Abstract
Staphylococcus pseudintermedius is a veterinary pathogen that has seldom been described as an agent of human disease. Features of this probably underreported coagulase-positive Staphylococcus species are depicted here through the description of a graft-versus-host disease-related wound infection caused by a multidrug-resistant strain.
CASE REPORT
A 65-year-old male patient who had received (3 years previously) an allogeneic bone marrow transplant (BMT) for chronic lymphoblastic leukemia was admitted to the Pescara Civic Hospital (Italy) because of a wound infection, located in the periumbilical region and showing two different purulent discharges. The lesion was due to chronic graft-versus-host disease (GvHD) that complicated the BMT. The patient affirmed that he lived in proximity to a pet dog and farm cows.
Pus staining revealed Gram-positive cocci within leukocytes, while cultures yielded a massive growth of mannitol-nonfermenting coagulase-positive staphylococci (CPS). Based on colony aspect (brightness of the white-gray color), two different isolates (named S32 and S33) were recognized, sharing a double-zone hemolysis on the sheep blood agar plate (Fig. 1). They were identified as Staphylococcus intermedius by both the Vitek2 and the API system (bioMérieux, Marcy l'Etoile, France). However, S. intermedius is the only species belonging to the so-called Staphylococcus intermedius group included in these systems' databases. The S. intermedius group includes S. intermedius, cultured from a wide variety of animals, Staphylococcus pseudintermedius, which recent studies indicate is the prevalent S. intermedius group species harbored by dogs and cats, and Staphylococcus delphini, first isolated from dolphins but later collected from several terrestrial animals (1, 2). Thus, we did not consider this identification as conclusive. Moreover, since the absence of mannitol fermentation, along with the double-zone hemolysis, was highly suggestive for S. intermedius/S. pseudintermedius, the patient was screened for S. intermedius group nasal and skin colonization. Five phenotypically similar organisms were grown from the nasal and the hand skin swabs. Again, they were identified as S. intermedius by the Vitek2 and the API system.
Fig 1.
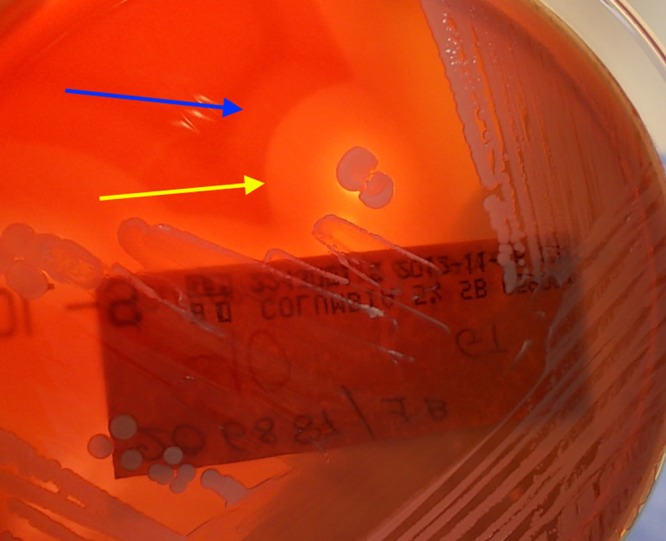
Double-zone hemolysis produced by strain S32. The yellow arrow indicates the inner area (first zone), a completely hemolytic band (β-hemolysis), while the blue arrow indicates the external area (second zone), an incompletely hemolytic band (α-hemolysis).
Different tests (both phenotypic and genotypic) were performed to confirm the identification. Phenotypically, all the strains were colistin resistant and showed a slow positivity to the Voges-Proskauer reaction (suggesting that the isolates were S. pseudintermedius rather than S. intermedius). Among genotypic assays, 16S rRNA sequencing did not provide a definitive discrimination among S. intermedius, S. pseudintermedius, and S. delphini. The isolates were then analyzed through automated ribotyping (RiboPrinter; Qualicon DuPont, USA). The instrument provided identical fingerprints for all of the strains and identified them as S. intermedius. A specific multiplex PCR was performed, as previously described (3). The isolates were identified as S. pseudintermedius, since a clear band was obtained at 926 bp (Fig. 2). This finding was finally confirmed by analyzing the tuf (4) and rpoB (5) genes, the latter allowing better discrimination than the tuf analysis alone. The isolates showed a high degree of similarity with the fully sequenced genome of Staphylococcus pseudintermedius ED99 (GenBank accession number CP002478.1); the rpoB sequence we obtained was submitted to GenBank.
Fig 2.
