Abstract
Tumor necrosis factor alpha (TNF-α)-induced NF-κB activation has been believed to depend on TRAF2- and cIAP1-mediated RIP1 ubiquitination. However, recent findings have challenged the notion that these proteins play essential roles in NF-κB activation. Here, by assessing the kinetics and amplitude of IκB kinase (IKK) activation, we report that TNF-α-induced immediate and robust activation of IKK requires K63-linked and linearly linked ubiquitination of RIP1 and that in the absence of RIP1 expression, TRAF2 and cIAP1 cooperatively induce delayed IKK activation by recruiting LUBAC to TNFR1. Knockdown of HOIP (a component of LUBAC) in RIP1-deficient cells completely impairs the recruitment and activation of IKK but does not affect K63-linked ubiquitination of TRAF2 and recruitment of TAK1 to TNFR1, suggesting that the K63-linked ubiquitin chain is not capable of recruiting IKK in vivo. We also demonstrate that TRAF2 and cIAP1 together, but not either one alone, directly catalyze linearly linked ubiquitination of RIP1. Importantly, in embryonic hepatocytes, TNF-α activates NF-κB through a RIP1-independent pathway. Thus, our findings clarify molecular details of this important signaling mechanism by providing evidence for the existence of two phases of IKK activation: the immediate phase, induced by TRAF2/cIAP1-mediated ubiquitination of RIP1, and the delayed phase, activated by TRAF2/cIAP1-dependent recruitment of LUBAC.
INTRODUCTION
Tumor necrosis factor alpha (TNF-α) is a pleiotropic cytokine that can exert opposing biological effects—proinflammatory and proapoptotic—by activating the NF-κB and caspase 8 pathways, respectively (1, 2). The cytokine acts through two distinct cell surface receptors, TNFR1 and TNFR2, and gene knockout studies have revealed that TNFR1 mediates both types of TNF-α activity (3, 4). Although numerous studies initially led to a good consensus on the molecular mechanisms underlying TNFR1-mediated NF-κB activation, a recent accumulation of apparently contradictory results has led to significant complication in those models (5–8).
In the field of TNFR1 study, the prevailing model for the signaling pathways whereby TNF-α leads to NF-κB activation has been as follows: activated TNFR1 recruits TNFR-associated death domain protein (TRADD), receptor-interacting protein 1 (RIP1), TNFR-associated factor 2 (TRAF2), and TRAF5; the RING domains of TRAF2 and TRAF5 then work in conjunction with the ubiquitin (Ub)-conjugating enzyme Ubc13/Uev1A to catalyze K63-linked polyubiquitination of RIP1 (RIP1-K63-pUb) at K377 (5, 9); the RIP1-K63-pUb chain then recruits the TAK1 (consisting of TAK1, TAB1, and TAB2 subunits) and IκB kinase (IKK) (consisting of IKKα, IKKβ, and IKKγ/NEMO subunits) complexes, binding directly to the Ub-binding domains present on TAB2 and NEMO; once recruited by RIP1-K63-pUb chains, TAK1 directly activates IKKβ through proximity-mediated phosphorylation (5). Recently, several studies suggested that TRAF2 lacks E3 ligase activity and that it recruits cIAP1/2 to promote RIP1 ubiquitination (10–13). On the other hand, Alvarez et al. reported more recently that TRAF2 does possess an E3 ligase function but that sphingosine-1-phosphate (S1P) binding directly to the TRAF2 RING domain is required for its activation (14). Nevertheless, TNF-α-induced NF-κB activation is believed to be impaired in RIP1 knockout (KO), TRAF2 and TRAF5 double-knockout (DKO), and cIAP1 and cIAP2 (cIAP1/2) double-knockdown and -knockout (10, 11, 15–19) cells.
We reported previously that TNF-α-induced activation of JNK, but not that of IKK, depends on the integrity of the TRAF2 RING domain and on Ubc13 expression (20). Notably, Yamamoto et al. reported that in B cells and mouse embryonic fibroblasts (MEFs), conditional ablation of Ubc13 could considerably impair JNK activation, without affecting IKK activation, in response to all tested stimuli except TNF-α (21). We reported recently that in both TRAF2 KO and TRAF2/5 DKO MEFs the classical IKK complex is constitutively activated to a certain level due to accumulation of NIK and that stimulating these cells with TNF-α further increases IKK activity, albeit weakly (8). Surprisingly, Xu et al. reported that IKK activation by TNF-α requires UbcH5 (rather than Ubc13), whose polyubiquitination of RIP1 is not restricted to the K63-pUb chain (7). On the other hand, Dynek et al. reported that cIAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 (RIP1-K11-pUb) and that RIP1-K11-pUb efficiently recruits NEMO to TNFR1 (22). More surprisingly, Wong et al. showed that TNF-α can activate NF-κB normally in both primary and immortalized RIP1 KO cells (6). Adding to these complexities, linear ubiquitination of NEMO by the HOIL-1L–HOIP complex (also known as LUBAC, for linear ubiquitin chain assembly complex) was also reported to stabilize the TNFR1 complex and allow TNF-α to efficiently activate NF-κB (23, 24). Collectively, these studies suggest that there might be redundant pathways that are capable of translating TNF-α signaling into NF-κB activation.
Western blot analysis of stimulation-induced IκBα degradation has been used as a standard method for assessing NF-κB activation. However, our extensive analyses of correlations among IKK activation, IκBα degradation, and NF-κB-dependent gene expression in TRAF2 KO and TRAF2/5 DKO cells revealed that the IKK immunokinase assay was a more sensitive means of assessing the strength and duration of signaling output (8, 25). In the current study, we used the latter approach to examine TNF-α-induced NF-κB activation in RIP1 KO and cIAP1/2-depleted cells. We found that TRAF2- and cIAP1-mediated RIP1 ubiquitination is essential for TNF-α-induced immediate IKK activation and that TRAF2- and cIAP1-dependent recruitment of LUBAC to TNFR1 mediates the delayed phase of IKK activation. Surprisingly, in HOIP knockdown RIP1 KO MEFs, TRAF2 and cIAP1 underwent increased autoubiquitination through K63 and K48 linkage following TNF-α stimulation, yet they were able to recruit TAK1, but not IKK, to TNFR1, suggesting that the K63-pUb chain is not capable of recruiting IKK in vivo. In addition, we demonstrate that TRAF2 and cIAP1 together, but not either one alone, directly conjugate RIP1 with linear-linkage Ub chains in vivo and in vitro, suggesting that linearly linked pUb chains recruit IKK in vivo. Thus, our findings shed new light on the mechanisms underlying TNF-α-induced NF-κB activation and also provide explanations for the conflicting conclusions that were drawn based on analysis of IκBα degradation.
MATERIALS AND METHODS
Cell lines, plasmids, and reagents.
Wild-type (WT), RIP1 KO, TRAF2 KO, and TRAF2/5 DKO MEFs were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% bovine calf serum (BCS) and antibiotics, and WT and RIP1-deficient T cells were maintained in Iscove's modified Dulbecco's medium (IMDM) supplemented with 10% fetal calf serum (FCS) and antibiotics. Antibodies (Abs) and reagents were purchased as follows: anti-TRAF2, anti-IKKγ, anti-IKKβ Abs and mouse HOIP-specific small interfering RNA (siRNA) from Santa Cruz Biotechnology (Santa Cruz, CA); panspecific anti-cIAP1/2, agonistic anti-TNFR1 and anti-TNFR2 Abs, and purified recombinant cIAP1 protein from R&D Systems (Minneapolis, MN); anti-TAK1 and anti-IκBα Abs from Cell Signaling (Danvers, MA); mouse and human TNF-α from Roche (Indianapolis, IN); anti-Flag Ab from Sigma Chemicals (St. Louis, MO); anti-polyubiquitin and anti-K63-polyubiquitin Abs from Enzo Life Sciences (Plymouth Meeting, PA); anti-HOIP and anti-HOIL-1 Abs from Abcam Inc. (Cambridge, MA); and a cocktail of inhibitors of proteases and phosphatases from Pierce (Rockford, IL). The anti-linear-pUb Ab was generated as described previously (26). siRNAs for mouse TRAF2, cIAP1, and cIAP2 were purchased from Thermo Scientific Dharmacon (Lafayette, CO); purified recombinant TRAF2 protein from SignalChem (Vancouver, Canada); and E1, UbcH5b, Ub-WT, Ub-K63, and Ub-K0 from Boston Biochem (Cambridge, MA). The retroviral plasmid PEAK12-RIP1 was generously provided by Adrian Ting (Mount Sinai Medical Center, NY), and pBabe-Flag-RIP1 and pQCXIN-Flag-RIP1 were then generated by subcloning the RIP1 cDNA into the pBabe-puro and pQCXIN plasmids.
TNFR1 immunoprecipitation and protein ubiquitination assays in vivo.
MEFs (in five 100-mm petri dishes per treatment) were treated with 2× Flag–TNF-α (1.0 μg/ml), and the cell pellets were lysed in TNE lysis buffer (20 mM HEPES, pH 7.4, 250 mM NaCl, 1% Triton X-100, 1 mM dithiothreitol [DTT], 1 mM EDTA, 20% glycerol, and a cocktail of protease and phosphatase inhibitors) containing 5 mM N-ethylmaleimide (NEM) on ice for 30 min, followed by centrifugation at 13,000 × g for 20 min at 4°C. The TNFR1 complex was then immunoprecipitated with 40 μl of anti-Flag-M2 beads (Sigma) at 4°C for 6 h. The precipitates were washed four times with the same lysis buffer, and recruitment of RIP1, TRAF2, cIAP1, HOIP, TAK1, and IKKβ to TNFR1 was monitored by Western blotting using corresponding Abs. In the cases of RIP1 and TRAF2 ubiquitination within the TNFR1 complex, the immunoprecipitated TNFR1 complex was first eluted from M2 beads with the TNE buffer containing 0.2 mg/ml 3× Flag peptide or 0.5% SDS, 0.5 M LiCl, and 5 mM NEM and then immunoprecipitated with anti-RIP1 or TRAF2 Ab after diluting the eluates 5-fold with TNE buffer. After SDS-PAGE and the membrane transfer, the membranes were sandwiched between filter papers and autoclaved for 30 min or subjected to treatment in denaturing solution (20 mM Tris, pH 7.5, 6 M guanidine chloride, and 5 mM β-mercaptoethanol) for 20 min at room temperature. After extensive washing, the membranes were blocked overnight at 4°C in Tris-buffered saline with Tween (TBS-T) containing 5% bovine serum albumin (BSA) and 10% goat serum. After blocking, the membranes were incubated with anti-linear-pUb or anti-K63-pUb Ab for 90 min at room temperature, washed extensively, and then incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit or anti-mouse Ab for 60 min at room temperature.
Western blot analysis.
MEFs and T cells were mock treated or treated with mouse TNF-α (mTNF-α) (10 ng/ml) and human TNF-α (hTNFα) (20 ng/ml) unless stated otherwise. Total protein samples were extracted with TNE buffer for 30 min on ice, followed by centrifugation at 12,500 × g for 15 min at 4°C. Twenty-five micrograms of each protein sample was separated by SDS-PAGE and transferred onto nitrocellulose membranes. For analysis of p65 phosphorylation, blots were blocked in 3% BSA in TBS-T for 4 h before incubation with phosphoantibodies overnight at 4°C. For analysis of the expression of other proteins, blots were blocked in 5% fat-free milk in TBS-T for 2 h and then incubated with the indicated antibody overnight at 4°C. Protein expression was then detected using enhanced chemiluminescence (ECL) solution.
