Abstract
Conserved from yeast to mammals, phosphorylation of the heptad repeat sequence Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7 in the carboxy-terminal domain (CTD) of the largest RNA polymerase II (RNA Pol II) subunit, RPB1, mediates the enzyme's promoter escape and binding of RNA-processing factors, such as the m7G capping enzymes. The first critical step, Ser5 phosphorylation, is carried out by cyclin-dependent kinase 7 (CDK7), a subunit of the basal transcription factor TFIIH. Many early-diverged protists, such as the lethal human parasite Trypanosoma brucei, however, lack the heptad repeats and, apparently, a CDK7 ortholog. Accordingly, characterization of trypanosome TFIIH did not identify a kinase component. The T. brucei CTD, however, is phosphorylated and essential for transcription. Here we show that silencing the expression of T. brucei cdc2-related kinase 9 (CRK9) leads to a loss of RPB1 phosphorylation. Surprisingly, this event did not impair RNA Pol II transcription or cotranscriptional m7G capping. Instead, we observed that CRK9 silencing led to a block of spliced leader (SL) trans splicing, an essential step in trypanosome mRNA maturation, that was caused by hypomethylation of the SL RNA's unique cap4.
INTRODUCTION
In eukaryotes, RNA polymerase II (Pol II) transcription of protein coding and small nuclear RNA (snRNA) genes is a highly conserved process involving a plethora of transcription and cotranscriptionally active RNA-processing factors. A key regulatory structure in this process is the carboxy-terminal domain (CTD) of the largest RNA Pol II subunit, RPB1. Conserved from some protists to humans, the CTD consists of 5 to 52 repeats of the heptad sequence Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7, whose regulated serine (de)phosphorylation cycle is essential for RNA Pol II function (1–3). RNA Pol II that is recruited to the core promoter has hypophosphorylated heptad repeats. Transcription initiation and RNA Pol II escape from the promoter then essentially depend on phosphorylation of Ser5 by cyclin-dependent kinase 7 (CDK7; the yeast ortholog is termed Kin28), which is a component of the basal transcription factor TFIIH. The same phosphorylation event leads to binding of the m7G capping enzyme to the CTD and cotranscriptional capping of pre-mRNA (4). Further downstream of the transcription initiation site, phosphorylation of Ser2 by CDK9 (Bur1) and CDK12/13 (Ctk1) transform RNA Pol II into an actively elongating enzyme (5–7). Finally, Ser7 phosphorylation appears to be particularly important for transcription of snRNA genes, since it mediates the interaction of the integrator 3′-end RNA processing complex with RNA Pol II. Ser7 is primarily phosphorylated by CDK7 (8–10), although CDK9 (10, 11) and DNA-activated protein kinase (12) may play roles as well.
The repetitive CTD structure, however, is not a universal feature of eukaryotes (13). Notably, protistan organisms, such as amoebozoa, kinetoplastids, trichomonads, and diplomonads, which appear to have diverged very early from other eukaryotic lineages, lack repetitive CTD motifs. Moreover, comparative genomics suggested that these organisms also lack a CDK7 ortholog (13). It therefore remains to be determined whether phosphorylation of nonrepetitive CTDs is important for RNA Pol II-mediated gene expression or whether the essential coordinating function of the canonical CTD evolved later in evolution.
The kinetoplastid Trypanosoma brucei is a lethal, vector-borne parasite of humans and livestock in Saharan Africa. It has emerged as a model organism for deviating gene expression mechanisms found in early-diverged eukaryotes. Its protein coding genes are arranged in long directional tandem arrays that are transcribed polycistronically. The resulting precursor RNAs are processed into individual mRNAs by spliced leader (SL) trans splicing and polyadenylation. The SL is derived from the small nuclear SL RNA and trans spliced onto each and every mRNA (14). The SL is 39 nucleotides (nt) long, pseudouridylated at position 28, and carries the most extensively modified cap found in any eukaryote. The cap structure is m7G(5′)ppp(5′)m6,6AmpAmpCmpm3Ump, and has been termed cap4 (15). Trypanosome RNA Pol II transcribes the protein coding gene arrays and the ∼100 tandem copies of the SL RNA gene (SLRNA) on chromosome 9, whereas other snRNAs in trypanosomes are synthesized by RNA Pol III. SLRNA transcription is monocistronic, initiates at a distinct initiation site, and requires the assembly of a conventional, albeit extremely divergent, preinitiation complex (16–18). The T. brucei genome encodes a full set of RNA Pol II subunits, termed RPB1 to RPB12 (19), and biochemical characterization of the enzyme complex identified all subunits except RPB10 (20–22). In T. brucei, there are two slightly different RPB1 genes (relevant T. brucei gene accession numbers are listed in Table S1 in the supplemental material) that appear to be functionally redundant, since deletion of three RPB1 alleles did not impair trypanosome proliferation in culture (23). All RPB1 homologs possess eight highly conserved regions, termed A to H, and the sequence downstream of region H is generally considered the CTD (1). The conserved regions can be clearly recognized in trypanosome RPB1, defining the CTD from residue 1481 to residue 1765. Despite the lack of the heptad repeat motif, this region is serine rich, phosphorylated, and essential for transcription (24, 25).
Characterization of the trypanosome phosphoproteome revealed 14 phosphorylated RPB1 residues that all reside in the CTD (26), yet the functional significance of CTD phosphorylation in trypanosomes remains unclear. In accordance with the notion that organisms with a noncanonical CTD lack a CDK7 ortholog, biochemical and structural characterization of trypanosome TFIIH, which is a basal factor for SLRNA transcription, revealed a full core complex of seven subunits but no kinase component (17, 27). These findings suggested that RNA Pol II transcription initiation in trypanosomes does not depend on TFIIH-based CTD phosphorylation as it does in eukaryotes with canonical CTDs. Moreover, CTD phosphorylation seems not to be important for cotranscriptional RNA processing either. Since SL trans splicing uncouples mRNA capping from RNA Pol II transcription, trypanosomes can utilize RNA Pols other than RNA Pol II to produce functional mRNA. T. brucei has evolved a multifunctional RNA Pol I that transcribes the large ribosomal gene units as in other eukaryotes and, in addition, gene units that encode their major cell surface proteins, procyclin and variant surface glycoprotein (28). In addition, genetically modified trypanosome cell lines that express T3 or T7 RNA polymerase are able to produce functional mRNA from the corresponding phage promoters (29). Thus, the use of promoters for different RNA Pols showed that processing of a reporter pre-mRNA was not coupled to RNA Pol II (30). On the other hand, there is strong evidence that m7G capping and cap4 formation of the SL RNA occurs cotranscriptionally (31, 32).
Canonical CTDs are primarily phosphorylated by CDKs. T. brucei harbors 11 enzymes of this class, termed cdc2-related kinase 1 (CRK1) to -4 and CRK6 to -12. Sequence divergence prevents an unambiguous assignment of these enzymes to putative counterparts in other eukaryotes, although CRK3 appears to be the functional homolog of eukaryotic CDK1 (33, 34). Here, we show that silencing of CRK9 expression leads to both a loss of RPB1 phosphorylation and a trans-splicing defect due to hypomethylation of the first four SL RNA nucleotides. Most interestingly and consistent with a CDK7-less TFIIH, loss of RPB1 phosphorylation did not impair RNA Pol II transcription or m7G capping of SL RNA.
MATERIALS AND METHODS
DNAs.
For CRK9, CRK1, CRK3, and CRK7 silencing, the coding regions from position 9 to position 508, from position 5 to position 505, from position 431 to position 930, and from position 6 to 506, respectively, were integrated in a stem-loop arrangement (sense-stuffer-antisense) into the pT7-stl construct (35). The RNA interference (RNAi) vector targeting the CRK9 3′ untranslated region (3′-UTR) was generated by inserting 501 bp of the CRK9 3′-UTR from positions 2326 to 2827 relative to the translation initiation codon into pT7-stl. For hemagglutinin (HA) tagging of RPB1, 1,077 bp of the 3′-terminal RPB1 coding sequence was inserted into the pC-HA-BLA vector (27) by utilizing the vector's ApaI and NotI restriction sites. The resulting vector was named pRPB1-HA-BLA. pCRK9-PTP-NEO was generated by inserting 1,532 bp of the CRK9 coding region (position 791 to position 2322) into the ApaI and NotI sites of pC-PTP-NEO (36). pCRK9-HA-BLA and pCRK9-HAT533A-BLA were generated by inserting 1,532 bp of the CRK9 coding region (position 791 to position 2322) into the ApaI and NotI sites of pC-HA-BLA. An A1597G transition was generated through overlapping PCR, which changed the threonine codon 533 to an alanine codon. For transfection, pRPB1-HA-BLA, pCRK9-PTP-NEO, pCRK9-HA-BLA, and pCRK9-HAT533A-BLA were linearized with restriction enzymes NarI, XhoI, SphI, and SgrA1, respectively. DNA oligonucleotides that were used in semiquantitative and quantitative reverse transcription-PCR (RT-PCR) and in primer extension assays are specified in Table S2 of the supplemental material. Plasmids used in dot blot assays were described previously and contained the complete coding regions of GPEET procyclin, α-tubulin, HSP70, and 18S rRNA or the tRNAThr/U6 snRNA gene association (37).
Cells.