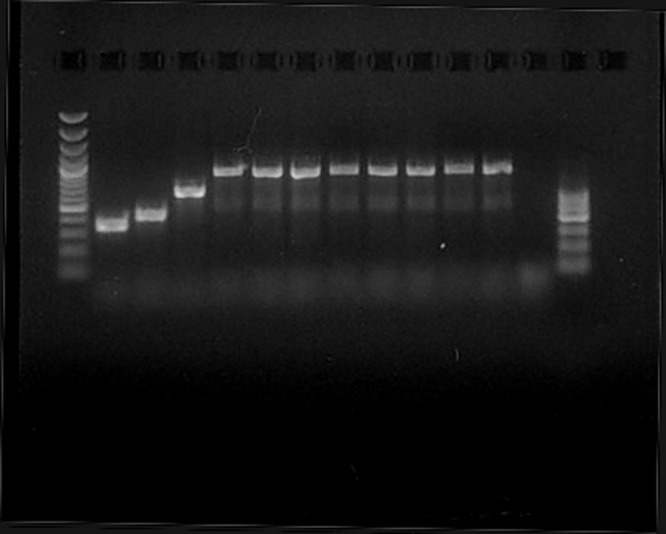
Multiplex PCR was performed according to the protocol in reference 3. Lanes: 1st, 100- to 3,000-bp molecular marker; 2nd, 359-bp product from a Staphylococcus aureus clinical isolate; 3rd, 430-bp product from Staphylococcus intermedius DSM 20373T (ATCC 29663); 4th, 661-bp product from Staphylococcus delphini DSM 20771T (ATCC 49171); 5th, 926-bp product from a Staphylococcus pseudintermedius isolated from veterinary sources; 6th to 12th, amplification products from the seven different isolates from the patient; 13th, negative control; 14th, 100- to 600-bp molecular marker.
The antimicrobial susceptibility profiles, obtained through the Vitek2 system, were then confirmed using both a disc diffusion method and MIC test strips (Liofilchem, Roseto degli Abruzzi, Italy). According to the EUCAST (version 2.0) interpretive breakpoints for Staphylococcus aureus, all of the isolates were resistant to benzylpenicillin, oxacillin, fluoroquinolones, macrolides, clindamycin, and cotrimoxazole and susceptible to tetracycline, chloramphenicol, rifampin, tigecycline, daptomycin, glycopeptides, and linezolid. Again, S32 and S33 showed gentamicin susceptibility, whereas all the other isolates displayed resistance to the aminoglycoside.
As cefoxitin sensitivity was observed in spite of the oxacillin resistance, the mecA gene was searched with an in-house method using the mecA primers and the conditions described in the paper of Oliveira and de Lencastre (6). We confirmed the results using the Gene X-pert system (Cepheid, Sunnyvale, CA, USA). Both techniques showed that the genetic element was harbored by all isolates.
To evaluate the genome relatedness between wound strains and those from nose and skin, random amplification of polymorphic DNA (RAPD) was performed, using a previously described protocol (7). Four different arbitrary primers were used, along with a control strain of veterinary origin. All the strains gave the same fingerprint with the different primers. Although it was clear then that we had a single S. pseudintermedius clone (confirmed by RAPD analysis and ribotyping), both isolate S32 and isolate S33 were deposited to the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen, GmbH) collection, Germany, under accession numbers DSM 25713 and DSM 25714, respectively.
A 10-day course of topical gentamicin (twice a day) led to sterilization of the ulcer; afterward, complete lesion healing was obtained by employing platelet-rich plasma gel (3 applications at 5-day intervals).
(A preliminary version of this clinical case was presented at the Gram Award [9 to 10 February 2012, Rome, Italy], a scientific event promoted by Novartis Farma S.p.A. The presentation was sponsored by Novartis Farma S.p.A. on behalf of IntraMed Communications S.r.L.)
The CPS of medical interest are mainly represented by S. aureus, which is highly prevalent in humans although increasingly isolated from different animals species (most of all, pets), and by the microorganisms belonging to the S. intermedius group.
Recently, CPS other than S. aureus have gained importance in veterinary medicine, as molecular techniques have allowed a better understanding of them, leading to a deep revision of their taxonomy and pathogenicity. In particular, the spread of multidrug-resistant (MDR) strains, mostly within the S. pseudintermedius species, has been reported more and more frequently, and the role of the mutual transmission of bacteria and/or virulence determinants between animals and their owners has been emphasized (2, 8, 9). In animals, S. intermedius group organisms are frequently isolated as colonizers, although they may cause diseases. In humans, it is not known whether S. delphini shows a zoonotic potential; conversely, S. intermedius has been related to bacteremia, pneumonia, brain abscesses, and wound infections (mostly of canine-inflicted wounds). S. pseudintermedius has been recognized on a few occasions as a pathogen (a case of rhinosinusitis, a catheter-related bacteremia, and an implantable cardioverter-defibrillator infection were previously published) (10–12).
