Abstract
Bone morphogenetic protein (BMP) signaling is critical for cerebellum development. However, the details of receptor regulated-Smad (R-Smad) and common partner Smad (Co-Smad, or Smad4) involvement are unclear. Here, we report that cerebellum-specific double conditional inactivation of Smad1 and Smad5 (Smad1/5) results in cerebellar hypoplasia, reduced granule cell numbers, and disorganized Purkinje neuron migration during embryonic development. However, single conditional inactivation of either Smad1 or Smad5 did not result in cerebellar abnormalities. Surprisingly, conditional inactivation of Smad4, which is considered to be the central mediator of canonical BMP-Smad signaling, resulted only in very mild cerebellar defects. Conditional inactivation of Smad1/5 led to developmental defects in the anterior rhombic lip (ARL), as shown by reduced cell proliferation and loss of Pax6 and Atoh1 expression. These defects subsequently caused the loss of the nuclear transitory zone and a region of the deep cerebellar nuclei. The normal maturation of the remaining granule cell precursors in the external granular layer (EGL) suggests Smad1/5 signaling is required for the specification process in ARL but not for the subsequent EGL development. Our results demonstrate functional redundancy for Smad1 and Smad5 but functional discrepancy between Smad1/5 and Smad4 during cerebellum development.
INTRODUCTION
The cerebellum is important for fine tuning body movements and maintaining balance and posture (1, 2). Cerebellar neuronal development involves lengthy and complex cellular events, including cell proliferation, specification, differentiation, and migration, that require precise genetic regulation (1, 3). Any defects during these developmental processes can lead to various pathologies, including cerebellar hypoplasia or neoplasias (2, 4). In mice, cerebellum development begins after the formation of the mid brain-hindbrain boundary, from which the two primary germinating zones of the cerebellum, the anterior rhombic lip (ARL) and the ventricular zone (VZ), arise (5). Between embryonic day 11.5 (E11.5) and E13.5, the VZ produces different types of GABAergic neurons, including the Purkinje cells, which are responsible for the sole output of the cerebellar cortex (6, 7). At similar time intervals, the neural progenitors in the ARL generate the nuclear transitory zone (NTZ) (8–10). This population of cells later becomes part of the deep cerebellar nuclei (DCN), which are connected to Purkinje cells and are responsible for sending the final neuronal output from the cerebellum (10, 11). At around E13.5, the ARL also starts to generate the granule cell precursors, which migrate tangentially along the cerebellar pial surface and form the external granular layer (EGL) (9, 12). The granule cell precursors then undergo proliferation and differentiation to generate the granule cell population in the developing cerebellum (13, 14). The granule cells, together with Purkinje cells and DCN, eventually build the fundamental neural circuit of the cerebellar cortex (1, 15). Thus, the specification program of progenitors in the ARL is critical for subsequent cerebellar functions. Although the lineage of the ARL is well characterized (10), the molecular pathways controlling the specification program of the progenitors in the ARL are still unclear.
Bone morphogenetic proteins (BMPs) are members of the transforming growth factor β superfamily that have been shown to play a crucial role in cerebellar granule cell development. In vitro experiments showed that BMP can induce the generation of cerebellar granule cells from nongranular cell lineages or embryonic stem cells (16–18). Bmp6, Bmp7, and Gdf7 are the major contributing ligands that are expressed in the roof plate of the neural tube or adjacent tissues before the formation of the cerebellum primordium (18). During cerebellum development, the ARL continues to express Bmp6 (19), whereas the choroid plexus expresses Bmp6, Bmp7, and Gdf7 (20, 21). Bmp7 can maintain the Atoh1 promoter activity that is important for granule cell specification (20). In addition, Gdf7-null mutants develop abnormal cerebellar foliation (21). Although Bmp3 is expressed in the cerebellar cortical transitory zone and Purkinje cells (22), its in vivo function has not been characterized. BMP signals can transduce through Smad-dependent or Smad-independent pathways (23–25), but the exact intracellular components of BMP signaling during cerebellar development are unclear. To confine our study, we focused here on canonical BMP signaling via Smad (here designated canonical BMP signaling [26]). In canonical BMP signaling, BMP ligands bind to membrane-bound serine/threonine kinase type I and II receptors, which results in the activation of receptor-regulated Smad proteins (R-Smads) through phosphorylation. Activated Smad1, -5, and -8 form complexes with common partner Smad (Co-Smad, or Smad4) and translocate into the nucleus, where they regulate the transcription of target genes (23–25). Both R-Smads and Co-Smad are expressed in the embryonic cerebellum (27). Bmpr1a and Bmpr1b encode the type I receptors for BMP signaling. Although Bmpr1a-Bmpr1b double conditional knockout in the cerebellum results in a severe cerebellar phenotype (28), surprisingly, conditional inactivation of Smad4 in the mouse cerebellum by Nestin-Cre does not result in observable cerebellar granule cell abnormality (29). Current dogma for canonical BMP signaling states that Smad4 is required for forming heterotrimers with R-Smads for R-Smad nuclear accumulation and transcriptional regulation of target genes (23–25, 30). These discrepancies prompted us to investigate the role and use of R-Smads and Smad4 during cerebellum development.
We utilized the Cre-loxP approach (31, 32) to inactivate Smad1, Smad5, and also Smad4 in the early embryonic cerebellum using conditional null (floxed) alleles of Smad1 (33), Smad5 (34), and Smad4 (35), together with an Engrailed1-Cre knock-in allele (En1Cre/+) (36), to circumvent the embryonic lethality caused by general knockout of these Smad genes (37–39). Our data showed that conditional inactivation of either Smad1 or Smad5 alone in the cerebellum did not result in cerebellar abnormality. However, the Smad1/5 double-conditional-knockout mutants showed cerebellar hypoplasia, a reduced number of cerebellar granule cells, and the loss of some parts of the DCN. These defects resulted from abnormal specification of the ARL, which failed to generate the early granule cell precursors and the NTZ. Interestingly, the remaining granule cell precursors that formed in the EGL could undergo normal differentiation and maturation. Our results demonstrate that two R-Smads (Smad1 and Smad5) are required and function redundantly during cerebellum development. The severe Smad1/5 mutant cerebellar phenotype stands in sharp contrast to the conditional inactivation of Smad4, which resulted in very mild cerebellar abnormalities. These results suggest a Co-Smad-independent BMP signaling pathway for cerebellum development that is inconsistent with the current understanding of canonical BMP signaling (24, 25).
MATERIALS AND METHODS
Generation and genotyping of conditional-knockout mice.
The generation and genotyping of mice with conditional alleles of Smad1 floxed (Smad1fx) (33), Smad5 floxed (Smad5fx) (34), and Smad4 floxed (Smad4fx) (35) have been described previously. The En1Cre allele has been described previously for the study of cerebellum development (36, 40, 41). The En1Cre mouse line expresses the Cre recombinase in the mid-hindbrain region of the early neural tube from which the cerebellum is derived. To generate the cerebellum-specific Smad1-Smad5 double-conditional-knockout mutant (here referred to as the Smad1/5 mutant), we crossed En1Cre/+ mice with Smad1fx/fx Smad5fx/fx mice. Their En1Cre/+ Smad1fx/+ Smad5fx/+ female offspring were then crossed with Smad1fx/fx Smad5fx/fx males to produce En1Cre/+ Smad1fx/+ Smad5fx/fx or En1Cre/+ Smad1fx/fx Smad5fx/+ males. To generate En1Cre/+ Smad1fx/fx Smad5fx/fx mutant embryos for analysis, Smad1fx/fx Smad5fx/fx females were then crossed with either En1Cre/+ Smad1fx/+ Smad5fx/fx or En1Cre/+ Smad1fx/fx Smad5fx/+ males. Their En1Cre/+ Smad1fx/+ Smad5fx/fx or En1Cre/+ Smad1fx/fx Smad5fx/+ littermates, which developed normally, were used as controls. For the generation of the cerebellum-specific Smad4 conditional-knockout mutant (here referred as the Smad4 mutant), mice with homozygous Smad4 floxed alleles (Smad4fx) were crossed with female mice harboring Zp3-Cre, which expresses Cre recombinase in oocytes (42), to generate mice with the deletion allele of Smad4 (Smad4Δ). We then crossed Smad4Δ/+ mice with En1Cre/+ mice to generate En1Cre/+ Smad4Δ/+ males. To generate En1Cre/+ Smad4Δ/f mutant embryos for analysis, Smad4fx/fx homozygous females were crossed with En1Cre/+ Smad4Δ/+ males. Their En1Cre/+ Smad4Δ/+ littermates, which developed normally, were used as controls. Due to the complexity of the genotypes required in the generation of the conditional mutants, wild-type littermates were not produced. Therefore, we used littermates with the En1Cre/+ allele as controls for the possible heterozygous null genotype effect of the En1 knock-in allele on the phenotypic analysis. Because the pedigrees have been maintained by intercross breeding, the background strain is a mixture of strains, including C57BL/6J, ICR, and 129/Sv.
Tails or yolk sac tissues were collected for DNA extraction and subjected to PCR analysis for genotyping. The primers used for genotyping were as follows: for the Cre allele, Cre-F (5′-GGACATGTTCAGGGATCGCCAGGCG-3′) and Cre-R (5′-CGACGATGAAGCATGTTTAGCTG-3′; for the Smad1 allele, Smad1-F (5′-GTTCCCATTTGGTTCCAAGC-3′), Smad1-R (5′-GAGCTCTGCTCCGCCACTCA-3′), and Smad1-rec (5′-CACCTGTGCCCCCTCCAAGT-3′); for the Smad5 allele, Smad5-F (5′-GAGCGTCTTCCTTAGCTAATGTG-3′), Smad5-R (5′-CACTGGCAAAGCAGAGGTTCAGA-3′), and Smad5-rec (5′-AAAAATCAGCGCTCGACACG-3′); for the Smad4 allele, Smad4-F (5′-CTTTTATTTTCAGATTCAGGGGTTC-3′), Smad4-rec (5′-AAAATGGGAAAACCAACGAG-3′), and Smad4-R (5′-TACAAGTGCTATGTCTTCAGCG-3′); for the Rap allele as an internal control, RAP-F (5′-AGGACTGGGTGGCTTCCAACTCCCAGACAC-3′) and RAP-R (5′-AGCTTCTCATTGCTGCGCGCCAGGTTCAGG-3′).