IKK immunokinase assays.
MEFs, T cells, and hepatocytes were treated with mTNF-α (10 ng/ml), hTNF-α (20 ng/ml), and anti-TNFR1 or anti-TNFR2 agonistic Ab (1.0 μg/ml), and protein samples were extracted using TNE lysis buffer (20 mM HEPES, pH 7.4, 250 mM NaCl, 1% Triton X-100, 1 mM DTT, 1 mM EDTA, 10% glycerol, and a cocktail of protease and phosphatase inhibitors). The IKK complex was immunoprecipitated using an anti-IKKγ antibody and then subjected to in vitro kinase assays in the presence of [γ-32P]ATP, and glutathione S-transferase (GST)–IκBα1-55 served as a substrate. The reaction mixtures were separated by SDS-PAGE, transferred onto nitrocellulose membranes, and exposed to X-ray film for 4 to 6 h to detect phosphorylation. The same membranes were stained with Ponceau S to monitor total GST-IκBα levels and then immunoblotted with an anti-IKKβ antibody.
Smac-mimetic and siRNA-mediated gene knockdown.
MEFs were treated with Smac-mimetic (SM) (200 ng/ml) for 4 h to deplete cIAP1/2. In the case of siRNA-mediated knockdown, MEFs were transfected with siRNA duplexes for mouse TRAF2 or HOIP (40 nM) using Lipofectamine RNAiMax reagent (Invitrogen) according to the manufacturer's instructions. Forty-eight hours after transfection, cells were treated with TNF-α, and IKK activity was then assessed by kinase assays.
Preparation of retroviral supernatants and infection of MEFs.
293T cells at 60 to 70% confluence were cotransfected with 2 μg of pMD.OGP (encoding gag-pol), 2 μg of pMD.G (encoding vesicular stomatitis virus G protein), and 2 μg of pBabe-puro-Flag-RIP1 or pQCXIN-neo-Flag-RIP1 by the standard calcium phosphate precipitation method. Forty-eight hours after transfection, the viral supernatant was collected and filtered through a 0.45-μm filter. The retroviral supernatants were diluted 3-fold with 10% fetal bovine serum (FBS)-DMEM and then used immediately for the infection of MEFs and T cells in the presence of 4 μg/ml Polybrene for 6 h (MEFs) or 24 h (T cells). Forty-eight hours after infection, cells were selected with puromycin (2.0 μg/ml) for 6 days or with neomycin (800 μg/ml) for 14 days, and resistant cells were pooled. Cells that stably express Flag-RIP1 at the physiological level were used for functional experiments within 1 month of establishment.
Multiplex and real-time RT-PCR.
For multiplex reverse transcription (RT)-PCR analysis, MEFs and T cells were untreated or treated with mTNF-α (10 ng/ml) and hTNF-α (20 ng/ml) for 2 h, and total RNA was extracted using the RNeasy minikit (Qiagen). Five micrograms of total RNA was treated with RQ1 RNase-free DNase for 30 min at 37°C and then reverse transcribed using an oligo(dT) primer in a 50-μl volume. Five microliters of the resulting cDNA was first subjected to conventional PCR amplification for 11 cycles in the presence of IP-10, interleukin 6 (IL-6), or RANTES primers, after which GAPDH (glyceraldehyde-3-phosphate dehydrogenase) primers were added to the same PCR mixtures and the PCR amplifications were continued for an additional 20 cycles. The thermocycling program was as follows: 94°C for 2 min; 16 and 20 cycles at 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min; and a final extension step at 72°C for 10 min. After cycling, the PCR products were separated on 1% agarose gels, stained with ethidium bromide, and visualized under UV light. For real-time RT-PCR analysis, cDNA was subjected to real-time PCR using the Power SYBR green AB Master Mix and an ABI Prism 7700 Sequence Detector (Applied Biosystems). Mouse GAPDH-specific primers were used to generate an internal control, and the average threshold cycle (CT) for samples in triplicate was used in the subsequent calculations. Relative expression levels of NF-κB target genes were calculated as the ratio with respect to the GAPDH expression level. The mean ± standard error (SE) of four independent experiments was considered to be statistically significant at a P value of <0.05. For PCR analysis of mouse genes, the same primer pairs reported in a previous publication were used (27). For RT-PCR of human genes in Jurkat T cells, the following primer pairs were used: GAPDH, 5′-ACCACAGTCCATGCCATCAC-3′ (forward) and 5′-TCCACCACCCTGTTGCTGTA-3′; IP-10, 5′-GCATCAGCATTAGTAATCAACC-3′ (forward) and 5′-GCTCCCCTCTGGTTTTAAGGAG-3′.
In vitro ubiquitination assay.
Bacterially expressed His-RIP1-Myc was first affinity purified with Ni-nitrilotriacetic acid (NTA) beads and then immunoprecipitated with anti-Myc antibody. In vitro-translated 35S-RIP1-Flag was immunoprecipitated with anti-Flag-M2 beads. After extensive washes with TNE lysis buffer containing 0.5 M NaCl, the beads were washed again with ubiquitination buffer (50 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 0.5 mM DTT, 2 mM NaF, and 3 μM okadaic acid) and then subjected to ubiquitination reactions in ubiquitination buffer supplemented with nontagged ubiquitin (1.5 μg), ATP (2 mM), E1 (150 ng), UbcH5b (0.5 μg), TRAF2 (0.15 μg), cIAP1 (0.15 μg), and/or TRADD (0.15 μg) for 45 min at 37°C. The beads were then washed extensively with TNE lysis buffer and subjected to Western blot and autoradiography analyses.
Primary hepatocyte isolation.
The livers from nine embryonic day 18 (E18) embryos and three 6-day-old CL57BL/6 mice were crudely sliced and digested at 37°C for 35 min in HEPES-buffered saline supplemented with 2 mM CaCl2, 2 mM MgCl2, 5% FBS, 1.0 mg/ml collagenase IV (Sigma), and 150 U/ml DNase I. Single-cell suspensions were obtained by gentle pipetting, followed by passing through a cell strainer with a 40-μm nylon mesh. Red blood cells were lysed in ammonium-chloride-potassium (ACK) lysis buffer at room temperature for 5 min, and then the remaining hepatocytes were washed twice with phosphate-buffered saline (PBS) containing 2% FBS and 2 mM EDTA. Dead cells were removed by Ficoll gradient centrifugation. Viable cells were then equilibrated at 37°C for 45 min under 5% CO2 in IMDM supplemented with 0.5% FBS and treated with mTNF-α.
Cytotoxicity assay.
Hepatocytes were plated on 12-well plates at a density of 2 × 104 cells/well and equilibrated at 37°C for 45 min under 5% CO2 in IMDM supplemented with 5% FBS, after which the cells were treated with TNF-α or with TNF-α plus cycloheximide (CHX) as indicated. At 36 h after treatment, cell death was assessed via the trypan blue exclusion assay. The data shown represent the results of three experiments performed in triplicate.
RESULTS
RIP1 is essential for TNF-α-induced immediate, but not delayed, IKK activation.
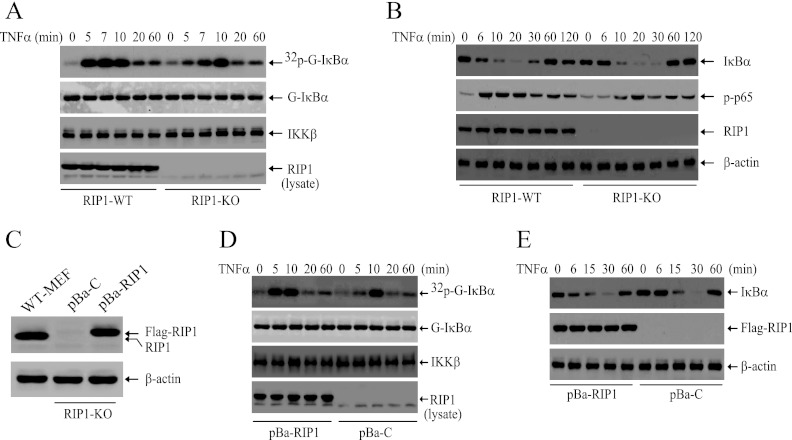
To better understand the role of RIP1 in TNF-α-induced NF-κB activation, we examined the strength and duration of IKK activity by immunokinase assays in WT and RIP1 KO MEFs following TNF-α stimulation. As shown in Fig. 1A, whereas in WT cells, TNF-α caused immediate and robust IKK activation within 5 min of stimulation, this immediate IKK activation is completely impaired in RIP1 KO MEFs. Interestingly, TNF-α still activated IKK in RIP1 KO MEFs 10 min after stimulation. In line with these observations, TNF-α-induced IκBα degradation and p65 phosphorylation were delayed in RIP1 KO MEFs compared to those in WT MEFs (Fig. 1B). Nevertheless, consistent with a previous study (6), TNF-α triggered complete degradation of IκBα in both WT and RIP1 KO MEFs 20 min after stimulation. To confirm our findings, we established RIP1 KO cell lines that stably express either empty vector (pBa-C) or Flag-RIP1 (pBa-RIP1) at a physiological level (Fig. 1C). As expected, stable expression of Flag-RIP1 in RIP1 KO MEFs restored TNF-α-induced immediate IKK activation and IκBα degradation (Fig. 1D and E). These data suggest that RIP1 expression is essential for TNF-α-induced immediate IKK activation and that TNF-α can still induce delayed IKK activation in the absence of RIP1.
Fig 1.
RIP1 is essential for TNF-α-induced immediate, but not delayed, IKK activation in MEFs. (A) WT and RIP1 KO MEFs were treated with TNF-α (10 ng/ml) as indicated, and IKK activation was then examined by kinase assay. 32p-G-IκBα, phosphorylated GST-IκBα; G-IκBα, total GST-IκBα. (B) WT and RIP1 KO MEFs were treated with TNF-α (10 ng/ml) as indicated, and degradation of IκBα and phosphorylation of p65 were monitored by Western blotting. (C) RIP1 KO MEFs were stably transfected with pBabe-puro vector control (pBa-C) and pBabe-puro-Flag-RIP1 (pBa-RIP1) by retroviral infection, and the expression level of RIP1 was analyzed by Western blotting using anti-RIP1 antibody. (D and E) pBa-C and pBa-RIP1 cells were treated with TNF-α (10 ng/ml) as indicated, after which IKK activity (D) and IκBα degradation (E) were determined by kinase assay and Western blotting.
TNF-α-induced delayed IKK activation in RIP1 KO MEFs is mediated by TNFR1.
