Abstract
Human endothelial nitric oxide synthase (eNOS) mRNA is highly stable in endothelial cells (ECs). Posttranscriptional regulation of eNOS mRNA stability is an important component of eNOS regulation, especially under hypoxic conditions. Here, we show that the human eNOS 3′ untranslated region (3′ UTR) contains multiple, evolutionarily conserved pyrimidine (C and CU)-rich sequence elements that are both necessary and sufficient for mRNA stabilization. Importantly, RNA immunoprecipitations and RNA electrophoretic mobility shift assays (EMSAs) revealed the formation of heterogeneous nuclear ribonucleoprotein E1 (hnRNP E1)-containing RNP complexes at these 3′-UTR elements. Knockdown of hnRNP E1 decreased eNOS mRNA half-life, mRNA levels, and protein expression. Significantly, these stabilizing RNP complexes protect eNOS mRNA from the inhibitory effects of its antisense transcript sONE and 3′-UTR-targeting small interfering RNAs (siRNAs), as well as microRNAs, specifically, hsa-miR-765, which targets eNOS mRNA stability determinants. Hypoxia disrupts hnRNP E1/eNOS 3′-UTR interactions via increased Akt-mediated serine phosphorylation (including serine 43) and increased nuclear localization of hnRNP E1. These mechanisms account, at least in part, for the decrease in eNOS mRNA stability under hypoxic conditions. Thus, the stabilization of human eNOS mRNA by hnRNP E1-containing RNP complexes serves as a key protective mechanism against the posttranscriptional inhibitory effects of antisense RNA and microRNAs under basal conditions but is disrupted under hypoxic conditions.
INTRODUCTION
The signaling molecule nitric oxide (NO), produced in endothelial cells (ECs) by endothelial nitric oxide synthase (eNOS), plays a pivotal role in the maintenance of homeostasis in the blood vessel wall (1). The appropriate expression and function of eNOS is critically important in blood vessel wall function and represents a sensitive and highly effective system for maintaining local blood flow to an organ. Targeted inactivation of the murine eNOS locus and physiologic assessment of eNOS−/− animals has reinforced this view (2, 3) and indicated that eNOS is important in the remodeling of vessel walls in response to changes in flow or distending pressure (4). Indeed, decreased eNOS expression and function has been implicated in the pathology of a number of cardiovascular diseases, including atherosclerosis and hypertension (1, 5), and is a key defining feature of the clinical entity termed endothelial dysfunction. Significantly, hypoxia is a major cellular stress that has a profound impact on endothelial cell biology, including critical changes in gene expression (6, 7). Thus, understanding the molecular regulation of eNOS gene expression is a high-priority area.
Importantly, several pathophysiological factors, especially hypoxia, and important models of endothelial activation lower eNOS expression. In these cases, a major contributing factor to downregulation of eNOS expression appears to be a reduction in the stability of the mature eNOS mRNA. Under normal conditions, the eNOS mRNA is highly stable in human ECs, with a half-life in excess of 24 h, as assessed by actinomycin D transcription arrest experiments (8). Notably, we and others have shown that hypoxia significantly downregulates eNOS gene expression in ECs, with major contributions from the posttranscriptional downregulation of eNOS mRNA expression (9–11). In addition, models of proliferation/injury (12), tumor necrosis factor alpha (TNF-α) treatment (13, 14), and exposure to lipopolysaccharide (15) or high levels of oxidized low-density lipoprotein (16) all decrease eNOS steady-state mRNA expression in cultured ECs in a manner dependent, in major part, on changes in eNOS mRNA stability.
We recently reported the existence of a cis-natural antisense transcript to eNOS called sONE (also know as ATG9B, NOS3AS, and APG9L2) that regulates eNOS via posttranscriptional mechanisms (17, 18). The genes for eNOS and sONE are arranged in a tail-to-tail orientation on human chromosome 7q36, and the transcripts for the 2 genes are complementary for a total of 662 nucleotides, including significant exon/exon overlap. The RNA product of the ATG9B gene has both noncoding (17) and putative coding functions (18). Under basal conditions, sONE transcripts are expressed at very low levels in ECs due to posttranscriptional regulation, whereas eNOS is highly abundant. Importantly, exposure of ECs to hypoxia, which downregulates eNOS mRNA and protein expression, markedly upregulates steady-state levels of sONE RNA (11). The decrease in eNOS mRNA abundance is attributed, at least in part, to the destabilization of eNOS mRNAs, and sONE depletion rescued eNOS expression. Thus, these findings implicate sONE as a posttranscriptional inhibitor of eNOS mRNA and protein expression, especially under hypoxic conditions. In addition to antisense RNAs, other posttranscriptional regulators also exist. A prime example is represented by microRNAs, which are ∼22-nucleotide (nt) endogenous small RNAs that function as potent posttranscriptional regulators of mRNA stability and translation (19–21). A recent estimate suggests that at least 60% of all human protein-coding genes are subject to regulation by microRNAs (22). However, it is not known whether eNOS is significantly regulated by microRNAs.
Given that mRNA stability contributes significantly to overall eNOS expression, we were interested to determine whether eNOS mRNA is actively stabilized in order to protect it from the inhibitory effects of the antisense transcript sONE and microRNAs. Thus, we undertook detailed functional analyses of eNOS 3′-untranslated region (UTR) regulatory cis elements. Here, we show that eNOS mRNA stability is determined by multiple evolutionarily conserved pyrimidine (C and CU)-rich elements in the 3′ UTR (referred to below as P1 through P5 below). Furthermore, eNOS mRNA is basally stabilized by the formation of ribonucleoprotein RNP complexes on these 3′-UTR cis elements. Importantly, we identified heterogenous nuclear RNP (hnRNP) E1, which is a ubiquitous, multifunctional RNA-binding protein with important nuclear and cytoplasmic roles (23), as a major component of these eNOS-stabilizing RNP complexes. Significantly, the stabilization of human eNOS mRNA by hnRNP E1-containing RNP complexes serves as a key protective mechanism against the posttranscriptional inhibitory effects of the eNOS antisense transcript sONE and 3′-UTR-targeting small interfering RNAs (siRNAs), as well as functionally important microRNAs, specifically, hsa-miR-765, under basal conditions. However, hypoxia disrupts hnRNP E1/eNOS 3′-UTR interactions via increased Akt-mediated serine phosphorylation (including serine 43) and nuclear localization of hnRNP E1. These mechanisms contribute significantly to the decrease in eNOS mRNA stability and overall expression in hypoxic ECs.
MATERIALS AND METHODS
Cell culture and hypoxia treatment.
Human umbilical vein endothelial cells (HUVEC) were harvested, used at passages two to five, and maintained as described previously (17, 24). Findings were replicated in HUVEC derived from independent cultures prepared from multiple unrelated individuals. HepG2 cells (human hepatocellular carcinoma cell line with epithelial morphology) were obtained from American Type Culture Collection (ATCC) and maintained in Dulbecco's modified Eagle's medium containing 10 mM HEPES. K562 human erythroleukemia cells (ATCC) were maintained in RPMI 1640 containing 10 mM HEPES. For hypoxia treatment, cells were subjected to 1% O2 in a temperature- and humidity-controlled incubator within a sealed anaerobic system (Thermo Forma model 1025). The hypoxic environment (1% O2) was achieved and maintained using a high-purity anaerobic gas mixture (5% CO2, 10% H2, 85% N2; Linde).
Cloning of the rabbit and mouse eNOS 3′-UTR cDNA.
Recombinant bacteriophage murine cDNA clones were isolated by plaque hybridization from an oligo(dT)- and random-primed murine fetal cardiac λZAP II expression library established at day 12.5 of embryonic life (25). A full-length, 4,052-nt human eNOS probe was labeled with [α-32P]dCTP and used to screen the amplified library as described previously (26). Similarly, a cDNA encoding rabbit eNOS was isolated by plaque hybridization from a rabbit inferior vena caval endothelial cell lambda gt11 cDNA library (27), using a full-length human eNOS cDNA as the hybridization probe. cDNA sequences were confirmed on both strands.
Plasmid construction and stable transfection.
Stable DNA expression cassettes were created based on the full-length, 4,052-nt eNOS cDNA using standard recombinant DNA techniques (27, 28). Transcription was directed by a 654-bp cytomegalovirus (CMV) promoter encoded in the pcDNA3 plasmid (Invitrogen). The vector-encoded 3′-mRNA processing signals were removed from the parental vector and replaced with human eNOS sequences representing 87 nucleotides of 3′-end-flanking genomic sequence. Luciferase (LUC) reporter constructs used the firefly luciferase cDNA from pSP Luc+ (Promega) ligated into the HindIII/XhoI sites of pcDNA3, followed by the terminal 714 base pairs of eNOS cDNA and 87 nucleotides of eNOS 3′-flanking genomic sequence. The point mutation of TGA to TCA was introduced within the eNOS 3′ UTR using the QuikChange mutagenesis kit (Stratagene) and the primer sequences 5′-GCC AGG CGG CTC TCA GGG GCT GTT GG-3′ and 5′-CCA ACA GCC CCT GAG AGC CGC CTG GC-3′ according to the manufacturer's recommendations.
