Abstract
Staphylococcus epidermidis is a major cause of catheter-related bloodstream infections (CRBSIs). Recent studies suggested the existence of well-adapted, highly resistant, hospital-associated S. epidermidis clones. The molecular epidemiology of S. epidermidis in Belgian hospitals and the Belgian community has not been explored yet. We compared a set of 33 S. epidermidis isolates causing CRBSI in hospitalized patients with a set of 33 commensal S. epidermidis isolates. The factors analyzed included resistance to antibiotics and genetic diversity as determined by pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), and SCCmec typing. Additionally, the presence of virulence-associated mobile genetic elements, the ica operon and the arginine catabolic mobile element (ACME), was assessed and compared against clinical data. CRBSI S. epidermidis isolates were significantly resistant to more antibiotics than commensal S. epidermidis isolates. The two populations studied were very diverse and genetically distinct as only 23% of the 37 PFGE types observed were harbored by both CRBSI and commensal isolates. ACME was found in 76% of S. epidermidis strains, regardless of their origin, while the ica operon was significantly more prevalent in CRBSI isolates than in commensal isolates (P < 0.05). Nine patients presented a clinically severe CRBSI, eight cases of which were due to an ica-positive multiresistant isolate belonging to sequence type 2 (ST2) or ST54. S. epidermidis isolates causing CRBSI were more resistant and more often ica positive than commensal S. epidermidis isolates, which were genetically heterogeneous and susceptible to the majority of antibiotics tested. Clinically severe CRBSIs were due to isolates belonging to two closely related MLST types, ST2 and ST54.
INTRODUCTION
Staphylococcus epidermidis is a common commensal bacterium of the human skin and mucosa. While S. epidermidis has long been considered nonpathogenic, it is now recognized as a relevant opportunistic pathogen (1). Most S. epidermidis-related infections are acquired in hospitals and are associated with the use of medical devices (2).
Certain S. epidermidis strains are able to form on polymer surfaces a multicellular agglomeration with a characteristic three-dimensional structure and physiology called biofilm (3). Compared to the planktonic state, biofilm confers to S. epidermidis increased mechanical, metabolic, immune, and antibiotic resistance and its production is essentially mediated by the production of a polysaccharide intercellular adhesin (PIA) encoded by the accessory intercellular adhesion (ica) operon (4). Another genetic locus, the arginine catabolic mobile element (ACME), is thought to play an important role in the pathogenicity of S. epidermidis. ACME comprises a six-gene arc operon that encodes an arginine deiminidase. The role of ACME has not yet been precisely determined, but various studies have showed that ACME increases the ability of S. epidermidis strains to colonize the skin and mucous membranes (2, 5).
Antibiotic resistance genes are widespread among the S. epidermidis population. One of the most common is the mec(A) gene, which confers resistance to oxacillin and is carried by a mobile genetic element called staphylococcal cassette chromosome mec (SCCmec). There are several types of SCCmec, which vary in size and structure. Until now, more than 11 different SCCmec structures have been identified for S. epidermidis (available on the website http://www.sccmec.org). Additionally, S. epidermidis strains often present high resistance rates, making their eradication very difficult. The common source of catheter-related bloodstream infections (CRBSIs) due to S. epidermidis is the endogenous flora of the patient. However, it has been reported that infections due to coagulase-negative staphylococci (CNS) can also be transmitted between hospitalized patients and health care workers (6, 7). Accordingly, studies conducted until now suggest the existence of well-adapted, highly resistant, hospital-associated S. epidermidis clones.
Using multilocus sequence typing (MLST), a typing method based on the sequence polymorphism of fragments of seven housekeeping genes designed to reveal long-term global epidemiology and population structure of S. epidermidis, several epidemic clonal lineages disseminated worldwide have been identified. The most predominant of these lineages, clonal complex 2 (CC2), is composed of a large number of sequence types (STs), due to an unusually high rate of recombination within the lineage (8). One of these STs, ST2, was recognized as representing the majority of methicillin-resistant S. epidermidis (MRSE) clinical strains (9, 10). ST2 isolates usually harbor the ica operon (11) and can carry different SCCmec types (12).
The sequence types and molecular characteristics of S. epidermidis strains circulating in Belgian hospitals as well as in the Belgian community have never been explored.
The objectives of this study were to compare a set of S. epidermidis isolates responsible for CRBSIs in hospitalized patients with a set of commensal S. epidermidis isolates from healthy volunteers (HVs) in terms of resistance to antibiotics, genetic diversity, and the presence of virulence-associated genetic loci (ica operon and ACME) and to compare these microbiological characteristics to the clinical data from patients with CRBSIs in order to identify factors that may be associated with a more severe clinical presentation.
MATERIALS AND METHODS
Study population.
The study was conducted at the Erasme Hospital, a university hospital with 864 beds. Sixty-six S. epidermidis isolates were studied: (i) 33 S. epidermidis isolates from patients who presented with a CRBSI from January 2006 to May 2011, selected among a collection of 48 S. epidermidis CRBSI isolates retrospectively identified via the database of the infection control unit, and (ii) 33 commensal S. epidermidis isolates, prospectively collected in 2011 from the skin of healthy volunteer students.
For each monomicrobial episode of CRBSI, we included at least one blood culture and one central venous catheter tip positive for S. epidermidis with an identical resistance profile and only one episode per patient. We excluded episodes not treated with antibiotics.