T. brucei cell culture, transfection, and the generation of stable cell lines by selection and limiting dilution were carried out as described previously (27, 38). In RNAi experiments, double-stranded RNA (dsRNA) synthesis was induced with 2 μg/ml of doxycycline. Cells were counted and diluted daily to 2 × 106 cells/ml for procyclic cell culture and to 2 × 105 cells/ml for bloodstream-form culture. A procyclic clonal cell line that exclusively expressed CRK9 with a C-terminally fused PTP tag was generated by targeted integration of linearized pCRK9-PTP-NEO in one CRK9 allele and by knocking out the remaining allele with a PCR product in which 101 bp of CRK9 5′ and 3′ gene flanks were fused to the hygromycin phosphotransferase coding region. Clonal CRK9 RNAi cell lines that expressed RPB1 with a C-terminal HA tag were generated by transfection of linearized pRPB1-HA-BLA.
RNA analysis.
To determine the relative abundances of RNAs in RNAi cells, total RNA prepared from 8 × 107 cells using TRIzol reagent (Invitrogen) was analyzed by semiquantitative and quantitative RT-PCR. Reverse transcriptions were carried out with SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's protocol and with oligo(dT) and random hexanucleotides (Roche) for the analysis of mature and pre-mRNAs, respectively. For each semiquantitative PCR, the number of cycles for the linear amplification range was determined empirically. For RNA quantifications, cDNA preparations from two independent RNAi experiments were analyzed by quantitative PCR (qPCR) assays using the SsoFast EvaGreen Supermix (Bio-Rad) on a CFX96 cycler (Bio-Rad) according to the manufacturer's recommendations. For each amplification and RNAi experiment, triplicate qPCR samples were analyzed using the CFX Manager software package (Bio-Rad). Each amplification product was evaluated for specificity by both agarose gel electrophoresis and melting curve analysis. Standard curves for oligonucleotide pairs were obtained from serial dilutions of noninduced cDNA samples and ranged in their coefficient of determination (R2) from 0.98 to 1.0. Samples were standardized with the 18S rDNA result from random hexamer-derived cDNA.
Labeling and analysis of nascent RNA by using a permeabilized cell system were carried out as described previously (37). For steady-state analysis of SL RNA and U2 snRNA abundances, 10 μg of total RNA prepared from induced or noninduced trypanosomes was assayed by primer extension using the 5′-32P-end-labeled oligonucleotides SLf and U2f (39). The methylation status of SL RNA cap4 was analyzed in a modified primer extension assay in which 4 μg of total RNA was reverse transcribed by using unmodified Moloney's murine leukemia virus (MMLV) reverse transcriptase (Invitrogen) at concentrations of individual deoxynucleoside triphosphates (dNTPs) that were reduced from 0.5 mM to 0.1 mM. DNA sequence ladders were generated by using the Thermo Sequenase cycle sequencing kit (USB) and radiolabeled SLf according to the manufacturer's specifications. Sequencing ladder and cap4-specific primer extension products were separated on high-resolution 50% urea–6% polyacrylamide gels and visualized by autoradiography.
The m7G capping status of SL RNA was analyzed by immunoprecipitation with a mouse monoclonal anti-2,2,7-trimethylguanosine antibody coupled to agarose beads that also recognizes the m7G cap nucleotide but not an unmethylated guanosine cap (Calbiochem). In immunoprecipitation mixtures, 5 μg of total RNA was diluted in 220 μl of buffer PA-150 (20 mM Tris-HCl [pH 7.7], 150 mM NaCl, 3 mM MgCl2, 0.5 mM dithiothreitol, 0.1% Tween 20), mixed with 30 μl of a 1:1 slurry of beads, and incubated for 1 h at 4°C. Immunoprecipitates were washed six times with 0.8 ml of PA-150 buffer. Bound RNA was extracted from the beads by using the TRIzol reagent and resuspended in 20 μl of RNase-free water. Ten-microliter aliquots of bound and unbound fractions (equal amounts) were used for primer extension analysis, using Superscript II reverse transcriptase as described above.
Protein analysis.
For the generation of a polyclonal anti-RPB1 immune serum, the CTD portion of RPB1, comprising the C-terminal 228 amino acids, was expressed with an N-terminal glutathione S-transferase (GST) tag in Escherichia coli, purified by glutathione affinity chromatography, and used as a GST fusion protein to immunize rats according to a previously published protocol (37).
RPB1 was detected on immunoblots by use of a 1:2,000 dilution of the anti-RPB1 immune serum followed by a 1:5,000 dilution of a monoclonal, peroxidase-labeled anti-rat IgG secondary antibody (Vector Laboratories). HA-tagged protein was detected with a commercial monoclonal rat anti-HA antibody (Roche), and RPA1 was detected with a polyclonal rabbit immune serum as described previously (40). Blots were developed with BM chemiluminescence blotting substrate (Roche) according to the manufacturer's protocol.
To demonstrate that the upper RPB1 band detected in immunoblots with anti-HA or anti-RPB1 antibodies corresponded to phosphorylated RPB1, 8 × 107 parasites were resuspended in 25 μl of lysis buffer (2% SDS, 100 mM Tris-HCl [pH 6.8]) and boiled for 5 min. To enable alkaline phosphatase treatment, the lysate was diluted 20-fold with Tryp wash solution (100 mM NaCl, 3 mM MgCl2, 20 mM Tris-HCl [pH 7.5]) to reduce the SDS concentration to 0.05% and subsequently reduced to a volume of 100 μl by using an Amicon Ultra 0.5-ml filter device (Millipore). The phosphatase reaction was carried out in a volume of 30 μl containing 20 μl of sample, 1× NEB3 buffer, and 10 units of calf intestinal phosphatase (New England BioLabs), and incubation at 37°C for 1 h. To block the phosphatase, 0.5 μl of phosphatase inhibitor cocktail 2 (Sigma) was added to the reaction mixture.
RESULTS
CRK9 silencing blocks SL trans splicing.
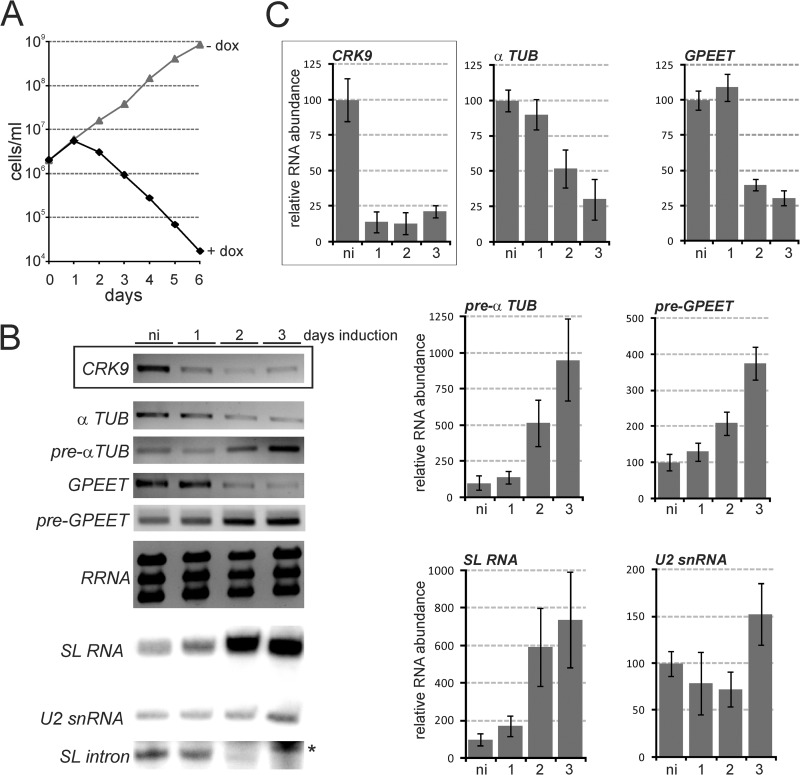
The research group of C. C. Wang (University of California, San Francisco) had previously studied all 11 T. brucei CRKs by RNAi-mediated gene knockdown. Their data indicated that CRK1, CRK3, and CRK9 were the most important CRKs for trypanosome viability (41, 42). Moreover, based on sequence similarity, it was previously proposed that CRK3 (43) and CRK7 (33) may represent CDK7 orthologs. To explore the possibility that these CRKs regulate gene expression and/or are involved in CTD phosphorylation, we generated clonal cell lines of the insect stage, procyclic form of T. brucei, in which each of these four CRK genes could be conditionally silenced by doxycycline-induced dsRNA expression (44). The knockdown of CRK9 (Fig. 1A) and of CRK1 and CRK3 (see Fig. S1 in the supplemental material) were lethal, halting culture growth after the first or second day of induction. While these growth defects were consistent with previous results (41, 42), the much stronger growth inhibition in our experiments was likely the consequence of our use of the well-regulated pT7-stl RNAi construct (35), which caused a rapid drop of the target mRNA levels within 24 h of induction (Fig. 1B, boxed panels; see also Fig. S1). Analysis of RNAs synthesized by all three RNA Pols showed that the CRK1 or CRK3 knockdown had no strong effect on RNA abundances (see Fig. S1), whereas CRK9 silencing consistently resulted in a clear reduction of α-tubulin and GPEET procyclin mRNA abundances and a concomitant increase of the corresponding pre-mRNAs. Since procyclin and α-tubulin genes are transcribed by RNA Pols I and II, respectively, it was unlikely that these effects were connected to the transcription of these genes. In addition to these changes in pre-mRNA and mature mRNA levels, we found that SL RNA abundance sharply increased 48 h after induction of CRK9 silencing, whereas there was no consistent effect on the abundances of rRNA and U2 snRNA which, in trypanosomes, are synthesized by RNA pols I and III, respectively (Fig. 1B). According to reverse transcription-quantitative real-time PCR experiments, these were strong effects: after 3 days of induction, the pools of mature α-tubulin and GPEET mRNAs were reduced to approximately one-quarter, while the levels of SL RNA and pre-mRNA were increased up to ∼9-fold (Fig. 1C). Since SL RNA is consumed in the first transesterifcation step of the splicing process, these findings strongly indicated that CRK9 silencing leads to inhibition of SL trans splicing before the first step. In accordance with this notion, a primer extension assay showed that the SL intron, the product of the first splicing step, disappeared specifically upon CRK9 silencing (Fig. 1B, bottom panel; compare with the corresponding panels in Fig. S1 in the supplemental material). CRK9 appears to have the same important functional role in gene expression in the mammalian-infective bloodstream trypanosome, because CRK9 knockdown in this life cycle stage was as lethal as in the procyclic form and also led to a decrease of α-tubulin mRNA and a concomitant increase of the corresponding pre-mRNA (see Fig. S2 in the supplemental material). Importantly, this result is in direct contrast to a previous report that found CRK9 to be essential only in the procyclic form (42). Again, this discrepancy may be explained by our use of the more suitable pT7-stl RNAi vector (35).