Translation read-through was confirmed by translation of 5′-capped synthetic transcripts synthesized using mMessage mMachine kits (Ambion) in rabbit reticulocyte lysate (Invitrogen) according to the manufacturer's suggestions in the presence of 20 μCi of [35S]methionine (1,000 Ci/mmol) (ICN Biomedicals). A 5-μl amount of each in vitro translation reaction product was size fractionated on 10% SDS polyacrylamide gels under reducing conditions. Translation products were visualized with fluorography using EN3HANCE autoradiography enhancer (Dupont), followed by autoradiographic analysis of dried gels using a Storm phosphorimager and ImageQuant software version 1.2MAC (Molecular Dynamics). Linker mutations were introduced into the eNOS 3′ UTR using modified PCR-based linker mutagenesis, as described by Gustin and Burk (29), with sequences unrelated to known mRNA regulatory element sequences. The final mutated sequences were 5′-GCA CAA GCT TCC AGG-3′ for P1 through P5 and 5′-ACA AGC TTC C-3′ for ΔARE. Briefly, two PCR amplicons were generated (a 5′ fragment and a 3′ fragment). The primers used to create the mutants are listed in Table S1 in the supplemental material. The 5′ amplicon was generated using primers Main-S and XXX-MAS (mutagenic antisense). Main-S is a gene-specific primer corresponding to nucleotides 3281 to 3298 in exon 25 of the eNOS cDNA. In XXX-MAS, XXX indicates that the 15-bp target of the mutation is a 36-mer antisense primer containing 22 nucleotides identical to eNOS cDNA and a 14-nt mutation at the 5′ end of the primer heterologous to eNOS cDNA. The 3′ amplicon was generated in a similar manner, using primers Main-AS and XXX-MS (mutagenic sense); Main-AS is an antisense primer homologous to base pairs 65 to 87 3′ to the site of cleavage in the 3′ flanking genomic region of eNOS. 5′ and 3′ amplicons were subcloned into pCR II (Invitrogen) and sequenced on an ABI Prism 377 sequencer (Applied Biosystems). The amplicons were then subcloned between the XhoI and NotI sites of the wild-type (WT) expression vector, creating 15-bp mutations in the targeted regions. In the case of 3′-UTR deletions, ligation of the 2 amplicons resulted in an internal deletion of sequences between the primers. For the triple P2/P3/P4 mutant, primers PYRIII-MS-Acl I and PYRIII-MAS-Acl I were used to amplify 3′ and 5′ amplicons using the mutant P3 and mutant P4 constructs, respectively. The final constructs were verified by sequence analysis.
Plasmid DNA was prepared using two rounds of gradient sedimentation ultracentrifugation in ethidium bromide-saturated cesium chloride cushions. A 1-μg amount of PvuI-linearized plasmid was transfected, using Lipofectin (Invitrogen), into 60-mm dishes that had been seeded 18 to 24 h earlier at a density of 7 × 105 cells/ml (3.5 ml). Cells were passaged 48 h after transfection into selection/maintenance medium containing 800 μg/ml G418 (Invitrogen). G418-resistant clones were pooled to avoid the potential effects of site-specific integration events. Multiple independent transfections were carried out in HepG2 cells with a minimum of three transfections per cell line. Estimation of integrated plasmid copy number was performed by slot blot Southern hybridization as described previously (30), using genomic DNA harvested at the time of mRNA half-life determination and a 1.1-kb midregion eNOS cDNA probe.
mRNA half-life determination using RNase protection assay and quantitative RT-PCR.
Cells were grown to confluence, and the medium replaced. Forty-eight hours later, actinomycin D was added to a final concentration of 5 μg/ml. At appropriate time points, total RNA was harvested using the method of Chomczynski and Sacchi (31). eNOS RNA levels were quantitated by RNase protection assay using the RPAII or RPAIII kit according to the manufacturer's suggestions (Ambion). Briefly, 10 to 20 μg of total cellular RNA from each time point was hybridized for 16 h at 42°C to [α-32P]CTP antisense riboprobes (approximately 1 × 109 cpm/μg) complementary to the terminal 292 nucleotides of the final exon, 350 nucleotides of exons 13 to 15 of the human eNOS cDNA, or 251 nucleotides of the firefly luciferase open reading frame generated using T7 or T3 MaxiScript in vitro transcription kits (Ambion). Human eNOS riboprobe cross-reactivity with bovine eNOS was limited to fragments less than 25 nt in length. Sample loading was monitored by the inclusion of a probe complementary to 98 nucleotides of the human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA in the hybridization reaction mixtures. Protected fragments were size fractionated through 5% acrylamide-7 M urea gels, followed by exposure to phosphorimager screens. Signal intensities were quantitated using ImageQuant version 1.2 (Molecular Dynamics), and mRNA half-lives calculated assuming exponential decay kinetics by nonlinear regression of observed data points (32, 33). RNA half-lives are expressed as the mean half-life and range representing the standard error of the mean (SEM) of the half-life calculated from the SEM of decay constants. Decay constants were tested for significant differences using F-tests. Alternatively, RNA levels were determined using quantitative reverse transcription (RT)-PCR at specific time points following actinomycin treatment.
Cytoplasmic extract preparation and RNA electrophoretic mobility shift assay (EMSA).
S100 cytoplasmic extracts were prepared from K562 cells or HUVEC as described previously (34). Briefly, approximately 1 × 109 HUVEC were harvested by trypsinization followed by inactivation using trypsin inhibitor (Invitrogen) and were washed three times in Dulbecco's phosphate-buffered saline (DPBS). Approximately 1 × 1010 K562 cells were harvested by centrifugation and washed three times in DPBS. Cells were resuspended (2 × 108 cells/ml) in hypotonic lysis buffer containing 20 mM HEPES (pH 7.5), 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol (DTT), and 1× Complete protease inhibitor cocktail, EDTA-free (Roche), at 4°C. Resuspended cells were lysed with 25 strokes of a type B pestle in a 15-ml Dounce homogenizer. Nuclei and cell debris were pelleted by centrifugation at 5,000 × g for 5 min at 4°C. The supernatant was adjusted to 40 mM KCl and centrifuged at 100,000 × g for 1 h at 4°C (38,000 rpm in a Beckman type 70.1 Ti rotor). The S100 supernatant was collected and adjusted to 5% glycerol (vol/vol). The protein contents of the extracts were determined using Bio-Rad protein assay reagent and bovine serum albumin standards. The extracts were aliquoted and stored at −80°C until use.
Templates for in vitro transcription of RNA electrophoretic mobility shift assays were constructed one of two ways. The first used partially double-stranded synthetic oligonucleotides as described previously (35), using the oligonucleotides 5′-CTC AAG AGG GGA GGA GCG GGG CTG ATC CTG GTC GGG CGG ACC TGA GTC GGG CAG CCG CTT CCC TAT AGT GAG TCG TAT TA-3′, which correspond to nucleotides +34 to +93 relative to the first nucleotide of the human eNOS translation stop codon, and 5′-TAA TAC GAC TCG CTA TCG-3′, which is complementary to this oligonucleotide, creating a double-stranded T7 promoter. Alternatively, wild-type and mutant eNOS expression cassettes were used as PCR templates in reaction mixtures designed to add synthetic T7 promoter sequences 5′ to the amplified region. The primer sequences 5′-TAA TAC GAC TCA CTA TAG GGT GGT GCC TTC TCA CAT C-3′ or 5′-TAA TAC GAC TCA CTA TAG GGA AGG AGC AAA ACG C-3′ and 5′-AGG CCC GAG GCA ACA GGC-3′ were used to amplify nucleotides 95 to 201 or 149 to 201 (containing P2) relative to the first nucleotide of the human eNOS translation stop codon. The primer sequences 5′-TAA TAC GAC TCA CTA TAG GAG CAA AAC GCC TCT TTC C-3′ and 5′-TGA GAG AGG CAA GAG GAA TCT AAC-3′ were used to amplify nucleotides 157 to 333 (containing P2, P3, and P4). Following amplification, PCR products were subcloned into pCRII (Invitrogen) and their sequences verified. Internally [α-32P]CTP-labeled sense riboprobes were synthesized using T7 Maxiscript kits (Ambion) and isolated by preparative gel electrophoresis through denaturing polyacrylamide gels. The full-length transcription products (specific activities of 1 × 109 to 5 × 109 cpm/μl) were eluted overnight in probe elution buffer (RPAII kit; Ambion) and ethanol precipitated. Approximately 0.5 ng (5,000 cpm) of labeled probe was incubated with 40 μg of extract in a final volume of 15 μl containing 1% 2-mercaptoethanol. The binding reactions took place at 4°C for 20 min following a 20-min preincubation with 0.125 U/μl SUPERase•In RNase inhibitor (Ambion) and antibodies/competitors as indicated below. Following a 10-min RNase treatment with 10 ng RNase A and 1 U RNase T1 at room temperature, nonspecific protein interactions were suppressed by a further 10 min of competition with heparin (LEO Pharmaceutical) at a final concentration of 5 μg/μl. The reaction products were electrophoresed through 6% nondenaturing acrylamide (60:1 acrylamide/bis)–0.5× Tris-borate-EDTA gels at 8 V/cm. RNA EMSA experiments were replicated a minimum of 3 times using at least 2 independently prepared cell extracts. For competition studies, unlabeled competitor RNAs were synthesized using T7 Megascript kits (Ambion) and quantified by trace labeling with [α-32P]CTP. Homoribopolymer competitors were from Sigma-Aldrich. Polyclonal antibodies FF1, FF2, and FF3, directed against hnRNP E1, E2, and E2-KL, respectively, were kindly provided by Stephen A. Liebhaber, University of Pennsylvania (36). FF1 is specific for hnRNP E1, and FF2 recognizes hnRNP E2 but not E2-KL or E1, while FF3 recognizes both hnRNP E2 and E2-KL but not E1.
hnRNP E knockdown.
Eighty-five to 95% confluent HUVEC were transfected with hnRNP E1- or hnRNP E2-specific ShortCut siRNAs (NEB) at a final concentration of 15 nM to 20 nM using Oligofectamine (Invitrogen) for 72 h as previously described (37). Following transfection, total cellular protein and RNA were extracted using the mirVana PARIS kit (Ambion).
RNA RIP.
RNA immunoprecipitations (RIPs) were performed using Magna RIP RNA-binding protein immunoprecipitation kits (Millipore) according to the manufacturer's recommendations. Approximately 1.5 × 107 cells were used for each RIP reaction mixture. Anti-hnRNP E1 rabbit polyclonal antibody (Cell Signaling), anti-hnRNP E2 (H-15) goat polyclonal antibody (Santa Cruz Biotechnology), and anti-DYKDDDDK tag (9A3) mouse monoclonal antibody (Cell Signaling) were used at a 1:50 dilution.
ChIP.
Chromatin immunoprecipitations (ChIPs) were performed in HUVEC as previously described (38).