The demographic data collected included age and gender. Each volunteer completed a questionnaire confirming the absence of intake of antibiotics in the previous 3 months and the absence of contact with a hospital or health care environment.
The clinical data collected for the 33 patients included the following: the major underlying disease, year of the CRBSI episode, hospitalization unit, length of hospital stay before CRBSI, time between catheter insertion and removal, indication for usage of the catheter (parenteral nutrition or not), prior hospitalization within 3 months, prior antibiotic therapy within the previous month, attributable death, presence of severe sepsis or septic shock, and antibiotic treatment. Several microbiological data were also registered, including the number and dates of blood and catheter cultures. The patients came from gastrointestinal surgery (n = 14), intensive care (n = 5), hematology (n = 5), pulmonary oncology (n = 5), and nephrology-urology (n = 4) units.
Definitions.
In accordance with CDC definitions of bloodstream infection (BSI) surveillance, confirmation of an S. epidermidis BSI requires that the patient presents with fever (temperature above 38°C) or hypothermia (temperature below 35°C) and/or hypotension and has undergone at least two pairs of blood cultures collected at different times (13). We defined CRBSI as a BSI with a central venous catheter positive for S. epidermidis presenting an identical susceptibility profile and treated with antibiotics. Clinically, the condition was considered severe if the patient presented with severe sepsis or septic shock at the time the blood culture samples were obtained. Sepsis associated with organ dysfunction, hypotension, or systemic manifestations of hypoperfusion constituted severe sepsis. Septic shock was defined as sepsis associated with hypotension unresponsive to intravenous fluid challenge or requiring a vasopressor agent (14). Persistent S. epidermidis bacteremia was defined as 2 or more consecutive positive blood cultures after removal of the catheter and despite 48 h of adequate antibiotic therapy.
A death was considered attributable to the CRBSI if it was reported during the episode (before the completion of antibiotic treatment) and no other cause was identified.
Bacterial isolate identification, characterization, and susceptibility testing.
Commensal isolates were obtained by swabbing of the neck skin (one isolate per volunteer) of randomly selected students (first-year undergraduate, Université Libre de Bruxelles). Clinical isolates (CRBSI) were isolated from blood cultures. Isolates were conserved at −80°C in pure glycerin and then subcultured onto Columbia 5% sheep blood agar for 18 to 24 h at 35°C in an aerobic atmosphere and then identified by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF). Susceptibility to 15 antimicrobial agents (penicillin, cefoxitin, erythromycin, clindamycin, ciprofloxacin, gentamicin, kanamycin, tobramycin, tetracycline, minocycline, rifampin, trimethoprim-sulfamethoxazole, fusidic acid, linezolid, and chloramphenicol) was determined by disk diffusion (Bio-Rad) according to CLSI criteria (15). For fusidic acid, Comité de l'Antibiogramme de la Société Française de Microbiologie (CASFM) criteria were used (http://www.sfm-microbiologie.org/UserFiles/file/CASFM/casfm_2010.pdf). Regarding vancomycin susceptibility, all strains were tested by the vancomycin screen agar method according to CLSI criteria. To ease data analysis, the sum of cases of resistance to non-β-lactam drugs observed for each isolate was calculated. In this calculation, the three aminoglycosides were regarded as a whole and isolates resistant to at least one of the three drugs were considered resistant to aminoglycosides.
Bacterial DNA was extracted as described by ünal et al. (16), and then all isolates were tested for the presence of the mec(A) gene as previously described (17). The presence of the antibiotic resistance genes aph(3′)-IIIa, ant(4′)-Ia, aac(6′)-Ie plus aph(2″), erm(A), erm(B), erm(C), msr(A), tet(K), and tet(M) was searched by PCR for all isolates nonsusceptible to kanamycin, tobramycin, gentamicin, erythromycin, and tetracycline, respectively (18–20).
Additionally, all isolates were tested for the presence of arc(A) and ica(A), to assess the presence of the ACME and the ica operon, respectively (4, 21).
Molecular typing techniques.
For all isolates, SmaI restriction fragments of genomic DNA were separated by pulsed-field gel electrophoresis (PFGE) as previously described (22). Bands in a size range between 36 and 700 kb were analyzed using the Bionumerics software version 6.5 by the Dice similarity coefficient, with an optimization set to 0.8% and a position tolerance set to 1%. A dendrogram of similarity was built using the unweighted pair-group method using arithmetic averages (UPGMA). Patterns were examined both by visual inspection and by computer matching. According to criteria established by Miragaia et al. (23), patterns differing by less than 79% (corresponding to a difference of less than seven bands) were considered to belong to the same type (represented by a letter). Multilocus sequence typing (MLST) was performed as described by Thomas et al. (24) on one randomly selected isolate of each PFGE type represented by more than one isolate. The sequences and allelic profiles assignments as well as the eBURST clustering (8) were performed via the S. epidermidis MLST database (http://www.mlst.net).
SCCmec typing was performed on all isolates by determination of the ccr and mec gene complex using multiplex PCR 1 (M-PCR1) and M-PCR2 as described by Kondo et al. (25).
Confidentiality and ethics committee approval.
The demographic, clinical, microbiological, and molecular epidemiology data were collected anonymously. The ethics committee of the Erasme Hospital approved the protocol before the beginning of the study (no. P2011/268).