All animal procedures were conducted with the approval of the Animal Ethics Committee of the Chinese University of Hong Kong.
SDS-PAGE and Western blot analysis.
E13.5 cerebella were dissected and placed in lysis buffer. After SDS-PAGE, Western blotting was performed using the Trans-Blot Turbo Transfer System (Bio-Rad). Standard immunodetection was performed to detect protein expression using the ECL detection system (GE Healthcare).
Histology, immunohistochemistry, and immunofluorescence.
Embryos dissected from the uterus at different stages were washed in phosphate-buffered saline (PBS) at 4°C, followed by fixation in 4% paraformaldehyde in PBS (PFA-PBS) at 4°C overnight. The fixed tissues were incubated in 20% sucrose at 4°C for 6 h and 30% sucrose at 4°C overnight, embedded in OCT compound (Tissue-Tek; Pelco International, Redding, CA), and cryosectioned at 10 μm. For paraffin embedding, fixed tissues were dehydrated, processed in paraffin, embedded using standard procedures, and sectioned at 5 μm. For histological analyses, paraffin sections were stained with hematoxylin and eosin. For immunohistochemistry, deparaffinized sections were subjected to antigen retrieval by heating them in a microwave for 10 min in 10 mM sodium citrate buffer (pH 6.0) and subsequently incubating them in 3% hydrogen peroxide for 10 min. After blocking in 10% sheep serum in PBS at room temperature for 1 h, the sections were incubated with primary antibodies diluted in 10% sheep serum in PBS overnight at 4°C. Horseradish peroxidase (HRP)-conjugated antibodies were used as the secondary antibodies using standard immunohistochemistry procedures. After color development by incubation with 3,3′-diaminobenzidine (DAB), nuclei were counterstained with hematoxylin. For immunofluorescence, deparaffinized sections were blocked in 10% sheep serum, 1% bovine serum albumin (BSA), and 1% Triton X in Tris-buffered saline (TBS) (pH 7.6) at room temperature for 1 h. The sections were then incubated in primary antibody diluted in 10% sheep serum, 1% BSA, and 0.1% Triton X in TBS at 4°C overnight. After three washes with TBS, fluorescently labeled secondary antibodies were applied. After final washes with TBS, nuclei were counterstained by incubating the sections in Hoechst (Invitrogen) in TBS (1:1,000) for 10 min. The sections were finally mounted with glass coverslips using ProLong Gold antifade reagent (Invitrogen) after two TBS 10-min washes. Images were captured using a fluorescence microscope equipped with a charge-coupled device (CCD) camera (DP72; Olympus.).
Floating-section immunostaining.
Sections (100 μm) of freshly dissected tissues were generated using a vibratome (Leica). The sections were fixed in 4% PFA-PBS for 2 h and then blocked in 10% horse serum, 1% BSA, and 1% Triton X in TBS at 4°C over two nights. The sections were then incubated in the primary antibodies diluted in TBS containing 10% horse serum, 1% BSA, and 0.1% Triton X at 4°C for 2 days. After three washes in TBS, fluorescently labeled secondary antibodies were diluted in TBS containing 10% horse serum, 1% BSA, and 0.1% Triton X at 4°C for 2 days. After three washes in TBS, nuclei were counterstained by incubating sections in Hoechst (33342; 1:1,000; Invitrogen) in TBS for 8 min, followed by two washes in TBS for 10 min each. The sections were mounted with coverslips using ProLong Gold antifade reagent (P36930; Invitrogen). Images were captured using a confocal microscope (FV1000; Olympus). Confocal images of a single optical section (0.6 μm) were acquired for each sample.
Quantitative analyses of proliferating and apoptotic cells.
Quantitative analyses of proliferating cells and apoptotic cells were performed by counting 30 alternative 5-μm paraffin sections from the medial region of the developing cerebellum. To quantify the proliferating cells within the ARL and the EGL, the total number of phospho-histone H3-immunoreactive cells was divided by the area with Pax6 immunoreactivity in the respective areas to calculate the mitotic index for comparison. In the case of apoptotic cells, the total number of cleaved-caspase 3-immunoreactive cells was divided by the area of the cerebellum. At least three animals were analyzed for each genotype. Statistical comparisons were tested for significance using a one-tailed Student's t test.
Quantitative analyses of the EGL.
To quantify the EGLs of mutant and control animals, the length of coverage of the EGL on the cerebellar pial surface was measured on 12 consecutive 5-μm paraffin sections from the medial region of E18.5 developing cerebella. EGL thickness was measured on five alternative 5-μm paraffin sections from the medial region of E18.5 developing cerebella. Regional measurements of the EGL were determined by dividing the EGL into three regions, anterior, dorsal, and posterior, according to the anatomical positions. At least three animals were analyzed for each genotype. Statistical comparisons were tested for significance using a two-tailed Student's t test.
Section in situ hybridization.
Ten-micrometer paraffin sections were hybridized with digoxigenin (DIG)-labeled RNA probes of Atoh1 (43) (provided by Jane E. Johnson) or Msx2 (44) synthesized, using a transcription kit (Roche), from linearized cDNA templates using T7 RNA polymerase (Promega) according to the manufacturer's protocol. In brief, the paraffin sections were dewaxed and hybridized with the DIG-labeled anti-sense RNA probe at 65°C overnight. After several washes, the sections were incubated with the alkaline phosphatase-conjugated anti-DIG antibody (Roche), and the expression signals were detected by standard alkaline phosphatase color development procedures.
Antibodies.
The primary antibodies and dilutions used for immunostaining were as follows: rabbit anti-Brn2 (1:500; Santa Cruz sc-28594), rabbit anti-calbindin (1:250; Millipore AB1778), rabbit anti-DAB-1 (1:100; Sigma D1569), rabbit anti-Lhx1 (1:200; Millipore AB3200), rabbit anti-Pax2 (1:500; Invitrogen 71-6000), rabbit anti-Pax6 (1:500; Millipore AB2237), mouse anti-phospho-histone H3 (1:100; Cell Signaling 9706), rabbit anti-phospho-Smad1/5 (1:50; Cell Signaling 9516), rabbit anti-Smad4 (1:100; Millipore 04-1033), mouse anti-TAG1 (1:40; Hybridoma bank 4D7), rabbit anti-Tbr1 (1:150; Abcam ab31940), rabbit anti-Tbr2 (1:500; Millipore AB2283), mouse anti-TUJ1 (1:5,000; Covance MMS-435P), and rabbit anti-Zic1 (1:400; Rockland 7838).
For the secondary antibodies used for immunostaining, HRP-conjugated affinity-purified goat anti-rabbit IgG (Millipore AP132P) and HRP-conjugated affinity-purified goat anti-mouse IgG (Millipore AP308P) were used at a dilution of 1:500. Alexa Fluor 488–goat anti-mouse IgG (Invitrogen A11029), Alexa Fluor 568–goat anti-mouse IgG (Invitrogen A11031), Alexa Fluor 488–goat anti-rabbit IgG (Invitrogen A11034), and Alexa Fluor 568–goat anti-rabbit IgG (Invitrogen A11036) were used at a dilution of 1:1,000.
The primary antibodies used for Western blotting were as follows: rabbit anti-phospho-Smad1/5 (1:1,000; Cell signaling 9516), rabbit anti-phospho-Smad1/5/8 (1:1,000; Cell Signaling 9511), rabbit anti-Smad4 (1:1,000; Abcam ab40759), and rabbit anti-beta actin (1:1,000; Cell Signaling 4967). The secondary antibodies used for Western blotting were ECL donkey anti-rabbit IgG and HRP-linked whole antibody (1:10,000; GE Healthcare NA934V).
RESULTS
Ablation of Smad1/5 and Smad4 in the mutant cerebella.
