Abstract
Transmission of methicillin-resistant Staphylococcus aureus (MRSA) between animals and humans is widely recognized. In this study, we describe the first case of infection of a goat and suspected transmission of MRSA ST398 to a human, which resulted in colonization of animal owners by MRSA sequence type 398.
CASE REPORT
A goat 1 month and 17 days old (mixed breed) was submitted for necropsy because of the sudden deaths of three goats in a week from the same stable box, as reported by the animal owners. After a thorough gross examination, pathohistological and bacteriological examinations were performed. In the foreground were the changes in the lungs. The lungs showed high-grade alveolar edema and moderate alveolar emphysema and, in some places, far-reaching atelectasis. Histologically, the lungs showed a focal, low-moderate desquamative pneumonia with focal purulent components and subtle fibrin in the alveoli. At one location, several coccoid bacteria presented in the alveoli. Bacteriological examination yielded Mycoplasma mycoides subsp. capri and Staphylococcus aureus (isolate 414_12). S. aureus species identification was confirmed by PCR (1). Antimicrobial susceptibility testing by the disk diffusion method according to guidelines of the Clinical and Laboratory Standards Institute (2, 3) showed that this isolate was resistant to oxacillin, cefoxitin, penicillin, amoxicillin-clavulanic acid, ceftiofur, cefovecin, cefquinome, tetracycline, doxycycline, and enrofloxacin and susceptible to marbofloxacin, ciprofloxacin, amikacin, gentamicin, chloramphenicol, azithromycin, erythromycin, clindamycin, teicoplanin, vancomycin (as determined by a MIC evaluator test), trimethoprim-sulfamethazole, linezolid, rifampin, and mupirocin. The production of β-lactamase was confirmed using a nitrocefin assay. After incubation on BBL CHROMagar MRSA II, the colonies showed a colony appearance typical of methicillin-resistant S. aureus (MRSA). The isolate was confirmed as MRSA by mecA PCR (4). The infected goat resided in a small goat farm in lower Austria, shared a stable box with 10 other goats, and was in frequent close contact with eight goats from a neighboring box, as reported by the owner. To establish the MRSA nasal colonization status of the goats and humans that had been in contact with the affected goat, diagnostic samples were requested. Two weeks after first isolation of MRSA, nasal swabs were collected from owners (female and male), from fellow workers (female and male), and from goats which shared the same box with the infected animal. Six out of 10 investigated goats were colonized with MRSA (isolates 5Z, 7Z, 8Z, 9Z, 11Z, 13Z), whereas both owners and fellow workers were negative for MRSA. Subsequently, 2 weeks later, the same persons were screened again for the presence of MRSA, as were eight goats from the neighboring box, a household dog, and three cats, resulting in two MRSA-positive samples (isolates 15H and 28Z), one originating from a goat and the other one from the female owner. Four weeks after isolation of human MRSA, nasal swabs from the person who tested positive were screened and remained positive (isolate 16H). All MRSA isolates were confirmed as described above and further investigated by PCR targeting Panton-Valentine leukocidin (PVL) genes (5), staphylococcal cassette chromosome mec (SCCmec) typing using the multiplex PCR method (6), staphylococcal protein A (spa) typing (7) (results of spa typing were interpreted by using BioNumerics version 5.1 software), and two different multilocus variable-number tandem-repeat analyses (MLVA): MLVA-14Orsay (8) and multiplex PCR MLVA (9). Multilocus sequence typing (MLST) was performed on all isolates. MLST was carried out by PCR amplification and sequencing of seven housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi, yqiL) as previously described (10). The allelic profiles and sequence types (ST) were assigned by the MLST website (http://saureus.mlst.net/). In order to determine the antibiotic profile of each isolate, they were tested against the above-mentioned antibiotics.
All MRSA isolates were PVL negative, carried an SCCmec type V cassette, belonged to spa type t011, and shared the same MLVA-14Orsay genotype (7 7 −1 5 1 1 5 0 4.5 4 5 3 3 2). All but one isolate utilized the same genotype obtained after multiplex PCR MLVA. For one goat isolate (11Z), primer pair clfB generated a band that was approximately 100 bp smaller than the bands generated by the same primer pair in all other isolates (Fig. 1). Therefore, on the basis of a difference of this band, this MRSA isolate could be assigned to another MLVA genotype. MLST revealed that all tested MRSA isolates belonged to the clonal lineage ST398. All isolates tested in this study shared the same antibiotic profile.
Fig 1.
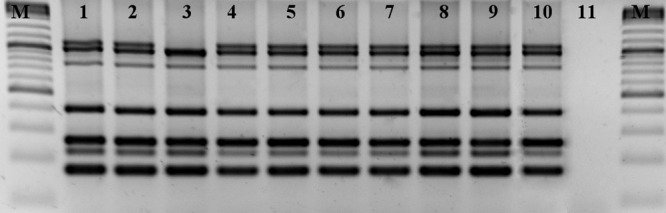
MLVA fingerprints of the 9 MRSA isolates from the goats and owner. M, 100-bp marker; lane 1, isolate 414_12; lane 2, isolate 9Z; lane 3, isolate 11Z; lane 4, isolate 8Z; lane 5, isolate 5Z; lane 6, isolate 7Z; lane 7, isolate 13Z; lane 8, isolate 28Z; lane 9, isolate 15H; lane 10, isolate 16H; lane 11, negative control.