Data analysis.
Stata data analysis and statistical software (version 10, copyright 1996 to 2012, StataCorp LP) was used for data analysis. The mean age and the sum of cases of resistance for S. epidermidis isolates were compared using the t test for equal or unequal variances. We used log10 with the length (days) of hospital stay before CRBSI and the duration (days) of the catheterization of the two groups to obtain a normal distribution before using the t test for equal or unequal variances. These results are presented using the geometric mean ± 1 standard deviation (SD). Fisher's exact tests were used to compare the proportion of persistent bacteremia, sex, the previous hospitalization, attributable death, underlying disease, presence of ica and ACME, and previous antibiotic treatment with severe clinical presentation and mixed isolates. The chi-square test was used to compare antimicrobial resistance. Statistical significance was achieved with 2-sided P value of <0.05.
RESULTS
All isolates tested (n = 66) were confirmed by MALDI-TOF to belong to the species Staphylococcus epidermidis. The 33 isolates of S. epidermidis responsible for CRBSI were distributed as follows: 11 strains in 2006, 7 in 2007, 7 in 2008, 4 in 2009, 2 in 2010, and 2 in 2011.
Resistance.
The resistance results are shown in Table 1. The majority (76%) of isolates were resistant to penicillin, although there was a marked difference between the two groups. Regarding the resistance to oxacillin, only one isolate from a volunteer carried the mec(A) gene, while 25 out of the 33 clinical isolates (76%) did so. Commensal isolates were, on average, resistant to 1 out of the 12 antibiotics other than β-lactams tested, with a maximum number of 2 antibiotics. Clinical strains were, on average, resistant to five non-β-lactam antibiotics (range, 0 to 7).
Table 1.
Antimicrobial resistance profiles and resistance-encoding genes of S. epidermidis isolates causing CRBSI versus S. epidermidis isolates collected from the skin of healthy volunteers
| Antimicrobial | Resistance-encoding gene(s) | No. of isolates with profile |
P value | ||
|---|---|---|---|---|---|
| HVs (n = 33) | CRBSIs (n = 33) | Overall (n = 66) | |||
| Penicillin | 20 | 30 | 50 | 0.04 | |
| Oxacillin (cefoxitin) | 1 | 25 | 26 | <0.001 | |
| mec(A) | 1 | 25 | 26 | ||
| Erythromycin | 15 | 25 | 40 | 0.012 | |
| erm(C) | 6 | 15 | 21 | ||
| msr(A) | 8 | 6 | 14 | ||
| erm(C) + erm(A) | 1 | 1 | |||
| erm(C) + msr(A) | 1 | 1 | 2 | ||
| erm(B) + msr(A) | 1 | 1 | |||
| Clindamycina | 1 (6) | 15 (2) | 16 (8) | <0.001 | |
| Ciprofloxacin | 0 | 19 | 19 | <0.001 | |
| Fusidic acid | 1 | 15 | 16 | <0.001 | |
| Trimethoprim-sulfamethoxazole | 0 | 16 | 16 | <0.001 | |
| Tetracycline | 3 | 8 | 11 | 0.099 | |
| tet(K) | 3 | 8 | 11 | ||
| Aminoglycosides (at least 1) | 0 | 13 | 13 | <0.001 | |
| Kanamycin | 11 | 11 | |||
| Tobramycin | 10 | 10 | |||
| Gentamicin | 6 | 6 | |||
| aac(6″)-aph(2″) | 2 | 2 | |||
| aph(3′) | 2 | 2 | |||
| ant(4′) | 3 | 3 | |||
| aac(6′)-aph(2″) + ant(4′) | 5 | 5 | |||
| Rifampin | 0 | 2 | 2 | 0.49 | |
| Chloramphenicol | 0 | 2 | 2 | 0.49 | |
| Minocycline | 0 | 0 | 0 | ||
| Vancomycin | 0 | 0 | 0 | ||
| Linezolid | 0 | 0 | 0 | ||
For clindamycin, the values in parentheses represent the number of isolates [erm(C) positive] with a clindamycin-inducible resistance phenotype.
Regarding the genes encoding these resistance profiles, 60% of the 40 erythromycin-resistant isolates possessed the erm(C) gene: one was erm(C) and erm(A) positive, and two were erm(C) and msr(A) positive. Fourteen isolates were msr(A) positive; one was msr(A) and erm(B) positive. All 11 strains resistant to tetracycline presented the tet(K) gene. When present, aminoglycoside resistance was mainly mediated by aac(6′)-aph(2″) or ant(4′) alone (two and three isolates, respectively) or in combination (five isolates). Two isolates carried aph(3′).
Molecular epidemiology. (i) PFGE.