Fig 1.
CRK9 expression silencing caused a decrease of mature mRNA levels and concomitant accumulations of SL RNA and pre-mRNAs. (A) A representative growth curve from one of three independently derived clonal cell lines was obtained in the absence and presence of the dsRNA synthesis-inducing reagent doxycycline (+dox versus −dox). (B) Relative CRK9 mRNA levels were determined by semiquantitative RT-PCR of total RNA prepared from noninduced (ni) cells and in cells that were silenced for 1, 2, or 3 days (boxed panel). In the same RNA preparations, the relative abundances of α-tubulin (α TUB) and of GPEET procyclin mature and pre-mRNAs were determined by semiquantitative RT-PCR, those of the large ribosomal RNAs by ethidium bromide staining, and those of the SL RNA, the U2 snRNA, and the SL intron by primer extension assays. Note that the SL intron signals were obtained through overexposure of the same gels; the asterisk indicates overexposed U2 signal just above the SL intron signal. (C) Corresponding RT-qPCR analyses. The relative level of the analyzed RNA in noninduced cells for each analysis was arbitrarily set to 100. The quantitative data were calculated from two independent experiments, each of which was analyzed three times by qPCR. The boxed panel shows the silenced CRK9 mRNA levels.
In accordance with previously published results (42), silencing of CRK7 did not impair trypanosome proliferation in culture, and abundances of rRNA, GPEET mRNA, and α-tubulin mRNA remained unaffected by the knockdown (see Fig. S3 in the supplemental material). Since these results strongly suggested that CRK7 is not a functional homolog of eukaryotic CDK7, we did not further analyze this kinase. It should be noted, though, that these results are in contrast to a genome-wide RNAi screen that found CRK7 to be important for trypanosome viability across different life cycle stages (45).
CRK9 silencing leads to a loss of RPB1 phosphorylation.
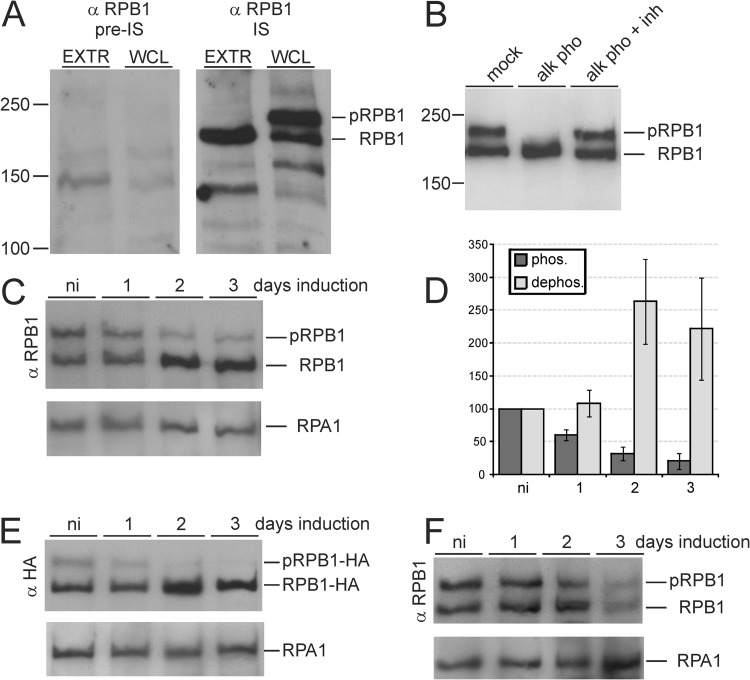
Since we had speculated that RPB1 CTD phosphorylation is mediated by a CDK, we wanted first to know whether CRK9 silencing affected the phosphorylation status of the CTD. We expressed a GST-CTD fusion protein in E. coli and raised a rat polyclonal anti-CTD immune serum against the purified recombinant protein. On immunoblots, the antibody specifically detected two specific protein bands in the range of 200 kDa in whole-cell lysates. However, the larger band was consistently absent in cell extract, suggesting that in the extract RPB1 is rapidly dephosphorylated (Fig. 2A). This finding was in accordance with previous studies in which detection of RPB1 phosphorylation in extract proved to be difficult (24, 25). Although we tested several phosphatase inhibitor cocktails, the loss of the upper band in extract could not be prevented (data not shown). To prove that the upper band detected with our anti-CTD antibody represented phosphorylated RPB1, we lysed trypanosomes in an SDS buffer and then reduced the detergent concentration to enable alkaline phosphatase treatment. As shown in Fig. 2B, the upper band was retained after mock incubation without enzyme while addition of alkaline phosphatase converted the upper band into the lower band, and this conversion was completely blocked with phosphatase inhibitor. Therefore, we concluded that the upper and lower bands in our immunodetections represented phosphorylated RPB1 (pRPB1) and dephosphorylated RPB1, respectively. Immunoblotting of whole-cell lysates from noninduced and induced cells then clearly showed that the pRPB1 signal became diminished upon CRK9 silencing, whereas dephosphorylated RPB1 increased (Fig. 2C). Quantification of immunoblot signals revealed that the pRPB1 signal decreased to 31% and 21% of noninduced levels on days 2 and 3, respectively, whereas the signal of dephosphorylated RPB1 increased ∼2.5-fold during the same period (Fig. 2D). To confirm this result, we generated a clonal procyclic cell line that expressed RPB1 with a C-terminal HA tag. Anti-HA immunoblotting detected the same two RPB1 bands as our polyclonal antibody, and CRK9 silencing again led to a signal decrease of the upper band and a concomitant signal increase of the lower band (Fig. 2E). To make sure that loss of RPB1 phosphorylation is a specific CRK9-dependent effect and not a general phenotype of dying trypanosomes, we analyzed RPB1 in CRK3-silenced cells that, like CRK9-silenced cells, ceased to proliferate after the first day of induction (see Fig. S1 in the supplemental material). Although we noted a general loss of RPB1 on day 3 of induction, the relative amounts of upper and lower bands remained at a similar level during the course of this experiment (Fig. 2F) and allowed us to conclude that CRK9 is essential for the majority, if not all, of RPB1 phosphorylations.
Fig 2.
CRK9 silencing caused a loss of RPB1 phosphorylation. (A) Cell extract (EXTR) or whole-cell lysate (WCL) was separated by SDS-PAGE, blotted, and probed with anti-RPB1 (α-RPB1) preimmune (pre-IS) or immune serum (IS). The immune serum specifically detected phosphorylated (pRPB1) and dephosphorylated RPB1. (B) Diluted whole-cell lysate was mock treated or incubated with alkaline phosphatase (alk pho) in the absence or presence (+inh) of a phosphatase inhibitor cocktail. (C) Anti-RPB1 immunoblot analysis of whole-cell lysates prepared from CRK9-silenced cells. Detection of the similar-sized RNA Pol I subunit RPA1 served as a loading control. (D) Densitometric quantification of the phosphorylated (phos.) and dephosphorylated (dephos.) RPB1 bands upon CRK9 silencing from three independent experiments. The signal of noninduced cells was arbitrarily set to 100. (E) Anti-HA immunoblot of a corresponding experiment with CRK9 RNAi cells that expressed RPB1-HA. (F) Anti-RPB1 immunoblot analysis of whole-cell lysates prepared from CRK3-silenced cells.
Although our data showed that RPB1 phosphorylation depended on CRK9, our results did not show whether CRK9 directly phosphorylates the CTD. To detect a physical link between CRK9 and RNA Pol II, we generated a cell line that exclusively expressed CRK9 C-terminally tagged with the composite PTP tag (36). Since CRK9 is essential for trypanosome viability, we concluded that the tag did not interfere with the function of the kinase. A pulldown of CRK9 under low-stringency conditions, however, did not coprecipitate detectable amounts of RPB1 (data not shown). Since this result may have been due to a transient or low-affinity interaction between the kinase and RPB1, we speculated that if RPB1 was phosphorylated at SLRNA, then the kinase should be cross-linkable to SLRNA chromatin. However, chromatin immunoprecipitation (ChIP) of PTP-tagged CRK9 or equivalently tagged RPB9, a specific subunit of RNA Pol II, showed high occupancy of the SLRNA promoter by RPB9, whereas a CRK9 association with this promoter could not be detected (see Fig. S4 in the supplemental material). Although these experiments do not rule out that CRK9 directly phosphorylates RPB1, they raise the possibility that a different, CRK9-dependent kinase carries out these phosphorylations.
CRK9 silencing does not impair RNA Pol II transcription.