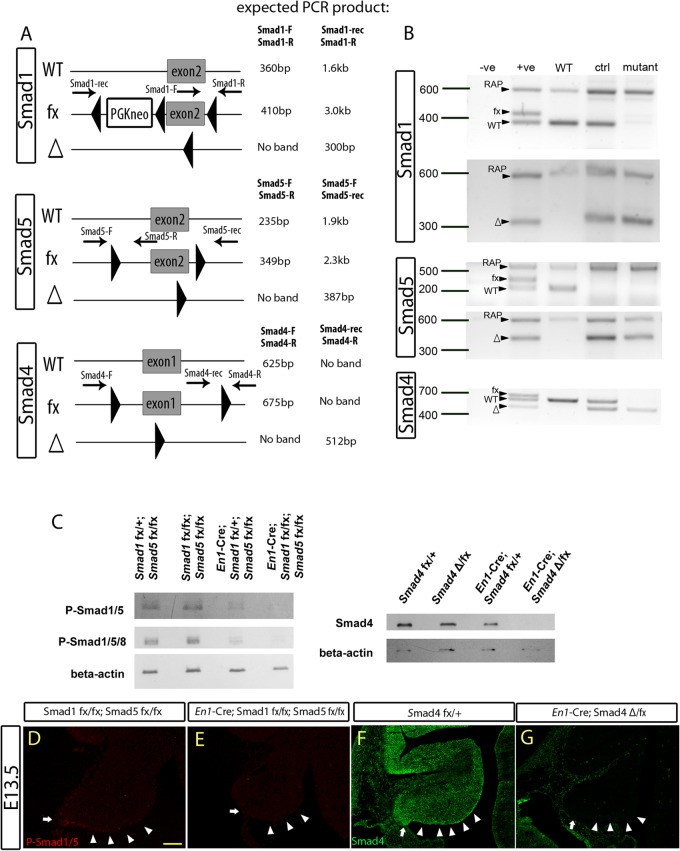
To investigate the role and use of Smads during cerebellum development, a conditional-knockout approach using the Cre-loxP system (31, 32) was employed to circumvent the early lethality limitation of the Smad1, Smad5, and Smad4 knockout mice (35, 37–39). We used the En1-driven Cre with strong activity in the mid-hindbrain boundary at early embryonic stages to ensure complete knockout of the targeted genes in the developing cerebellum (36, 45). Smad1/5 En1-Cre and Smad4 En1-Cre mice are referred to as Smad1/5 mutants and Smad4 mutants, respectively. The controls are defined in Materials and Methods. To determine whether the Smad1, Smad5, and Smad4 floxed alleles were recombined in the mutant cerebella, DNA from control and mutant cerebella at E13.5 were isolated and subjected to PCR analysis. The data demonstrated complete Cre-mediated recombination of the Smad1, Smad5, and Smad4 floxed alleles in the mutant cerebella (Fig. 1A and B). Successful inactivation of the targeted Smad alleles was further supported by the loss of Smad proteins in the mutant E13.5 cerebella assessed by Western blot analysis (Fig. 1C). In addition, immunofluorescent staining showed no expression of phospho-Smad1/5 and Smad4 in the ARL and the VZ of the Smad1/5 and Smad4 mutant E13.5 cerebella, respectively, upon Cre-mediated recombination (Fig. 1D to G).
Fig 1.
En1-Cre-mediated recombination and inactivation of the conditional Smad alleles in the embryonic cerebellum. (A) Schematic diagrams showing the wild-type (WT) allele, floxed allele (fx), and Cre-recombined allele (Δ), with the locations of PCR primers and their expected product sizes indicated. (B) The PCR results for genomic DNA from E13.5 cerebella using specific primers (indicated in panel A) showed Cre-mediated in vivo recombination activities of the different targeted conditional Smad alleles in the cerebellum. (C) Western blot analysis detecting the expression of phospho-Smad1/5/8 (P-Smad1/5/8), P-Smad1/5, and Smad4 in the E13.5 cerebellum, with the genotype annotated, showing the loss of Smad protein in the corresponding mutant cerebella. (D and E) Immunofluorescence staining detecting phospho-Smad1/5 in E13.5 cerebellar sagittal sections from control (Smad1fx/fx Smad5fx/fx) (D) and Smad1/5 mutant (En1Cre/+ Smad1fx/fx Smad5fx/fx) (E) cells. Phospho-Smad1/5 expression was detected in the ARL (arrows) and parts of the ventricular zone (arrowheads) of control E13.5 cerebellum but not in the Smad1/5 mutant cerebellum. (F and G) Immunofluorescence staining detecting Smad4 on E13.5 cerebellar sagital sections from control (Smad4fx/+) (F) and Smad4 mutant (En1Cre/+ Smad4Δ/fx) (G) cells. Smad4 expression was detected in the E13.5 whole cerebellum, including the ARL (arrows) and parts of the ventricular zone (arrowheads) of the control but not in the Smad4 mutant cerebellum. Scale bar, 100 μm.
Cerebellar hypoplasia and a reduced population of granule cells in the Smad1/5 mutant.
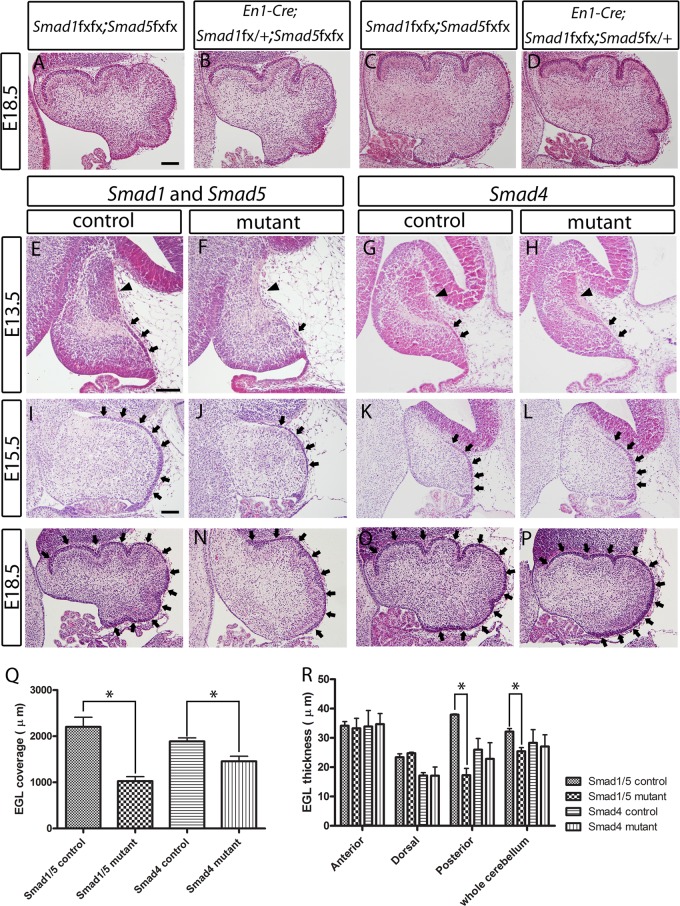
Conditional inactivation of either Smad1 or Smad5 alone in the cerebellum (En1Cre/+ Smad1fx/fx Smad5fx/+ or En1Cre/+ Smad1fx/+ Smad5fx/fx) did not result in abnormalities in cerebellum development (Fig. 2A to D), and both of these single conditional mutant mice were fertile with a normal life span. However, the double conditional inactivation of both Smad1 and Smad5 resulted in severe cerebellar hypoplasia with reduced foliation at E18.5 (Fig. 2M and N). The length of coverage and thickness of the EGL of the Smad1/5 mutants were reduced during embryonic development (E13.5 to E18.5) compared with controls at the corresponding stages (Fig. 2E, F, I, J, M, N, Q, and R). In addition, the NTZ was absent in the Smad1/5 mutant cerebella at E13.5 (Fig. 2E and F).
Fig 2.
Histological analyses of the developing cerebella of Smad1/5 double-conditional-knockout mutant mice and Smad4 conditional-knockout mice. (A to D) Hematoxylin and eosin (H&E)-stained sagittal sections showed no defect and significant difference between E18.5 cerebella of the no-Cre control (Smad1fx/fx Smad5fx/fx) (A and C), Smad1 single-conditional-knockout mice (En1Cre/+ Smad1fx/+ Smad5fx/fx) (B), and Smad5 single-conditional-knockout mice (En1Cre/+ Smad1fx/fx Smad5fx+x) (D). (E and F) H&E-stained sagittal sections revealed the loss of NTZ (arrowheads) and the reduced EGL (arrows) in the E13.5 cerebella of the Smad1/5 mutant compared with the control. (I, J, M, and N) H&E-stained sagittal sections revealed the reduced EGL (arrows) in the E15.5 and E18.5 cerebella of the Smad1/5 mutant compared with the control. (G, H, K, L, O, and P) H&E-stained sagittal sections showed normal NTZ (arrowheads) and EGL (arrows) in the cerebellum of the Smad4 mutant at E13.5, E15.5, and E18.5. (Q) Measurement of the length of cerebellar EGL coverage. EGL coverage was significantly reduced by 50% and 20% in the Smad1/5 mutant and the Smad4 mutant, respectively (*, P < 0.05). (R) Measurement of EGL thickness. Significant reductions of EGL thickness were detected in the posterior region (by 50%) and the whole cerebellum (by 20%) of the Smad1/5 mutant (*, P < 0.05). No significant reduction of EGL thickness was detected in the anterior region and dorsal region of the Smad1/5 mutant cerebellum. No significant difference in EGL thickness was detected in the anterior, dorsal, or posterior region or whole cerebellum between the Smad4 mutant and the control. The error bars indicate standard deviations. Scale bars = 100 μm.
Interestingly, the Smad4 conditional mutants showed very mild cerebellar morphological abnormalities during embryonic development compared with the Smad1/5 double mutants. The EGL and NTZ were comparable between the Smad4 mutants and controls (Fig. 2G, H, K, L, O, and P). Histologically, no observable phenotype was detected in the Smad4 mutants except a mild reduction in cerebellum size and EGL area at E18.5 (Fig. 2O, P, Q, and R).
Specification of the ARL progenitors requires Smad1/5 but not Smad4.