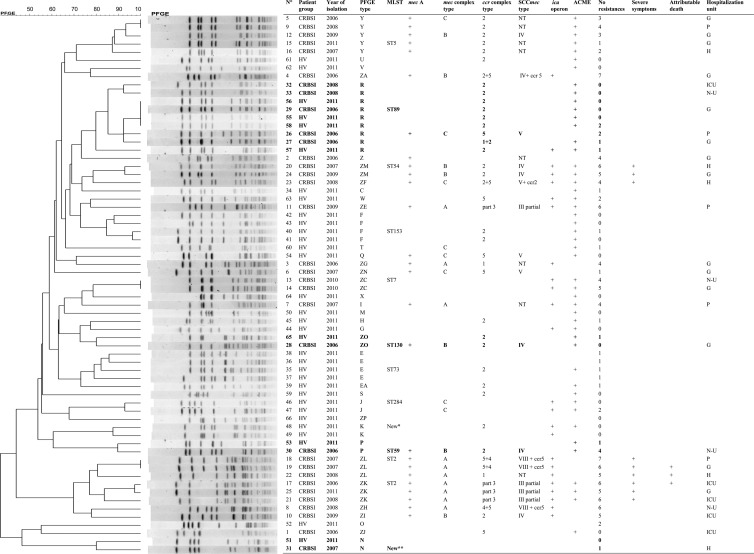
The 66 isolates of S. epidermidis produced a broad range of restriction patterns, divided into 37 PFGE types (Fig. 1). Twenty-four PFGE types were represented by a single isolate. The group of CRBSI isolates showed as much genetic diversity (19 PFGE types) as the group of commensal isolates (22 PFGE types). Four PFGE types were shared by clinical and commensal isolates (mixed PFGE types): PFGE type R (4 commensal and 5 clinical isolates) and PFGE types N, P, and ZO (two isolates each). It is interesting to note that among the eight clinical isolates involved in these mixed PFGE types, only three were carriers of the mec(A) gene, and their average resistance to antibiotics other than β-lactams was 1.
Fig 1.
Dendrogram of PFGE profiles of 66 S. epidermidis isolates causing CRBSI (n = 33) or collected from the skin of healthy volunteers (n = 33). For each isolate represented, the year of isolation and the presence of the mecA gene, the ica operon, and the ACME are listed, as well as the number of cases of resistance observed, the MLST ST (when tested), and the SCCmec type (mecA-positive isolates). Additionally, for isolates causing CRBSI, severe clinical presentation, hospitalization unit, and attributable death are indicated. PFGE types shared by CRBSI and commensal isolates are represented in boldface. Abbreviations: HV, healthy volunteer; G, gastrointestinal surgery units; H, hematology unit; ICU, intensive care units; N-U, nephrology-urology units; P, pulmonary oncology. *, close to ST86; **, close to ST19; °, number of cases of resistance observed among the 12 non-β-lactam antimicrobials tested (Table 1).
(ii) MLST sequencing.
Thirteen isolates were typed by MLST (one randomly selected per PFGE type composed of more than one isolate). In total, 12 different MLST types were identified. PFGE type R (no. of isolates [n] = 9) was found to belong to ST89, PFGE type E (n = 4) to ST73, PFGE type F (n = 4) to ST153, PFGE type Y (n = 5) to ST5, and PFGE types ZL (n = 3) and ZK (n = 3) to ST2 (Fig. 1).
The other identified MLST types were ST54 (n = 2), ST7 (n = 2), ST130 (n = 2), ST59 (n = 2), ST284 (n = 2), and two new STs, single-locus variants of ST86 (n = 2) and ST19 (n = 2), respectively.
Using the default group definition (minimum number of identical loci for group definition of 6), the eBURST algorithm classified ST2, ST5, ST7, ST54, and ST130 into the same clonal complex (CC7). Remarkably, this CC was thus composed almost exclusively of methicillin-resistant CRBSI isolates. Two PFGE ZC-ST7 isolates were methicillin-sensitive S. epidermidis (MSSE), and one MSSE PFGE type ZO-ST130 isolate was commensal.
However, when the eBURST analysis was rerun with a less stringent group definition (minimum number of identical loci for group definition of 5), all but three STs (ST73 and the two new STs) clustered into a single CC, corresponding to the CC2 described by Miragaia et al. (8).
(iii) SCCmec.
Among the 26 mec(A)-positive isolates (of which 25 were from CRBSI), eight different combinations of ccr and mec complexes were found. The most frequent type was type IV (n = 7). Type III with a partial deletion and type V were each carried by four isolates. Type VIII flanked by an additional ccr5 was carried by three isolates. Finally, SCCmec types of eight isolates were classified as nontypeable (NT). In most cases, this was due to an “unclassified” combination of ccr and mec complexes types (1A and 2C, for example) or was due to the absence of amplification for one of the two complexes. The community MRSE isolate had SCCmec type V.
Although a few clinical epidemic MRSE PFGE types were associated with a single SCCmec type (ZK with the partial SCCmec III and ZM with SCCmec IV), others (PFGE types Y and ZL) were associated with up to 3 different combinations of ccr and mec complexes.
Surprisingly, among the 40 mec(A)-negative isolates, up to 22 isolates (5 from CRBSI, 17 from volunteers) were positive for either a ccr gene complex (ccr type 2; 16 isolates) or a mec gene complex (class C mec; 3 isolates).
Virulence-associated genes.
We observed ica-positive isolates in both volunteers and patients with a ratio of 21% (7/33) versus 52% (17/33) in favor of the clinical isolates (P < 0.05). Most strains (76%) were arc(A) positive and were distributed evenly between the two studied groups (26/33 commensal isolates and 24/33 CRBSI isolates; P = 0.566).
Clinical data.
The clinical data for the 33 patients with CRBSI due to S. epidermidis are shown in Table 2. There were 10 women and 23 men, with a mean age of 57 ± 13 years.
Table 2.