Transient plasmid transfection.
sONE overexpression: HepG2 cells stably expressing wild-type and mutant eNOS were transfected with pMACS Kk.II plasmids (Miltenyi Biotec) expressing a 1,555-nt sequence derived from sONE exons 10 to 12 that contain the 662 nucleotides that overlap eNOS. Transfected cells were enriched as described in reference 17, and verification of immunomagnetic enrichment of transfected cells was performed by monitoring the expression of the red fluorescent protein DsRed2 encoded by the pDsRed2 N1 plasmid. Enrichment was observed to be >90%. Forty-eight hours after enrichment, total cellular protein and RNA were extracted using the mirVana PARIS kit (Ambion). Plasmids encoding the FLAG-tagged wild-type hnRNP E1 (pCMV14-3X Flag-hnRNP E1) and the S43A mutant (with an S-to-A change at position 43) (pCMV14-3X Flag-hnRNP E1-S43A) (kindly provided by Philip H. Howe, Medical University of South Carolina) were transfected by electroporation into 1 to 2 million HUVEC resuspended in 400 μl of 5% fetal bovine serum in Opti-MEM (Invitrogen) using a BTX ECM 830 (Harvard Apparatus), with a single pulse of 200 V for 70 ms (20 μg plasmid per transfection).
Manipulation of Dicer and microRNA expression.
Dicer knockdowns in HUVEC were performed using Oligofectamine as previously described (37). Transfection of synthetic hsa-miR-765 mimics (Dharmacon), antagomirs (i.e., hairpin inhibitors; Dharmacon), and corresponding negative controls were performed using Oligofectamine (Invitrogen) as previously described (37). Transfection efficiencies of >95% were achieved.
eNOS 3′-UTR-targeting siRNAs.
Transfection of a pool of three siRNAs (Dharmacon, Thermo Scientific) that target the eNOS 3′ UTR in HUVEC was performed as previously described (37), at a total final concentration of 5 nM. The siRNA sequences were 5′-CUG CAA GGA UUC AGC AUU AUU-3′, 5′-GGU CCG CCU UAA UCU GGA AUU-3′, and 5′-GUU UCU UAG UCG AAU GUU AUU-3′.
Akt and Pak1 kinase inhibition.
Cellular Akt activity was inhibited by the addition of Akt inhibitor IV (EMD Millipore) at a final concentration of 5 μM. Cellular Pak1 activity was inhibited by the addition of Pak inhibitor IPA-3 (EMD Millipore) at a final concentration of 5 μM. Cells were treated with the corresponding inhibitor for 1 h before being subjected to hypoxia.
Immunofluorescence and confocal microscopy.
HUVEC were seeded onto 0.2% gelatin-coated, 22-mm by 22-mm glass coverslips. Following treatment, cells were fixed in 4% paraformaldehyde and rinsed with PBS, followed by permeabilization and blocking with 1% BSA, 10% normal goat serum, 0.3 M glycine 0.1% PBS-Tween. Cells were stained using an anti-hnRNP E1 rabbit polyclonal (Abcam) antibody and Dylight 488-conjugated goat anti-rabbit polyclonal secondary antibody (Abcam). Coverslips were mounted on glass slides using Vectashield mounting medium with DAPI (4′,6-diamidino-2-phenylindole; Vector Laboratories) and visualized using a Zeiss Axio LSM 700 confocal microscope. Images were analyzed using the Zeiss ZEN and ImageJ softwares.
Quantitative RT-PCR.
First-strand synthesis and quantitative real-time RT-PCR were performed as previously described (37). The results were normalized to the results for 18S rRNA. Primer sequences for eNOS, endogenous sONE, 18S, and CYPA were previously described in reference 11. Specifically, the sONE amplicon spans the exon 7/8 boundary of the gene. Primer sequences for hnRNP E1 were 5′-AGC CGC CAA AGA CTT GAC C-3′ (forward) and 5′-AGC CGA ATG GTG AGA GTC ACA-3′ (reverse). Primer sequences for hnRNP E2 were 5′-GAA GGT GGA TTA AAT GTC ACT-3′ (forward) and 5′-TCC CTT CTG AGA TGT TGA TAC-3′ (reverse). Primer sequences for total sONE were 5′-TCG CCG CAG ACA AAC AT-3′ (forward) and 5′-TGA GGG CGC AAT GGT AAC-3′ (reverse).
Immunoblotting.
Protein concentrations were determined using Bio-Rad protein assay kit II (Bio-Rad). Fifteen micrograms of total cellular protein was size fractionated on NuPAGE Novex 4 to 12% Bis-Tris gels (Invitrogen) and transferred onto nitrocellulose membranes. hnRNP E1 protein was detected using an anti-hnRNP E1 mouse polyclonal antibody (Novus Biologicals) (immunoblot) or an anti-hnRNP E1 rabbit polyclonal antibody (Cell Signaling) (immunoprecipitation and immunoblot). hnRNP E2 protein was detected using an anti-hnRNP E2 (5F12) mouse monoclonal antibody (Novus Biologicals). eNOS protein was detected using an anti-NOS3 (C-20) rabbit polyclonal antibody (Santa Cruz Biotechnology). Dicer protein was detected using an anti-Dicer (13D6) mouse monoclonal antibody (Abcam). Total Akt (Akt1, Akt2, and Akt3) protein was detected using an anti-Akt rabbit polyclonal antibody (Cell Signaling). Phospho-Akt protein was detected using an anti-phospho-Akt (Ser 473) rabbit polyclonal antibody (Cell Signaling). Serine-phosphorylated proteins were detected using an anti-phospho-serine (PSR-45) mouse monoclonal antibody (Abcam). Total Pak1 protein was detected using an anti-Pak1 rabbit polyclonal antibody (Cell Signaling). Phospho-Pak1 protein was detected using an anti-phospho-Pak1/Pak2/Pak3 (Thr 423) rabbit polyclonal antibody (Abcam). Threonine-phosphorylated proteins were detected using an anti-phospho-threonine rabbit polyclonal antibody (Abcam). Lamin A/C protein was detected using an anti-lamin A/C (346) mouse monoclonal antibody (Santa Cruz Biotechnology). α-Tubulin protein was detected using an anti-α-tubulin mouse monoclonal antibody (Sigma-Aldrich). Horseradish peroxidase (HRP)-linked goat anti-rabbit IgG (Santa Cruz Biotechnology) and HRP-linked rabbit anti-mouse IgG (Abcam) were used as secondary antibodies. Signal detection and analysis were performed using an ECL plus/prime instrument (GE Healthcare) and ImageJ (NIH), respectively.
Statistical analysis.
All results represent the means ± SEM of at least three independent experiments. Statistical analysis was performed using a two-tailed t test or analysis of variance (ANOVA) as appropriate. A P value of <0.05 was considered to be statistically significant.
RESULTS
The 3′ UTR confers exceptional stability to eNOS mRNA.
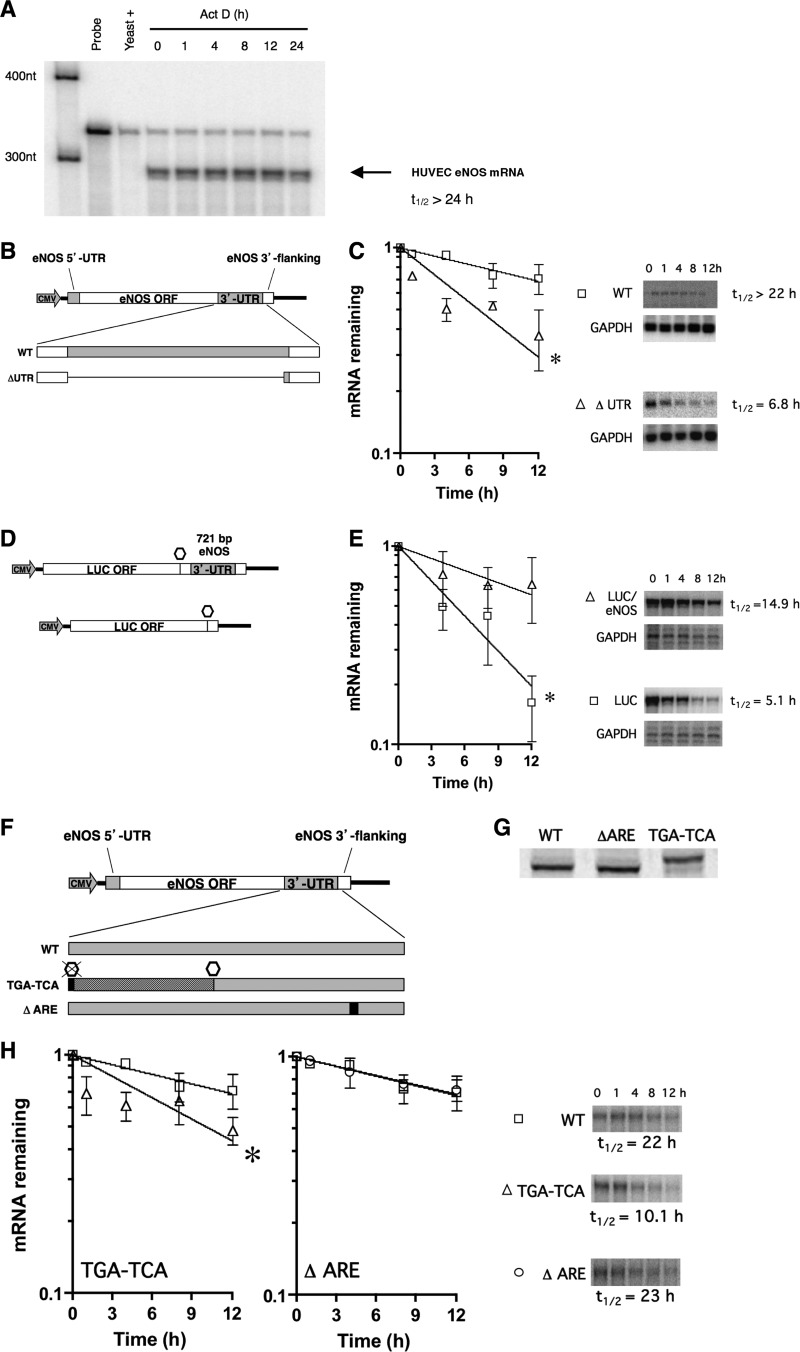
mRNA stability is directed by complex interactions between a variety of RNA binding factors and mRNA regulatory cis elements, an important number of which are found in 3′ UTRs. The human eNOS mRNA is highly stable in ECs, with a half-life in excess of 24 h (Fig. 1A). To address the functional contribution of the 421-nt human 3′ UTR to eNOS mRNA stability, we expressed full-length eNOS mRNAs either with (WT) or without the 3′ UTR (ΔUTR). Stable transfections of eNOS expression constructs were performed in HepG2 cells, which do not express eNOS. Wild-type and ΔUTR expression cassettes contained eNOS 3′ polyadenylation signals and 3′ flanking genomic sequences (Fig. 1B). In HepG2 stable cell lines, the removal of the 3′ UTR from eNOS resulted in a decrease in mRNA half-life from 22 h to 6.8 h (Fig. 1C), thus indicating that cis elements contained therein confer enhanced stability to the human eNOS mRNA.