MRSA was not detectable after the first examination of the female owner, but she became positive 2 weeks afterward and remained positive for another 4 weeks. She reported that she would daily feed and hold the goats, especially the young animals. During the time between the first isolation of MRSA and the last sample collection, neither the tested animals nor the humans had received any antibiotic therapy nor showed any clinical signs. As reported by the owner, none of the examined persons had been exposed to pigs and veal calves, which is a well-documented risk factor associated with acquisition of MRSA ST398 (11). Based on the data presented in the current case, we believe that the owner most likely became a carrier of MRSA ST398 after acquisition of MRSA by the goat, because the human isolate was indistinguishable from eight out of nine goat isolates by the methods used.
MRSA is a frequent pathogen of humans and many animal species. In the past years, the transfer of MRSA isolates between animals and humans gained specific attention, especially in the case of livestock-associated MRSA ST398 belonging to clonal complex 398 (CC398), which has been the most commonly reported MRSA strain found in association with livestock (12). However, there is still a scarcity of information on infections with MRSA ST398 in goats. Colonization of humans with ST398 in contact with infected colonized animals has been widely documented (12). Consequential infection of humans have also been reported (13), and in some areas with high livestock density, the prevalence is rising (14). Two Austrian regions, upper Austria and southeast Styria, with intensive pig farming account for practically all human MRSA ST398 infections (15, 16). Although pig farming is as common as in upper Austria, there are no reports of MRSA ST398 in lower Austria. We recognize that the results of our study have limitations, because it was not possible to completely determine the source of MRSA. Despite this limitation, this study is the first to report MRSA ST398 in Austrian goats and, to our knowledge, the first to suggest transmission from goats to human.
In conclusion, our study contributes to the growing evidence that MRSA ST398 could be transmitted from animal to human. At this point, it could be highly recommended that unnecessary close contact with positive animals should be avoided.
ACKNOWLEDGMENT
There are no financial declarations or conflicts of interest to disclose.
Footnotes
Published ahead of print 27 February 2013
REFERENCES
- 1. Hirotaki S, Sasaki T, Kuwahara-Arai K, Hiramatsu K. 2011. Rapid and accurate identification of human-associated staphylococci by use of multiplex PCR. J. Clin. Microbiol. 49:3627–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clinical and Laboratory Standards Institute 2008. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals—3rd ed. Approved standard M31-A3. CLSI, Wayne, PA [Google Scholar]
- 3. Clinical and Laboratory Standards Institute 2012. Performance standards for antimicrobial susceptibility testing: 21st informational supplement. M100-22 CLSI, Wayne, PA [Google Scholar]
- 4. Strommenger B, Kettlitz C, Werner G, Witte W. 2003. Multiplex PCR assay for simultaneous detection of nine clinically relevant antibiotic resistance genes in Staphylococcus aureus. J. Clin. Microbiol. 41:4089–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, Vandenesch F, Etienne J. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128–1132 [DOI] [PubMed] [Google Scholar]
- 6. Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, Vogel U. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pourcel C, Hormigos K, Onteniente L, Sakwinska O, Deurenberg RH, Vergnaud G. 2009. Improved multiple-locus variable-number tandem-repeat assay for Staphylococcus aureus genotyping, providing a highly informative technique together with strong phylogenetic value. J. Clin. Microbiol. 47:3121–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rasschaert G, Vanderhaeghen W, Dewaele I, Janez N, Huijsdens X, Butaye P, Heyndrickx M. 2009. Comparison of fingerprinting methods for typing methicillin-resistant Staphylococcus aureus sequence type 398. J. Clin. Microbiol. 47:3313–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Cleef BA, Monnet DL, Voss A, Krziwanek K, Allerberger F, Struelens M, Zemlickova H, Skov RL, Vuopio-Varkila J, Cuny C, Friedrich AW, Spiliopoulou I, Pászti J, Hardardottir H, Rossney A, Pan A, Pantosti A, Borg M, Grundmann H, Mueller-Premru M, Olsson-Liljequist B, Widmer A, Harbarth S, Schweiger A, Unal S, Kluytmans JA. 2011. Livestock-associated methicillin-resistant Staphylococcus aureus in humans, Europe. Emerg. Infect. Dis. 17:502–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Smith TC, Pearson N. 2011. The emergence of Staphylococcus aureus ST398. Vector Borne Zoonotic Dis. 11:327–339 [DOI] [PubMed] [Google Scholar]
- 13. Fluit AC. 2012. Livestock-associated Staphylococcus aureus. Clin. Microbiol. Infect. 18:735–744 [DOI] [PubMed] [Google Scholar]
- 14. Verkade E, Bergmans AM, Budding AE, van Belkum A, Savelkoul P, Buiting AG, Kluytmans J. 2012. Recent emergence of Staphylococcus aureus clonal complex 398 in human blood cultures. PLoS One 7:e41855 doi:10.1371/journal.pone.0041855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grisold AJ, Zarfel G, Hoenigl M, Krziwanek K, Feierl G, Masoud L, Leitner E, Wagner-Eibel U, Badura A, Marth E. 2010. Occurrence and genotyping using automated repetitive-sequence-based PCR of methicillin-resistant Staphylococcus aureus ST398 in Southeast Austria. Diagn. Microbiol. Infect. Dis. 66:217–221 [DOI] [PubMed] [Google Scholar]
- 16. Krziwanek K, Metz-Gercek S, Mittermayer H. 2009. Methicillin-resistant Staphylococcus aureus ST398 from human patients, upper Austria. Emerg. Infect. Dis. 15:766–769 [DOI] [PMC free article] [PubMed] [Google Scholar]