Patient and isolate characteristics of 33 CRBSI episodes according to the clinical presentation and the genotype of the isolate
| Parameter | Result bya: |
|||||
|---|---|---|---|---|---|---|
| Clinical presentation |
PFGE type |
|||||
| Severe (n = 9) | Not severe (n = 24) | P valueb | Mixed (n = 8) | Other (n = 25) | P valueb | |
| Patients | ||||||
| Age, mean yr ± SD | 52 ± 15 | 59 ± 12 | 0.218 | 61 ± 12 | 56 ± 14 | 0.378 |
| No. male/no. female | 7/2 | 16/8 | 0.686 | 6/2 | 17/8 | 1.000 |
| No. (%) with underlying major comorbidity | 9 (100) | 20 (83) | 0.555 | 8 (100) | 21 (84) | 0.550 |
| No. (%) with previous hospitalization | 6 (67) | 5 (21) | 0.033 | 2 (25) | 9 (36) | 0.687 |
| No. (%) with previous antibiotic treatment | 9 (100) | 6 (25) | <0.001 | 1 (13) | 14 (56) | 0.046 |
| Length of hospital stay before CRBSI, geometric mean days (−1 SD; +1 SD) | 26 (14; 50) | 13 (7; 22) | 0.003* | 12 (8; 18) | 16 (8; 34) | 0.182* |
| Time between catheter insertion and removal, geometric mean days (−1 SD; +1 SD) | 14 (7; 27) | 9 (6; 15) | 0.052* | 8 (7; 10) | 11 (6; 20) | 0.037* |
| No. (%) with parenteral nutrition | 2 (22) | 16 (66) | 0.047 | 6 (75) | 12 (48) | 0.242 |
| No. (%) with attributable death | 3 (33) | 0 (0) | 0.015 | 0 (0) | 3 (12) | 0.560 |
| Isolates | ||||||
| Mean sum of cases of resistance ±SD | 6 ± 1 | 3 ± 2 | <0.001 | 1 ± 1 | 4 ± 2 | <0.001 |
| No. (%) with ica operon | 9 (100) | 8 (33) | <0.001 | 0 (0) | 17 (68) | <0.001 |
| No. (%) in mixed-PFGE group | 0 (0) | 8 (33) | 0.073 | NAb | NA | NA |
| No. (%) with ACME | 7 (77) | 17 (71) | 1.000 | 6 (75) | 18 (7) | 1.000 |
*, use of log10 to normalize the variable. NA, not applicable.
Statistically significant values (P < 0.05) are highlighted in boldface.
The most frequent comorbidity was an oncologic tumor, which was present in 18 patients; 4 patients presented with a hematologic tumor. Fifty-five percent (18/33) of patients received parenteral nutrition through the incriminated central venous catheter.
Within the subgroup of the nine patients who presented severe clinical symptoms, 67% had been hospitalized during the previous 3 months, compared to 21% for the other patients (P < 0.05) and all had received antibiotic therapy during the month prior to the CRBSI episode compared to 25% of the other patients (P < 0.05). Patients with severe clinical symptoms presented a much longer mean delay between admission and CRBSI than the other patients (P < 0.05). Among this subgroup, we observed also three deaths attributable to CRBSI and associated with severe clinical profiles: one liver cirrhosis patient, one renal transplant patient, and one neutropenic patient (acute myeloid leukemia). Furthermore, eight isolates implicated were multidrug resistant (MDR) and belonged to two closely related STs (single-locus variants of each other): ST2 and ST54.
Finally, when comparing the groups of CRBSIs due to an S. epidermidis isolate belonging to a mixed PFGE type to the group of CRBSIs due to an S. epidermidis isolate belonging to a PFGE type exclusively composed of CRBSI isolates (“CRBSI-specific” PFGE type), several significant discrepancies were observed: all isolates from the group belonging to a mixed PFGE type were ica negative and were resistant to an average of one antibiotic compared to four for the other group (P < 0.05). Twenty-five percent of them had previously been hospitalized, compared with 36% for patients who presented with CRBSI due to an S. epidermidis strain belonging to a CRBSI-specific PFGE type. The average duration on the catheter at the time of removal was 8 days for S. epidermidis isolates with mixed PFGE types versus 11 days for the other group (P < 0.05). No patient in the mixed-PFGE group developed severe clinical symptoms.
DISCUSSION
In this study, we compared 33 S. epidermidis isolates responsible for CRBSIs in hospitalized patients to 33 commensal S. epidermidis isolates from healthy volunteers in terms of resistance to antibiotics, genetic diversity, and the presence of virulence-associated gene loci; we also compared these microbiological characteristics to the clinical data from patients with CRBSIs. Our study has several limitations: most importantly, the sample size was small (n = 66). Additionally, the sampling of healthy volunteers (2011) was offset in time compared with that for patients with CRBSIs (2006 to 2011), which renders the comparison somewhat artificial. Furthermore, only one S. epidermidis strain per patient and per volunteer was studied: given the frequent polyclonality (26) of S. epidermidis infections and colonization, important information was possibly missed.