Fig 1.
The eNOS 3′-UTR contains cis elements necessary for active stabilization. (A) Half-life (t1/2) measurements of native eNOS mRNA in HUVEC using RNA protection assays. (B) Schematic of constructs used to create cell lines in HepG2 cells stably expressing full-length eNOS and eNOS mRNA lacking 3′-UTR sequences (ΔUTR). ORF, open reading frame. (C) Half-life measurements of wild-type eNOS mRNA (WT) and ΔUTR mRNA from stably transfected expression cassettes in HepG2 cells. Representative gels are shown. (D) Schematic of constructs used to created stable HepG2 cell lines expressing luciferase or chimeric luciferase/human eNOS mRNA. Open octagons in the schemata indicate luciferase coding stop codon. Black lines represent vector sequence. (E) Half-life measurements of luciferase mRNA (LUC) and chimeric luciferase/eNOS mRNA (LUC/eNOS) from stably transfected HepG2 cells after actinomycin D treatment. Representative gels are shown. (F) Mutation of the eNOS wild-type stop codon (TGA to TCA), represented by a black box, allows translating ribosomes to read 180 nucleotides into the eNOS 3′ UTR (dark grey box) before reaching the next in-frame stop codon (open hexagon). Mutation of the ARE nonamer is depicted as a black box. (G) Translational read-through was confirmed in the TGA-to-TCA mutant by in vitro translation of synthetic RNA in rabbit reticulocyte lysates. Representative gels are shown. (H) Half-life measurements of wild-type eNOS mRNA (WT), the translation antitermination mutant (TGA-TCA), and the ARE nonamer sequence mutant (ΔARE) mRNAs from stably transfected expression cassettes in HepG2 cells. Representative gels are shown. Data represent means ± SEM (n = 3). *, P < 0.05.
In the converse experiment, we tested the ability of the eNOS 3′ UTR to stabilize a heterologous firefly luciferase reporter mRNA. We expressed luciferase mRNAs from stably transfected expression constructs containing nucleotides 3338 to 4052 of the human eNOS cDNA (293 nucleotides of the eNOS open reading frame and the entire 3′ UTR) (Fig. 1D). Inclusion of the full-length eNOS 3′ UTR stabilized luciferase reporter mRNA compared to the stability of the reporter mRNA that did not contain the 3′ UTR (Fig. 1E). The half-lives of the reporter mRNAs were 5.1 h and 14.9 h for LUC and LUC/eNOS mRNA, respectively. Given that the addition of the eNOS 3′ UTR prolonged the half-life of heterologous reporter mRNA, we concluded that the eNOS mRNA 3′ UTR contains stabilizing cis elements that can be transferred to heterologous sequences.
Next, to further examine the functional significance of the eNOS 3′ UTR, we employed a biologically relevant screen of the proximal half of the eNOS 3′ UTR. This was done by mutating the wild-type eNOS translation termination codon (TGA) to a serine codon (TCA), allowing translating ribosomes to read 180 nucleotides further into the 3′ UTR before reaching the next in-frame stop codon (TAG, positions 178 to 180) (Fig. 1F). The rationale for this experiment is based upon the molecular mechanism underlying the human α2-globin Constant Spring mutation, which is the most common form of nondeletional α-thalassemia (39). This mutation prevents the active stabilization of α2-globin mRNAs by disrupting the formation of stabilizing complexes between α-complex proteins (αCPs), also known as hnRNP E proteins or poly(C) binding proteins (PCBPs), and important pyrimidine (C and CU)-rich sequences in the α2-globin mRNA (40, 41). Thus, by mutating a single nucleotide and making a mutation equivalent to that observed in human α2-globin Constant Spring, we functionally screened approximately half of the eNOS 3′ UTR. In addition, we created a 10-nt mutation to target a conserved AU-rich element (ARE)-like nonamer [UUAUUUA(A/U)(A/U)] (42–45) in the distal 3′ UTR, just upstream of the cleavage and polyadenylation site (Fig. 1F) (28). This sequence is followed very closely by an ARE-like pentamer (AUUUA) that is also conserved among the species we compared. AREs often play a central role in the rapid turnover of labile mRNAs (reviewed in references 46 and 47). Translational read-through of proximal eNOS sequences was confirmed by in vitro translation, which revealed the extended open reading frame (Fig. 1G). When translating ribosomes were allowed to read through the proximal 180 nucleotides of the eNOS 3′ UTR in the TGA-to-TCA mutant, the mRNAs were considerably less stable than wild-type mRNAs (22-h versus 10.1-h half-life) (Fig. 1H). It is of interest that the P1 and P2 but not the P3 or P4 cis elements (see next section) are affected by the TGA-to-TCA mutation. In contrast, mutation of the nonamer component of the ARE did not significantly change the eNOS mRNA half-life (23 h) (Fig. 1H). From these data, we concluded that mRNA sequences in the proximal 180 nucleotides of the eNOS 3′ UTR are necessary for the mRNA stabilization, whereas ARE-like sequences in the distal eNOS 3′ UTR do not contribute significantly to basal mRNA stability.
Multiple conserved 3′-UTR cis elements contribute to eNOS mRNA stability.
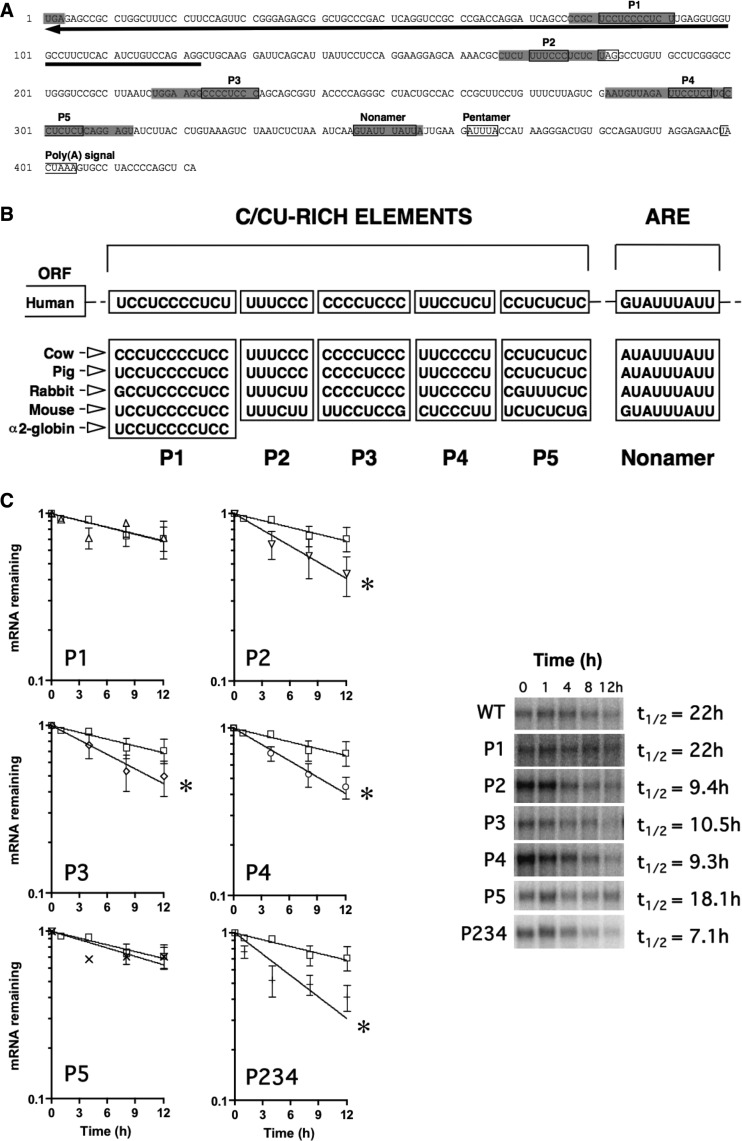
We determined, in silico, potential regulatory cis elements in the eNOS mRNA 3′ UTR (Fig. 2A and B). We cloned cDNAs from rabbit and mouse endothelial cell cDNA libraries and extracted data from publicly available databases. Cross-species comparison of eNOS 3′-UTR sequences revealed that overall nucleotide identities with the human eNOS 3′ UTR were 57%, 59%, 66%, and 37% for bovine, porcine, rabbit, and mouse eNOS 3′ UTRs, respectively. In contrast with this overall lack of sequence conservation, we noted several highly conserved regions. In particular, multiple pyrimidine (C and CU)-rich elements are evolutionarily conserved and shared, in part, with sequences responsible for the formation of the α2-globin mRNA stability complex (Fig. 2A and B) (48). Pyrimidine-rich elements have been associated with highly stable mRNAs (48). We will refer to these pyrimidine-rich sequence elements in eNOS as P1 through P5, respectively. The first of the conserved pyrimidine-rich elements (P1) is located in a region complementary to the eNOS antisense transcript sONE. The conserved nonamer ARE (which was mutated as described in the legend to Figure 1F) is also indicated (Fig. 2A and B).
Fig 2.
Conserved cis elements in the eNOS 3′ UTR. (A) Conserved sequence elements (boxed) of the human eNOS 3′ UTR. The stop codon (UGA) is shaded, as are regions that were mutated for functional analysis. The next in-frame stop codon, UAG (boxed), is also indicated. The region of complementarity to sONE is denoted by a black arrow. (B) Cross-species sequence alignment of conserved elements in the eNOS 3′ UTR. (C) Half-life measurements of wild-type (WT) and mutant (P1 to P5) eNOS mRNAs in HepG2 cells. P234 represents a construct containing targeted mutations of P2/P3/P4 in combination. Representative gels are shown. Data represent means ± SEM (n = 3 or 4). *, P < 0.05.