To our knowledge, we are reporting the second methicillin-resistant S. pseudintermedius (MRSP) infection in a human host (10), although it is clear from the published literature that current knowledge of the S. intermedius group is fragmentary, requiring further revisions. In particular, it may be hypothesized that a large number of strains previously identified as S. intermedius may be S. pseudintermedius, and a wide majority of reported S. intermedius human infections might actually be due to S. pseudintermedius (1, 2).
Based on current knowledge, however, this case expands the spectrum of S. pseudintermedius diseases to wound infections and emphasizes the risk of zoonoses in compromised subjects; in particular, the underlying GvHD-related immunosuppression and skin damage reasonably played a pivotal role in the development of the disease.
From a laboratory point of view, clinical microbiologists should not hastily label and dismiss mannitol-nonfermenting microorganisms as coagulase-negative staphylococci but carefully perform a coagulase test if the hemolysis on blood agar plates is suggestive for an S. intermedius group organism. Typically, a complete inner band (β-hemolysis) is observed, while the outer one is incomplete (α-hemolysis), although it becomes complete at 4°C (hot-cold hemolysis). Among the mannitol-nonfermenting staphylococci, this double zone has been known to be pathognomonic of S. intermedius and S. pseudintermedius (Fig. 1) (13).
Although some phenotypic features have been employed in the past decades to discriminate within the S. intermedius group, these do not actually appear to be conclusive (2, 14). Also, in the context of the newest approaches to microbial identification, matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) libraries have recently been shown not to be adequate, as yet, to reliably characterize S. intermedius group strains (15). Therefore, molecular methodologies are mandatory to provide a correct species identification.
Similarly, we did not obtain a conclusive characterization either by 16S rRNA sequencing or through the automated ribotyping; it is known in fact that the first method cannot always distinguish among such closely related species (those belonging to the S. intermedius group), while S. pseudintermedius profiles are not included in the RiboPrinter reference databases. Ribotyping data for these organisms should be obtained, along with MALDI-TOF MS data.
Like other authors (15), we finally achieved a good identification through the multiplex PCR proposed by Sasaki et al. (3) and the rpoB gene analysis (5).
Concerning the antibiotic susceptibility testing, it has to be emphasized that the isolates were susceptible to cefoxitin but resistant to oxacillin; this result can be misleading, as cefoxitin screening is widely used in clinical laboratories as a marker for methicillin resistance in S. aureus. The mecA gene was detected through molecular methods, and this finding was not surprising, since the spread of MRSP has been previously and widely reported, although in the veterinary context (2, 8). In our opinion, it is critical that methicillin/oxacillin resistance in CPS (other than S. aureus) should as a routine practice be screened through different methods, including molecular tools, pending specific criteria for in vitro testing and interpretation of the activities of drugs against S. intermedius group members.
It is likely that human and veterinary S. pseudintermedius isolates have been misidentified as S. aureus, S. intermedius, and S. delphini in the past (1, 2). Nevertheless, when a patients' history includes contact with pets (mostly dogs or cats), the potential role of S. pseudintermedius (rather than S. intermedius) as the agent of zoonoses has to be taken into account, and a correct identification may be performed only by using molecular techniques.
The real prevalence and incidence of S. pseudintermedius (as a colonizer or pathogen) in the human population is surely underestimated, and more accurate phenotype- and genome-based investigations are warranted to further highlight the behavior of this organism as an emerging human pathogen in the future.
Nucleotide sequence accession number.
The rpoB sequence of strain S32 was deposited in GenBank and assigned accession number KC680229.
ACKNOWLEDGMENTS
The project was partially supported by grant no. N N401 017740 from the Polish Ministry of Science and Education (to J.M.) and by a grant from the Scientific Committee of the IRCCS Arcispedale Santa Maria Nuova, Reggio Emilia (to E.C.).
We thank Cecilia Passeri and Ornella Iuliani (Department of Transfusion Center, Spirito Santo Hospital, Pescara, Italy) and Giorgia Mancini (Department of Hematology, Ospedali Riuniti of Ancona, Ancona, Italy) for providing us with details about the patient's history and follow-up and for their interest in this case.