Using PFGE, a typing method known to provide reliable information on the short-term epidemiology, the two groups of isolates observed here (commensal versus CRBSI isolates) were very diverse and genetically distinct, as only 23% of PFGE types were common to both populations. This large genetic variability is in accordance with previous studies conducted on the genetic diversity of S. epidermidis in the hospital environment and/or in the community (27–29). However, these two distinct populations were found to be phylogenetically related, as MLST (a typing method designed to study long-term global epidemiology and population structure) revealed that all but three STs (ST73 and two new STs, close to ST19 and ST86, respectively) clustered into a unique CC that corresponds to the major CC, CC7 (previously CC2), described by Miragaia and coworkers (8, 29).
CRBSI S. epidermidis isolates were more resistant to antibiotics than commensal strains obtained from the skin of healthy volunteers, which is in concordance with other studies (29). Oxacillin and other β-lactams were the most affected antibiotics, followed by erythromycin, ciprofloxacin, and trimethoprim-sulfamethoxazole. These differences are probably the result of an adaptation to the significant pressure caused by the intensive use of antibiotics in hospitals, as has been particularly demonstrated by others for β-lactams and quinolones (30). Li et al. (11), for example, observed 96% methicillin resistance among nosocomial S. epidermidis isolates in China.
It has been reported that the majority of nosocomial S. epidermidis strains contain the ica operon in their genome (4). In our study, the ica operon was far more prevalent in strains from clinical settings than in commensal strains from the community, and all isolates responsible for clinically severe CRBSIs were ica positive, confirming that the presence of the ica operon plays an important role in the pathogenesis of CRBSI (biofilm formation). In contrast, the ACME islet, which confers increased virulence and pathogenicity to the USA300 methicillin-resistant Staphylococcus aureus (MRSA) clone (2, 9), was harbored by most isolates in this study (76%). This percentage, which is higher than the 50 to 70% reported in other studies (31, 32), was found in both commensal and CRBSI S. epidermidis isolates, which is in accordance with current opinion that in S. epidermidis, ACME actually plays a more important role in colonization than in virulence (5).
CRBSI S. epidermidis isolates either belonged to CRBSI-specific PFGE types or shared their PFGE types with commensal isolates (mixed PFGE types). Several significant discrepancies were observed between these two groups of isolates, suggesting that these CRBSI-specific S. epidermidis lineages are more adapted to the hospital environment than the others: they were resistant to more antibiotics and were more often ica positive than the isolates belonging to the mixed PFGE group. The patients' clinical histories were also different in the two groups, as patients from the CRBSI-specific PFGE group were more often exposed to antibiotics within the month preceding the CRBSI episode, more often had a severe clinical presentation, and developed their CRBSI episode on a catheter 3 days “older” on average. ST2 isolates accounted for nearly 20% of all CRBSIs isolates and were thus not as frequently recovered as in other studies conducted in the United States, Australia, or Germany, where the majority of S. epidermidis isolates responsible for nosocomial infections belonged to ST2 (9, 10, 33). However, all of the ST2 isolates belonged to the CRBSI-specific PFGE group. Furthermore, all were implicated in severe CRBSI episodes (attributable death, persistent bacteremia, and sepsis), as were isolates belonging to the closely related (single-locus variant) ST54. Additionally, these ST2 and ST54 isolates were all multidrug resistant and ica positive. All of these findings strongly suggest that certain S. epidermidis lineages, by combining virulence and resistance, are well adapted to the hospital environment and are able to cause more severe diseases. These ST2 and ST54 CRBSI-specific PFGE types were observed causing CRBSI in patients otherwise epidemiologically unrelated (different units, different years), suggesting the persistence of these clones over time in our institution, as observed by others (34). Identification more precisely of the reservoir and the routes of colonization of venous catheters by these S. epidermidis clones would be useful to develop efficient prevention strategies, as several methods are possible and are commonly described: via the flora of the patient, who would have been progressively colonized with these hospital-adapted S. epidermidis isolates (autoinfection) or via the hands of colonized health care workers (cross-infection) (35–37). Some authors even evoke the role of ambient air in nosocomial MRSE transmission (38).
Regarding SCCmec type distribution, diversity was also high among our CRBSI MRSE isolates. Up to eight different combinations of ccr and mec complex types were found, two of which were “unclassified” (1A and 2C). SCCmec type IV was the most abundant (36%), as reported in the literature (1, 14). Note that isolates belonging to the same PFGE type could carry up to 3 different combinations of ccr and mec complexes, and several isolates were harboring SCCmec with multiple ccr complexes. Additionally, half of the MSSE isolates seemed to carry remnants of SCCmec. All of these factors indicate that the loss and acquisition of mobile genetic elements in S. epidermidis are probably extremely frequent, even in the community setting (8).
In conclusion, S. epidermidis isolates that cause CRBSIs in hospitals are much more resistant to antibiotics and are more frequently carriers of the ica operon than commensal S. epidermidis isolates found in the community. Although PFGE typing indicates that the two populations are very diverse and genetically distinct, the majority of isolates seems to belong to the same genetic lineage. Two closely related MLST types (ST2 and ST54) were associated with ica-positive, MRSE isolates responsible for clinically severe CRBSIs, suggesting the possibility of an association between the genetic background of the isolate causing the CRBSI and the severity of the disease.
ACKNOWLEDGMENTS
We thank Sylvie Arias for help with statistical analysis of the data and for support. We thank Chaïma El Batik and Madjid Taguemount for skilled technical assistance.
This project was conducted with support no. 2012-820690-005 from the Iris Research Fund (Fonds Iris Recherche) awarded through the King Baudouin Foundation.