To further define the functional importance of the pyrimidine-rich cis elements in the eNOS mRNA 3′ UTR, specifically as to their potential role in the active stabilization of the eNOS mRNA, a series of 15-nt linker mutations were used to target the highly conserved P1 through P5 elements (Fig. 2A). The mutant sequences (5′-GCA CAA GCT TCC AGG-3′) are unrelated to any known mRNA regulatory element sequences. Importantly, the results indicated that the P2, P3, and P4 mutant eNOS mRNAs were unstable compared to wild-type eNOS mRNA. Specifically, P2, P3, and P4 mRNAs had half-lives of 9.4 h, 10.5 h, and 9.3 h, respectively, versus 22 h for the wild type (Fig. 2C). Mutation of the P1 and P5 elements alone failed to decrease eNOS mRNA stability, resulting in half-lives of 22 h, and 18.1 h, respectively. Finally, mutant eNOS mRNAs containing the combined triple mutation of the P2, P3, and P4 elements (i.e., P2/P3/P4, abbreviated as the P234 mutant in figures) had a significantly shorter half-life (i.e., 7.1 h) than wild-type eNOS mRNAs (Fig. 2C). Importantly, the effects of the P3 and P4 mutations indicate that regions in the distal region of the eNOS 3′ UTR, beyond those affected by the TGA-to-TCA mutation, also contribute functionally to eNOS mRNA stability. From these results, we concluded that the conserved P2, P3, and P4 elements play a functional role in the active stabilization of eNOS mRNA.
RNP complexes form on eNOS 3′-UTR cis elements.
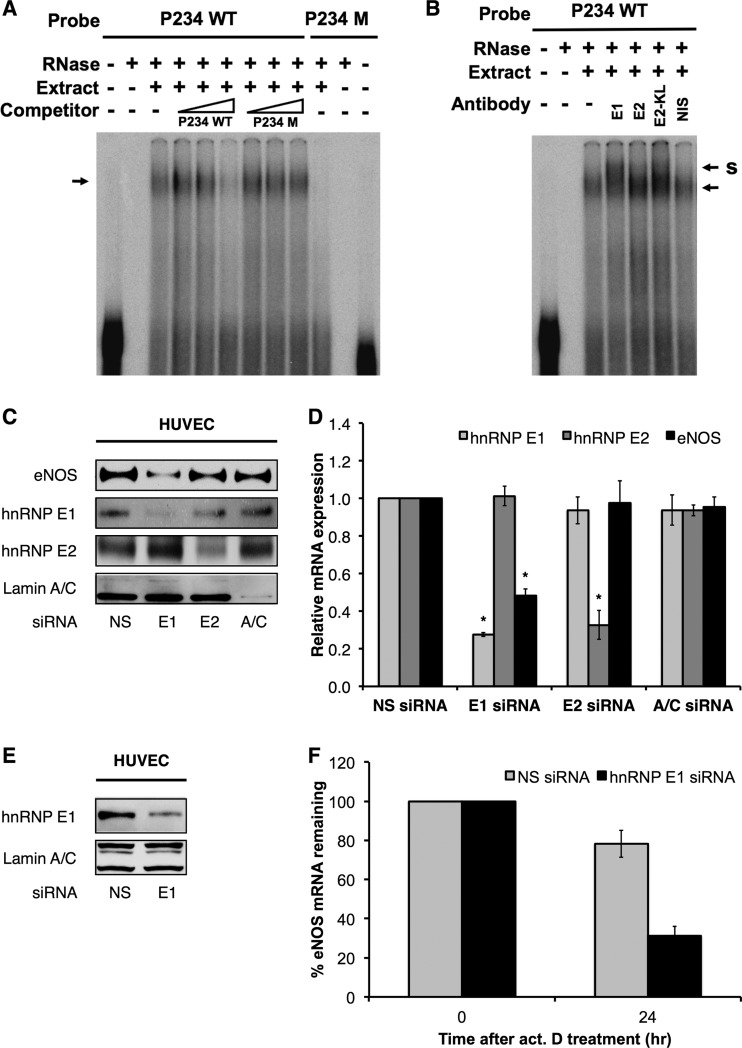
The pyrimidine-rich elements identified above that contribute to the exceptional stability of the eNOS mRNA bear strong homology to cis elements that recruit stabilizing RNP complexes (e.g., α2-globin mRNA) (Fig. 2B). Thus, we examined the interaction of these elements with cellular trans factors in HUVEC and K562 human erythroleukemia cells and compared the RNP complex formation with the formation of α2-globin RNP complexes that have been well characterized in K562 cells (49). The findings were comparable in both cell types. Importantly, we observed the formation of eNOS RNP complexes on probes comprising nucleotides 157 to 333 of the eNOS 3′ UTR, which span the P2/P3/P4 elements (Fig. 3A). Complex formation on the P2/P3/P4 sequences was inhibited by adding an increasing (10-, 100-, 250-fold) molar excess of unlabeled probe (lanes 4 to 6). Complex formation was not inhibited by the addition of an increasing molar excess of unlabeled mutant probe (lanes 7 to 9). Importantly, RNP complex formation was not observed on mutant P2/P3/P4 probes (lanes 10 to 12) (Fig. 3A).
Fig 3.
eNOS-stabilizing RNP complexes contain hnRNP E1. (A) In vitro-transcribed, 32P-labeled riboprobe containing the wild-type eNOS P2/P3/P4 sequences (P234 WT) was incubated with or without S100 cytoplasmic extract from HUVEC and unlabeled competitors (10-, 100-, and 250-fold molar excess) as indicated. Formation of eNOS RNP complexes in vitro (indicated by an arrow) was performed in the absence (3rd lane) and presence of 10-, 100-, and 250-fold molar excess of unlabeled P234 WT probe (4th to 6th lanes) or an unlabeled P234 M probe containing 15-nt mutations of eNOS P2, P3, and P4 sequences in the context of otherwise wild-type sequence (7th to 9th lanes). 32P-labeled P234 M riboprobes failed to form eNOS RNP complexes in vitro with HUVEC S100 extract (10th lane). (B) eNOS RNP complexes were formed as described for panel A. Supershift position is indicated by S. (C) Representative immunoblots of HUVEC under hnRNP E1- or hnRNP E2-specific siRNA knockdown conditions. Lamin A/C knockdown was performed as an internal control for siRNA specificity. (D) hnRNP E1, E2, and eNOS mRNA levels in HUVEC under hnRNP E1 or E2 knockdown conditions. (E) Representative immunoblots of HUVEC under hnRNP E1 (E1)-specific siRNA knockdown conditions. (F) Half-life measurements of eNOS mRNA in HUVEC under hnRNP E1 knockdown (hnRNP E1 siRNA) or nonsilencing (NS siRNA) conditions. Data represent means ± SEM (n = 3). *, P < 0.05 compared to results for corresponding nonsilencing siRNA. act. D treatment, actinomycin D transcription arrest.
Complex formation was also observed on probes containing the P2 element (comprising nucleotides 95 to 201 of the eNOS 3′ UTR) with trans factors in HUVEC (see Fig. S1A in the supplemental material) and K562 cell S100 cytoplasmic extracts (data not shown). In addition, the formation of RNP complexes on the P2 element was sensitive to competition by poly(CU) and, to some extent, poly(U) but not poly(C), poly(A), or poly(G) (see Fig. S1B), which is consistent with previous reports (40, 50). Importantly, complex formation was also observed on probes containing P1, P3, or P4 elements in isolation (data not shown). Significantly, these findings indicate that eNOS- and α2-globin-stabilizing RNP complexes contain common components.
Together, our findings strongly indicate that the formation of stabilizing RNP complexes on pyrimidine-rich elements (especially P2 to P4) in the eNOS 3′ UTR are functionally linked to the active stabilization of eNOS mRNA. These findings strongly suggest that eNOS 3′-UTR RNP complexes contain protein components of the α-complex, i.e., hnRNP E proteins.
hnRNP E1 is a major component of the eNOS-stabilizing 3′-UTR RNP complex.
To confirm the involvement of hnRNP E proteins in eNOS-stabilizing RNP complex formation, we performed RNA EMSAs using antisera directed against the major isoforms of hnRNP E proteins, i.e., hnRNP E1, E2, and E2-KL. Importantly, the results indicated that the addition of antiserum directed against hnRNP E1 produced supershifts of eNOS RNP complexes in HUVEC extracts (Fig. 3B). Supershifts were also observed occasionally and to a lesser extent with E2-KL antiserum. In contrast, supershifts were not evident with the addition of antisera directed against hnRNP E2 or nonimmune serum (NIS). Taken together, these data strongly implicate hnRNP E1 as a key protein component of the RNP complex that is responsible for the active stabilization and extraordinary basal stability of eNOS mRNA.
hnRNP E proteins are an integral part of the stabilizing α-complex and are similarly recruited to other highly stable mRNAs (48). Thus, to test the functional relevance of hnRNP E for eNOS expression, we performed siRNA-mediated knockdowns of hnRNP E1 and E2. Steady-state measurements showed that eNOS protein (Fig. 3C) and mRNA expression (Fig. 3D) were decreased by ∼60% and ∼50%, respectively, under E1 but not E2 knockdown conditions. In addition, eNOS mRNA stability measurements using actinomycin D showed that eNOS mRNA stability is significantly decreased under E1 knockdown conditions (i.e., ∼15 h, compared to the basal half-life of >48 h) (Fig. 3E and F). Taken together, these results strongly support hnRNP E1 as a major component of the 3′-UTR RNP complex that actively stabilizes eNOS mRNA.