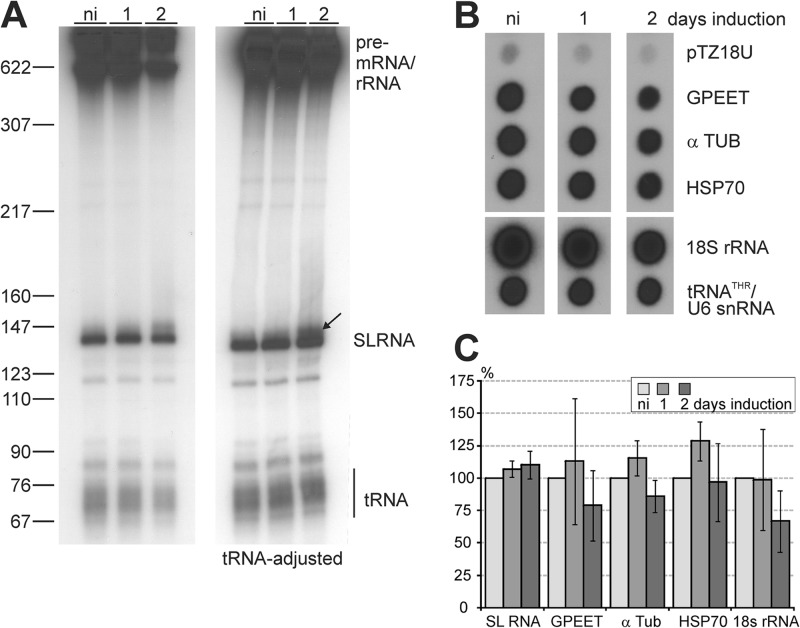
Since CTD phosphorylation is essential for transcription initiation in yeast and mammals, we explored whether CRK9-dependent RPB1 phosphorylation is required for RNA Pol II transcription in trypanosomes. We employed a permeabilized cell system that supports labeling of nascent RNA in a linearly increasing fashion for a period of 15 to 20 min (46). Upon separation of labeled RNA by denaturing PAGE, the 142-nt-long SL RNA could be readily detected due to its strong overall synthesis rate (Fig. 3A). The amount of SL RNA was roughly the same in noninduced cells and in cells in which the CRK9 knockdown was induced for 1 day. After 2 days of induction, the SL RNA was reduced by approximately one-half, but the signals for tRNA and larger RNAs were similarly reduced, most likely due to an increasing number of dying cells. Accordingly, when we used densitometry to adjust the tRNA signal, labeled SL RNA remained at the same high level 2 days after induction (Fig. 3A, right panel). Signal quantification from independent experiments confirmed that nascent SL RNA synthesis was not perturbed upon CRK9 silencing (Fig. 3C). Due to a large number of dead or dying cells, the experiment could not be performed meaningfully with cells that were CRK9 silenced for 3 days. However, since trypanosome growth ceased already after 1 day of CRK9 silencing and since the levels of pre-mRNA, mature mRNA, and SL RNA were already clearly perturbed after 2 days of induction (Fig. 1), we concluded that the loss of CRK9-dependent RPB1 phosphorylation did not impair SL RNA synthesis. Importantly, the SLRNA promoter is the only characterized RNA Pol II promoter in trypanosomes that assembles a transcription preinitiation complex, including TFIIH, the factor responsible for initial CTD phosphorylation in other eukaryotes. Therefore, this finding strongly indicated that, in trypanosomes, TFIIH-mediated CTD phosphorylation is not required for RNA Pol II escape from the SLRNA promoter.
Fig 3.
CRK9 silencing does not affect RNA Pol II transcription. (A) Nascent RNA of cells that were noninduced (ni) or CRK9 silenced for 1 or 2 days was radiolabeled in a permeabilized cell system, extracted, separated on a 50% urea–6% polyacrylamide gel, and visualized by autoradiography. For the left panel, the loading of RNA was standardized to the number of cells, and for the right panel, the loading was standardized according to the tRNA signal, as indicated. The arrow points to an RNA band that most likely represents a previously described SL RNA that is extended at the 3′ end. (B) The same labeled RNA preparations were used to probe dot blots of plasmids containing the coding regions of T. brucei GPEET procyclin, α-tubulin (α TUB), heat shock protein 70 (HSP70), and 18S rRNA, as well as the associated threonine tRNA/U6 snRNA genes (tRNATHR/U6 snRNA). The plasmid without insert (pTZ18U) served as a negative control. For each lane, upper and lower panels are from the same image. (C) The radioactive signals from four independent experiments were quantified by scintillation counting and normalized to the RNA Pol III-mediated tRNATHR/U6 snRNA signal, because CRK9 silencing did not affect the abundance of the RNA Pol III-synthesized U2 transcript. The SL RNA signal was quantified by densitometry and normalized to the tRNA signal as shown in panel A. The error bars represent standard deviations.
To explore the possibility that RPB1 phosphorylation is important for transcription of protein coding genes, we hybridized labeled nascent RNA to dot blots of plasmids carrying trypanosome gene inserts (Fig. 3B). The experiment was repeated three times and revealed that labeling efficiency was reduced throughout the array at 2 days after induction. For comparison, we normalized all signals with the signal of the RNA Pol III-transcribed threonine tRNA/U6 snRNA gene association (47), which on day 2 was reduced on average by ∼35%. Quantification of hybridization signals did not detect a specific defect in RNA Pol II-mediated transcription of protein coding genes (Fig. 3C). Two days after induction, the α-tubulin and HSP70 signals were reduced by 14% and 3%, respectively, and RNA Pol I-synthesized GPEET procyclin RNA was reduced by 21%. While none of these reductions proved to be significant (unpaired Student's t test, assuming equal variance and a two-tailed distribution), the strongest signal reduction of 33% was observed in rRNA gene transcription. Since it is well-known from other systems that inhibition of rRNA synthesis in stationary or stressed cells is often a first step in downregulating gene expression, this signal reduction could reflect the poor health of CRK9-silencend cells.
In summary, these experiments failed to detect a specific defect in RNA Pol II transcription and thus strongly indicated that CRK9-dependent RPB1 phosphorylation is neither required for transcription initiation at the SLRNA promoter nor an essential modification for transcription elongation within the protein coding gene arrays.
CRK9 silencing impairs cap4 formation of the SL RNA.
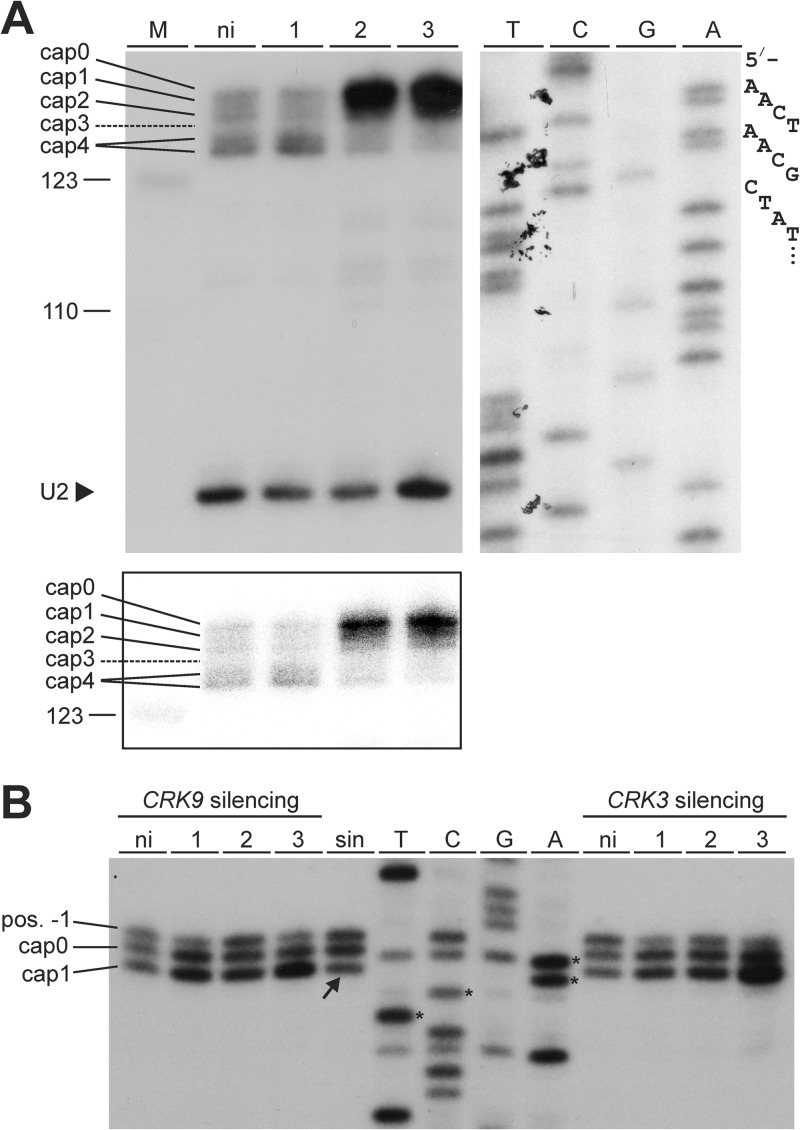
As detailed above, reporter gene expression by different RNA Pols demonstrated that pre-mRNA processing in trypanosomes is uncoupled from RNA Pol II transcription (30). In accordance with these results, CRK9 silencing affected RNA Pol I-synthesized GPEET procyclin RNA in the same way as it did RNA Pol II-synthesized α-tubulin mRNA (Fig. 1). However, these results did not exclude the possibility that SL RNA modification depended on RNA Pol II, because SL trans splicing is an essential maturation step for all trypanosome mRNAs independent of the transcribing RNA Pol. Since trans splicing depends on the methylation status of the SL RNA cap4 structure (48, 49) and since there is evidence that SL RNA cap4 formation occurs cotranscriptionally (31), it was possible that the trans-splicing block observed upon CRK9 silencing was due to inappropriate cap4 formation. To monitor cap4 methylation on the SL RNA, we employed a modified primer extension assay with a reduced concentration of deoxyribonucleotides and with unmodified MMLV reverse transcriptase, which terminates polymerization before the methylated nucleotide under these conditions (50, 51). In noninduced cells, the SL RNA-specific extension products spanned six positions: cap0 to cap4, in which, as previously noted (52), a cap3-specific extension product is typically not observed because MTR3 methylates positions 3 and 4, and cap4, which can generate two extension products (Fig. 4A). The cap4 signals were the strongest in these cells, and the pattern did not change after 1 day of CRK9 silencing. Conversely, on days 2 and 3, the cap4 signals faded, whereas the main extension product of the accumulating SL RNA was the cap0 product, indicating a complete loss of cap4 methylations (Fig. 4A). Furthermore, loss of cap1 modification was previously shown to correlate with ineffective trimming of the SL RNA 3′ end by the nuclease TbSNIP, leading to slightly larger SL RNAs (52). In accordance with this finding, we detected an accumulating signal above the correct SL RNA band in labeled nascent RNA (Fig. 3A, arrow). In summary, we concluded that CRK9 silencing caused a degree of cap4 hypomethylation that impaired SL trans splicing that led to SL RNA and pre-mRNA accumulations and decreasing levels of mature mRNAs.