Footnotes
Published ahead of print 13 March 2013
REFERENCES
- 1. Otto M. 2009. Staphylococcus epidermidis—the ‘accidental’ pathogen. Nat. Rev. Microbiol. 7:555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Widerstrom M, Wistrom J, Sjostedt A, Monsen T. 2012. Coagulase-negative staphylococci: update on the molecular epidemiology and clinical presentation, with a focus on Staphylococcus epidermidis and Staphylococcus saprophyticus. Eur. J. Clin. Microbiol. Infect. Dis. 31:7–20 [DOI] [PubMed] [Google Scholar]
- 3. Vadyvaloo V, Otto M. 2005. Molecular genetics of Staphylococcus epidermidis biofilms on indwelling medical devices. Int. J. Artif. Organs 28:1069–1078 [DOI] [PubMed] [Google Scholar]
- 4. Kozitskaya S, Cho SH, Dietrich K, Marre R, Naber K, Ziebuhr W. 2004. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect. Immun. 72:1210–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Diep B, Stone G, Bauino L, Graber C, Miller A, des Etages S, Jones AA, Pallazzolo-Balance A, Perdreau-Remington F, Sensabaugh G, DeLeo F, Chambers H. 2008. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 197:1523–1530 [DOI] [PubMed] [Google Scholar]
- 6. Milisavljevic V, Wu F, Cimmoti J, Haas J, Della-Latta P, Larson E, Saiman L. 2005. Genetic relatedness of Staphylococcus epidermidis from infected infants and staff in the neonatal intensive care unit. Am. J. Infect. Control 33:341–347 [DOI] [PubMed] [Google Scholar]
- 7. Liakopoulos V, Petinaki E, Efthimiadi G, Klapsa D, Giannapoulo M, Dovas S, Eleftheriadis T, Mertens PR, Stefanidis I. 2008. Clonal relatedness of methicillin-resistant coagulase-negative staphylococci in the haemodialysis unit of a single university centre in Greece. Nephrol. Dial. Transplant. 23:2599–2603 [DOI] [PubMed] [Google Scholar]
- 8. Miragaia M, Thomas JC, Couto I, Enright MC, de Lencastre H. 2007. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J. Bacteriol. 189:2540–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gordon RJ, Miragaia M, Weinberg AD, Lee CJ, Rolo J, Giacalone JC, Slaughter MS, Pappas P, Naka Y, Tector AJ, de Lencastre H, Lowy HF. 2012. Staphylococcus epidermidis colonization is highly clonal across US cardiac centers. J. Infect. Dis. 205:1391–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Widerstrom M, McCullough CA, Coombs GW, Monsen T, Christiansen KJ. 2012. A multidrug-resistant Staphylococcus epidermidis clone (ST2) is an ongoing cause of hospital-acquired infection in a Western Australian hospital. J. Clin. Microbiol. 50:2147–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li M, Wang X, Gao Q, Lu Y. 2009. Molecular characterization of Staphylococcus epidermidis strains isolated from a teaching hospital in Shangai, China. J. Med. Microbiol. 58:456–461 [DOI] [PubMed] [Google Scholar]
- 12. Garza-Gonzalez E, Lopez D, Pezina C, Muruet W, Bocanegra-Garcia V, Munoz I, Ramirez C, Llaca-Diaz JM. 2010. Diversity of staphylococcal cassette chromosome mec structures in coagulase-negative staphylococci and relationship to drug resistance. J. Med. Microbiol. 59:323–329 [DOI] [PubMed] [Google Scholar]
- 13. National Nosocomial Infections Surveillance System 2004. National Nosocomial Infections Surveillance (NNIS) System report, data summary from January 1992 through June 2004. Am. J. Infect. Control 32:470–485 [DOI] [PubMed] [Google Scholar]
- 14. American College of Chest Physicians/Society of Critical Care Medicine 1992. American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference: definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Crit. Care Med. 20:864–874 [PubMed] [Google Scholar]
- 15. Clinical Laboratory Standards Institute 2009. Performance standards for antimicrobial susceptibility testing: 19th informational supplement. M100S19. CLSI, Wayne, PA [Google Scholar]
- 16. Ünal S, Hoskins J, Flokowitsch JE, Wu CYE, Preston DA, Skatrud PL. 1992. Detection of methicillin-resistant staphylococci by using the polymerase chain reaction. J. Clin. Microbiol. 30:1685–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maes N, Magdalena J, Rottiers S, De Gheldre Y, Struelens MJ. 2002. Evaluation of a triplex PCR assay to discriminate Staphylococcus aureus from coagulase-negative staphylococci and determine methicillin resistance from blood cultures. J. Clin. Microbiol. 40:1514–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vanhoof R, Godard C, Content J, Nyssen HJ, Hannecart-Pokorni E. 1994. Detection by polymerase chain reaction of genes encoding aminoglycoside-modifying enzymes in methicillin-resistant Staphylococcus aureus isolates of epidemic phage types. Belgian Study Group of Hospital Infections (GDEPIH/GOSPIZ). J. Med. Microbiol. 41:282–290 [DOI] [PubMed] [Google Scholar]
- 19. Ng LK, Martin I, Alfa M, Mulvey M. 2001. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 15:209–215 [DOI] [PubMed] [Google Scholar]