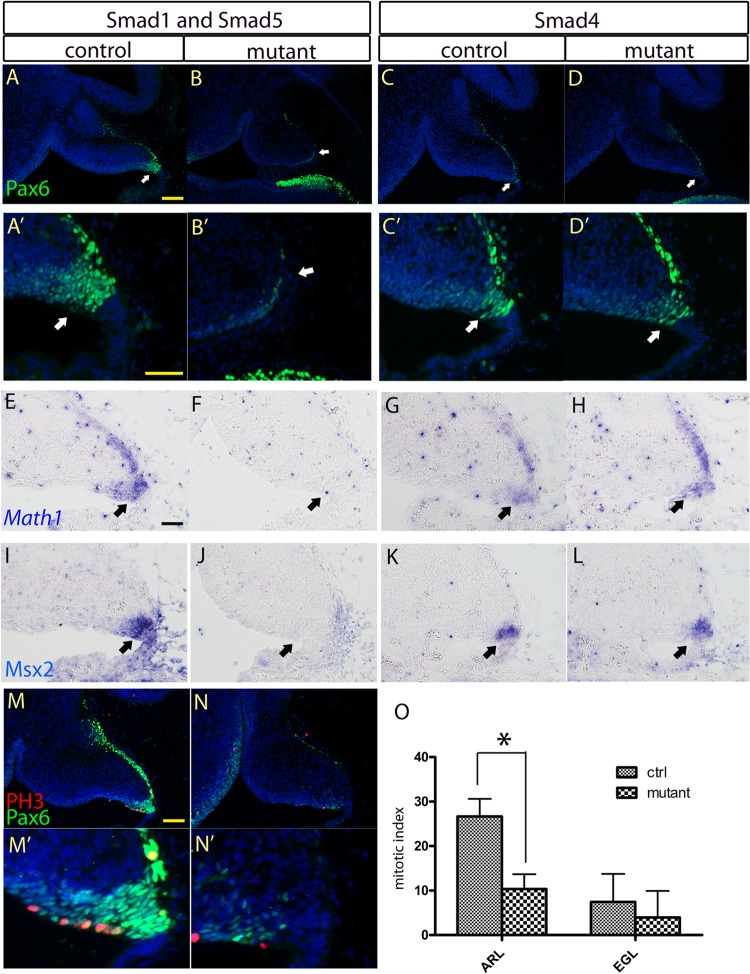
To determine whether the morphological alterations in the EGL and NTZ of the Smad1/5 mutant cerebellum were due to defects in the specification program of the progenitor cells in the ARL, we examined the expression of two transcription factors, Pax6 and Atoh1, important for the ARL specification program (46, 47). Pax6 and Atoh1 expression was detected by immunostaining and in situ hybridization, respectively. Both Pax6 and Atoh1 were not expressed in the ARL of the Smad1/5 mutants at E13.5 (Fig. 3A, B, A′, B′, E, and F), indicating that there were defects in the ARL specification program in the absence of canonical BMP Smad signaling. In contrast, Pax6 and Atoh1 were detected in the ARL of the Smad4 mutants (Fig. 3C, D, C′, D′, G, and H). We next investigated whether canonical BMP Smad signaling was activated in the ARL of the Smad4 mutants. The expression of a well-known BMP downstream target, Msx2, was examined (48, 49). Msx2 expression similar to that of controls was detected in the ARL of the Smad4 mutant (Fig. 3K and L), whereas there was loss of Msx2 expression in the Smad1/5 mutant ARL (Fig. 3I and J).
Fig 3.
Investigation of the specification program and cell proliferation of the progenitor cells in the ARL. (A, B, E, and F) Immunostaining and in situ hybridization of cerebellar sagittal sections revealed the loss of Pax6 and Atoh1 expression, respectively, in the ARL (arrows) of the E13.5 Smad1/5 mutant cerebellum compared with the control. (C, D, G, and H) Expression of Pax6 and Atoh1 was detected in the ARL (arrows) of the E13.5 Smad4 mutant cerebellum using immunostaining and in situ hybridization, respectively. (A′ to D′) Magnified views of the ARLs in panels A to D. (I to L) In situ hybridization of E13.5 cerebellar sagittal sections detecting Msx2 expression indicated the loss of Msx2 signal and hence canonical BMP signaling activities in the ARL (arrows) of the Smad1/5 mutant (J), while Msx2 expression was maintained in the Smad4 mutant (L). (M, M′, N, and N′) Co-immunofluorescence staining of Pax6 and phospho-histone H3 (PH3) in sagittal sections of E13.5 cerebella showed the cell-proliferating activities of the ARL from the control (M and M′) and Smad1/5 mutant (N and N′). (M′ and N′) Magnified views of the ARLs in panels M and N. (O) Cell proliferation of the ARL from the E13.5 Smad1/5 mutant showed significant reduction by 61% compared with the control (*, P < 0.005). The error bars indicate standard deviations. Scale bars: 100 μm (A and M) and 50 μm (A′ and E).
To investigate whether the reduction of the granule cell population in the Smad1/5 mutants was caused by reduced cell proliferation in the ARL, immunostaining of phospho-histone H3 was performed. There was an approximately 60% reduction in phospho-histone H3-positive cells in the Smad1/5 mutant ARL at E13.5 compared with controls (P = 0.0027) (Fig. 3M to O). Cell death in the ARL was examined by immunostaining of cleaved caspase 3, a hallmark of apoptotic cells. There were only very few apoptotic cells in the ARLs of both the Smad1/5 mutants and controls. Thus, no increase in apoptosis was observed in the Smad1/5 mutant (data not shown).
Smad1/5 and Smad4 are not required for the differentiation and maturation of granule cell precursors in the EGL.
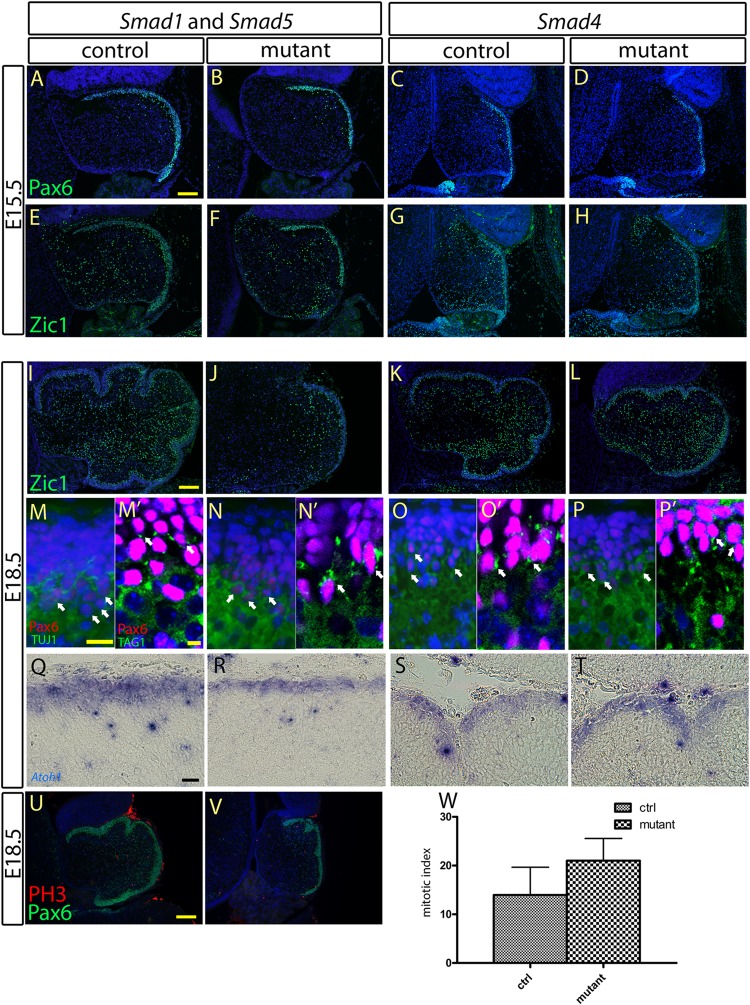
After their specification from the ARL, the development of cerebellar granule neurons involves many subsequent steps, including proliferation, differentiation, and maturation of granule cell precursors within the EGL (9, 13, 14, 50). We asked whether Smad1/5 and Smad4 are required for the differentiation and maturation of the granule cell precursors in the EGL of the cerebellum. We analyzed the expression of Pax6 and Atoh1, which are involved in early granule cell differentiation and EGL development (46, 47). At E15.5 and E18.5, Pax6- and Atoh1-positive cell populations could be clearly detected in the EGL of the Smad1/5 mutants and Smad4 mutants (Fig. 4A to D, M to P, and Q to T). In addition, expression of Zic1, another marker of granule cells that plays an important role in granule cell development (28, 51), was also detected in the EGLs of E15.5 and E18.5 Smad1/5 mutants and Smad4 mutants (Fig. 4E to H and I to L). Furthermore, the expression of TAG-1, a marker of granule cell maturation (13, 50), was expressed normally in the E18.5 granule cells of the Smad1/5 mutants and Smad4 mutants (Fig. 4M′ to P′). To confirm that granule neurons were postmitotic, coimmunostaining of Pax6 and TUJ1 was performed to label granule neurons and postmitotic neurons, respectively (52). Coexpression of Pax6 and TUJ1 was detected in the EGLs of Smad1/5 mutants and Smad4 mutants, indicating the normal maturation of granule cells (Fig. 4M to P). Because there was a severe reduction in the granule cell population in the Smad1/5 mutants, quantitative analysis of phospho-histone H3-positive cells was performed to detect if alterations in cell proliferation were occurring in the Smad1/5 mutant EGL at E18.5. No significant change in the number of phospho-histone H3-positive cells in the EGL at E18.5 were observed between the Smad1/5 mutants and controls (Fig. 4U to W). Altogether, these results suggest that Smad1/5 or Smad4 signaling is not required for the differentiation and maturation of granule cell precursors in the EGL during embryonic cerebellum development.
Fig 4.