Fig 4.
CRK9 silencing resulted in specific hypomethylation of SL RNA cap4. (A) Primer extension analysis with MMLV reverse transcriptase and a reduced dNTP concentration (100 nM) of total RNA prepared from noninduced (ni) or CRK9-silenced cells using 32P-5′-end-labeled oligonucleotides SLf and U2f, which are complementary to SL RNA and U2 snRNA, respectively. The extension products were separated on a high-resolution 6% sequencing gel and visualized by autoradiography. The lower exposure of the SL RNA extension products at the bottom reveals that CRK9 silencing led to accumulation of larger extension products, particularly that of the cap0 product. For comparison, a SL RNA-specific sequencing ladder was generated by linear amplification sequencing with labeled SLf; the nucleotide specification for each lane corresponds to the coding strand sequence. The sequencing ladder was coseparated on the same gel but required a longer exposure. (B) A corresponding analysis of the U1 snRNA cap1 in CRK9- and CRK3-silenced cells. The first four U1 nucleotides of the sequencing ladder are marked by asterisks, and the partial loss of cap1 in sinefungin-treated cells (sin) is denoted by the arrow.
Next, we addressed the question of whether cap4 hypomethylation is linked to the loss of RPB1 dephosphorylation. We first analyzed the U1 snRNA cap. While the T. brucei U1 gene appears to be transcribed by RNA Pol III (53), the U1 snRNA has a cap1 structure consisting of a trimethylguanosine (TMG) cap and a methylated adenosine at position 1 that leads to a premature primer extension stop at position 2 (52, 53). Importantly, cap1 methylation of both SL RNA and U1 snRNA is carried out by the same methyltransferase, MTR1 (52). Hence, if CRK9 silencing led to a direct deactivation of MTR1, we would expect to see a loss of cap1 in U1 snRNA. To test this, we carried out a modified primer extension assay with total RNA prepared from CRK9- and CRK3-silenced cells using a U1-complementary oligonucleotide (Fig. 4B). We reproducibly detected three U1-specific products in our assay: the two lower bands were specific for cap1 and unmodified cap0, whereas the fainter product with an extranucleotide (position −1) was possibly generated due to the TMG cap (as opposed to the SL RNA m7G cap). As expected, treating the cells for 6 h with sinefungin resulted in a signal shift from cap1 to cap0/pos-1 products. This shift was not observed in CRK9- or CRK3-silenced cells, even after RNAi induction for 3 days, indicating that the MTR1 enzyme activity remained active. Consequently, the specific loss of SL RNA cap1 methylation upon CRK9 silencing may be due to a failure in MTR1 recruitment to the SL RNA 5′ end.
To support this notion, we evaluated pseudouridylation of SL RNA at position 28, which is carried out by the SLA1 ribonucleoprotein particle (RNP) that comprises several proteins, including the pseudouridine synthase CBF5 (54) and MTR1 (55). We hypothesized that if MTR1 is not recruited to the SL RNA cap in CRK9-silenced cells, then pseudouridylation may also be impaired, because MTR1 and CBF5 belong to the same RNA-protein complex. However, the accumulating SL RNA in CRK9-silenced cells was clearly pseudouridylated (see Fig. S5 in the supplemental material), indicating that the SLA1 RNP can interact with SL RNA independently of CRK9-controlled RPB1 phosphorylations. As follows, our data do not unequivocally support the notion that RPB1 phosphorylation mediates cap4 formation. Testing this hypothesis is beyond the scope of this study because it requires the simultaneous knockdown of the two T. brucei RPB1 genes and the ectopic expression of a tagged, RNAi-resistant version of the large RPB1 gene carrying mutations of phosphorylated CTD residues. Nevertheless, the results presented here strongly indicate that the unique methylations of cap4 that are essential for trans splicing depend on CRK9.
CRK9 silencing does not affect m7G capping of SL RNA.
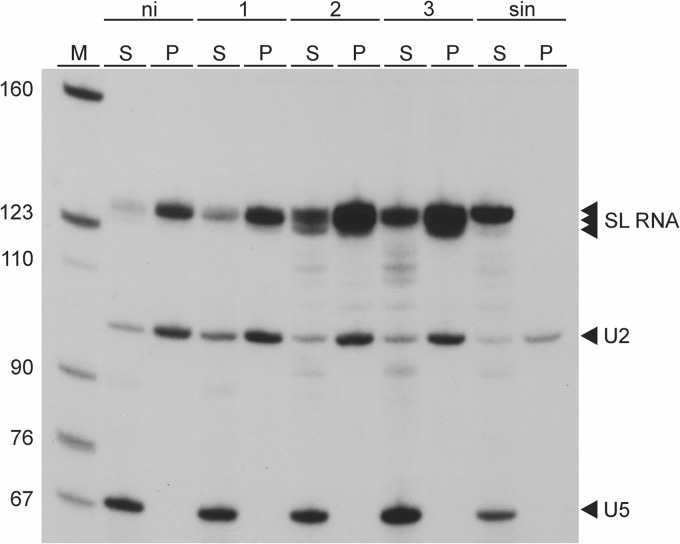
The previous demonstration, that prematurely terminated SL RNA transcripts as short as 30 nt were capped with an m7G residue, led to the conclusion that m7G capping of T. brucei SL RNA occurs cotranscriptionally (31). It was therefore possible that CRK9 silencing and the resulting CTD dephosphorylation abrogated m7G capping of the SL RNA. To analyze the SL RNA capping status in CRK9-silenced cells, we performed immunoprecipitations with an antibody directed against a TMG cap that also recognizes the m7G cap but does not interact with an unmethylated guanosine cap or uncapped RNAs. As shown in Fig. 5, the SL RNA was efficiently precipitated from total RNA of noninduced cells as well as of cells in which CRK9 was silenced for up to 3 days. In contrast, SL RNA from sinefungin-treated cells that lacked methylations and the uncapped U5 snRNA, which served as an internal negative control, were not precipitated. These results clearly demonstrated that CRK9-dependent CTD phosphorylation is not required for SL RNA m7G capping, indicating that a potential interaction between trypanosome RNA Pol II and the capping enzyme is independent of RPB1 phosphorylation.
Fig 5.
CRK9 silencing does not affect m7G capping of SL RNA. Total RNA preparations from noninduced cells (ni), cells in which CRK9 was silenced for 1, 2, or 3 days, or cells which were treated with the methylation inhibitor sinefungin (sin) were immunoprecipitated with an antibody that recognizes both m2,2,7G and m7G caps. Equal amounts of supernatants (S) and immunoprecipitates (P) were analyzed by primer extension with oligonucleotides complementary to the SL RNA (m7G cap), the U2 snRNA (m2,2,7G cap), and the uncapped U5 snRNA.
RPB1 phosphorylation and SL RNA cap4 formation depend on CRK9 kinase activity.