Footnotes
Published ahead of print 13 March 2013
REFERENCES
- 1. Sasaki T, Kikuchi K, Tanaka Y, Takahashi N, Kamata S, Hiramatsu K. 2007. Reclassification of phenotypically identified Staphylococcus intermedius strains. J. Clin. Microbiol. 45:2770–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bond R, Loeffler A. 2012. What's happened to Staphylococcus intermedius? Taxonomic revision and emergence of multi-drug resistance. J. Small Anim. Pract. 53:147–154 [DOI] [PubMed] [Google Scholar]
- 3. Sasaki T, Tsubakishita S, Tanaka Y, Sakusabe A, Ohtsuka M, Hirotaki S, Kawakami T, Fukata T, Hiramatsu K. 2010. Multiplex-PCR method for species identification of coagulase-positive staphylococci. J. Clin. Microbiol. 48:765–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Heikens E, Fleer A, Paauw A, Florijn A, Fluit AC. 2005. Comparison of genotypic and phenotypic methods for species-level identification of clinical isolates of coagulase-negative staphylococci. J. Clin. Microbiol. 43:2286–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Drancourt M, Raoult D. 2002. rpoB gene sequence-based identification of Staphylococcus species. J. Clin. Microbiol. 40:1333–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oliveira DC, de Lencastre H. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Damiani G, Telecco S, Comincini S, Sironi M, Carretto E, Marone P. 1996. Comparison of an improved RAPD fingerprinting with different typing methods for discriminating clinical isolates of Staphylococcus spp. Eur. J. Epidemiol. 12:163–169 [DOI] [PubMed] [Google Scholar]
- 8. van Duijkeren E, Catry B, Greko C, Moreno MA, Pomba MC, Pyorala S, Ruzauskas M, Sanders P, Threlfall EJ, Torren-Edo J, Torneke K, Scientific Advisory Group on Antimicrobials 2011. Review on methicillin-resistant Staphylococcus pseudintermedius. J. Antimicrob. Chemother. 66:2705–2714 [DOI] [PubMed] [Google Scholar]
- 9. Walther B, Hermes J, Cuny C, Wieler LH, Vincze S, Abou Elnaga Y, Stamm I, Kopp PA, Kohn B, Witte W, Jansen A, Conraths FJ, Semmler T, Eckmanns T, Lubke-Becker A. 2012. Sharing more than friendship—nasal colonization with coagulase-positive staphylococci (CPS) and co-habitation aspects of dogs and their owners. PLoS One 7:e35197 doi:10.1371/journal.pone.0035197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stegmann R, Burnens A, Maranta CA, Perreten V. 2010. Human infection associated with methicillin-resistant Staphylococcus pseudintermedius ST71. J. Antimicrob. Chemother. 65:2047–2048 [DOI] [PubMed] [Google Scholar]
- 11. Chuang CY, Yang YL, Hsueh PR, Lee PI. 2010. Catheter-related bacteremia caused by Staphylococcus pseudintermedius refractory to antibiotic-lock therapy in a hemophilic child with dog exposure. J. Clin. Microbiol. 48:1497–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Van Hoovels L, Vankeerberghen A, Boel A, Van Vaerenbergh K, De Beenhouwer H. 2006. First case of Staphylococcus pseudintermedius infection in a human. J. Clin. Microbiol. 44:4609–4612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Devriese LA, Vancanneyt M, Baele M, Vaneechoutte M, De Graef E, Snauwaert C, Cleenwerck I, Dawyndt P, Swings J, Decostere A, Haesebrouck F. 2005. Staphylococcus pseudintermedius sp. nov., a coagulase-positive species from animals. Int. J. Syst. Evol. Microbiol. 55:1569–1573 [DOI] [PubMed] [Google Scholar]
- 14. Savini V, Polilli E, Polakowska K, Marrollo R, Białecka A, Kasprowicz A, Fazii P, D'Antonio D, Carretto E, Międzobrodzki J. 2012. Arginine dehydrolase and beta-gentiobiose cannot discriminate within the Staphylococcus intermedius group. Vet. Microbiol. 161:236–237 [DOI] [PubMed] [Google Scholar]
- 15. Dmitrenko O, Balbutskaya A, Voronina O, Ankirskaya A, Lubasovskaya L, Voytenko A, Lammier C. 2012. Species identification of Staphylococcus intermedius group by multiplex-PCR and Gap gene amplification and sequencing compared to matrix-assisted laser desorption ionization time-of-fight mass spectrometry, poster 1518. ECCMID, London, United Kingdom, 31 March to 3 April 2012 [Google Scholar]