Characterization of the specification, differentiation, and maturation processes of the granule cell precursors at the EGL. (A to D, M to P, and Q to T) Immunostaining and in situ hybridization of the cerebellar sagittal sections at E15.5 and E18.5 showed normal expression of Pax6 and Atoh1, respectively, at the EGLs of the Smad1/5 mutant and the Smad4 mutant, indicating normal specification of the granule cell precursors at the EGL. (E to L) Immunostaining of Zic1 revealed no detectable abnormality in the differentiation process of granule cell precursors at the EGLs of the Smad1/5 mutant and the Smad4 mutant. Normal expression of Zic1 was detected in the cerebellar sagittal sections from both the controls and mutants at E15.5 (E to H) and E18.5 (I to L). (M to P′) Co-immunofluorescence staining of Pax6 with TUJ1 (M to P) and of Pax6 with TAG1 (M′ to P′) in the EGL sagittal sections of the controls, the Smad1/5 mutant, and the Smad4 mutant E18.5 cerebella showed coexpression of Pax6 with TUJ1 and TAG1 in granule cells, indicating normal maturation of granule cells from EGLs in both the Smad1/5 mutant and the Smad4 mutant. (U and V) Co-immunofluorescence staining of Pax6 and phospho-histone H3 (PH3) in sagittal sections of the control and Smad1/5 mutant E18.5 cerebella showed the cell-proliferating activity of the EGL. (W) No significant difference in cell proliferation of the EGL was detected between the control and Smad1/5 mutant at E18.5 (P = 0.0854). The error bars indicate standard deviations. Scale bars: 100 μm (A and I), 20 μm (M and Q), 5 μm (M′), and 150 μm (U).
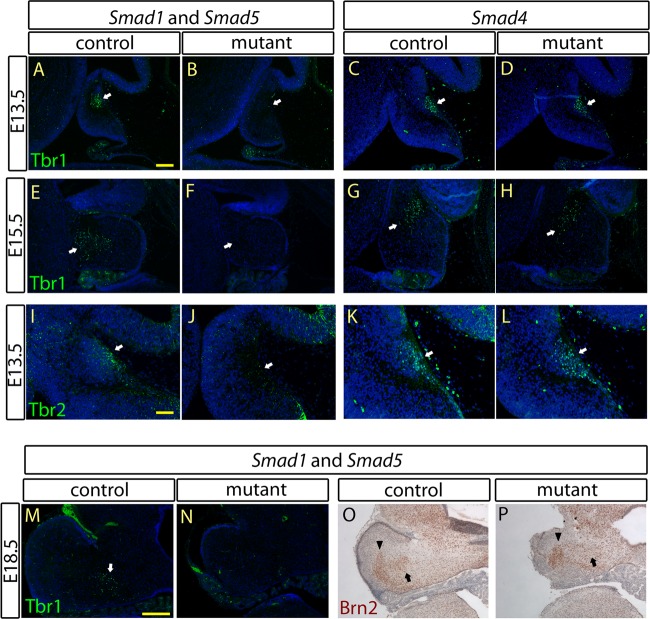
Impaired development of the nuclear transitory zone and deep cerebellar nuclei in the Smad1/5 mutant but not in the Smad4 mutant.
Progenitor cells originating from the ARL contribute to the formation of the NTZ from E13.5, which then contribute to the DCN (8–10). Because the specification program in the ARL was affected in the Smad1/5 mutant, we investigated whether there were any defects in NTZ and DCN development in the absence of Smad1/5 or Smad4 signaling. In addition to the histological studies on the loss of the NTZ observed in the Smad1/5 mutant cerebella (Fig. 2E and F), immunostaining for Tbr1 and Tbr2, markers of the NTZ at E13.5 (9, 10), was performed. Loss of both Tbr1 and Tbr2 expression was observed in the Smad1/5 mutants at E13.5 (Fig. 5A, B, I, and J). Expression of Tbr1 at E15.5, which marks the downwardly migrating NTZ (10, 53), was also lost in the Smad1/5 mutants compared with controls (Fig. 5E and F). Defects in NTZ development of the Smad1/5 mutants prompted us to investigate DCN development. The expression domain of Tbr1 in coronal cerebellar sections at E18.5, a molecular marker for the fastigial nuclei (FN) (10), was lost in the Smad1/5 mutants compared to controls (Fig. 5M and N). In addition, other subtypes of the DCN, including interpositus nuclei (IN) and dentate nuclei (DN), which arise from the ARL and ventricular zone, respectively, were investigated (10, 11). We performed immunostaining for Brn2, which is a molecular marker of IN and DN (10). Brn2 expression was absent in the IN but detected in the DN of the Smad1/5 mutant cerebella at E18.5 (Fig. 5O and P). Together, these results suggest that Smad1/5 signaling is required for the neuronal specification program in the ARL but may not be required for the specification program in the ventricular zone. In contrast, normal expression of Tbr1 and Brn2 at E13.5 and Tbr1 at E15.5 was detected in the Smad4 mutants (Fig. 5C, D, G, H, K, and L). These results further indicate that there are no detectable defects in the generation and migration of the NTZ in the Smad4 mutants.
Fig 5.
Defects of NTZ and DCN development were observed in the Smad1/5 mutant but not in the Smad4 mutant. Immunostaining of Tbr1 (A to D) and Tbr2 (I to L) in the cerebellar sagittal sections at E13.5 revealed loss of the NTZ (arrows) in Smad1/5 mutants but normal formation of the NTZ (arrows) in Smad4 mutants compared to the controls. (E to H) Immunostaining of Tbr1 in the E15.5 cerebellar sagittal sections showed loss of the NTZ (arrows) in the Smad1/5 mutant at the stage of NTZ downward migration, but normal NTZ downward migration was observed in the Smad4 mutant. (M and N) Immunostaining of Tbr1 in the E18.5 cerebellar coronal sections showed loss of FN (arrow) in the Smad1/5 mutant cerebellum, indicating a defect of DCN development in the Smad1/5 mutant. (O and P) Immunostaining of Brn2 in the E18.5 cerebellar coronal sections showed loss of IN (arrow) but the presence of DN (arrowhead) in the Smad1/5 mutant but the presence of both DN (arrowhead) and IN (arrow) in the control. Scale bars: 100 μm (A), 50 μm (I), and 200 μm (M).
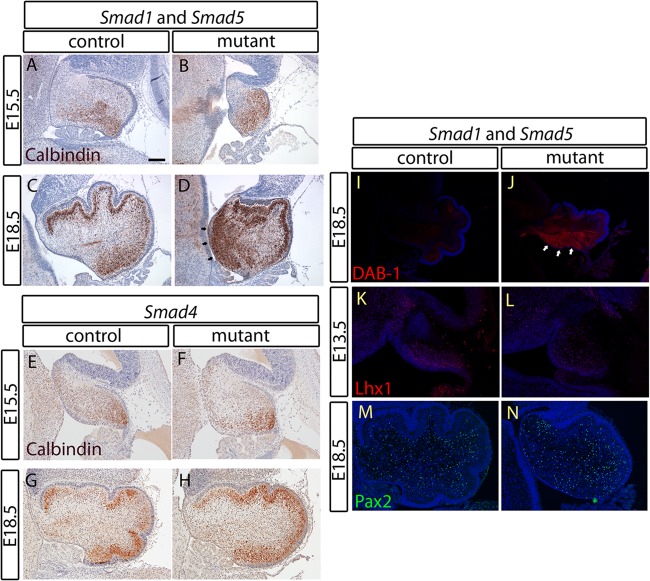
Purkinje cell migration is affected in the Smad1/5 mutant cerebella.
The Purkinje cell is another major neuronal type in the cerebellar cortex, and its migration depends on the reelin signal secreted by granule cells (54, 55). To determine whether the inactivation of Smad1/5 in the cerebellum would affect Purkinje cell migration, immunostaining of calbindin, a marker of Purkinje cells in the cerebellum (6, 17), was performed. Purkinje cells should migrate to the Purkinje cell plate just under the EGL during embryonic development; however, in the Smad1/5 double-mutant cerebellum, a large population of the Purkinje cells remained in the region near the ventricular zone (Fig. 6A to D). On the other hand, ectopically located Purkinje cells were not detected in the Smad4 mutant (Fig. 6E to H). The mislocated Purkinje cells in the Smad1/5 mutants could be due to defective migration caused by changes in the reelin signaling pathway. Therefore, we performed immunostaining for Dab-1, a downstream mediator of the reelin signaling pathway (56). There was an obvious upregulation of Dab-1 in the Smad1/5 mutant cerebella, particularly in the Purkinje cells that remained near the ventricular zone (Fig. 6I and J). This suggests that the amount of secreted reelin in the Smad1/5 mutant cerebellum was reduced and, thus, Dab-1 expression was upregulated due to a feedback mechanism (56, 57). In addition, the expression of Lhx1 and Pax2 was investigated to determine if there was a defect in the specification and differentiation of Purkinje and GABAergic neuronal cells, respectively, arising from the ventricular zone in the Smad1/5 mutants (6, 11, 58–60). Lhx1 and Pax2 expression levels were comparable between the Smad1/5 mutant and control cerebella at E13.5 and E18.5 (Fig. 6K to N). These results suggest that the specification and generation of neurons arising from the ventricular zone were not affected in the absence of Smad1/5 signaling.
Fig 6.
A migration defect of Purkinje neurons in the cerebellum of the Smad1/5 mutant is related to reelin deficiency. (A to H) Immunostaining of calbindin in cerebellar sagittal sections at E15.5 and E18.5 from both the Smad1/5 mutant and the Smad4 mutant with their respective controls showed a migration defect in a population of Purkinje neurons of the Smad1/5 mutant that remains in the region near the VZ (D, arrows). Normal Purkinje cell migration was observed in the Smad4 mutant. (I and J) Immunostaining of DAB-1 in the E18.5 cerebellar sagittal sections showed overexpression of DAB-1 in the Purkinje cell population remaining near the VZ (arrows), indicating a defect in cerebellar reelin signaling of the Smad1/5 mutant. (K and L) Immunostaining of Lhx1 in E13.5 cerebellar sagittal sections showed a normal Lhx1 expression domain in the VZ of the Smad1/5 mutant, indicating a normal specification process of the VZ in the Smad1/5 mutant cerebellum. (M and N) Normal GABAergic neuron generation from the VZ of the Smad1/5 mutant shown by immunostaining of Pax2 in the E18.5 cerebellar sagittal sections. Scale bar, 100 μm.