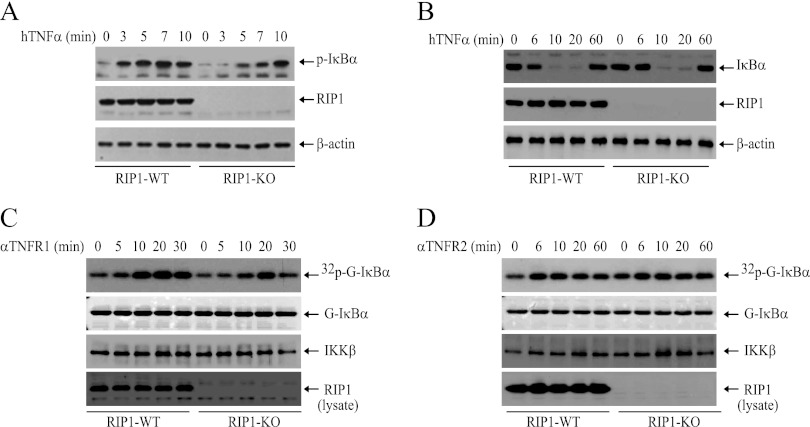
TNFR2 directly recruits TRAF2 and activates NF-κB in response to stimulation by membrane-bound TNF-α (28). To exclude the possibility that delayed IKK activation in RIP1 KO MEFs is regulated by TNFR2, we stimulated WT and RIP1 KO MEFs with hTNF-α, which binds only to mouse TNFR1 and not to TNFR2 (29). As expected, Western blotting revealed that hTNF-α triggers IκBα phosphorylation and degradation immediately in WT, but not in RIP1 KO, MEFs and that hTNF-α can still cause IκBα degradation in RIP1 KO MEFs 15 min after treatment (Fig. 2A and B). To further confirm that ligation of TNFR1 activates NF-κB though RIP1-dependent and -independent pathways, we stimulated WT and RIP1 KO MEFs with either a TNFR1- or a TNFR2-specific agonistic Ab. Interestingly, TNFR1 Ab-induced IKK activation was kinetically delayed in both cell lines, yet it still occurred about 10 min earlier in WT MEFs than in RIP1 KO MEFs (Fig. 2C). On the other hand, the TNFR2 Ab weakly induced IKK activation in both WT and RIP1 KO MEFs at the same strength and kinetics (Fig. 2D). Collectively, these data confirm that ligation of TNFR1 activates IKK through RIP1-dependent and -independent pathways.
Fig 2.
TNF-α induces delayed IKK activation in RIP1 KO MEFs downstream of TNFR1. (A and B) WT and RIP1 KO MEFs were treated with hTNF-α (20 ng/ml) as indicated, after which IκBα phosphorylation and degradation were monitored by Western blotting. (C and D) WT and RIP1 KO MEFs were treated with anti-mouse TNFR1 (αTNFR1) or TNFR2 (αTNFR2) agonistic antibody (1 μg/ml) as indicated, after which IKK activation was determined by kinase assays.
TNF-α-induced IKK activation is impaired in RIP1-deficient Jurkat T cells.
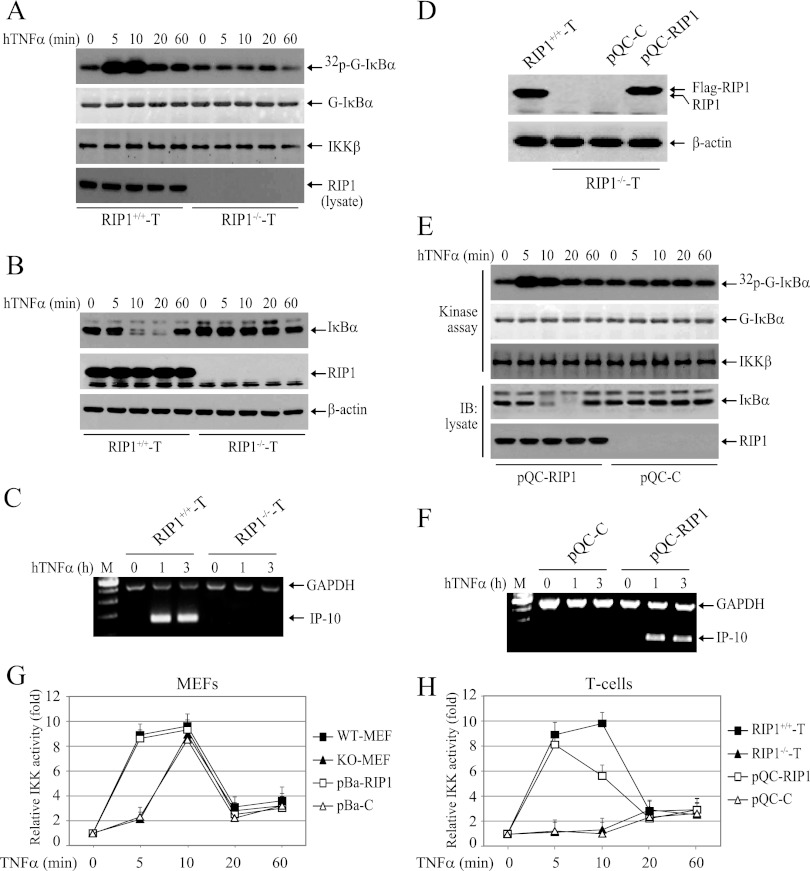
TNF-α-induced NF-κB activation has been shown to be impaired in RIP1-deficient Jurkat T cell mutants (RIP1−/− T cells) (17). Therefore, we also examined the strength and duration of IKK activation in these cells by immunokinase assay following stimulation of the cells with TNF-α. Interestingly, both the immediate and delayed phases of IKK activation were completely impaired in RIP1−/− T cells (Fig. 3A). Consistently, TNF-α-induced degradation of IκBα and expression of IP-10 (an NF-κB target gene) were also completely impaired (Fig. 3B and C). As expected, stable expression of Flag-RIP1 in RIP1−/− T cells efficiently restored TNF-α-induced immediate IKK activation, IκBα degradation, and IP-10 expression (Fig. 3D, E, and F). Next, we repeated the IKK kinase assay three times in these MEFs and T cell mutants, as well as in RIP1-deficient MEFs and T cell mutants reconstituted with Flag-RIP1. As shown in Fig. 3G and H, whereas stable expression of Flag-RIP1 in RIP1 KO MEFs completely restored the temporal profile of IKK activity to a level comparable to that seen in WT MEFs, stable expression of Flag-RIP1 in RIP1−/− T cells failed to do so (IKK activity declined at a 10-min time point to a level clearly lower than that seen in RIP1+/+ T cells). Notably, it has been shown that primary T cells of RIP1 KO thymus isolated from E18 embryos exhibit clear degradation of IκBα within 20 min of TNF-α treatment to a level similar to that observed in WT counterparts (6). A possible explanation for the lack of delayed IKK activation in RIP1−/− T cells is that RIP1−/− T cells were created from human Jurkat T cells by exposing the cells to the mutagen ICR191 and by selectively enriching cell clones that are defective in TNF-α-induced NF-κB activation (17). In addition, RIP1 KO MEFs and RIP1−/− T cells are different types of cells; one is of fibroblast origin, whereas the other is of T lymphocyte origin. Nevertheless, the RIP1 reconstitution experiments in RIP1-deficient cells demonstrated that RIP1 expression is indeed essential for TNF-α-induced rapid IKK activation in both human and murine cells.
Fig 3.
RIP1 is essential for TNF-α-induced IKK activation in Jurkat T cells. (A and B) WT (RIP1+/+ T) and RIP1-deficient (RIP1−/− T) Jurkat T cells were treated with hTNF-α (20 ng/ml) as indicated, and IKK activation and IκBα degradation were examined by kinase assay and Western blotting. (C) RIP1+/+ T cells and RIP1−/− T cells were treated with hTNF-α (20 ng/ml) as indicated, and expression of IP-10 was monitored by conventional multiplex RT-PCR. Lane M, DNA size markers. (D) RIP1−/− T cells were stably transfected with pQCXIN-neo vector control (pQC-C) or pQCXIN-neo-Flag-RIP1 (pQC-RIP1) by retroviral infection, and the expression level of RIP1 was then analyzed by Western blotting using anti-RIP1 antibody. (E) pQC-C and pQC-RIP1 cells were treated with hTNF-α (20 ng/ml) as indicated, and IKK activation and IκBα degradation were examined by kinase assay and Western blotting. (F) pQC-C and pQC-RIP1 cells were treated with hTNF-α (20 ng/ml) as indicated, and expression of IP-10 was monitored by conventional multiplex RT-PCR. (G and H) The IKK kinase assay was repeated three times in MEFs and T cells, and the average IKK activities are presented as means and standard deviations (SD).
RIP1 is essential for rapid and efficient recruitment of IKK to TNFR1.
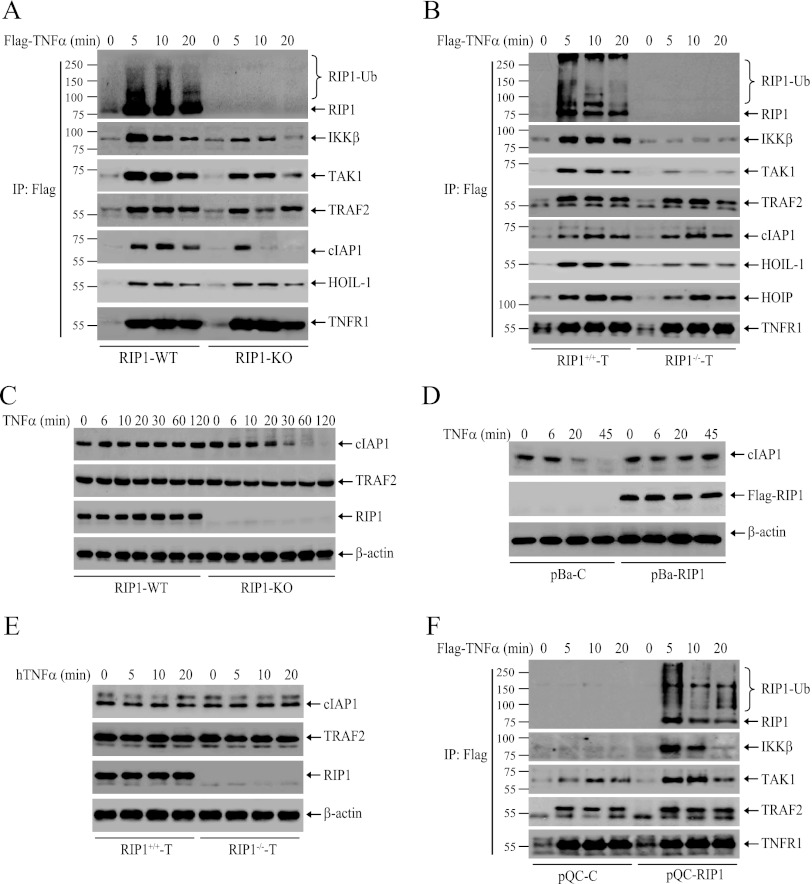
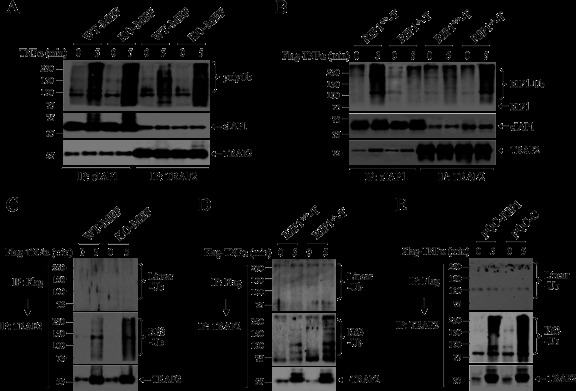
RIP1 ubiquitination has been believed to be essential for TNF-α-induced recruitment of TAK1 and IKK to the TNFR1 complex (5, 9). To understand the possible mechanism for delayed IKK activation in RIP1 KO MEFs and the lack of any IKK activation in RIP−/− T cells, we examined the recruitment of IKK, TAK1, TRAF2, cIAP1, and LUBAC (HOIP, HOIL-1, and SHARPIN) to TNFR1 in WT and RIP1-deficient MEFs and T cells. We initially expected that IKK and TAK1 recruitment would be delayed in RIP1 KO MEFs. However, in contrast to our expectation, the recruitment of IKK and TAK1 was not delayed but reduced dramatically in RIP1 KO MEFs compared to that in WT MEFs (Fig. 4A). It seems that immediate activation of IKK requires immediate recruitment of sufficient amounts of TAK1 and IKK to TNFR1. Interestingly, immediate recruitment of TRAF2, cIAP1, and HOIL-1 to TNFR1 was comparable between WT and RIP1 KO MEFs at the 5-min time point, which is entirely consistent with a recent report by Haas et al. (24). However, in RIP1 KO MEFs, the protein level of cIAP1 within the TNFR1 complex was reduced to an undetectable level 10 min after stimulation, whereas the level of TRAF2 protein was reduced slightly at 10 min, followed by a slight increase at the 20-min time point (Fig. 4A). On the other hand, in RIP−/− T cells, TNF-α-induced recruitment of IKK to TNFR1 was completely impaired, recruitment of TAK1 was severely reduced, and recruitment of HOIP and HOIL-1 was also reduced (Fig. 4B). In contrast, the recruitment of TRAF2 and cIAP1 to TNFR1 in RIP−/− T cells was comparable to that in RIP+/+ T cells. Nevertheless, these data suggest that RIP1 is essential for immediate and efficient recruitment of IKK to TNFR1.