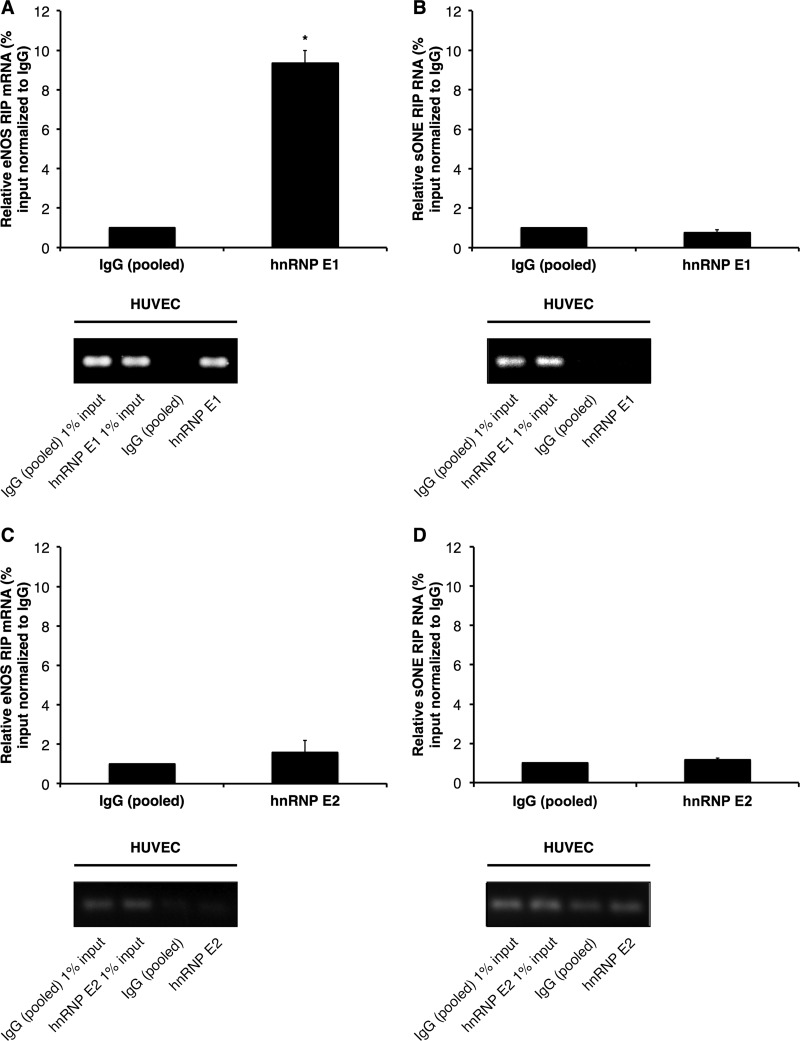
To confirm the interaction between the pyrimidine-rich elements in eNOS 3′ UTR and hnRNP E proteins, we performed RNA immunoprecipitation (RIP) experiments against hnRNP E1 in HUVEC that express native eNOS. The results indicated that hnRNP E1 associated specifically with endogenous wild-type eNOS mRNA, with a significant, ∼10-fold enrichment over control IgG pulldowns (Fig. 4A). In contrast, hnRNP E1 was not associated with sONE RNA (Fig. 4B). Furthermore, hnRNP E2-specific RIPs revealed no significant associations between either eNOS mRNA (Fig. 4C) or sONE RNA (Fig. 4D) and hnRNP E2. CYPA mRNA, used as a negative control, was not associated with either hnRNP E1 or E2 (see Fig. S2 in the supplemental material).
Fig 4.
hnRNP E1 but not E2 associates with eNOS mRNA. hnRNP E1 or E2 and associated RNAs were immunoprecipitated using an hnRNP E1-specific or hnRNP E2-specific antibody in HUVEC. Levels of hnRNP E1-associated eNOS mRNA (A) and sONE RNA (B) and of hnRNP E2-associated eNOS mRNA (C) and sONE RNA (D) are shown. Data are presented as percentage of input relative to measurements of control IgG (pooled) pulldown. Data represent means ± SEM (n = 3). *, P < 0.05 compared to results for control IgG (pooled). Bottom panels represent real-time PCR products obtained from the experiments whose measurements are shown in the respective top panels.
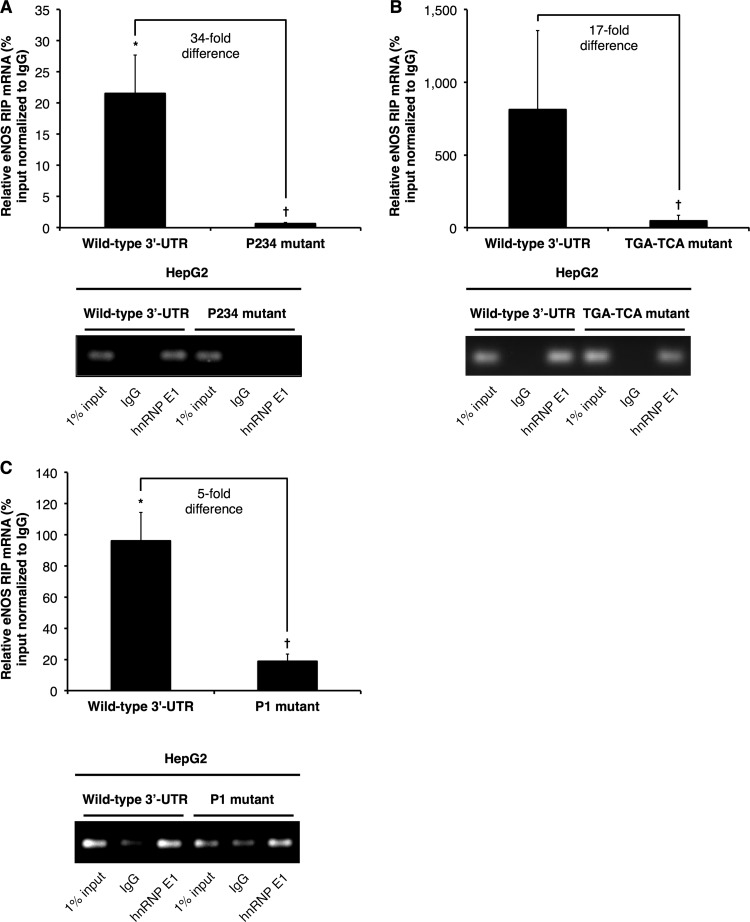
The specific association of eNOS mRNA with hnRNP E1 was recapitulated in HepG2 cells that stably express wild-type eNOS mRNA (Fig. 5), as the results indicated significant enrichment of eNOS mRNAs in hnRNP E1 versus control IgG pulldown samples (Fig. 5). Importantly, mutating the P2, P3, and P4 elements disrupted the interaction between hnRNP E1 and eNOS mRNAs (Fig. 5A and B). Specifically, the levels of association of P2/P3/P4 (Fig. 5A) and the TGA-to-TCA (Fig. 5B) mutant eNOS mRNAs with hnRNP E1 were ∼34-fold and ∼17-fold lower, respectively, than the level of association between wild-type eNOS mRNA and hnRNP E1. Similarly, we observed an ∼5-fold decrease in the association between eNOS mRNA and hnRNP E1 when the P1 element was mutated (Fig. 5C). These findings are consistent with the results of the RNA EMSAs and, taken together with the mRNA half-life measurements, strongly indicate that hnRNP E1, as part of a 3′-UTR RNP complex, plays an important function in the active stabilization of eNOS mRNA. hnRNP E proteins are known to shuttle between the nucleus and cytoplasm, with functional consequences for the translational efficiencies of their target mRNAs (e.g., α2-globin mRNA) (51–54). Indeed, chromatin immunoprecipitations (ChIPs) against hnRNP E1 revealed specific E1 enrichment at eNOS P2/P3/P4 elements compared to the amount at the eNOS promoter (data not shown). These results indicate that the interaction of hnRNP E1 with eNOS mRNA occurs as an early post- or cotranscriptional event. Future studies will be necessary to define the contribution of additional factors in the association of hnRNP E1 with eNOS mRNA.
Fig 5.
hnRNP E1 associates with wild-type but not mutant eNOS mRNA 3′ UTR. hnRNP E1 and associated RNAs were immunoprecipitated using an hnRNP E1-specific antibody in HepG2 stable cell lines. hnRNP E1-associated eNOS mRNA levels in HepG2 cells that stably express full-length eNOS mRNA with wild-type 3′ UTR, the P2/P3/P4 3′-UTR mutant (P234 mutant) (A), the TGA-to-TCA mutant (B), or the P1 3′-UTR mutant (P1 mutant) (C). Data are presented as percentage of input relative to measurements of control IgG pulldown. Data represent means ± SEM (n = 3 or 4). * and †, P < 0.05 compared to results for control IgG and wild-type 3′ UTR, respectively. Bottom panels represent real-time PCR products obtained from the experiments whose measurements are shown in the respective top panels.
Stabilization protects eNOS mRNA from sONE antisense-mediated inhibition.