DISCUSSION
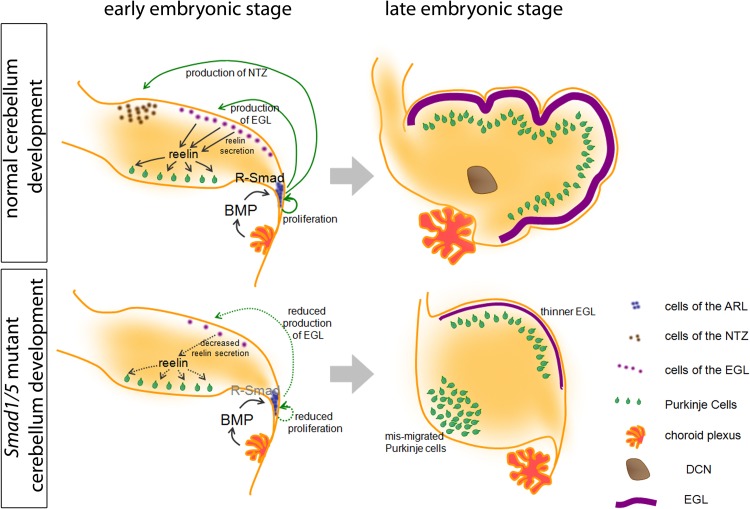
In this study, we investigated the role and usage of R-Smads and Co-Smad in BMP-mediated cerebellum development using a genetic approach. Through the use of conditional inactivation of different R-Smads and the Co-Smad Smad4, we have shown that canonical BMP Smad signaling acts through both Smad1 and Smad5 in a functionally redundant manner during cerebellar development. This Smad1/5 signaling is important for the specification of neuronal progenitor cells within the ARL but is not required for the differentiation or maturation of cerebellar granule cell precursors in the EGL, as illustrated in Fig. 7. Surprisingly, our data suggest that Smad4 is not essential for these processes.
Fig 7.
Schematic diagram showing the developmental alterations in the Smad1/5 mutant. The development of the normal and mutant cerebella is shown. In the normal cerebellum, the BMP ligands are secreted from different origins, including the choroid plexus. The BMP signals regulate the proliferation of ARL progenitors and the specification of ARL progenitors into NTZ and EGL populations. The granule cells of the EGL then secrete reelin, which signals Purkinje cell migration. At the end, the proper EGL, DCN, and Purkinje cell plate are formed. In the Smad1/5 mutant cerebellum, the ARL cannot respond to BMPs due to the loss of R-Smad, which results in the reduced proliferation of ARL progenitors, loss of the NTZ, and a reduced number of granule cell precursors in the EGL. The reduced EGL then leads to a lower level of reelin and results in a Purkinje cell migration defect. At the end, loss of DCN, a thinner EGL, and mismigration of Purkinje cells in the Smad1/5 mutant cerebellum resulted.
Co-Smad independence of the canonical BMP Smad signaling during embryonic cerebellum development.
We showed that Smad4 conditional-knockout mutant mice developed only a very mild cerebellar phenotype compared to that of the Smad1/5 mutants. The normal expression of Msx2 in the ARL of the Smad4 mutant clearly demonstrates that R-Smads are still functional in the absence of Co-Smad. Apparently, the R-Smad molecules accumulate in the nucleus independently of Smad4 to regulate the transcription of target genes. These results challenge the current model of the canonical BMP Smad signaling, which considers Smad4 a requisite mediator of this pathway by forming an obligate heterotrimer with R-Smad (23–25).
In fact, this Co-Smad independence is not restricted to nervous system development. Inactivation of Smad1/5 during lens development causes cell death, but inactivation of Smad4 does not (61). In addition, Smad1/5 are essential for bone development (26), but inactivation of Smad4 only results in mild bone defects (62). Furthermore, initial heart specification requires R-Smad to induce Nkx2.5 expression (63, 64); however, Nkx2.5 expression is not abolished when Smad4 is inactivated (35). This implies cell- or tissue-specific Co-Smad independence in canonical BMP signaling. Moreover, a similar phenomenon is found in Drosophila. Mutants for Mad (encoding the orthologue of mammalian R-Smad) have more severe defects than Medea (encoding the orthologue of mammalian Co-Smad) mutants (65). These findings, together with our current results, contrast with the current perception that Co-Smad is essential for canonical BMP signaling. In the Drosophila Medea mutant, the regions of the wing imaginal disc that normally receive low Dpp signal are most severely affected (65, 66). This suggests that Co-Smad dependence increases with a decrease in BMP signal. We propose that in some cellular processes that depend on low levels of BMP signaling, Co-Smad is essential for the R-Smads to maximize their full functions, probably by forming an obligate heterotrimer with R-Smads. However, in other cellular processes that have high levels of BMP signaling, abundant phosphorylated R-Smads can be formed. Under these conditions, R-Smads can still activate their downstream targets. In this situation, Co-Smad may be less important. Together, our study and previous findings add to the understanding of the role of R- and Co-Smad in BMP signaling. In contrast to our findings, a previous report showed abnormal cerebellum development and severe ARL specification defects when Smad4 was inactivated using the same En1-Cre mouse line as ours but with a different Smad4 conditional allele (27). However, in another report using Nestin-Cre to delete Smad4 in the nervous system, there were only mild cerebellar phenotypes resembling those in our study (29). These differences in phenotypes may be due to variation in the genetic backgrounds used in the different studies. Nevertheless, it is interesting to find that the dependency of Smad4, which was initially thought to be the central mediator of the canonical BMP signaling pathway (23–25), could vary between different genetic backgrounds. Thus, our finding provides new insight into the role and use of Co-Smad in the BMP signaling pathway, which was previously based on various biochemical studies using different cell line models.
Smad1 and Smad5 are functionally redundant for cerebellum development.
Although the involvement of BMP signaling in embryonic cerebellum development is known (16–18), the role and use of R-Smad are unclear. Previous dissociated cell culture and organotypic culture assays suggested the involvement of Smad1 (67) and Smad5 (68) use, respectively. However, severe cerebellar defects were observed only in our Smad1/5 mutants, consistent with the results of Bmpr1a and Bmpr1b inactivation (28). Hence, we suggest that both Smad1 and Smad5 indeed act in a functionally redundant manner during cerebellum development. In addition, this study provides further evidence to support the idea that functional redundancy between Smad1 and Smad5 may be a rather common phenomenon and is not restricted to bone development (26), eye development (61), limb development (44), and suppressing metastatic tumor development in gonads (69) but is also involved in the development of the central nervous system.
Although we have identified the R-Smads involved during cerebellar development, many details of the canonical BMP Smad signaling pathway remain unclear. The type II receptor and the coreceptor involved in this developmental process are not clearly identified (70). WNT signaling and fibroblast growth factor (FGF) signaling can regulate R-Smad degradation through the phosphorylation of its linker region (71). However, the details of the control of R-Smad degradation involved in cerebellum development remain unclear. More importantly, as we have demonstrated Co-Smad independence, the translocation of R-Smad into the nucleus may involve other proteins that are largely unknown (72). Further investigation of the canonical BMP Smad signaling pathway involvement in the cerebellum ARL specification program may allow better understanding of the Co-Smad-independent signaling pathway.
Co-Smad-independent canonical BMP Smad signaling is specifically required for the specification program of progenitor cells within the anterior rhombic lip.
To further understand the Co-Smad-independent canonical BMP Smad signaling, we precisely dissected the requirement for it in specific cellular processes. We have shown that this signaling is essential for the specification of the progenitors in the ARL. In the absence of Smad1/5, the ARL at E13.5 has shown reduced cell proliferation and lacked expression of Pax6 and Atoh1, which are essential for the specification of ARL (46, 47). The postmitotic NTZ originates from the ARL (8, 9, 12), and as a result of Smad1/5 inactivation, the NTZ and two subsequent DCN subtypes derived from NTZ (FN and IN) are lost at E13.5 and E18.5, respectively, in the Smad1/5 double mutant. Our results also reiterate the genetic analyses of the Tbr1 knockout mice that have DCN morphogenesis defects (53, 73).
On the other hand, our data show that Co-Smad-independent canonical BMP Smad signaling is important for the regulation of the granule cell precursor population derived from the ARL but is not essential for the generation of granule cells from the ARL. The reduced population of granule cell precursors in the EGL of the Smad1/5 mutant may be due to the defective specification program and reduced cell proliferation of the ARL in the absence of Smad1/5 signaling. Nevertheless, the remaining granule cell precursors that derived from the ARL can still proliferate within the EGL at the later stage, even in the absence of Smad1/5 signaling. Despite the reduced population of granule cell precursors in the EGL of the Smad1/5 mutants, the granule cell precursors undergo normal proliferation, early differentiation, and maturation. Taken together, there may be a distinct temporal and spatial requirement for canonical BMP Smad signaling during the granule cell development program within the ARL and EGL.
Since En1-Cre is not specific to the cerebellum, other possible defects in glial cell generation in the midbrain (74) or the early neural tube specification (75) may occur in the Smad1/5 mutants. These events may secondarily affect cerebellar development. However, the ARL abnormality is very likely to be the primary cause of the cerebellar defects observed in the Smad1/5 mutants, as those defects are related to NTZ and EGL development.
Our data also provide further insights into the molecular regulation of granule cell development by Atoh1. BMP is known to regulate Atoh1 expression during cerebellar granule cell development (16, 18, 47). We have shown that Atoh1 is not expressed in the Smad1/5 mutant ARL but is still expressed in the mutant EGL. Thus, Atoh1 expression in the embryonic EGL may either be independent of BMP signaling or depend on noncanonical BMP signaling. This implies the differential enhancer usage of Atoh1. Further investigations of the BMP regulation of Atoh1 expression in different cerebellar germinating zones may be required.