Fig 4.
RIP1 is required for immediate and efficient recruitment of IKK to TNFR1. (A and B) WT and RIP1-deficient MEFs (A) and T cells (B) were treated with Flag–TNF-α (1.0 μg/ml) as indicated, after which the TNFR1 complexes were pulled down with anti-Flag-M2 beads. Ubiquitination of RIP1 and recruitment of IKKβ, TAK1, TRAF2, cIAP1, HOIP, and HOIL-1 were then monitored by Western blotting. IP, immunoprecipitation. (C) RIP1 WT and KO MEFs were left untreated or treated with TNF-α (10 ng/ml) as indicated, after which cIAP1 degradation was detected by Western blotting. (D) pBa-C and pBa-RIP1 MEFs were left untreated or treated with TNF-α (10 ng/ml) as indicated, after which cIAP1 degradation was detected by Western blotting. (E) RIP1+/+ T cells and RIP1−/− T cells were treated with hTNF-α (20 ng/ml) as indicated, and expression of TRAF2 and cIAP1 was monitored by Western blotting. (F) pQC-C and pQC-RIP1 T cells were treated with Flag–TNF-α (1.0 μg/ml) as indicated, and ubiquitination of RIP1 and recruitment of IKKβ, TAK1, and TRAF2 to TNFR1 were then monitored as for panel A.
TNF-α triggers cIAP1 degradation in the absence of RIP1 expression in MEFs but not in T cells.
Gentle et al. have recently reported that TNF-α stimulation of RIP1 KO MEFs causes proteasomal degradation of cIAP1 and TRAF2 (30). Thus, we also examined the total protein levels of cIAP1 and TRAF2 in WT and RIP1 KO MEFs and T cells following TNF-α stimulation. Consistent with the report by Gentle et al., cIAP1 was degraded almost completely 30 min after TNF-α stimulation in RIP1 KO MEFs, whereas the total protein level of TRAF2 was reduced at the 30-min time point and then recovered 120 min after stimulation (Fig. 4C). Reconstitution of RIP1 KO MEFs with Flag-RIP1 completely blocked TNF-α-induced cIAP1 degradation (Fig. 4D), confirming that RIP1 is a bona fide substrate of cIAP1 and TRAF2, as the RING domain-containing E3 ligases undergo autoubiquitination in the absence of their substrates (31). On the other hand, the protein levels of TRAF2 and cIAP1 were not changed in RIP1−/− T cells following TNF-α stimulation (Fig. 4E). It is possible that in RIP1−/− T cells the mutagen ICR191 also introduced mutations to effector proteins that regulate cIAP1 ubiquitination and degradation. Nevertheless, stable expression of Flag-RIP1 in RIP1−/− T cells restored TNF-α-induced efficient recruitment of IKK and TAK1 to TNFR1 (Fig. 4F), confirming that RIP1 is not only essential but also sufficient for rapidly recruiting IKK and TAK1 to TNFR1.
K63-linked TRAF2 ubiquitination is not sufficient for recruiting IKK to TNFR1.
Next, we examined the ubiquitination of cIAP1 and TRAF2 in total cell lysates prepared from TNF-α-treated WT and RIP1-deficient MEFs and T cells. Consistent with the reduction in total protein levels, TNF-α stimulation induced increased ubiquitination of cIAP1 and TRAF2 in RIP1 KO MEFs compared to that in WT MEFs (Fig. 5A). On the other hand, in RIP1−/− T cells, the ubiquitination of cIAP1 was significantly reduced while the ubiquitination of TRAF2 was significantly augmented compared to that in WT T cells (Fig. 5B). Because TRAF2 underwent increased ubiquitination in both RIP1 KO MEFs and T cells after TNF-α stimulation, we further examined the topology of TRAF2 pUb chains within the TNFR1 complex. As shown in Fig. 5C and D, TNF-α-induced increased ubiquitination of TRAF2 in RIP1-deficient cells was, at least partially, achieved through K63 linkage but not through linear linkage. As expected, stable expression of Flag-RIP1 in RIP1−/− T cells efficiently suppressed TRAF2 ubiquitination (Fig. 5E). Given that TNF-α-induced IKK recruitment to TNFR1 was completely impaired in RIP1−/− T cells (Fig. 4B), our data suggest that K63-linked ubiquitination of TRAF2 alone is not sufficient for recruitment and activation of IKK.
Fig 5.
TRAF2 and cIAP1 undergo increased ubiquitination in the absence of RIP1 expression. (A and B) WT and RIP1-deficient MEFs (A) and T cells (B) were treated with TNF-α as indicated, and TRAF2 and cIAP1 were immunoprecipitated with corresponding Abs. Ubiquitination of TRAF2 and cIAP1 was then monitored by Western blotting using anti-pUb Ab. (C to E) RIP1-WT and -KO MEFs and T cells, as well as pQC-C and pQC-RIP1 T cells, were treated with Flag–TNF-α (1.0 μg/ml) as indicated, after which the TNFR1 complexes were immunoprecipitated with anti-Flag-M2 beads. The TNFR1 complex was then eluted, diluted, and reimmunoprecipitated with anti-TRAF2 Ab. Ubiquitination of TRAF2 was monitored by Western blotting using anti-linear-pUb and anti-K63-pUb Abs.
cIAP1/2 and TRAF2, but not TRAF5, regulate the immediate and delayed phases of IKK activation.
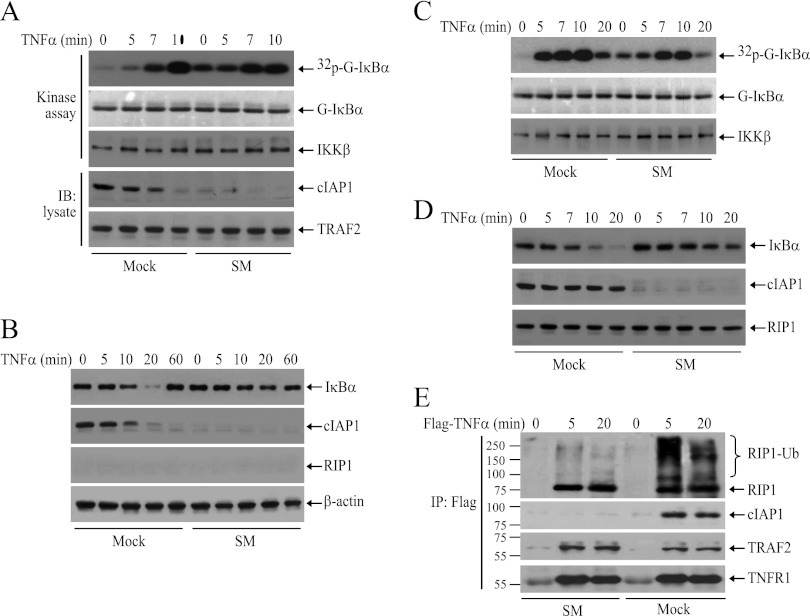
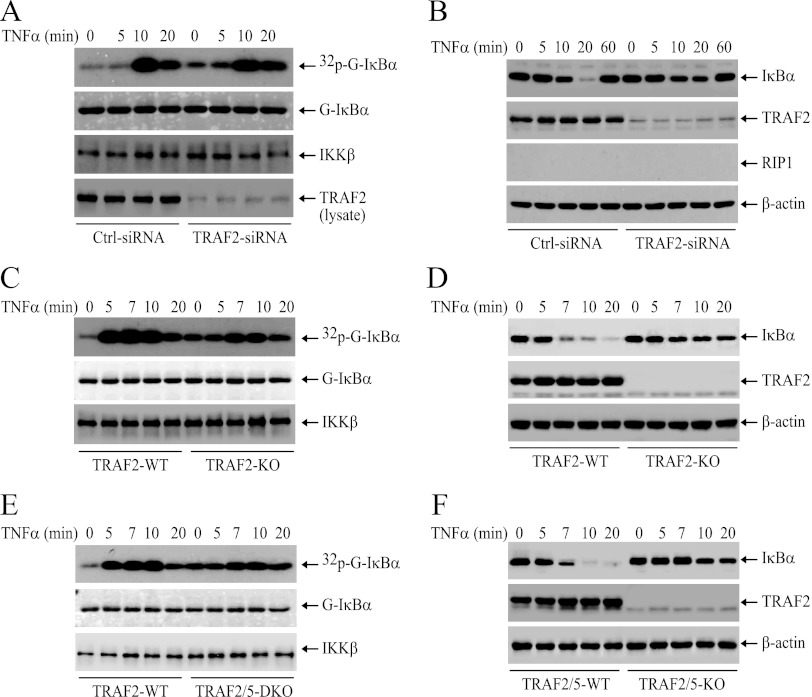
As the ubiquitination of cIAP1 and TRAF2 was increased in RIP1 KO MEFs, we speculated that ubiquitinated cIAP1 and TRAF2 trigger the delayed phase of IKK activation in RIP1 KO MEFs. To test this possibility, we first depleted cIAP1/2 by SM in RIP1 KO MEFs, as described by Wang et al. (32), and then examined TNF-α-induced delayed IKK activation. Interestingly, depletion of cIAP1/2 increased the basal IKK activity and attenuated, but did not completely abolish, the delayed phase of IKK activation (Fig. 6A). Notably, Western blotting revealed that IκBα degradation was severely reduced in cIAP1/2-depleted RIP1 KO MEFs (Fig. 6B). These data prompted us to examine the effect of cIAP1/2 depletion on TNF-α-induced IKK activation and IκBα degradation in WT MEFs. Interestingly, depletion of cIAP1/2 in WT MEFs also increased the basal IKK activity and impaired the immediate phase of IKK activation but failed to completely impair delayed IKK activation (Fig. 6C). Again, Western blotting revealed that IκBα degradation was severely reduced in cIAP1/2-depleted WT MEFs (Fig. 6D). In addition, consistent with published reports, in cIAP1/2-depleted MEFs, although both RIP1 and TRAF2 were recruited normally to the TNFR1 complex, RIP1 ubiquitination was severely impaired (Fig. 6E), indicating that TRAF2 by itself is not a strong E3 ligase toward RIP1. Next, we knocked down TRAF2 with siRNA in RIP1 KO MEFs and then examined IKK activation. Similarly, knockdown of TRAF2 also slightly increased basal IKK activity and severely reduced TNF-α-induced IκBα degradation but did not completely impair delayed IKK activation (Fig. 7A and B). As expected, in both TRAF2 KO and TRAF2/5 DKO MEFs, TNF-α-induced immediate IKK activation was also impaired and IκBα degradation was severely reduced, yet TNF-α still induced delayed IKK activation in these cells, albeit weakly (Fig. 7C, D, E, and F). We tried to knock down both TRAF2 and cIAP1/2 in RIP1 KO MEFs, but our attempts failed repeatedly because the majority of siRNA-transfected cells did not survive overnight culture. Altogether, these data suggest that TRAF2 and cIAP1/2 regulate both the immediate and delayed phases of IKK activation in response to TNF-α stimulation and that TRAF5 may not have a substantial role in TNF-α-induced immediate IKK activation.