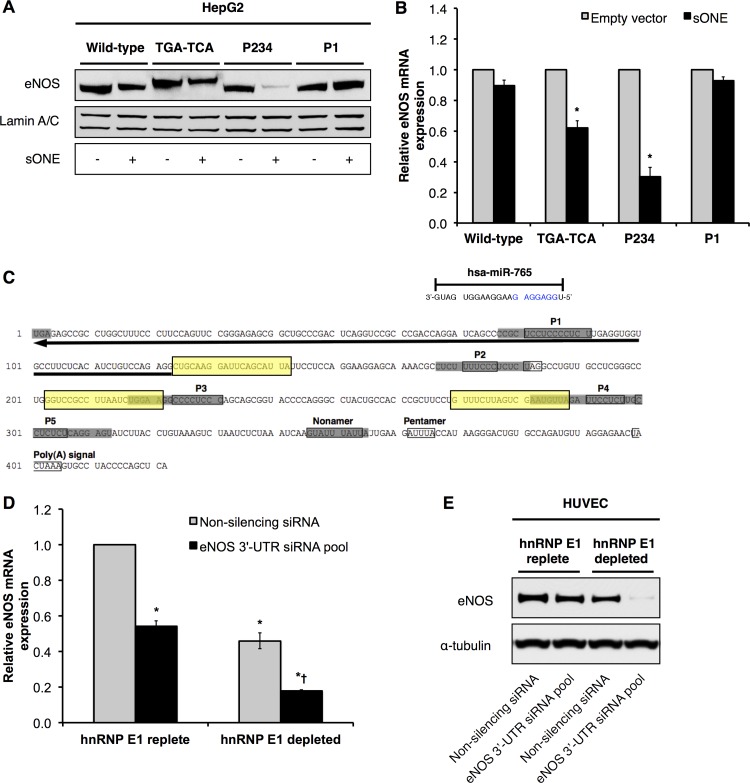
Next, we sought to determine the biological significance of active eNOS mRNA stabilization by hnRNP E1-containing RNP complexes. We have previously identified an inhibitory antisense transcript to eNOS, i.e., sONE, which is arranged in a tail-to-tail orientation to eNOS, and the mRNAs for the 2 genes are complementary for a total of 662 nucleotides, including significant exon/exon overlap (17). Importantly, sONE regulates eNOS expression via posttranscriptional mechanism(s) (17). Therefore, we hypothesized that the eNOS-stabilizing 3′-UTR RNP complex protects and allows eNOS mRNA to accumulate despite the inhibitory antisense effects of sONE. To test this hypothesis, we overexpressed a region of the sONE transcript containing the maximal (622-nucleotide) overlap with eNOS in HepG2 cells that stably express full-length wild-type (WT) and 3′-UTR mutant eNOS. We first confirmed that total sONE expression was increased by 10- to 12-fold following sONE overexpression (see Fig. S3A in the supplemental material). As expected, this increase in overall sONE expression was attributed to overexpression of the sONE overlap region, since the endogenous levels of sONE remain unchanged under these conditions in HepG2 cells (see Fig. S3B in the supplemental material). Next, we measured eNOS protein (Fig. 6A) and mRNA (Fig. 6B) levels from the wild-type and mutant eNOS constructs. Compared to the levels in empty-vector-transfected cells, we observed an ∼35% decrease in eNOS protein levels when sONE was overexpressed in cells that express the wild-type eNOS 3′ UTR (Fig. 6A), while the eNOS mRNA levels remained essentially unaffected (Fig. 6B). This is consistent with our previous findings with endogenous eNOS in HUVEC (17). In contrast, both the eNOS protein and mRNA levels from the TGA-to-TCA mutant were reduced, to ∼55% and 60%, respectively, in the presence of sONE (Fig. 6A and B). Significantly, the expression of eNOS protein from the P2/P3/P4 triple mutant was the most dramatically affected, as eNOS protein was reduced to ∼15% in the presence of sONE (Fig. 6A) and eNOS mRNA levels similarly decreased to ∼30% (Fig. 6B). Interestingly, the levels of expression of eNOS protein and mRNA from the P1 mutant were unaffected by exogenous expression of sONE overlap regions (Fig. 6A and B). Taken together, these findings suggest that the formation of the stabilizing RNP complex protects eNOS mRNA from sONE-mediated posttranscriptional/translational downregulation.
Fig 6.
hnRNP E1 protects eNOS mRNA and protein from the antisense transcript sONE and 3′-UTR-targeting siRNAs. (A) Representative immunoblots of HepG2 cells that stably express full-length eNOS mRNA or 3′-UTR mutants and which were transiently transfected with a plasmid encoding either 1,555 nucleotides of RNA corresponding to the region of sONE that overlaps with eNOS or empty vector. (B) Wild-type and mutant eNOS mRNA levels in HepG2 cells transiently transfected with the sONE overlapping region or empty vector. Data represent means ± SEM (n = 3). *, P < 0.05 compared to results for corresponding empty vector. (C) Target sites of eNOS 3′-UTR-targeting siRNAs (boxed in yellow). Hsa-miR-765 binding site in eNOS 3′UTR as predicted by TargetScan is also indicated. Seed sequence is highlighted in blue. (D and E) eNOS mRNA expression (D) and representative immunoblots (E) following siRNA treatment of HUVEC under nonsilencing knockdown (hnRNP E1-replete) versus hnRNP E1 knockdown (hnRNP E1-depleted) conditions. Data represent means± SEM (n = 3). * and †, P < 0.05 compared to results for nonsilencing siRNA and eNOS 3′-UTR siRNA pool, respectively, in hnRNP E1-replete HUVEC.
Regulation of eNOS expression is Dicer dependent.
RNA interference (RNAi) and microRNA regulation are well-established mechanisms of posttranscriptional gene regulation. Given the importance of posttranscriptional regulation of eNOS expression, as evidenced in this work and prior work from us and others (9–17), we sought to determine whether RNAi/microRNA pathways are implicated in the posttranscriptional regulation of eNOS and/or sONE, as well as the functional relevance of the stabilizing RNP complex in this context. Dicer is a key enzyme involved in the biogenesis and activity of microRNAs and siRNAs. We have recently shown that the downregulation of Dicer in hypoxia is an important adaptive mechanism that serves to maintain the cellular response to hypoxia (37).
Thus, we first examined the Dicer dependence of basal eNOS and sONE expression. Importantly, our results indicated that eNOS mRNA and protein levels, as well as sONE RNA levels (see Fig. S4 in the supplemental material), were upregulated under Dicer knockdown conditions in HUVEC. These findings indicate that Dicer-dependent mechanisms are involved in the regulation of eNOS and sONE expression. Given that sONE downregulates eNOS posttranscriptionally (Fig. 6A), an increase in sONE should lead to a decrease in basal eNOS protein expression (17). However, the opposite was observed (see Fig. S4A and B). This paradoxical increase in eNOS protein expression upon Dicer knockdown, which has also been previously reported (55), suggests that sONE-independent, Dicer-dependent microRNAs could be repressing eNOS under basal conditions.
Stabilization protects eNOS mRNA from siRNA-mediated inhibition.
We first tested whether hnRNP E1-containing stabilizing RNP complexes can protect eNOS from destabilizing RNAi mechanisms. To do so, we transfected into HUVEC siRNAs that targeted the eNOS 3′ UTR, specifically at regions between the P2, P3, and P4 elements (Fig. 6C), under hnRNP E1-replete or -depleted conditions. We chose to design siRNAs against 3′-UTR sequences because endogenous microRNAs predominantly target the 3′ UTR. Consistent with the protective function of hnRNP E1, the siRNAs were much more effective at downregulating eNOS mRNA (Fig. 6D) and, especially, eNOS protein (Fig. 6E) in hnRNP E1-depleted cells. Thus, these results strongly support our model that hnRNP E1 is involved in the active stabilization and protection of eNOS mRNAs from siRNAs/microRNAs that target the eNOS 3′ UTR.
Stabilization protects eNOS mRNA from microRNA-mediated inhibition.
We performed in silico analyses to identify endogenous, biologically relevant microRNAs that target eNOS. The results indicated that eNOS is potentially targeted by hsa-miR-765, which is an abundant species (within the top 20% of all detected microRNAs) in HUVEC (37). Importantly, a putative target site of hsa-miR-765 was located within the P1 element in the eNOS mRNA 3′ UTR (as predicted by TargetScan) (Fig. 6C).
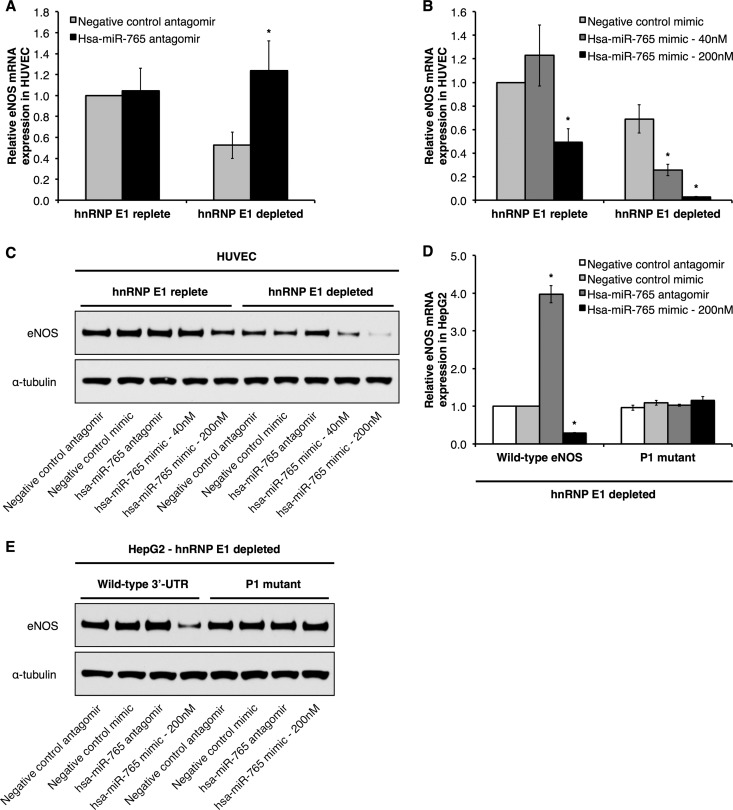
Next, we addressed the functional relevance of hsa-miR-765 for eNOS expression. To do so, we manipulated hsa-miR-765 expression in HUVEC by transfecting synthetic inhibitors (i.e., antagomirs) and mimics of hsa-miR-765 and measured steady-state levels of eNOS mRNA and protein. In hnRNP E1-replete HUVEC, we observed no effect on eNOS mRNA (Fig. 7A and B) or protein (Fig. 7C) levels when we inhibited hsa-miR-765 expression/activity with hsa-miR-765 antagomirs or overexpressed hsa-miR-765 mimics. Only when hsa-miR-765 mimics were expressed at very high levels (200 nM) did we observe modest decreases of ∼50% and ∼15% in eNOS mRNA (Fig. 7B) and protein expression (Fig. 7C), respectively. In contrast, however, hsa-miR-765 had a much more potent effect on eNOS mRNA and protein expression in cells depleted of hnRNP E1 (via siRNA-mediated knockdown of hnRNP E1). Here, hsa-miR-765 inhibition resulted in a significant, ∼2.5-fold increase in eNOS mRNA compared to the level in cells treated with a negative-control antagomir (Fig. 7A), and hsa-miR-765 overexpression at 40 nM and 200 nM final concentrations resulted in significant downregulation of eNOS mRNA, by ∼60% and ∼95%, respectively (Fig. 7B). Likewise, we observed in hnRNP E1-depleted HUVEC that eNOS protein levels were significantly elevated, by ∼1.3-fold, and reduced by ∼50% (at 40 nM) and ∼70% (at 200 nM) under hsa-miR-765 inhibition and overexpression conditions, respectively (Fig. 7C).