The Purkinje cell population is limited by the number of granule cells.
Generation of GABAergic neurons and the specification process of the ventricular zone are not affected but Purkinje cell migration is disorganized in the Smad1/5 mutant. The migration defect is most probably caused by the reduction in granule cell numbers (55, 56), which leads to a decreased amount of reelin molecule secreted (54). This explains the upregulation of Dab-1 expression in those nonmigrating Purkinje cells and the variability in migration and alignment between different individual Purkinje cells in the Smad1/5 mutants. Therefore, precise regulation between the numbers of Purkinje cells and granule cells is essential for proper cerebellar development and function. Importantly, reduced granule cell numbers limit the number of functional Purkinje cells because the Purkinje cells generated cannot migrate properly when reelin signal is insufficient. As Purkinje cells send the sole output of the cerebellar cortex, its distribution and population are directly related to cerebellar function (1, 6). Thus, this probably implies that during the evolution of cerebellar complexity among chordates (11), increasing the number of granule cells or the production of reelin molecules is a critical step for the expansion of the Purkinje cell population and thus the size or complexity of the cerebellum.
ACKNOWLEDGMENTS
We thank Richard R. Behringer (University of Texas M. D. Anderson Cancer Center, Houston, TX) for providing the Smad1/5 conditional mouse line, helpful advice, and comments on the manuscript; Elizabeth J. Robertson (University of Oxford, Oxford, United Kingdom) for providing the Smad4 conditional mutant mouse line; Jane E. Johnson (University of Texas Southwestern Medical Center, Dallas, TX) for providing the Math1 probe; and Sze-Nee Lim, Yuk-Lau Wong, and Wang-Chi Lau for technical assistance.
This research was supported by the General Research Fund of the Research Grants Council of Hong Kong Special Administrative Region, China (project no. CUHK 466708) and Focused Investments Scheme, Scheme B, of the Chinese University of Hong Kong (BL08650).
Footnotes
Published ahead of print 4 March 2013
REFERENCES
- 1. Millen KJ, Gleeson JG. 2008. Cerebellar development and disease. Curr. Opin. Neurobiol. 18:12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Villanueva R. 2012. The cerebellum and neuropsychiatric disorders. Psychiatry Res. 198:527–532 [DOI] [PubMed] [Google Scholar]
- 3. ten Donkelaar HJ, Lammens M, Wesseling P, Thijssen HO, Renier WO. 2003. Development and developmental disorders of the human cerebellum. J. Neurol. 250:1025–1036 [DOI] [PubMed] [Google Scholar]
- 4. Garel C, Fallet-Bianco C, Guibaud L. 2011. The fetal cerebellum: development and common malformations. J. Child Neurol. 26:1483–1492 [DOI] [PubMed] [Google Scholar]
- 5. Dastjerdi FV, Consalez GG, Hawkes R. 2012. Pattern formation during development of the embryonic cerebellum. Front. Neuroanat. 6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao Y, Kwan KM, Mailloux CM, Lee WK, Grinberg A, Wurst W, Behringer RR, Westphal H. 2007. LIM-homeodomain proteins Lhx1 and Lhx5, and their cofactor Ldb1, control Purkinje cell differentiation in the developing cerebellum. Proc. Natl. Acad. Sci. U. S. A. 104:13182–13186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Glasgow SM, Henke RM, MacDonald RJ, Wright CV, Johnson JE. 2005. Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development 132:5461–5469 [DOI] [PubMed] [Google Scholar]
- 8. Wang VY, Rose MF, Zoghbi HY. 2005. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron 48:31–43 [DOI] [PubMed] [Google Scholar]
- 9. Machold R, Fishell G. 2005. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron 48:17–24 [DOI] [PubMed] [Google Scholar]
- 10. Fink AJ, Englund C, Daza RA, Pham D, Lau C, Nivison M, Kowalczyk T, Hevner RF. 2006. Development of the deep cerebellar nuclei: transcription factors and cell migration from the rhombic lip. J. Neurosci. 26:3066–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hashimoto M, Hibi M. 2012. Development and evolution of cerebellar neural circuits. Dev. Growth Differ. 54:373–389 [DOI] [PubMed] [Google Scholar]
- 12. Machold R, Klein C, Fishell G. 2011. Genes expressed in Atoh1 neuronal lineages arising from the r1/isthmus rhombic lip. Gene Expr. Patterns 11:349–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Behesti H, Marino S. 2009. Cerebellar granule cells: insights into proliferation, differentiation, and role in medulloblastoma pathogenesis. Int. J. Biochem. Cell Biol. 41:435–445 [DOI] [PubMed] [Google Scholar]
- 14. Gao WO, Heintz N, Hatten ME. 1991. Cerebellar granule cell neurogenesis is regulated by cell-cell interactions in vitro. Neuron 6:705–715 [DOI] [PubMed] [Google Scholar]
- 15. Marr D. 1969. A theory of cerebellar cortex. J. Physiol. 202:437–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Salero E, Hatten ME. 2007. Differentiation of ES cells into cerebellar neurons. Proc. Natl. Acad. Sci. U. S. A. 104:2997–3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Su HL, Muguruma K, Matsuo-Takasaki M, Kengaku M, Watanabe K, Sasai Y. 2006. Generation of cerebellar neuron precursors from embryonic stem cells. Dev. Biol. 290:287–296 [DOI] [PubMed] [Google Scholar]
- 18. Alder J, Lee KJ, Jessell TM, Hatten ME. 1999. Generation of cerebellar granule neurons in vivo by transplantation of BMP-treated neural progenitor cells. Nat. Neurosci. 2:535–540 [DOI] [PubMed] [Google Scholar]
- 19. Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. 2003. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425:917–925 [DOI] [PubMed] [Google Scholar]
- 20. Krizhanovsky V, Ben-Arie N. 2006. A novel role for the choroid plexus in BMP-mediated inhibition of differentiation of cerebellar neural progenitors. Mech. Dev. 123:67–75 [DOI] [PubMed] [Google Scholar]
- 21. Lee KJ, Mendelsohn M, Jessell TM. 1998. Neuronal patterning by BMPs: a requirement for GDF7 in the generation of a discrete class of commissural interneurons in the mouse spinal cord. Genes Dev. 12:3394–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thomadakis G, Ramoshebi LN, Crooks J, Rueger DC, Ripamonti U. 1999. Immunolocalization of Bone Morphogenetic Protein-2 and -3 and Osteogenic Protein-1 during murine tooth root morphogenesis and in other craniofacial structures. Eur. J. Oral Sci. 107:368–377 [DOI] [PubMed] [Google Scholar]
- 23. Attisano L, Lee-Hoeflich ST. 2001. The Smads. Genome Biol. 2:REVIEWS3010 doi:10.1186/gb-2001-2-8-reviews3010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miyazono K, Maeda S, Imamura T. 2005. BMP receptor signaling: transcriptional targets, regulation of signals, and signaling cross-talk. Cytokine Growth Factor Rev. 16:251–263 [DOI] [PubMed] [Google Scholar]
- 25. Sieber C, Kopf J, Hiepen C, Knaus P. 2009. Recent advances in BMP receptor signaling. Cytokine Growth Factor Rev. 20:343–355 [DOI] [PubMed] [Google Scholar]
- 26. Retting KN, Song B, Yoon BS, Lyons KM. 2009. BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development 136:1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fernandes M, Antoine M, Hebert JM. 2012. SMAD4 is essential for generating subtypes of neurons during cerebellar development. Dev. Biol. 365:82–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qin L, Wine-Lee L, Ahn KJ, Crenshaw EB., III 2006. Genetic analyses demonstrate that bone morphogenetic protein signaling is required for embryonic cerebellar development. J. Neurosci. 26:1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhou YX, Zhao M, Li D, Shimazu K, Sakata K, Deng CX, Lu B. 2003. Cerebellar deficits and hyperactivity in mice lacking Smad4. J. Biol. Chem. 278:42313–42320 [DOI] [PubMed] [Google Scholar]
- 30. Makkar P, Metpally RP, Sangadala S, Reddy BV. 2009. Modeling and analysis of MH1 domain of Smads and their interaction with promoter DNA sequence motif. J. Mol. Graph. Model. 27:803–812 [DOI] [PubMed] [Google Scholar]
- 31. Kwan KM. 2002. Conditional alleles in mice: practical considerations for tissue-specific knockouts. Genesis 32:49–62 [DOI] [PubMed] [Google Scholar]
- 32. Nagy A. 2000. Cre recombinase: the universal reagent for genome tailoring. Genesis 26:99–109 [PubMed] [Google Scholar]
- 33. Huang S, Tang B, Usoskin D, Lechleider RJ, Jamin SP, Li C, Anzano MA, Ebendal T, Deng C, Roberts AB. 2002. Conditional knockout of the Smad1 gene. Genesis 32:76–79 [DOI] [PubMed] [Google Scholar]
- 34. Umans L, Vermeire L, Francis A, Chang H, Huylebroeck D, Zwijsen A. 2003. Generation of a floxed allele of Smad5 for cre-mediated conditional knockout in the mouse. Genesis 37:5–11 [DOI] [PubMed] [Google Scholar]
- 35. Chu GC, Dunn NR, Anderson DC, Oxburgh L, Robertson EJ. 2004. Differential requirements for Smad4 in TGFbeta-dependent patterning of the early mouse embryo. Development 131:3501–3512 [DOI] [PubMed] [Google Scholar]