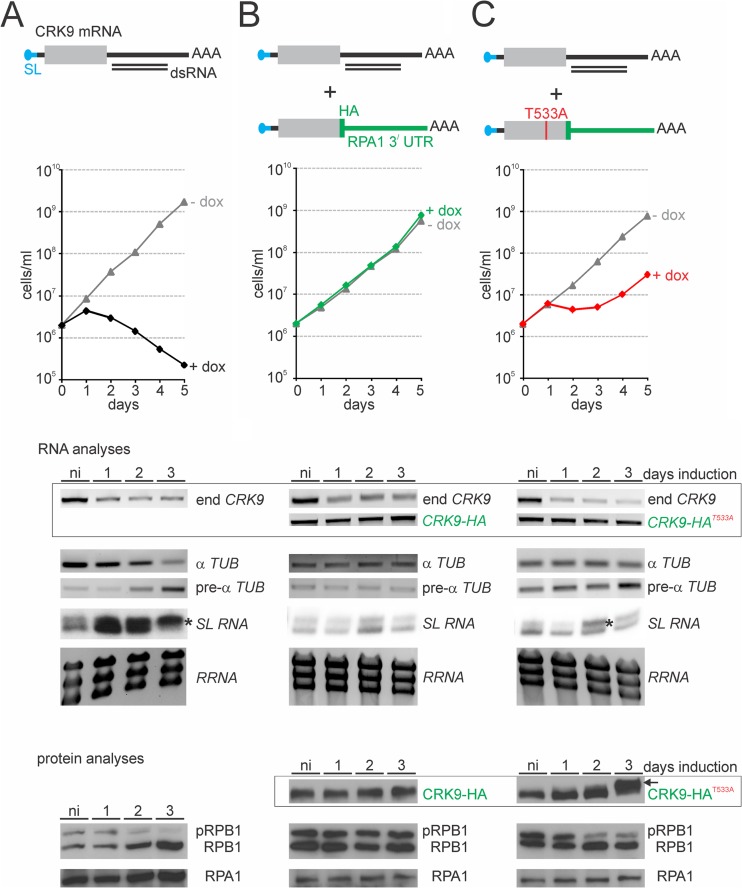
Thus far, we have shown that inducible expression of dsRNA that targets the 5′-terminal CRK9 coding region leads to an efficient knockdown of CRK9 mRNA and to RPB1 dephosphorylation, SL RNA cap4 hypomethylation, and the described RNA processing defects. To unequivocally demonstrate that these effects depend on CRK9 and its enzymatic activity and not to an off-target effect of the 500-bp-long dsRNA, we generated a new procyclic RNAi cell line in which we targeted the 3′-UTR of the CRK9 mRNA (Fig. 6A). The corresponding knockdown led to a similar, rapid decline of cell numbers as observed previously and, as expected, the levels of mature and premature α-tubulin mRNA decreased and increased upon induction of dsRNA synthesis, respectively. The cap4 methylation defect and RPB1 dephosphorylation were similarly obvious (Fig. 6A). Moreover, when we integrated a plasmid into an endogenous CRK9 allele that fused an HA tag sequence and the RPA1 3′-UTR to the CRK9 coding region, we could rescue culture growth and all the defects (Fig. 6B). The corresponding RNA analysis showed that endogenous CRK9 mRNA was effectively knocked down, whereas CRK9-HA mRNA was resistant to the RNAi response. Correspondingly CRK9-HA protein abundance remained unaffected upon doxycycline induction. These results unequivocally demonstrated that RPB1 phosphorylation and SL RNA cap4 formation are controlled by CRK9. In a final step, we wanted to make sure that the kinase activity of CRK9 is important for this control. In general, phosphorylation of the T loop threonine residue (T161 in human CDK1) leads to activation of a CDK/cyclin complex. The corresponding threonine residue in CRK9 of T. brucei had been mapped to position 533 (34). We therefore repeated the rescue experiment, but this time we mutated the T533 to an alanine in the CRK9-HA protein (Fig. 6C). RNAi induction showed a growth defect after 24 h, similar to the other (nonrescued) CRK9-silencing experiments. However, these cells recovered after 2 to 3 days to normal growth, and this was observed with three independently obtained clonal cell lines. Concomitant with recovery, we reproducibly noticed a slightly higher migrating band of CRK9-HAT553A 3 days after induction (Fig. 6C, arrow in the CRK9-HAT553A immunoblot), suggesting that compensatory phosphorylations or other posttranslational modifications led to an unconventional activation of CRK9-HAT553A. Nevertheless, while only a minor perturbation of α-tubulin (pre-)mRNA was apparent 3 days after induction, cap4 hypomethylation and RPB1 dephosphorylation were clearly detectable 2 days after induction. These data strongly indicated that CRK9 enzyme activity is required for RPB1 phosphorylation and SL RNA cap4 formation.
Fig 6.
RPB1 phosphorylation and SL RNA cap4 formation depend on CRK9 kinase activity. (A and B) A CRK9 RNAi parent cell line (A) was generated in which the inducibly expressed dsRNA targets the 3′-UTR of CRK9 mRNA. This cell line was further modified by targeted integration of a plasmid into an endogenous allele that fused an HA tag sequence and the 3′-UTR of the largest RNA Pol I subunit, RPA1, to the 3′ end of the gene (B). (C) A third cell line was generated analogously, except that the threonine codon at position 533 was mutated to an alanine codon. For each cell line, culture growth curves were obtained in the presence or absence of doxycyline, and RNA and protein analyses were conducted with noninduced cells (ni) and with cells that were induced for 1, 2, or 3 days. Semiquantitative RT-PCR analyses demonstrated efficient knockdown of endogenous (end) CRK9 RNA and RNAi resistance of CRK9-HA RNA (boxed). The CRK9 silencing effect on α-tubulin mRNA (α Tub) and pre-mRNA (pre-α Tub) was assessed by semiquantitative RT-PCR, and on SL RNA by primer extension assays. Dectection of rRNA (RRNA) by ethidium bromide staining served as a control for RNA amount. Asterisks identify increased hypomethylated cap4 signals. Anti-HA and anti-RPB1 immunoblot analyses confirmed RNAi resistance of CRK9-HA (boxed) and revealed the effects of CRK9 silencing on RBP1 phosphorylation. Detection of RPA1 served as a loading control. The arrow on the right identifies a slower-migrating band of CRK9-HAT553A protein.
DISCUSSION
We have analyzed the role in gene expression of four out of the 11 trypanosome CDKs. Our hypothesis was that, in line with phosphorylation of heptad repeats, this type of kinase phosphorylates the noncanonical RPB1 of trypanosomes and is of central importance for gene expression in this early-diverged eukaryote. Accordingly, we demonstrated that silencing of trypanosome CRK9 led to a loss of RPB1 phosphorylation. Since it was shown that the RPB1 CTD is phosphorylated (24, 25) and that phosphoproteomics exclusively detected phosphorylated residues in the CTD of the large RPB1 protein (26), it is likely that most if not all RPB1 phosphorylations are located in the CTD. It seems that all of these phosphorylations depend on CRK9, because silencing of its gene led to a loss of the pRPB1 band in immunoblots and to a concomitant increase of unphosphorylated RPB1 without the appearance of an intermediate band. Although we have not been able to show so far that CRK9 directly phosphorylates the CTD, our results are informative about the function of the noncanonical CTD. We found that reducing phosphorylated RPB1 to 30% on day 2 of CRK9 silencing did not impact transcription of protein coding and SLRNA genes, indicating a mechanistic difference in transcription initiation between trypanosomes and eukaryotes with a canonical CTD. In the latter, RNA Pol II recruitment into the preinitiation complex involves the mediator complex that binds to hypophosphorylated RNA Pol II. TFIIH/CDK7-mediated CTD phosphorylation disrupts this interaction, presumably facilitating promoter escape of the polymerase (56). Trypanosomes do possess a mediator complex that is essential for SLRNA transcription, interacts with TFIIH, and is required for the formation of a transcription preinitiation complex at the SLRNA promoter (18). However, whereas the eukaryotic mediator consists of head, middle, tail, and CDK8 modules, electron microscopic structure analysis of the trypanosome complex indicated that it is confined to the head module (18). Thus, the absence of a TFIIH kinase (17) and the presence of a minimal mediator complex suggest that, in trypanosomes, a mediator-CTD interaction does not occur and a release of RNA Pol II from the mediator is not a prerequisite for promoter escape of the enzyme. Accordingly, tandem affinity purification of the trypanosome mediator did not copurify detectable amounts of RNA Pol II subunits (18), and vice versa (20, 21).
Furthermore, RPB1 dephosphorylation did not impact m7G capping of SL RNA. In higher eukaryotes, the capping enzyme binds to the CTD after Ser5 of the heptad repeat motif is phosphorylated by CDK7 (4); capping then occurs cotranscriptionally. Since in trypanosomes, SL RNA capping does occur cotranscriptionally as well (31), our results strongly suggest that the trypanosome capping enzyme interacts with the transcription machinery in a trypanosome-specific manner independent of CTD phosphorylation.
Nevertheless, CRK9 silencing resulted in a striking defect in trypanosome gene expression, namely, a block of trans splicing before the first transesterification reaction caused by hypomethylation of SL RNA cap4. Formation of the unique cap4 structure involves seven methylations on the first four nucleotides by three or more different methyltransferases. Thus, the most intriguing aspect of our work is the demonstration that cap4 formation is controlled by a single enzyme. Since the trypanosome cap4 structure is unique, essential for trans splicing (48, 49), and important for translation (57), identification of the methyltransferases has been a priority and, thus far, the three 2′-O-ribose methyltransferases MTR1, MTR2 (also termed COM or MT48), and MTR3 (also termed MT57), which methylate positions 1, 2, and 3/4, respectively, have been identified (51, 52, 58–60). Surprisingly, however, elimination of individual MTR genes left trypanosomes viable and did not impair trans splicing. This functional redundancy of the known MTRs has dampened enthusiasm for these enzymes as potential chemotherapeutic targets. However, the dependence of a critical number of cap4 methylations on CRK9 alone reinstates cap4 formation as a target. CRK9 appears to be particularly promising in this respect. First, CRK9 has an essential central role in trypanosome gene expression, controlling RPB1 phosphorylation and SL RNA cap4 formation. Second, SL trans splicing does not occur in mammals and, thus, the block of SL RNA maturation is a parasite-specific defect. Third, since SL RNA is consumed in trans splicing of every mRNA, partial inhibition of SL RNA biogenesis may already be lethal. This notion is backed up by independent observations that a reduction of SL RNA synthesis to ∼30% is already lethal to cultured trypanosomes (23, 37). Fourthly, cyclin-dependent kinases in general are considered to be among the most promising drug targets, due to their key function in cell proliferation (61). Accordingly, CRK9 silencing has been shown to affect trypanosome mitosis and cytokinesis (42, 62).