- 20. Lina G, Quaglia A, Reverdy ME, Leclercq R, Vandenesch F, Etienne J. 1999. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob. Agents Chemother. 43:1062–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kozitskaya S, Olson ME, Fey PD, Witte W, Ohlsen K, Ziebuhr W. 2005. Clonal analysis of Staphylococcus epidermidis isolates carrying or lacking biofilm-mediating genes by multilocus sequence typing. J. Clin. Microbiol. 43:4751–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deplano A, Witte W, van Leeuwen WJ, Brun Y, Struelens MJ. 2000. Clonal dissemination of epidemic methicillin-resistant Staphylococcus aureus in Belgium and neighboring countries. Clin. Microbiol. Infect. 6:239–245 [DOI] [PubMed] [Google Scholar]
- 23. Miragaia M, Carriço JA, Thomas JC, Couto I, Enright MC, de Lencastre H. 2008. Comparison of molecular typing methods for characterization of Staphylococcus epidermidis: proposal for clone definition. J. Clin. Microbiol. 46:118–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thomas JC, Vargas MR, Miragaia M, Peacock SJ, Archer GL, Enright MC. 2007. Improved multilocus sequence typing scheme for Staphylococcus epidermidis. J. Clin. Microbiol. 45:616–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Hiramatsu K. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Van Eldere J, Peetermans WE, Struelens M, Deplano A, Bobbaers H. 2000. Polyclonal staphylococcal endocarditis caused by genetic variability. Clin. Infect. Dis. 31:24–30 [DOI] [PubMed] [Google Scholar]
- 27. Nunes AP, Teixeira LM, Bastos CC, Silva MG, Ferreira RB, Fonseca LS, Santos KR. 2005. Genomic characterization of oxacillin-resistant Staphylococcus epidermidis and Staphylococcus haemolyticus isolated from Brazilian medical centres. J. Hosp. Infect. 59:19–26 [DOI] [PubMed] [Google Scholar]
- 28. Bogado J, Limansky A, Sutich E, Marchiaro P, Marzi M, Putero J, Viale A. 2002. Molecular chracterization of methicillin-resistant coagulase-negative staphylococci from a neonatal intensive care unit. Infect. Control. Hosp. Epidemiol. 23:447–451 [DOI] [PubMed] [Google Scholar]
- 29. Rolo J, de Lencastre H, Miragaia M. 2012. Strategies of adaptation of Staphylococcus epidermidis to hospital and community: amplification and diversification of SCCmec. J. Antimicrob. Chemother. 67:1333–1341 [DOI] [PubMed] [Google Scholar]
- 30. Raad I, Alrahwan A, Rolston K. 1998. Staphylococcus epidermidis: emerging resistance and need for alternative agents. Clin. Infect. Dis. 26:1182–1187 [DOI] [PubMed] [Google Scholar]
- 31. Barbier F, Lebeaux D, Hernandez D, Delannoy AS, Caro V, Francois P, Schrenzel J, Ruppe E, Gaillard K, Wolff M, Brisse S, Andremont A, Ruimy R. 2011. High prevalence of the arginine catabolic mobile element in carriage isolates of methicillin-resistant Staphylococcus epidermidis. J. Antimicrob. Chemother. 66:29–36 [DOI] [PubMed] [Google Scholar]
- 32. Miragaia M, de Lencastre H, Perdreau-Remington F, Chambers HF, Higashi J, Sullam PM, Lin J, Wong KI, King KA, Otto M, Sensabaugh GF, Diep BA. 2009. Genetic diversity of arginine catabolic mobile element in Staphylococcus epidermidis. PLoS One 4:e7722 doi:10.1371/journal.pone.0007722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Widerstrom M, Monsen T, Karlsson C, Edebro H, Johansson A, Wistrom J. 2009. Clonality among multidrug-resistant hospital-associated Staphylococcus epidermidis in northern Europe. Scand. J. Infect. Dis. 41:642–649 [DOI] [PubMed] [Google Scholar]
- 34. Muldrew KL, Tang Li Y-WH, Stratton CW. 2008. Clonal dissemination of Staphylococcus epidermidis in an oncology ward. J. Clin. Microbiol. 46:3391–3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Livesley MA, Tebbs SE, Moss HA, Faroqui MH, Lambert PA, Elliott TSJ. 1998. Use of pulsed field gel electrophoresis to determine the source of microbial contamination of central venous catheters. Eur. J. Clin. Microbiol. Infect. Dis. 17:108–112 [DOI] [PubMed] [Google Scholar]
- 36. Müller-Premru M, Cernelc P. 2004. Molecular epidemiology of catheter-related bloodstream infections caused by coagulase-negative staphylococci in haematological patients with neutropenia. Epidemiol. Infect. 132:921–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cimiotti JP, Wu F, Della-Latta P, Nesin M, Larson E. 2004. Emergence of resistant staphylococci on the hands of new graduate nurses. Infect. Control. Hosp. Epidemiol. 25:431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Botelho AM, Nunes Z, Asensi MD, Gomes MZ, Fracalanzza SE, Figueiredo AM. 2012. Characterization of coagulase-negative staphylococci isolated from hospital indoor air and a comparative analysis between airborne and inpatient isolates of Staphylococcus epidermidis. J. Med. Microbiol. 61:1136–1145 [DOI] [PubMed] [Google Scholar]