- 36. Kimmel RA, Turnbull DH, Blanquet V, Wurst W, Loomis CA, Joyner AL. 2000. Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation. Genes Dev. 14:1377–1389 [PMC free article] [PubMed] [Google Scholar]
- 37. Yang X, Castilla LH, Xu X, Li C, Gotay J, Weinstein M, Liu PP, Deng CX. 1999. Angiogenesis defects and mesenchymal apoptosis in mice lacking SMAD5. Development 126:1571–1580 [DOI] [PubMed] [Google Scholar]
- 38. Tremblay KD, Dunn NR, Robertson EJ. 2001. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development 128:3609–3621 [DOI] [PubMed] [Google Scholar]
- 39. Sirard C, de la Pompa JL, Elia A, Itie A, Mirtsos C, Cheung A, Hahn S, Wakeham A, Schwartz L, Kern SE, Rossant J, Mak TW. 1998. The tumor suppressor gene Smad4/Dpc4 is required for gastrulation and later for anterior development of the mouse embryo. Genes Dev. 12:107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chi CL, Martinez S, Wurst W, Martin GR. 2003. The isthmic organizer signal FGF8 is required for cell survival in the prospective midbrain and cerebellum. Development 130:2633–2644 [DOI] [PubMed] [Google Scholar]
- 41. Sgaier SK, Millet S, Villanueva MP, Berenshteyn F, Song C, Joyner AL. 2005. Morphogenetic and cellular movements that shape the mouse cerebellum; insights from genetic fate mapping. Neuron 45:27–40 [DOI] [PubMed] [Google Scholar]
- 42. Lewandoski M, Wassarman KM, Martin GR. 1997. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr. Biol. 7:148–151 [DOI] [PubMed] [Google Scholar]
- 43. Lai HC, Klisch TJ, Roberts R, Zoghbi HY, Johnson JE. 2011. In vivo neuronal subtype-specific targets of Atoh1 (Math1) in dorsal spinal cord. J. Neurosci. 31:10859–10871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wong YL, Behringer RR, Kwan KM. 2012. Smad1/Smad5 signaling in limb ectoderm functions redundantly and is required for interdigital programmed cell death. Dev. Biol. 363:247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ellisor D, Koveal D, Hagan N, Brown A, Zervas M. 2009. Comparative analysis of conditional reporter alleles in the developing embryo and embryonic nervous system. Gene Expr. Patterns 9:475–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Engelkamp D, Rashbass P, Seawright A, van Heyningen V. 1999. Role of Pax6 in development of the cerebellar system. Development 126:3585–3596 [DOI] [PubMed] [Google Scholar]
- 47. Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. 1997. Math1 is essential for genesis of cerebellar granule neurons. Nature 390:169–172 [DOI] [PubMed] [Google Scholar]
- 48. Machold RP, Kittell DJ, Fishell GJ. 2007. Antagonism between Notch and bone morphogenetic protein receptor signaling regulates neurogenesis in the cerebellar rhombic lip. Neural Dev. 2:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rodriguez-Carballo E, Ulsamer A, Susperregui AR, Manzanares-Cespedes C, Sanchez-Garcia E, Bartrons R, Rosa JL, Ventura F. 2011. Conserved regulatory motifs in osteogenic gene promoters integrate cooperative effects of canonical Wnt and BMP pathways. J. Bone Miner. Res. 26:718–729 [DOI] [PubMed] [Google Scholar]
- 50. Powell SK, Rivas RJ, Rodriguez-Boulan E, Hatten ME. 1997. Development of polarity in cerebellar granule neurons. J. Neurobiol. 32:223–236 [DOI] [PubMed] [Google Scholar]
- 51. Aruga J, Minowa O, Yaginuma H, Kuno J, Nagai T, Noda T, Mikoshiba K. 1998. Mouse Zic1 is involved in cerebellar development. J. Neurosci. 18:284–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Menezes JR, Luskin MB. 1994. Expression of neuron-specific tubulin defines a novel population in the proliferative layers of the developing telencephalon. J. Neurosci. 14:5399–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bulfone A, Smiga SM, Shimamura K, Peterson A, Puelles L, Rubenstein JL. 1995. T-brain-1: a homolog of Brachyury whose expression defines molecularly distinct domains within the cerebral cortex. Neuron 15:63–78 [DOI] [PubMed] [Google Scholar]
- 54. Goldowitz D, Cushing RC, Laywell E, D'Arcangelo G, Sheldon M, Sweet HO, Davisson M, Steindler D, Curran T. 1997. Cerebellar disorganization characteristic of reeler in scrambler mutant mice despite presence of reelin. J. Neurosci. 17:8767–8777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Miyata T, Ono Y, Okamoto M, Masaoka M, Sakakibara A, Kawaguchi A, Hashimoto M, Ogawa M. 2010. Migration, early axonogenesis, and Reelin-dependent layer-forming behavior of early/posterior-born Purkinje cells in the developing mouse lateral cerebellum. Neural Dev. 5:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rice DS, Sheldon M, D'Arcangelo G, Nakajima K, Goldowitz D, Curran T. 1998. Disabled-1 acts downstream of Reelin in a signaling pathway that controls laminar organization in the mammalian brain. Development 125:3719–3729 [DOI] [PubMed] [Google Scholar]
- 57. Honda T, Kobayashi K, Mikoshiba K, Nakajima K. 2011. Regulation of cortical neuron migration by the Reelin signaling pathway. Neurochem Res. 36:1270–1279 [DOI] [PubMed] [Google Scholar]
- 58. Hori K, Hoshino M. 2012. GABAergic neuron specification in the spinal cord, the cerebellum, and the cochlear nucleus. Neural Plast. 2012:921732 doi:10.1155/2012/921732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Leto K, Bartolini A, Yanagawa Y, Obata K, Magrassi L, Schilling K, Rossi F. 2009. Laminar fate and phenotype specification of cerebellar GABAergic interneurons. J. Neurosci. 29:7079–7091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Leto K, Carletti B, Williams IM, Magrassi L, Rossi F. 2006. Different types of cerebellar GABAergic interneurons originate from a common pool of multipotent progenitor cells. J. Neurosci. 26:11682–11694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rajagopal R, Huang J, Dattilo LK, Kaartinen V, Mishina Y, Deng CX, Umans L, Zwijsen A, Roberts AB, Beebe DC. 2009. The type I BMP receptors, Bmpr1a and Acvr1, activate multiple signaling pathways to regulate lens formation. Dev. Biol. 335:305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang J, Tan X, Li W, Wang Y, Wang J, Cheng X, Yang X. 2005. Smad4 is required for the normal organization of the cartilage growth plate. Dev. Biol. 284:311–322 [DOI] [PubMed] [Google Scholar]
- 63. Liberatore CM, Searcy-Schrick RD, Vincent EB, Yutzey KE. 2002. Nkx-2.5 gene induction in mice is mediated by a Smad consensus regulatory region. Dev. Biol. 244:243–256 [DOI] [PubMed] [Google Scholar]
- 64. Shi Y, Katsev S, Cai C, Evans S. 2000. BMP signaling is required for heart formation in vertebrates. Dev. Biol. 224:226–237 [DOI] [PubMed] [Google Scholar]
- 65. Wisotzkey RG, Mehra A, Sutherland DJ, Dobens LL, Liu X, Dohrmann C, Attisano L, Raftery LA. 1998. Medea is a Drosophila Smad4 homolog that is differentially required to potentiate DPP responses. Development 125:1433–1445 [DOI] [PubMed] [Google Scholar]
- 66. Tendeng C. 2012. Experiments, measurements, and mathematical modeling to decipher time signals in development. Birth Defects Res. C Embryo Today 96:121–131 [DOI] [PubMed] [Google Scholar]
- 67. Angley C, Kumar M, Dinsio KJ, Hall AK, Siegel RE. 2003. Signaling by bone morphogenetic proteins and Smad1 modulates the postnatal differentiation of cerebellar cells. J. Neurosci. 23:260–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rios I, Alvarez-Rodriguez R, Marti E, Pons S. 2004. Bmp2 antagonizes sonic hedgehog-mediated proliferation of cerebellar granule neurones through Smad5 signalling. Development 131:3159–3168 [DOI] [PubMed] [Google Scholar]
- 69. Pangas SA, Li X, Umans L, Zwijsen A, Huylebroeck D, Gutierrez C, Wang D, Martin JF, Jamin SP, Behringer RR, Robertson EJ, Matzuk MM. 2008. Conditional deletion of Smad1 and Smad5 in somatic cells of male and female gonads leads to metastatic tumor development in mice. Mol. Cell. Biol. 28:248–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zakin L, De Robertis EM. 2010. Extracellular regulation of BMP signaling. Curr. Biol. 20:R89–R92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Fuentealba LC, Eivers E, Ikeda A, Hurtado C, Kuroda H, Pera EM, De Robertis EM. 2007. Integrating patterning signals: Wnt/GSK3 regulates the duration of the BMP/Smad1 signal. Cell 131:980–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen X, Xu L. 2010. Specific nucleoporin requirement for Smad nuclear translocation. Mol. Cell. Biol. 30:4022–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bulfone A, Martinez S, Marigo V, Campanella M, Basile A, Quaderi N, Gattuso C, Rubenstein JL, Ballabio A. 1999. Expression pattern of the Tbr2 (Eomesodermin) gene during mouse and chick brain development. Mech. Dev. 84:133–138 [DOI] [PubMed] [Google Scholar]
- 74. Mecklenburg N, Garcia-Lopez R, Puelles E, Sotelo C, Martinez S. 2011. Cerebellar oligodendroglial cells have a mesencephalic origin. Glia 59:1946–1957 [DOI] [PubMed] [Google Scholar]
- 75. Wassef M, Bally-Cuif L, Alvarado-Mallart RM. 1993. Regional specification during cerebellar development. Perspect. Dev. Neurobiol. 1:127–132 [PubMed] [Google Scholar]