Fig 6.
cIAP1/2 are essential for TNF-α-induced immediate IKK activation. (A and B) RIP1 KO MEFs depleted of cIAP1/2 with SM were treated with TNF-α (10 ng/ml) as indicated, and then IKK activation and IκBα degradation were assessed by kinase assay and Western blotting (IB). (C and D) RIP1 WT MEFs depleted of cIAP1/2 with SM were treated with TNF-α (10 ng/ml) as indicated, and then IKK activation and IκBα degradation were assessed by kinase assay and Western blotting. (E) WT MEFs depleted of cIAP1/2 with SM were treated with Flag–TNF-α (1.0 μg/ml) as indicated, after which the TNFR1 complexes were immunoprecipitated with anti-Flag-M2 beads. Ubiquitination of RIP1 and recruitment of TRAF2 and cIAP1 were then monitored by Western blotting.
Fig 7.
TRAF2 is essential for TNF-α-induced immediate IKK activation. (A and B) RIP1 KO MEFs were transfected with a scrambled control or mouse TRAF2-specific siRNA. Forty-eight hours after transfection, the cells were treated with TNF-α (10 ng/ml) as indicated, and IKK activation and IκBα degradation were monitored by kinase assay and Western blotting. (C to F) WT, TRAF2 KO, and TRAF2/5 DKO MEFs were treated with mTNF-α (10 ng/ml) as indicated, and IKK activation and IκBα degradation were assessed by kinase assays and Western blotting.
LUBAC mediates TNF-α-induced delayed IKK activation in the absence of RIP1.
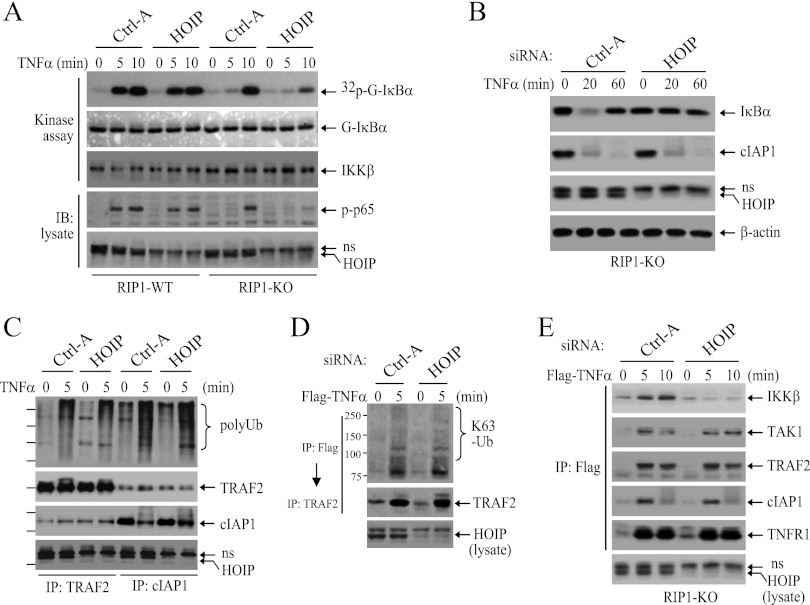
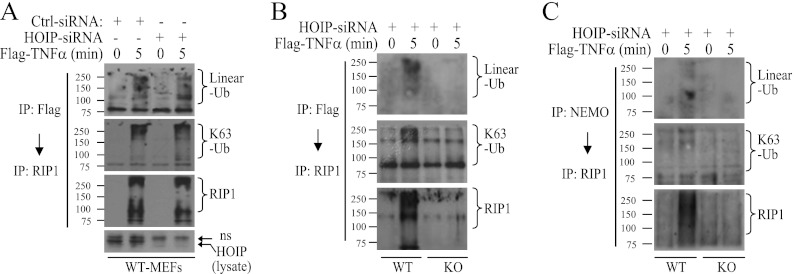
Haas et al. have reported that ubiquitinated TRAF2 and cIAP1/2 recruit LUBAC to TNFR1 and promote efficient TNF-α-induced NF-κB activation (24). Thus, to further define the mechanisms that underlie TNF-α-induced delayed IKK activation in RIP1 KO MEFs, we knocked down HOIP with siRNA in RIP1 WT and KO MEFs and then examined IKK activation. Interestingly, knockdown of HOIP almost completely inhibited delayed IKK activation in RIP1 KO MEFs, while it only slightly reduced IKK activation at the 10-min point in WT MEFs (Fig. 8A). In addition, IκBα degradation and p65 phosphorylation were also almost completely impaired in HOIP knockdown RIP1 KO MEFs (Fig. 8A and B). As expected, knockdown of HOIP in RIP1 KO MEFs had no effect on overall ubiquitination of cIAP1 and TRAF2 (Fig. 8C) or on K63-linked ubiquitination of TRAF2 within the TNFR1 complex (Fig. 8D). Surprisingly, knockdown of HOIP in RIP1 KO MEFs significantly reduced TNF-α-induced recruitment of IKK to TNFR1 but had no effect on the recruitment of TRAF2, cIAP1, and TAK1 (Fig. 8E). These data suggest that ubiquitinated TRAF2 and cIAP1/2 recruit LUBAC and induce delayed IKK activation in the absence of RIP1 expression.
Fig 8.
LUBAC is essential for activation of the delayed phase of IKK. (A and B) RIP1 WT and KO MEFs were transfected with a scrambled control (Ctrl-A) or mouse HOIP-specific siRNA. Forty-eight hours after transfection, the cells were treated with TNF-α (10 ng/ml) as indicated, and IKK activation, p65 phosphorylation, and IκBα degradation were then examined by kinase assay and Western blotting. ns, nonspecific band. (C to E) RIP1 KO MEFs were transfected with a scrambled control or mouse HOIP-specific siRNA. Forty-eight hours after transfection, the cells were stimulated with TNF-α as indicated, and the ubiquitination of TRAF2 and cIAP1 in the total lysate (C), TRAF2 K63 ubiquitination within the TNFR1 complex (D), and recruitment of IKKβ, TAK1, TRAF2, and cIAP1 to TNFR1 (E) were then analyzed as in Fig. 4A and 5A and C.
TNF-α induces linear ubiquitination of RIP1 independent of LUBAC.
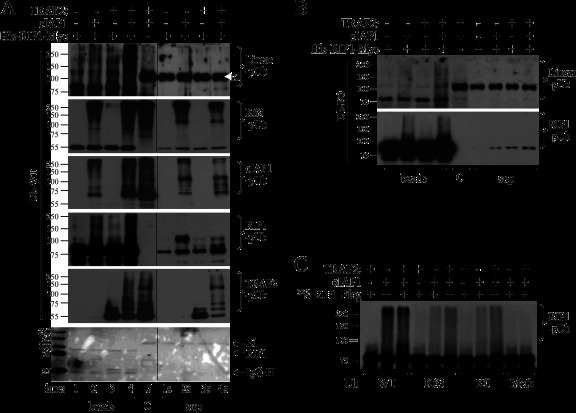
Our data demonstrate that K63-linked ubiquitination of TRAF2 is not capable of recruiting IKK directly to TNFR1. This prompted us to examine the topology of RIP1 ubiquitination within the TNFR1 complex. To do so, we first knocked down HOIP in WT MEFs, stimulated the cells with Flag–TNF-α, and then immunoprecipitated the TNFR1 complex with anti-Flag-M2 beads. The complexes were then eluted, diluted, and subjected to RIP1 reimmunoprecipitation. Western blotting using chain-specific antibodies revealed that RIP1 is ubiquitinated through both K63 and linear linkages (Fig. 9A and B). Interestingly, when the NEMO complex was first immunoprecipitated, followed by RIP1 reprecipitation, linear-pUb-specific antibody produced slightly stronger signals than did K63-pUb-specific antibody (Fig. 9C). These data led us to hypothesize that the other E3 ligases found in the signaling complex (e.g., TRAF2 and cIAP1) may mediate linear ubiquitination of RIP1 and immediate activation of IKK in WT MEFs. To test whether TRAF2 and cIAP1 cooperatively catalyze linear ubiquitination of RIP1, we subjected immunoprecipitated His-RIP1-Myc to in vitro ubiquitination assays in the presence of nontagged Ub, UbcH5b, TRAF2, and/or cIAP1 and then extensively washed the RIP1-bound protein G (Gp) beads after the reactions. Western blot analysis revealed that TRAF2 and cIAP1 together, but not either one alone, directly and efficiently conjugate RIP1 with linear-linkage Ub chains (Fig. 10A, lane 4). Notably, cIAP1 also strongly ubiquitinated TRAF2, as well as itself, and these two proteins, but not E1 and E2, bound to the RIP1 beads very tightly after the reactions and were not washed off the beads even with high-salt buffers. Nevertheless, in a control reaction that contained a 2-fold excess of TRAF2 and cIAP1, the linear ubiquitination was not detected under the same conditions (Fig. 10A, compare lanes 4 and 5). Interestingly, when lysine-free Ub (Ub-K0) was used for ubiquitination reactions, cIAP1 alone was able to catalyze linear ubiquitination of RIP1, albeit weakly (Fig. 10B). To confirm these data, we in vitro translated RIP1-Flag in the presence of [35S]Met, immunoprecipitated it with anti-Flag-M2 beads, and then carried out ubiquitination assays using Ub-WT, -K63, -K0, and -Meth (methylated Ub). As expected, TRAF2 and cIAP1 together more efficiently generated polyubiquitin chains on 35S-RIP1 from Ub-K63 and -K0 but not from Ub-Meth, confirming that the signals depict pUb chains but not multiple monoubiquitination, since methylation of Ub blocks all Lys and the α-amino group of the N-terminal Met (Fig. 10C). Collectively, these data suggest that TRAF2's function is not only to recruit cIAP1 to the TNFR1 complex, but also to promote cIAP1-mediated linear and K63-linked ubiquitination of RIP1.
Fig 9.