How does CRK9 enable cap4 formation? A plausible explanation is that cap4 methylations occur cotranscriptionally, requiring a phosphorylated CTD. This would be consistent with previous observations that cap4 methylations can be detected on newly synthesized, prematurely terminated SL RNA transcripts (31). Part of our results support this scenario: CRK9 silencing did not cause a detectable defect of cap1 formation in the RNA Pol III-synthesized U1 snRNA, suggesting that MTR1, the methyltransferase that forms cap1 in both U1 and SL RNA, remains functional during CRK9 knockdown. Thus, the loss of SL RNA cap1 may be explained by failed recruitment of MTR1 by dephosphorylated RNA Pol II. Although MTRs do not copurify with T. brucei RNA Pol II (20, 21), the interaction may have been lost due to rapid RPB1 dephosphorylation in extract. As our data showed (Fig. 2A), the phosphorylated form of RPB1 was only detected in whole-cell lysates and rapidly converted to the dephosphorylated form during extract preparation, possibly due to the presence of 15 putative homologs of the CTD phosphatase FCP1 in the T. brucei genome (www.TritrypDB.org).
On the other hand, there are findings that suggest that the T. brucei methyltransferases do not require RNA Pol II for recruitment. T. brucei MTR1 was found to be in a complex with the SLA1 RNP, which contains several subunits, including the pseudouridine synthase CBF5, and which guides pseudouridylation of SL RNA position 28 (55). While SL RNA pseudouridylation is not essential for trypanosome viability (32), CBF5 silencing affected cap4 formation, indicating that the SLA1 RNP is important for MTR1 function (54). Since SLA1 can base pair with the SL RNA around the pseudouridylation site, this interaction could bring MTR1 to the SL RNA 5′ end independently of RNA Pol II (55). Accordingly, we found that pseudouridylation was not impaired during CRK9 silencing, which indicates that the SLA1 RNP can interact with the SL RNA independent of phosphorylated RNA Pol II. Furthermore, MTR2 and MTR3 cap4 methylations depend on Sm protein binding to the SL RNA 3′-terminal domain (63, 64), suggesting that the SL RNP possesses RNA Pol II-independent determinants for MTR2 and MTR3 recruitment. However, these arguments are not conclusive, and there are other findings that are more in line with cotranscriptional cap4 formation. First, the interaction between SLA1 and SL RNA appears to be weak and transient, since purification of the SLA1 RNP did not copurify detectable amounts of SL RNA, indicating that base pairing alone may not be enough for SLA1 recruitment to the SL RNA (55). Second, SLA1 colocalizes with the SLRNA transcription factor SNAP2, also known as tSNAP42, suggesting that SLRNA transcription and cap1 formation occur at the same distinct site in the nucleus (32). Taken together, it will require a dedicated study to decipher whether CTD phosphorylation is directly linked to SL RNA cap4 formation or whether RPB1 phosphorylation and cap4 formation are independent processes.
Overall, our study has identified a cyclin-dependent kinase that is of central importance to trypanosome gene expression, being essential for SL RNA maturation and RPB1 phosphorylation. Together with previous TFIIH and mediator characterizations, we have provided compelling evidence for RNA Pol II transcription initiation and m7G capping functioning independently of a TFIIH-based, CDK7-mediated CTD phosphorylation event. Our data strongly suggest that the essential CTD function in RNA Pol II transcription initiation evolved after the divergence of major protistan lineages, because it appears to be coupled to the canonical CTD with heptad repeat motifs.
Supplementary Material
ACKNOWLEDGMENTS
We thank David A. Campbell and Nancy R. Sturm (UCLA) for discussions and Tu N. Nguyen and Justin K. Kirkham for critical reading of the manuscript.
This work was funded by NIH grant R01 AI073300 to A.G.
Footnotes
Published ahead of print 11 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00156-13.
REFERENCES
- 1. Chapman RD, Heidemann M, Hintermair C, Eick D. 2008. Molecular evolution of the RNA polymerase II CTD. Trends Genet. 24:289–296 [DOI] [PubMed] [Google Scholar]
- 2. Buratowski S. 2009. Progression through the RNA polymerase II CTD cycle. Mol. Cell 36:541–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hsin JP, Manley JL. 2012. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 26:2119–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fabrega C, Shen V, Shuman S, Lima CD. 2003. Structure of an mRNA capping enzyme bound to the phosphorylated carboxy-terminal domain of RNA polymerase II. Mol. Cell 11:1549–1561 [DOI] [PubMed] [Google Scholar]
- 5. Karagiannis J, Balasubramanian MK. 2007. A cyclin-dependent kinase that promotes cytokinesis through modulating phosphorylation of the carboxy terminal domain of the RNA Pol II Rpb1p sub-unit. PLoS One 2:e433 doi:10.1371/journal.pone.0000433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qiu H, Hu C, Hinnebusch AG. 2009. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol. Cell 33:752–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu Y, Warfield L, Zhang C, Luo J, Allen J, Lang WH, Ranish J, Shokat KM, Hahn S. 2009. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol. Cell. Biol. 29:4852–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Akhtar MS, Heidemann M, Tietjen JR, Zhang DW, Chapman RD, Eick D, Ansari AZ. 2009. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol. Cell 34:387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Glover-Cutter K, Larochelle S, Erickson B, Zhang C, Shokat K, Fisher RP, Bentley DL. 2009. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol. Cell. Biol. 29:5455–5464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim M, Suh H, Cho EJ, Buratowski S. 2009. Phosphorylation of the yeast Rpb1 C-terminal domain at serines 2, 5, and 7. J. Biol. Chem. 284:26421–26426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tietjen JR, Zhang DW, Rodriguez-Molina JB, White BE, Akhtar MS, Heidemann M, Li X, Chapman RD, Shokat K, Keles S, Eick D, Ansari AZ. 2010. Chemical-genomic dissection of the CTD code. Nat. Struct. Mol. Biol. 17:1154–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Egloff S, Szczepaniak SA, Dienstbier M, Taylor A, Knight S, Murphy S. 2010. The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. J. Biol. Chem. 285:20564–20569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guo Z, Stiller JW. 2004. Comparative genomics of cyclin-dependent kinases suggest coevolution of the RNAP II C-terminal domain and CTD-directed CDKs. BMC Genomics 5:69 doi:10.1186/1471-2164-5-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Günzl A. 2010. The pre-mRNA splicing machinery of trypanosomes: complex or simplified? Eukaryot. Cell 9:1159–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bangs JD, Crain PF, Hashizume T, McCloskey JA, Boothroyd JC. 1992. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 267:9805–9815 [PubMed] [Google Scholar]
- 16. Das A, Banday M, Bellofatto V. 2008. RNA polymerase transcription machinery in trypanosomes. Eukaryot. Cell 7:429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee JH, Jung HS, Günzl A. 2009. Transcriptionally active TFIIH of the early-diverged eukaryote Trypanosoma brucei harbors two novel core subunits but not a cyclin-activating kinase complex. Nucleic Acids Res. 37:3811–3820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee JH, Cai G, Panigrahi AK, Dunham-Ems S, Nguyen TN, Radolf JD, Asturias FJ, Günzl A. 2010. A TFIIH-associated mediator head is a basal factor of small nuclear spliced leader RNA gene transcription in early-diverged trypanosomes. Mol. Cell. Biol. 30:5502–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kelly S, Wickstead B, Gull K. 2005. An in silico analysis of trypanosomatid RNA polymerases: insights into their unusual transcription. Biochem. Soc. Trans. 33:1435–1437 [DOI] [PubMed] [Google Scholar]
- 20. Devaux S, Lecordier L, Uzureau P, Walgraffe D, Dierick JF, Poelvoorde P, Pays E, Vanhamme L. 2006. Characterization of RNA polymerase II subunits of Trypanosoma brucei. Mol. Biochem. Parasitol. 148:60–68 [DOI] [PubMed] [Google Scholar]
- 21. Das A, Li H, Liu T, Bellofatto V. 2006. Biochemical characterization of Trypanosoma brucei RNA polymerase II. Mol. Biochem. Parasitol. 150:201–210 [DOI] [PubMed] [Google Scholar]
- 22. Martinez-Calvillo S, Saxena A, Green A, Leland A, Myler PJ. 2007. Characterization of the RNA polymerase II and III complexes in Leishmania major. Int. J. Parasitol. 37:491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ruan JP, Arhin GK, Ullu E, Tschudi C. 2004. Functional characterization of a Trypanosoma brucei TATA-binding protein-related factor points to a universal regulator of transcription in trypanosomes. Mol. Cell. Biol. 24:9610–9618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chapman AB, Agabian N. 1994. Trypanosoma brucei RNA polymerase II is phosphorylated in the absence of carboxyl-terminal domain heptapeptide repeats. J. Biol. Chem. 269:4754–4760 [PubMed] [Google Scholar]
