying S. saprophyticus in urine (5). Only a small number of S. saprophyticus isolates have been tested in previous studies for assessment of the identification accuracy of the BBL CHROMagar Orientation system (3, 6). The results of our study clearly demonstrate that two distinct colony colors (white and pink) for S. saprophyticus on BBL CHROMagar Orientation agar were not in accordance with the interpretation instructions provided by the manufacturer.
In summary, our findings show that MALDI-TOF MS is more accurate than other commercial identification methods for identifying CoNS, particularly urinary isolates of S. saprophyticus.
Footnotes
Published ahead of print 6 February 2013
REFERENCES
- 1. Minardi D, d'Anzeo G, Cantoro D, Conti A, Muzzonigro G. 2011. Urinary tract infections in women: etiology and treatment options. Int. J. Gen. Med. 4:333–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chatzigeorgiou KS, Sergentanis TN, Tsiodras S, Hamodrakas SJ, Bagos PG. 2011. Phoenix 100 versus Vitek 2 in the identification of gram-positive and gram-negative bacteria: a comprehensive meta-analysis. J. Clin. Microbiol. 49:3284–3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. D'Souza HA, Campbell M, Baron EJ. 2004. Practical bench comparison of BBL CHROMagar Orientation and standard two-plate media for urine cultures. J. Clin. Microbiol. 42:60–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dupont C, Sivadon-Tardy V, Bille E, Dauphin B, Beretti JL, Alvarez AS, Degand N, Ferroni A, Rottman M, Herrmann JL, Nassif X, Ronco E, Carbonnelle E. 2010. Identification of clinical coagulase-negative staphylococci, isolated in microbiology laboratories, by matrix-assisted laser desorption/ionization-time of flight mass spectrometry and two automated systems. Clin. Microbiol. Infect. 16:998–1004 [DOI] [PubMed] [Google Scholar]
- 5. Grant CE, Sewell DL, Pfaller M, Bumgardner RV, Williams JA. 1994. Evaluation of two commercial systems for identification of coagulase-negative staphylococci to species level. Diagn. Microbiol. Infect. Dis. 18:1–5 [DOI] [PubMed] [Google Scholar]
- 6. Hengstler KA, Hammann R, Fahr AM. 1997. Evaluation of BBL CHROMagar orientation medium for detection and presumptive identification of urinary tract pathogens. J. Clin. Microbiol. 35:2773–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iorio NL, Ferreira RB, Schuenck RP, Malvar KL, Brilhante AP, Nunes AP, Bastos CC, Dos Santos KR. 2007. Simplified and reliable scheme for species-level identification of Staphylococcus clinical isolates. J. Clin. Microbiol. 45:2564–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim M, Heo SR, Choi SH, Kwon H, Park JS, Seong MW, Lee DH, Park KU, Song J, Kim EC. 2008. Comparison of the MicroScan, Vitek 2, and Crystal GP with 16S rRNA sequencing and MicroSeq 500 v2.0 analysis for coagulase-negative staphylococci. BMC Microbiol. 8:233 doi:10.1186/1471-2180-8-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Samra Z, Heifetz M, Talmor J, Bain E, Bahar J. 1998. Evaluation of use of a new chromogenic agar in detection of urinary tract pathogens. J. Clin. Microbiol. 36:990–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alatoom AA, Cunningham SA, Ihde SM, Mandrekar J, Patel R. 2011. Comparison of direct colony method versus extraction method for identification of Gram-positive cocci by use of Bruker Biotyper matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 49:2868–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Benagli C, Rossi V, Dolina M, Tonolla M, Petrini O. 2011. Matrix-assisted laser desorption ionization–time of flight mass spectrometry for the identification of clinically relevant bacteria. PLoS One 6:e16424 doi:10.1371/journal.pone.0016424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bergeron M, Dauwalder O, Gouy M, Freydiere AM, Bes M, Meugnier H, Benito Y, Etienne J, Lina G, Vandenesch F, Boisset S. 2011. Species identification of staphylococci by amplification and sequencing of the tuf gene compared to the gap gene and by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Eur. J. Clin. Microbiol. Infect. Dis. 30:343–354 [DOI] [PubMed] [Google Scholar]
- 13. Fox K, Fox A, Rose J, Walla M. 2011. Speciation of coagulase negative staphylococci, isolated from indoor air, using SDS PAGE gel bands of expressed proteins followed by MALDI TOF MS and MALDI TOF-TOF MS-MS analysis of tryptic peptides. J. Microbiol. Methods 84:243–250 [DOI] [PubMed] [Google Scholar]
- 14. Khot PD, Couturier MR, Wilson A, Croft A, Fisher MA. 2012. Optimization of matrix-assisted laser desorption ionization–time of flight mass spectrometry analysis for bacterial identification. J. Clin. Microbiol. 50:3845–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Murray PR. 2010. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry: usefulness for taxonomy and epidemiology. Clin. Microbiol. Infect. 16:1626–1630 [DOI] [PubMed] [Google Scholar]
- 16. Spanu T, De Carolis E, Fiori B, Sanguinetti M, D'Inzeo T, Fadda G, Posteraro B. 2011. Evaluation of matrix-assisted laser desorption ionization–time-of-flight mass spectrometry in comparison to rpoB gene sequencing for species identification of bloodstream infection staphylococcal isolates. Clin. Microbiol. Infect. 17:44–49 [DOI] [PubMed] [Google Scholar]
- 17. Ferreira L, Sánchez-Juanes F, González-Avila M, Cembrero-Fuciños D, Herrero-Hernández A, González-Buitrago JM, Muñoz-Bellido JL. 2010. Direct identification of urinary tract pathogens from urine samples by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J. Clin. Microbiol. 48:2110–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chuang CY, Yang YL, Hsueh PR, Lee PI. 2010. Catheter-related bacteremia caused by Staphylococcus pseudintermedius refractory to antibiotic-lock therapy in a hemophilic child with dog exposure. J. Clin. Microbiol. 48:1497–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Drancourt M, Raoult D. 2002. rpoB gene sequence-based identification of Staphylococcus species. J. Clin. Microbiol. 40:1333–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mellmann A, Becker K, von Eiff C, Keckevoet U, Schumann P, Harmsen D. 2006. Sequencing and staphylococci identification. Emerg. Infect. Dis. 12:333–336 [DOI] [PMC free article] [PubMed] [Google Scholar]