RIP1 is ubiquitinated through both K63 and linear linkages. (A to C) RIP1 WT and KO MEFs were transfected with a scrambled control or mouse HOIP-specific siRNA. Forty-eight hours after transfection, the cells were treated with Flag–TNF-α (1.0 μg/ml) as indicated, after which the signaling complexes were immunoprecipitated either with anti-Flag-M2 beads (A and B) or with anti-NEMO Ab (C). The complexes were then eluted, diluted, and reprecipitated with anti-RIP1 Ab. Ubiquitination of RIP1 was monitored by Western blotting using anti-linear-pUb and anti-K63-pUb Abs.
Fig 10.
TRAF2 and cIAP1 cooperatively catalyze K63-linked and linearly linked ubiquitination of RIP1. (A) Bacterially expressed and purified His-RIP1-Myc was immunoprecipitated with anti-Myc Ab and then subjected to in vitro ubiquitination reactions in the presence of nontagged Ub-WT, TRAF2, and/or cIAP1. Following the reaction, the Gp beads (lanes 1 to 4) and supernatants (sup) (lanes 1s to 4s) were separated, and the beads were washed before being subjected to Western blot analysis using anti-linear-pUb, anti-K63-pUb, anti-cIAP1, anti-RIP1, and anti-TRAF2 Abs. Lane 5 is a control ubiquitination reaction mixture containing 2-fold excess of TRAF2 and cIAP1 but without RIP1. At the bottom is shown Ponceau S staining of one of the membranes, and a thin black line in each gel indicates a blank lane cropped out of the image derived from one Western blot image. (B) Immunoprecipitated His-RIP1-Myc was subjected to in vitro ubiquitination reactions in the presence of nontagged, lysine-free Ub (Ub-K0), TRAF2, and/or cIAP1, and ubiquitination of RIP1 was then monitored by Western blotting using anti-linear-pUb and anti-RIP1 Abs as for panel A. (C) In vitro-translated and immunopurified 35S-RIP1-Flag was subjected to ubiquitination reactions as in the presence of WT, K63-only (K63), K0, and methylated (Meth) Ub, and ubiquitination of RIP1 was then monitored by autoradiography.
Both RIP1 and LUBAC are essential for efficient expression of NF-κB target genes.
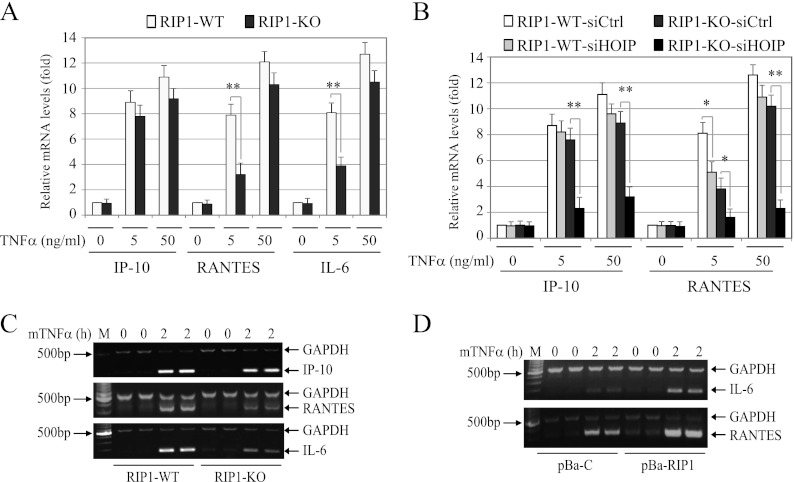
To determine the relevance of RIP1- and LUBAC-mediated activation of IKK in NF-κB-dependent gene expression, we first analyzed the expression of well-known NF-κB target genes (Ip-10, Rantes, and Il-6) by quantitative real-time RT-PCR in WT and RIP1 KO MEFs following stimulation of the cells with low (5-ng/ml) and high (50-ng/ml) doses of TNF-α. Interestingly, although the expression levels of IP-10 were similar between the two cell lines, the expression of RANTES and IL-6 was reduced significantly in RIP1 KO MEFs compared to that in WT MEFs when the cells were treated with a low, but not with a high, dose of TNF-α (Fig. 11A). Next, we knocked down HOIP and then examined IP-10 and RANTES expression in RIP1 WT and KO MEFs following TNF-α stimulation. As expected, knockdown of HOIP in RIP1 KO MEFs almost completely inhibited TNF-α-induced expression of both IP-10 and RANTES at both doses (Fig. 11B). Interestingly, knockdown of HOIP in WT MEFs also significantly reduced the expression of RANTES induced by a low dose of TNF-α treatment. To confirm these results, we performed conventional multiplex RT-PCR analysis. Consistently, transcription of RANTES and IL-6 was clearly lower in RIP1 KO MEFs than in WT MEFs (Fig. 11C). To further confirm these results, we examined the expression of RANTES and IL-6 in pBa-C and pBa-RIP1 cell lines. As expected, TNF-α-induced expression of RANTES and IL-6 was markedly higher in pBa-RIP1 cells than in pBa-C cells (Fig. 11D). These data suggest that both RIP1 and LUBAC play essential roles in efficient expression of a subset of NF-κB target genes in response to a pathophysiologically relevant dose of TNF-α stimulation.
Fig 11.
Both RIP1 and LUBAC are essential for TNF-α-induced efficient expression of a subset of NF-κB target genes. (A and B) WT and RIP1 KO MEFs were treated with mTNF-α for 2 h as indicated, and expression of IP-10, RANTES, and IL-6 was then determined by real-time RT-PCR. *, P < 0.05; **, P < 0.01. The error bars indicate SD. (C and D) RIP1 WT, KO, pBa-C, and pBa-RIP1 MEFs were treated with mTNF-α (10 ng/ml) as indicated, and expression of IP-10, IL-6, and RANTES was determined by multiplex RT-PCR.
Embryonic hepatocytes do not express RIP1 and exhibit delayed IKK activation.
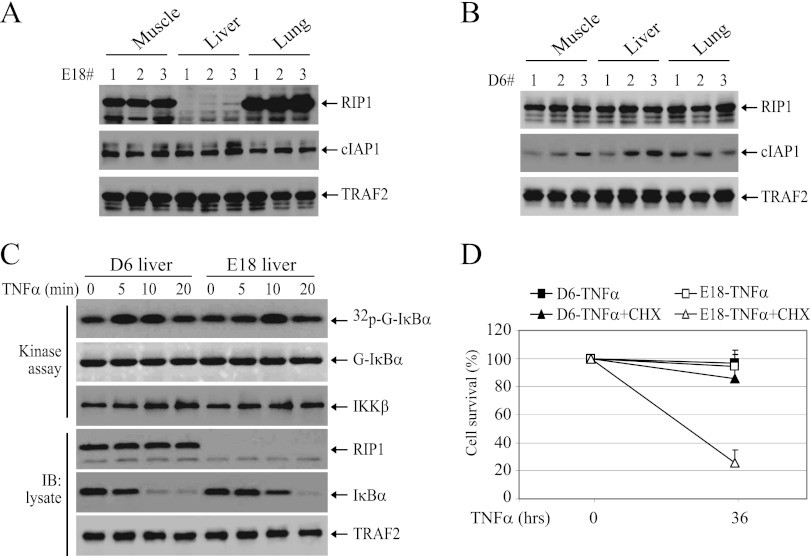
Recently, Wong et al. reported that embryonic hepatocytes do not express RIP1 protein, possibly due to degradation (6). We also found that embryonic hepatocyte do not express RIP1 protein, while hepatocytes derived from 6-day-old mice do (Fig. 12A and B). To assess the physiological relevance of the RIP1-independent pathway in regulating TNF-α-induced IKK activation, we isolated hepatocytes from E18 embryos and 6-day-old mice and examined IKK activation. As expected, TNF-α-induced IKK activation was delayed in embryonic hepatocytes but not impaired (Fig. 12C). Importantly, this delayed IKK activation is sufficient to confer resistance to TNF-α-induced cell death on embryonic hepatocytes (Fig. 12D). On the other hand, embryonic hepatocytes were found to be more susceptible to apoptosis induced by TNF-α in the presence of a low level of CHX (0.3 μg/ml), a condition that does not cause apparent cell death in RIP1-expressing cells (Fig. 12D). These data suggest that, during embryonic development, TNF-α activates IKK though a RIP1-independent pathway in hepatocytes.
Fig 12.
TNF-α activates NF-κB in embryonic hepatocytes through a RIP1-independent pathway. (A and B) The muscles, livers, and lungs were removed from E18 embryos (A) and 6-day-old mice (D6) (B), and protein extracts were prepared by homogenization in TNE lysis buffer. Expression of RIP1, cIAP1, and TRAF2 was then determined by Western blotting. (C) Hepatocytes prepared from E18 embryos and 6-day-old mice were treated with TNF-α (10 ng/ml) as indicated, and IKK activation and IκBα degradation were then detected by kinase assay and Western blotting. (D) Hepatocytes prepared from E18 embryos and 6-day-old mice were treated with TNF-α (10 ng/ml) or TNF-α (10 ng/ml) plus CHX (0.3 μg/ml), and cell viability was assessed 36 h after treatment by trypan blue staining. The error bars indicate SD.
DISCUSSION
TNF-α is a key modulator of inflammatory responses and is targeted in therapeutic strategies for a range of diseases involving chronic inflammation (33). Ironically, although TNF-α/TNFR1 signaling is among the most studied signal transduction pathways, our understanding of the molecular mechanisms underlying TNFR1-mediated activation of NF-κB has been significantly complicated by the recent accumulation of apparently contradictory results (2, 6, 8, 11, 21, 34). For example, early studies conducted in a mutant Jurkat T cell line lacking RIP1 and in Abelson murine leukemia virus-transformed pre-B cell lines derived from RIP1 KO mice revealed that RIP1 expression plays an essential role in TNF-α-induced NF-κB activation (16, 17). In contrast, Wong et al. reported recently that TNF-α can normally activate NF-κB in a variety of primary and immortalized cells derived from the same RIP1 KO mice (6). Our study demonstrates that different experimental approaches could result in different, if not conflicting, conclusions. For example, whereas Western blotting showed that TNF-α induces IκBα degradation normally in RIP1 KO MEFs, kinase assays revealed that TNF-α-induced immediate IKK activation is impaired (Fig. 1A and B). In the case of RIP1−/− T cells, although Western blotting revealed that reconstitution of these RIP1−/− T cells with Flag-RIP1 restores TNF-α-induced IκBα degradation, kinase assays revealed that the IKK activity declines faster than that in parental WT T cells (Fig. 3E and G).
Similarly, comparing results from gene expression further revealed that which target genes are analyzed can have an impact on the experimental outcome. Specifically, real-time RT-PCR analysis of IP-10 expression suggested that TNF-α-induced expression levels of NF-κB targets are comparable between WT and RIP1 KO MEFs, yet analysis of the expression of RANTES and IL-6 revealed a significant reduction in RIP1 KO cells (Fig. 11A). The expression of NF-κB target genes is regulated at multiple levels, including phosphorylation of the p65 subunit (by IKK, Akt, and protein kinase Cζ) and the immediate accessibility and modification of the target gene's promoter regions (1, 35, 36). Some NF-κB target genes (e.g., ikba and Ip-10) are particularly sensitive to TNF-α stimulation, as their promoters are always immediately accessible to transcription factors; even transient NF-κB activation is sufficient to induce their expression (37, 38). In contrast, other promoters require stimulus-dependent modifications to chromatin structure to become accessible to NF-κB (e.g., Rantes and IL-6); thus, they are activated only in the context of strong and prolonged NF-κB activation (37, 38). This explains why RIP1 KO MEFs express IP-10 and IκBα at levels similar to those of WT MEFs but express RANTES and IL-6 at significantly lower levels. Collectively, these data suggest that assessment of TNF-α-induced NF-κB activation by analyzing the degradation of IκBα and the expression of particular NF-κB target genes can lead to conflicting conclusions.