- 25. Das A, Bellofatto V. 2009. The non-canonical CTD of RNAP-II is essential for productive RNA synthesis in Trypanosoma brucei. PLoS One 4:e6959 doi:10.1371/journal.pone.0006959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nett IR, Martin DM, Miranda-Saavedra D, Lamont D, Barber JD, Mehlert A, Ferguson MA. 2009. The phosphoproteome of bloodstream form Trypanosoma brucei, causative agent of African sleeping sickness. Mol. Cell. Proteomics 8:1527–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee JH, Nguyen TN, Schimanski B, Günzl A. 2007. Spliced leader RNA gene transcription in Trypanosoma brucei requires transcription factor TFIIH. Eukaryot. Cell 6:641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Günzl A, Bruderer T, Laufer G, Schimanski B, Tu LC, Chung HM, Lee PT, Lee MG. 2003. RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei. Eukaryot. Cell 2:542–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wirtz E, Hartmann C, Clayton C. 1994. Gene expression mediated by bacteriophage T3 and T7 RNA polymerases in transgenic trypanosomes. Nucleic Acids Res. 22:3887–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stewart M, Haile S, Jha BA, Cristodero M, Li CH, Clayton C. 2010. Processing of a phosphoglycerate kinase reporter mRNA in Trypanosoma brucei is not coupled to transcription by RNA polymerase II. Mol. Biochem. Parasitol. 172:99–106 [DOI] [PubMed] [Google Scholar]
- 31. Mair G, Ullu E, Tschudi C. 2000. Cotranscriptional cap 4 formation on the Trypanosoma brucei spliced leader RNA. J. Biol. Chem. 275:28994–28999 [DOI] [PubMed] [Google Scholar]
- 32. Hury A, Goldshmidt H, Tkacz ID, Michaeli S. 2009. Trypanosome spliced-leader-associated RNA (SLA1) localization and implications for spliced-leader RNA biogenesis. Eukaryot. Cell 8:56–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parsons M, Worthey EA, Ward PN, Mottram JC. 2005. Comparative analysis of the kinomes of three pathogenic trypanosomatids: Leishmania major, Trypanosoma brucei and Trypanosoma cruzi. BMC Genomics 6:127 doi:10.1186/1471-2164-6-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hammarton TC. 2007. Cell cycle regulation in Trypanosoma brucei. Mol. Biochem. Parasitol. 153:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brandenburg J, Schimanski B, Nogoceke E, Nguyen TN, Padovan JC, Chait BT, Cross GA, Günzl A. 2007. Multifunctional class I transcription in Trypanosoma brucei depends on a novel protein complex. EMBO J. 26:4856–4866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schimanski B, Nguyen TN, Günzl A. 2005. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot. Cell 4:1942–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schimanski B, Brandenburg J, Nguyen TN, Caimano MJ, Günzl A. 2006. A TFIIB-like protein is indispensable for spliced leader RNA gene transcription in Trypanosoma brucei. Nucleic Acids Res. 34:1676–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Park SH, Nguyen TN, Kirkham JK, Lee JH, Günzl A. 2011. Transcription by the multifunctional RNA polymerase I in Trypanosoma brucei functions independently of RPB7. Mol. Biochem. Parasitol. 180:35–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Günzl A, Cross M, Bindereif A. 1992. Domain structure of U2 and U4/U6 small nuclear ribonucleoprotein particles from Trypanosoma brucei: identification of trans-spliceosomal specific RNA-protein interactions. Mol. Cell. Biol. 12:468–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schimanski B, Klumpp B, Laufer G, Marhöfer RJ, Selzer PM, Günzl A. 2003. The second largest subunit of Trypanosoma brucei's multifunctional RNA polymerase I has a unique N-terminal extension domain. Mol. Biochem. Parasitol. 126:193–200 [DOI] [PubMed] [Google Scholar]
- 41. Tu X, Wang CC. 2004. The involvement of two cdc2-related kinases (CRKs) in Trypanosoma brucei cell cycle regulation and the distinctive stage-specific phenotypes caused by CRK3 depletion. J. Biol. Chem. 279:20519–20528 [DOI] [PubMed] [Google Scholar]
- 42. Gourguechon S, Wang CC. 2009. CRK9 contributes to regulation of mitosis and cytokinesis in the procyclic form of Trypanosoma brucei. BMC Cell. Biol. 10:68 doi:10.1186/1461-2121-10-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, Anupama A, Apostolou Z, Attipoe P, Bason N, Bauser C, Beck A, Beverley SM, Bianchettin G, Borzym K, Bothe G, Bruschi CV, Collins M, Cadag E, Ciarloni L, Clayton C, Coulson RM, Cronin A, Cruz AK, Davies RM, De Gaudenzi J, Dobson DE, Duesterhoeft A, Fazelina G, Fosker N, Frasch AC, Fraser A, Fuchs M, Gabel C, Goble A, Goffeau A, Harris D, Hertz-Fowler C, Hilbert H, Horn D, Huang Y, Klages S, Knights A, Kube M, Larke N, Litvin L, Lord A, Louie T, Marra M, Masuy D, Matthews K, Michaeli S, Mottram JC, Muller-Auer S, Munden H, Nelson S, Norbertczak H, Oliver K, O'neil S, Pentony M, Pohl TM, Price C, Purnelle B, Quail MA, Rabbinowitsch E, Reinhardt R, Rieger M, Rinta J, Robben J, Robertson L, Ruiz JC, Rutter S, Saunders D, Schafer M, Schein J, Schwartz DC, Seeger K, Seyler A, Sharp S, Shin H, Sivam D, Squares R, Squares S, Tosato V, Vogt C, Volckaert G, Wambutt R, Warren T, Wedler H, Woodward J, Zhou S, Zimmermann W, Smith DF, Blackwell JM, Stuart KD, Barrell B, Myler PJ. 2005. The genome of the kinetoplastid parasite, Leishmania major. Science 309:436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wirtz E, Leal S, Ochatt C, Cross GAM. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89–101 [DOI] [PubMed] [Google Scholar]
- 45. Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, Hertz-Fowler C, Horn D. 2011. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 21:915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ullu E, Tschudi C. 1990. Permeable trypanosome cells as a model system for transcription and trans-splicing. Nucleic Acids Res. 18:3319–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nakaar V, Günzl A, Ullu E, Tschudi C. 1997. Structure of the Trypanosoma brucei U6 snRNA gene promoter. Mol. Biochem. Parasitol. 88:13–23 [DOI] [PubMed] [Google Scholar]
- 48. Ullu E, Tschudi C. 1991. Trans splicing in trypanosomes requires methylation of the 5′ end of the spliced leader RNA. Proc. Natl. Acad. Sci. U. S. A. 88:10074–10078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McNally KP, Agabian N. 1992. Trypanosoma brucei spliced-leader RNA methylations are required for trans splicing in vivo. Mol. Cell. Biol. 12:4844–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mandelboim M, Estrano CL, Tschudi C, Ullu E, Michaeli S. 2002. On the role of exon and intron sequences in trans-splicing utilization and cap 4 modification of the trypanosomatid Leptomonas collosoma SL RNA. J. Biol. Chem. 277:35210–35218 [DOI] [PubMed] [Google Scholar]
- 51. Zamudio JR, Mittra B, Zeiner GM, Feder M, Bujnicki JM, Sturm NR, Campbell DA. 2006. Complete cap 4 formation is not required for viability in Trypanosoma brucei. Eukaryot. Cell 5:905–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zamudio JR, Mittra B, Foldynova-Trantirkova S, Zeiner GM, Lukes J, Bujnicki JM, Sturm NR, Campbell DA. 2007. The 2′-O-ribose methyltransferase for cap 1 of spliced leader RNA and U1 small nuclear RNA in Trypanosoma brucei. Mol. Cell. Biol. 27:6084–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Djikeng A, Ferreira L, D'Angelo M, Dolezal P, Lamb T, Murta S, Triggs V, Ulbert S, Villarino A, Renzi S, Ullu E, Tschudi C. 2001. Characterization of a candidate Trypanosoma brucei U1 small nuclear RNA gene. Mol. Biochem. Parasitol. 113:109–115 [DOI] [PubMed] [Google Scholar]
- 54. Barth S, Hury A, Liang XH, Michaeli S. 2005. Elucidating the role of H/ACA-like RNAs in trans-splicing and rRNA processing via RNA interference silencing of the Trypanosoma brucei CBF5 pseudouridine synthase. J. Biol. Chem. 280:34558–34568 [DOI] [PubMed] [Google Scholar]
- 55. Zamudio JR, Mittra B, Chattopadhyay A, Wohlschlegel JA, Sturm NR, Campbell DA. 2009. Trypanosoma brucei spliced leader RNA maturation by the cap 1 2′-O-ribose methyltransferase and SLA1 H/ACA snoRNA pseudouridine synthase complex. Mol. Cell. Biol. 29:1202–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sogaard TM, Svejstrup JQ. 2007. Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J. Biol. Chem. 282:14113–14120 [DOI] [PubMed] [Google Scholar]
- 57. Zamudio JR, Mittra B, Campbell DA, Sturm NR. 2009. Hypermethylated cap 4 maximizes Trypanosoma brucei translation. Mol. Microbiol. 72:1100–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Arhin GK, Li H, Ullu E, Tschudi C. 2006. A protein related to the vaccinia virus cap-specific methyltransferase VP39 is involved in cap 4 modification in Trypanosoma brucei. RNA 12:53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Arhin GK, Ullu E, Tschudi C. 2006. 2′-O-methylation of position 2 of the trypanosome spliced leader cap 4 is mediated by a 48 kDa protein related to vaccinia virus VP39. Mol. Biochem. Parasitol. 147:137–139 [DOI] [PubMed] [Google Scholar]
- 60. Hall MP, Ho CK. 2006. Functional characterization of a 48 kDa Trypanosoma brucei cap 2 RNA methyltransferase. Nucleic Acids Res. 34:5594–5602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Malumbres M, Barbacid M. 2009. Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer 9:153–166 [DOI] [PubMed] [Google Scholar]
- 62. Li Z. 2012. Regulation of the cell division cycle in Trypanosoma brucei. Eukaryot. Cell 11:1180–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mandelboim M, Barth S, Biton M, Liang XH, Michaeli S. 2003. Silencing of Sm proteins in Trypanosoma brucei by RNA interference captured a novel cytoplasmic intermediate in spliced leader RNA biogenesis. J. Biol. Chem. 278:51469–51478 [DOI] [PubMed] [Google Scholar]
- 64. Zeiner GM, Foldynova S, Sturm NR, Lukes J, Campbell DA. 2004. SmD1 is required for spliced leader RNA biogenesis. Eukaryot. Cell 3:241–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.