Also, in TRAF2 KO, TRAF2/5 DKO, and cIAP1/2-depleted cells, while Western blotting showed that TNF-α-induced IκBα degradation is severely reduced, kinase assays revealed that basal IKK activity is elevated and TNF-α-induced IKK activation is attenuated but not impaired (Fig. 6A to D and 7C to F). Recent studies revealed the existence of extensive cross talk between the canonical and noncanonical NF-κB pathways. For example, accumulated NIK activates both the classical IKK complex and the IKKα homodimer (8, 39). NIK is accumulated in TRAF2 KO and cIAP1/2-depleted cells, and it is thus conceivable that basal IKK activity is elevated in TRAF2 KO and cIAP1/2 knockdown cells. The conclusion that depletion of both cIAP1 and cIAP2 or knockout of both TRAF2 and TRAF5 can abrogate TNF-α-induced NF-κB activation was based on analyses of IκBα protein levels in these cells (10, 11, 15). In WT MEFs, TNF-α-induced IκBα phosphorylation is robust and immediate, resulting in complete degradation of IκBα within 15 min. If, however, the IKK complex is constitutively activated to a certain degree, IκBα will likewise be constitutively phosphorylated, degraded, and resynthesized, which will partially mask stimulation-induced IκBα degradation. This is why TNF-α-induced IκBα degradation seems impaired in cIAP1/2-depleted and TRAF2/5 DKO cells, even though IKK activation is not abolished. Thus, we believe that an accurate assessment of TNF-α-induced NF-κB activation requires examination of the early versus late phases of IKK activation and of the expression of sets of genes that are regulated by transient versus prolonged NF-κB activation.
A growing body of evidence shows that TRAF2 recruits cIAP1/2 to TNFR1 and that the E3 ligase activity of cIAP1/2, but not of TRAF2, is responsible for attaching K63-linked pUb chains to RIP1 (10, 11, 13, 25). This form of RIP1 ubiquitination has been believed to serve as a platform to recruit both the TAK1 and IKK complexes (5, 9). Recently, LUBAC (containing HOIL-1, HOIP, and SHARPIN) was reported to work in conjunction with UbcH5 to catalyze linear ubiquitination of RIP1 and NEMO and to promote NF-κB activation (24, 26, 40). These notions, however, were challenged by Xu et al., who asserted that K63-linked ubiquitination is dispensable for IKK activation by TNFR1 and that LUBAC is not required for polyubiquitination of RIP1 and activation of IKK (7). Gerlach et al. reported, based on mass spectrometric analysis of the TNFR1 complex, that RIP1 can be ubiquitinated through K48, K63, K11, and linear linkages (40). In vitro binding and ubiquitination assays have revealed that cIAP1 can bind directly to and is capable of conjugating RIP1 with diverse types of ubiquitin chains (11, 32, 41). However, in the absence TRAF2, cIAP1 is not able to induce immediate IKK activation in response to TNF-α stimulation. Notably, TRAF2 and cIAP1 undergo increased autoubiquitination, at least partially, through K63 linkage in RIP1 KO MEFs following TNF-α stimulation, yet they are not able to directly recruit and activate IKK in the absence of HOIP expression (Fig. 8A to E). In contrast, TAK1 recruitment was not affected significantly under the same conditions. In addition, knockdown of HOIP had no significant effect on TNF-α-induced linear ubiquitination of RIP1 and immediate activation of IKK in WT MEFs (Fig. 8A and E and 9A). Interestingly, when nontagged Ub-WT was used in the ubiquitination reactions, cIAP1 conjugated RIP1 with linear-linkage Ub chains only in the presence of TRAF2 (Fig. 10A), whereas when Ub-K0 and -K63 were used in the reactions, cIAP1 alone was able to conjugate RIP1 with linear and K63 linkage chains, albeit less efficiently/weakly, and such cIAP1-mediated ubiquitination was clearly augmented in the presence of TRAF2 (Fig. 10B and C). Of note, when Ub acceptor sites of substrates are mutated, E3 ligases often conjugate Ub to alternative sites on substrates (5, 42). Remarkably, the affinity of TAB2 for the K63-pUb chain is much higher than for the linear pUb chain (43). On the other hand, NEMO has 100-fold-higher affinity for the linear pUb chain than for the K63-pUb chain (44). RIP1 is the most strongly ubiquitinated component of the TNFR1 signaling complex (2, 34). Thus, these data suggest that TRAF2's function is not only to recruit cIAP1, but also to regulate cIAP1-mediated RIP1 ubiquitination through linear and K63 linkages, and that linear-pUb chains recruit the IKK complex and K63-pUb chains recruit the TAK1 and LUBAC complexes (Fig. 13A). It is also likely that although NEMO binds to K63-pUb chains in vitro, it may not compete well with TAB2 and HOIP for binding to K63-pUb chains under physiological conditions in vivo.
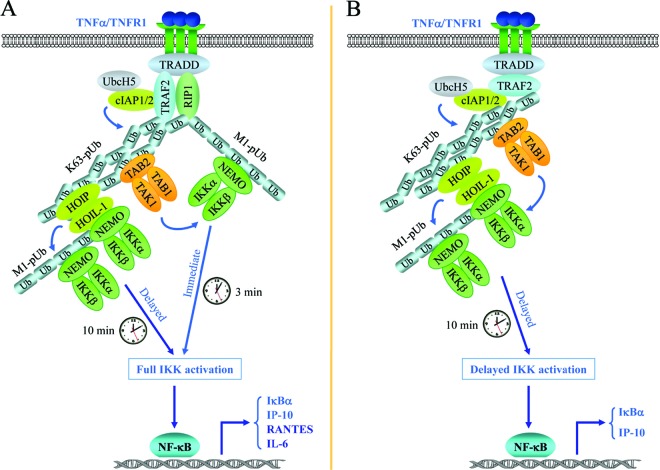
Fig 13.
TNF-α signaling pathways to IKK activation. (A) Signaling pathways to full IKK activation. Upon ligation, TNFR1 recruits TRADD, RIP1, TRAF2, and cIAP1/2. TRAF2 and cIAP1/2 then, in conjunction with UbcH5, catalyze linearly linked (M1-pUb) and K63-linked (K63-pUb) polyubiquitination of RIP1, as well as autoubiquitination of themselves (not shown for cIAPs). K63-pUb recruits the TAK1-TAB1-TAB2 complex, and M1-pUb recruits the IKKα-IKKβ-NEMO complex, leading to TAK1 activation and TAK1-mediated activation of the early phase of IKK. Thereafter, K63-pUb recruits the HOIL-1L–HOIP complex, which in turn catalyzes linear ubiquitination of NEMO, leading to more IKK recruitment and full activation of IKK. Although the early phase of IKK activation is sufficient for induction of some NF-κB target genes (e.g., IκBα and IP-10 genes), full activation of IKK is essential for the efficient expression of other genes (e.g., RANTES and IL-6 genes) that require strong and prolonged NF-κB activation. (B) RIP1-independent pathway to delayed IKK activation. In embryonic hepatocytes, RIP1 protein is not detectable, due most likely to posttranslational degradation. As a result, upon TNFR1 ligation, receptor-recruited TRAF2 and cIAP1/2 undergo increased autoubiquitination through K63 (K63-pUb) and K48 linkages. This K63-pUb can mediate the recruitment of the TAK1-TAB1-TAB2 and HOIP–HOIL-1 complexes but not the IKK complex. Consequently, the early phase of IKK activation is impaired. However, the HOIL-1L–HOIP complex (and possibly also TRAF2-K63-pUb) is still able to recruit the IKK complex with delayed kinetics compared to that of M1-pUb. This triggers HOIP–HOIL-1-mediated linear ubiquitination of NEMO and the subsequent recruitment of more IKK, resulting in the activation of the delayed phase of IKK. Such delayed IKK activation is sufficient for induction of some NF-κB target genes (e.g., IκBα and IP-10 genes).
Of note, cIAP1, in conjunction with UbcH5, seems very promiscuous in in vitro ubiquitination assays, conjugating almost all types of Ub chains. The best way to assess whether cIAP1 and TRAF2 together conjugate the linear Ub chain in vivo is to analyze RIP1 ubiquitination by liquid chromatography-tandem mass spectrometry (LC–MS-MS) using HOIP gene KO cells, as HOIP, but not HOIL-1 and SHARPIN, is the catalytic core unit of LUBAC, which has been firmly established to conjugate linear Ub chains (24, 26, 40). However, HOIP KO mice have not yet been generated, and siRNA-mediated knockdown of HOIP may not be appropriate for MS-MS analysis, because siRNA knockdown is usually not complete. Therefore, it is also possible that cIAP1/TRAF2-mediated RIP1 K63-pUb recruits the TAK1 complex and LUBAC-mediated RIP1 linear-pUb recruits the IKK complex in vivo in response to TNF-α stimulation, resulting in rapid and full activation of IKK in WT cells, or that RIP1-K63-pUb plus the RIP1 protein itself cooperatively efficiently recruit the IKK complex in vivo. In fact, some deubiquitinating enzymes (DUBs) display specificity for both substrates and particular Ub chain types (45); thus, like some DUBs, NEMO may bind directly to RIP1 and K63-pUb with a certain affinity.
One of the puzzling questions in the field of TNFR1 study is why so many E3 ligases and different types of pUb chains are required for NF-κB activation (2, 34). Our data demonstrate that RIP1 and LUBAC can independently mediate NF-κB activation in response to TNF-α stimulation and that ablation of either pathway significantly reduces the expression of IL-6 and RANTES when cells are stimulated with a low dose of TNF-α (Fig. 11A and B). TNF-α is a growth factor for hematopoietic cells, and its production is increased following physical trauma and pathogenic infection (33). It is most likely that involvement of several E3 ligases and different types of pUb chains in TNF-α signaling serves to increase the sensitivity of the cell's response to a slight increase in serum TNF-α levels to initiate an immediate and effective host response to infection and injury. In fact, unlike p65 or IKKβ KO mice, RIP1 KO mice survive embryonic development but die perinatally due to multiple morphological defects, and elimination of either TNF-α or TNFR1 rescues RIP1 KO mice, allowing them to grow nearly normally (4, 16). These published findings and our data indicate that during embryonic development, TNF-α can activate NF-κB in RIP1-deficient hepatocytes by the TRAF2-cIAP1/2-LUBAC pathway (Fig. 13B) and that during postnatal growth RIP1 is required for efficient activation of NF-κB and inhibition of TNF-α-induced cell death. Thus, our findings not only shed new light on the mechanisms underlying TNF-α-induced NF-κB activation, but also have significant implications for the interpretation of previous and future studies on TNF-α signaling.
ACKNOWLEDGMENTS
We thank Xiaodong Wang (University of Texas Southwestern Medical Center, Dallas, TX) for Smac-mimetic, Bruce Hostager (University of Iowa, Iowa City, IA) for anti-murine HOIP antibody (46), and John Silke (La Trobe University, Victoria, Australia) for RIP1 WT and KO MEFs.
Support by NCI grant CA138475 (to H.H.) is gratefully acknowledged.
None of us have conflicts of interest.
Footnotes
Published ahead of print 4 March 2013
REFERENCES
- 1. Hayden MS, Ghosh S. 2008. Shared principles in NF-kappaB signaling. Cell 132:344–362 [DOI] [PubMed] [Google Scholar]
- 2. Wajant H, Scheurich P. 2011. TNFR1-induced activation of the classical NF-kappaB pathway. FEBS J. 278:862–876 [DOI] [PubMed] [Google Scholar]
- 3. Vallabhapurapu S, Karin M. 2009. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 27:693–733 [DOI] [PubMed] [Google Scholar]
- 4. Gerondakis S, Grumont R, Gugasyan R, Wong L, Isomura I, Ho W, Banerjee A. 2006. Unravelling the complexities of the NF-kappaB signalling pathway using mouse knockout and transgenic models. Oncogene 25:6781–6799 [DOI] [PubMed] [Google Scholar]
- 5. Chen ZJ, Sun LJ. 2009. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33:275–286 [DOI] [PubMed] [Google Scholar]
- 6. Wong WW, Gentle IE, Nachbur U, Anderton H, Vaux DL, Silke J. 2010. RIPK1 is not essential for TNFR1-induced activation of NF-kappaB. Cell Death Differ. 17:482–487 [DOI] [PubMed] [Google Scholar]
- 7. Xu M, Skaug B, Zeng W, Chen ZJ. 2009. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol. Cell 36:302–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang L, Blackwell K, Thomas GS, Sun S, Yeh WC, Habelhah H. 2009. TRAF2 suppresses basal IKK activity in resting cells and TNFalpha can activate IKK in TRAF2 and TRAF5 double knockout cells. J. Mol. Biol. 389:495–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. 2006. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation. Nat. Cell Biol. 8:398–406 [DOI] [PubMed] [Google Scholar]
- 10. Mahoney DJ, Cheung HH, Mrad RL, Plenchette S, Simard C, Enwere E, Arora V, Mak TW, Lacasse EC, Waring J, Korneluk RG. 2008. Both cIAP1 and cIAP2 regulate TNFalpha-mediated NF-kappaB activation. Proc. Natl. Acad. Sci. U. S. A. 105:11778–11783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Varfolomeev E, Goncharov T, Fedorova AV, Dynek JN, Zobel K, Deshayes K, Fairbrother WJ, Vucic D. 2008. c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor alpha (TNFalpha)-induced NF-kappaB activation. J. Biol. Chem. 283:24295–24299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yin Q, Lamothe B, Darnay BG, Wu H. 2009. Structural basis for the lack of E2 interaction in the RING domain of TRAF2. Biochemistry 48:10558–10567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vince JE, Pantaki D, Feltham R, Mace PD, Cordier SM, Schmukle AC, Davidson AJ, Callus BA, Wong WW, Gentle IE, Carter H, Lee EF, Walczak H, Day CL, Vaux DL, Silke J. 2009. TRAF2 must bind to cellular inhibitors of apoptosis for tumor necrosis factor (TNF) to efficiently activate nf-{kappa}b and to prevent TNF-induced apoptosis. J. Biol. Chem. 284:35906–35915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alvarez SE, Harikumar KB, Hait NC, Allegood J, Strub GM, Kim EY, Maceyka M, Jiang H, Luo C, Kordula T, Milstien S, Spiegel S. 2010. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature 465:1084–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tada K, Okazaki T, Sakon S, Kobarai T, Kurosawa K, Yamaoka S, Hashimoto H, Mak TW, Yagita H, Okumura K, Yeh WC, Nakano H. 2001. Critical roles of TRAF2 and TRAF5 in tumor necrosis factor-induced NF-kappa B activation and protection from cell death. J. Biol. Chem. 276:36530–36534 [DOI] [PubMed] [Google Scholar]
- 16. Kelliher MA, Grimm S, Ishida Y, Kuo F, Stanger BZ, Leder P. 1998. The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity 8:297–303 [DOI] [PubMed] [Google Scholar]
- 17. Ting AT, Pimentel-Muinos FX, Seed B. 1996. RIP mediates tumor necrosis factor receptor 1 activation of NF-kappaB but not Fas/APO-1-initiated apoptosis. EMBO J. 15:6189–6196 [PMC free article] [PubMed] [Google Scholar]
- 18. Feltham R, Moulin M, Vince JE, Mace PD, Wong WW, Anderton H, Day CL, Vaux DL, Silke J. 2010. Tumor necrosis factor (TNF) signaling, but not TWEAK (TNF-like weak inducer of apoptosis)-triggered cIAP1 (cellular inhibitor of apoptosis protein 1) degradation, requires cIAP1 RING dimerization and E2 binding. J. Biol. Chem. 285:17525–17536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moulin M, Anderton H, Voss AK, Thomas T, Wong WW, Bankovacki A, Feltham R, Chau D, Cook WD, Silke J, Vaux DL. 2012. IAPs limit activation of RIP kinases by TNF receptor 1 during development. EMBO J. 31:1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Habelhah H, Takahashi S, Cho SG, Kadoya T, Watanabe T, Ronai Z. 2004. Ubiquitination and translocation of TRAF2 is required for activation of JNK but not of p38 or NF-kappaB. EMBO J. 23:322–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yamamoto M, Okamoto T, Takeda K, Sato S, Sanjo H, Uematsu S, Saitoh T, Yamamoto N, Sakurai H, Ishii KJ, Yamaoka S, Kawai T, Matsuura Y, Takeuchi O, Akira S. 2006. Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nat. Immunol. 7:962–970 [DOI] [PubMed] [Google Scholar]
- 22. Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L, Helgason E, Fairbrother WJ, Deshayes K, Kirkpatrick DS, Vucic D. 2010. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 29:4198–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Iwai K, Tokunaga F. 2009. Linear polyubiquitination: a new regulator of NF-kappaB activation. EMBO Rep. 10:706–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E, Feltham R, Vince J, Warnken U, Wenger T, Koschny R, Komander D, Silke J, Walczak H. 2009. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell 36:831–844 [DOI] [PubMed] [Google Scholar]
- 25. Zhang L, Blackwell K, Shi Z, Habelhah H. 2010. The RING domain of TRAF2 plays an essential role in the inhibition of TNFalpha-induced cell death but not in the activation of NF-kappaB. J. Mol. Biol. 396:528–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, Yamamoto M, Akira S, Takao T, Tanaka K, Iwai K. 2009. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat. Cell Biol. 11:123–132 [DOI] [PubMed] [Google Scholar]
- 27. Blackwell K, Zhang L, Thomas GS, Sun S, Nakano H, Habelhah H. 2009. TRAF2 phosphorylation modulates tumor necrosis factor alpha-induced gene expression and cell resistance to apoptosis. Mol. Cell. Biol. 29:303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wajant H, Henkler F, Scheurich P. 2001. The TNF-receptor-associated factor family: scaffold molecules for cytokine receptors, kinases and their regulators. Cell Signal 13:389–400 [DOI] [PubMed] [Google Scholar]
- 29. Lewis M, Tartaglia LA, Lee A, Bennett GL, Rice GC, Wong GH, Chen EY, Goeddel DV. 1991. Cloning and expression of cDNAs for two distinct murine tumor necrosis factor receptors demonstrate one receptor is species specific. Proc. Natl. Acad. Sci. U. S. A. 88:2830–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gentle IE, Wong WW, Evans JM, Bankovacki A, Cook WD, Khan NR, Nachbur U, Rickard J, Anderton H, Moulin M, Lluis JM, Moujalled DM, Silke J, Vaux DL. 2011. In TNF-stimulated cells, RIPK1 promotes cell survival by stabilizing TRAF2 and cIAP1, which limits induction of non-canonical NF-kappaB and activation of caspase-8. J. Biol. Chem. 286:13282–13291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Joazeiro CA, Weissman AM. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549–552 [DOI] [PubMed] [Google Scholar]
- 32. Wang L, Du F, Wang X. 2008. TNF-alpha induces two distinct caspase-8 activation pathways. Cell 133:693–703 [DOI] [PubMed] [Google Scholar]
- 33. Feldmann M, Maini RN. 2003. Lasker Clinical Medical Research Award. TNF defined as a therapeutic target for rheumatoid arthritis and other autoimmune diseases. Nat. Med. 9:1245–1250 [DOI] [PubMed] [Google Scholar]
- 34. Schmukle AC, Walczak H. 2012. No one can whistle a symphony alone: how different ubiquitin linkages cooperate to orchestrate NF-kappaB activity. J. Cell Sci. 125:549–559 [DOI] [PubMed] [Google Scholar]
- 35. Hoffmann A, Levchenko A, Scott ML, Baltimore D. 2002. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science 298:1241–1245 [DOI] [PubMed] [Google Scholar]
- 36. Okazaki T, Sakon S, Sasazuki T, Sakurai H, Doi T, Yagita H, Okumura K, Nakano H. 2003. Phosphorylation of serine 276 is essential for p65 NF-kappaB subunit-dependent cellular responses. Biochem. Biophys. Res. Commun. 300:807–812 [DOI] [PubMed] [Google Scholar]
- 37. Hoffmann A, Leung TH, Baltimore D. 2003. Genetic analysis of NF-kappaB/Rel transcription factors defines functional specificities. EMBO J. 22:5530–5539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saccani S, Pantano S, Natoli G. 2001. Two waves of nuclear factor kappaB recruitment to target promoters. J. Exp. Med. 193:1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zarnegar B, Yamazaki S, He JQ, Cheng G. 2008. Control of canonical NF-kappaB activation through the NIK-IKK complex pathway. Proc. Natl. Acad. Sci. U. S. A. 105:3503–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, Webb AI, Rickard JA, Anderton H, Wong WW, Nachbur U, Gangoda L, Warnken U, Purcell AW, Silke J, Walczak H. 2011. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471:591–596 [DOI] [PubMed] [Google Scholar]
- 41. Bertrand MJ, Lippens S, Staes A, Gilbert B, Roelandt R, De Medts J, Gevaert K, Declercq W, Vandenabeele P. 2011. cIAP1/2 are direct E3 ligases conjugating diverse types of ubiquitin chains to receptor interacting proteins kinases 1 to 4 (RIP1-4). PLoS One 6:e22356 doi:10.1371/journal.pone.0022356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. 2006. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol. Cell 22:245–257 [DOI] [PubMed] [Google Scholar]
- 43. Kulathu Y, Akutsu M, Bremm A, Hofmann K, Komander D. 2009. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nat. Struct. Mol. Biol. 16:1328–1330 [DOI] [PubMed] [Google Scholar]
- 44. Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, Randow F, Wakatsuki S, Dikic I. 2009. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell 136:1098–1109 [DOI] [PubMed] [Google Scholar]
- 45. Komander D, Clague MJ, Urbe S. 2009. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10:550–563 [DOI] [PubMed] [Google Scholar]
- 46. Hostager BS, Kashiwada M, Colgan JD, Rothman PB. 2011. HOIL-1L interacting protein (HOIP) is essential for CD40 signaling. PLoS One 6:e23061 doi:10.1371/journal.pone.0023061 [DOI] [PMC free article] [PubMed] [Google Scholar]