Fig 7.
hnRNP E1 protects eNOS mRNA and protein from hsa-miR-765. Relative eNOS mRNA expression in HUVEC that have been transfected with hsa-miR-765 antagomir (A) and hsa-miR-765 mimic (B) under hnRNP E1-replete versus hnRNP E1-depleted conditions. (C) Representative immunoblots of HUVEC transfected with hsa-miR-765 antagomir and mimic under hnRNP E1-replete versus hnRNP E1-depleted conditions. (D and E) eNOS mRNA levels (D) and representative immunoblots (E) of HepG2 cells that stably express either full-length eNOS mRNA with wild-type 3′ UTR or the P1 3′-UTR mutant (P1 mutant) and which have been transfected with hsa-miR-765 antagomir and mimic under hnRNP E1-depleted conditions. Data represent means ± SEM (n = 3). *, P < 0.05 compared to results with the corresponding negative-control antagomir or mimic.
Of great interest, the predicted target site for hsa-miR-765 in the eNOS 3′ UTR corresponds to the P1 element (Fig. 6C). Although mutation of the P1 element does not affect steady-state eNOS mRNA under basal conditions, the loss of microRNA binding could still be associated with stabilization of eNOS mRNA. Thus, we performed similar experiments in HepG2 cell lines that stably express either full-length eNOS mRNA with the wild-type 3′ UTR or the P1 3′-UTR mutant (P1 mutant) under hnRNP E1-depleted conditions. Similar to our findings in hnRNP E1-depleted ECs, wild-type eNOS mRNA expression was significantly increased, by ∼4-fold, when hsa-miR-765 was inhibited and significantly downregulated, by ∼70%, when hsa-miR-765 was overexpressed (Fig. 7D). Likewise, we observed a significant decrease in wild-type eNOS protein levels, by ∼65%, when hsa-miR-765 was overexpressed (Fig. 7E). Importantly, hsa-miR-765 inhibition and overexpression had no effect on the levels of eNOS mRNA and protein derived from the P1 mutant (Fig. 7D and E). Together, these results indicate that under basal conditions, stabilizing hnRNP E1-containing RNP complexes protect human eNOS from downregulation by hsa-miR-765. Thus, hnRNP E1 protects eNOS mRNA from the posttranscriptional inhibitory effects of both sONE and hsa-miR-765, thus serving as a protective mechanism that promotes overall eNOS mRNA and protein expression. Notably, hsa-miR-765 expression was significantly decreased in a Dicer-dependent manner in hypoxic HUVEC (see Fig. S5A in the supplemental material), with a corresponding accumulation of hsa-miR-765 precursors (see Fig. S5B). This is consistent with our recent report that chronic hypoxia leads to functional depletion of Dicer in ECs, which results in the downregulation of an important number of biologically relevant microRNAs, including hsa-miR-765 (37).
Hypoxia modulates the formation and function of the stabilizing complex on eNOS mRNA.
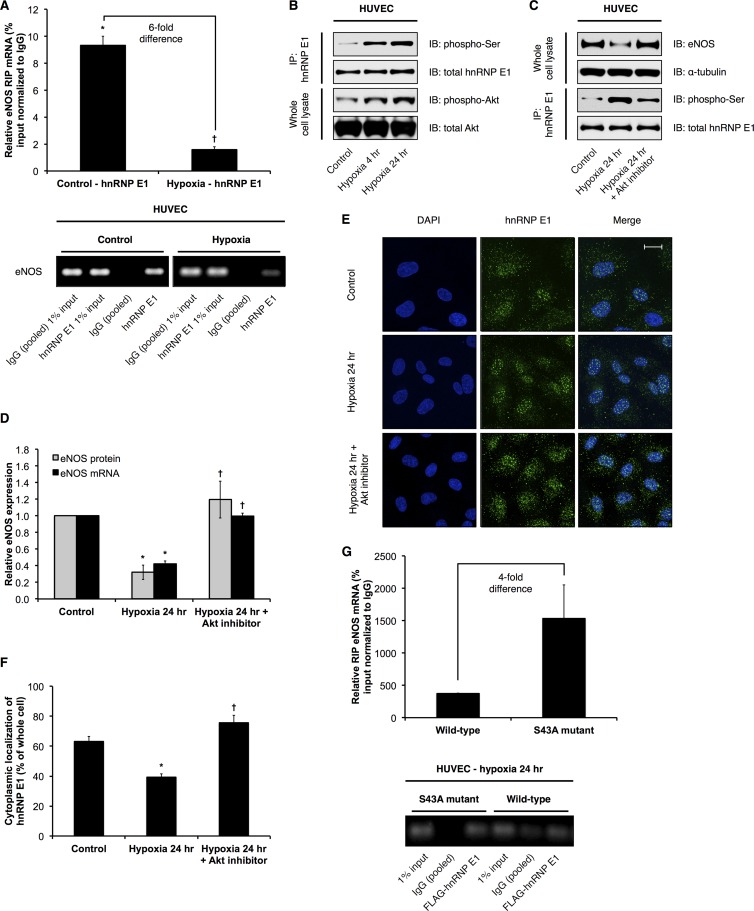
eNOS mRNA and protein expression are significantly decreased by hypoxia (see Fig. S5C to E in the supplemental material), in part due to posttranscriptional mechanisms (9, 11). In contrast, sONE is highly induced by hypoxia (see Fig. S5E); specifically, the increase in steady-state levels of sONE RNA is due to increased RNA stability rather than changes in the transcription rate of sONE (11). Given that the stabilizing RNP complex protects eNOS from sONE-mediated inhibition and that eNOS expression is downregulated by hypoxia, we wanted to determine if hnRNP E1/eNOS complex formation is disrupted in hypoxia. Notably, steady-state hnRNP E1 protein (see Fig. S5C and D) and mRNA (data not shown) expression are not affected by hypoxia (1% O2 for 24 h) in HUVEC. Thus, we performed RIPs against hnRNP E1 in control (21% O2) versus hypoxic (1% O2, 24 h) HUVEC and measured the associated eNOS mRNA levels. Consistent with our hypothesis, we observed a significantly reduced association of eNOS mRNA with hnRNP E1 under conditions of hypoxia relative to their association in the control (Fig. 8A). RIPs revealed that neither sONE RNA nor mRNA of the housekeeping CYPA (see Fig. S6 in the supplemental material) associated significantly with hnRNP E1 under either control or hypoxic conditions. Taken together, these results indicate that the posttranscriptional downregulation of eNOS in hypoxia is achieved, at least in part, by the inhibition of hnRNP E1-containing RNP complex formation on eNOS mRNAs. Importantly, we observed a significant increase in the serine phosphorylation of hnRNP E1 under hypoxic conditions (Fig. 8B). Notably, a recent study has shown that Akt-dependent serine 43 phosphorylation of hnRNP E1 inhibits its ability to bind to its target mRNAs (56). We found that Akt is phosphorylated and activated under hypoxic conditions in ECs (Fig. 8B). Importantly, inhibition of Akt activity in hypoxic HUVEC significantly rescued the hypoxia-mediated decrease in eNOS mRNA and protein expression, presumably due, at least in part, to the attenuation of the increase in hnRNP E1 serine phosphorylation (Fig. 8C and D). Significantly, we also observed, for the first time, that hypoxia led to increased nuclear retention of hnRNP E1 compared to its localization in the control, and Akt inhibition effectively abolished this increase in nuclear localization (Fig. 8E and F). Finally, we tested whether serine 43 is specifically involved in the reduced association of hnRNP E1 with eNOS mRNA. Thus, we performed RIPs against FLAG-tagged constructs containing either the wild-type or a phosphorylation-insensitive mutant hnRNP E1, whose serine 43 has been mutated to an alanine residue (S43A). Importantly, the results indicated that in hypoxic HUVEC, where wild-type hnRNP E1 is phosphorylated, the phosphorylation-insensitive S43A mutant exhibited a higher (i.e., ∼4-fold) degree of association with eNOS mRNA than did wild-type hnRNP E1 (Fig. 8G).
Fig 8.
Effects of hypoxia on hnRNP E1 posttranslational modification and subcellular localization and consequences for eNOS expression. (A) hnRNP E1-specific RNA immunoprecipitations (RIPs) were performed. hnRNP E1-associated eNOS mRNA levels in control (21% O2) versus hypoxic (1% O2, 24 h) HUVEC were measured. Data are presented as percentage of input relative to measurements of control IgG (pooled) pulldown. Data represent means ± SEM (n = 3). * and †, P < 0.05 compared to control IgG (pooled) and control hnRNP E1 levels, respectively. Bottom panel represents real-time PCR products obtained from experiments whose measurements are shown in the top panel. (B) Representative immunoblots (IB) of control versus hypoxic HUVEC hnRNP E1 immunoprecipitation (IP) samples and whole-cell lysates of control versus hypoxic HUVEC. (C) Representative immunoblots of whole-cell lysates and IP samples from control HUVEC, hypoxic HUVEC, and hypoxic HUVEC treated with 5 μM Akt inhibitor IV. (D) eNOS mRNA and protein levels (quantification of immunoblot results shown in panel C) in control HUVEC, hypoxic HUVEC, and hypoxic HUVEC treated with Akt inhibitor IV. Data represent means ± SEM (n = 3). * and †, P < 0.05 compared to results for corresponding control and 24-h hypoxia, respectively. (E) Subcellular localization of hnRNP E1 was determined by immunofluorescence using confocal microscopy in control HUVEC, hypoxic HUVEC, and hypoxic HUVEC treated with Akt inhibitor IV (representative image shown). The scale bar represents 20 μm. (F) Quantification of cytoplasmic hnRNP E1 based on images represented in panel E, expressed as percentage of total cellular hnRNP E1 immunofluorescence. Data represent means ± SEM (n = 3). * and †, P < 0.05 compared to results for control and 24-h hypoxia, respectively. (G) RNA immunoprecipitations (RIPs) were performed against FLAG-tagged wild-type hnRNP E1 versus the S43A mutant. Data are presented as percentage of input relative to measurements of control IgG (pooled) pulldown. Data represent means ± SEM (n = 3). Bottom panel represents real-time PCR products obtained from experiments whose measurements are shown in the top panel.
In addition to serine phosphorylation, hnRNP E1 can also be phosphorylated at threonine residues (e.g., threonine 60 and 127) in a Pak1-dependent manner, which also results in decreased association with target mRNAs, as well as nuclear retention (57). Indeed, increased hnRNP E1 threonine phosphorylation and Pak1 phosphorylation/activation were observed in hypoxic HUVEC (see Fig. S7A in the supplemental material). However, inhibition of Pak1 activity did not have any significant functional effects on hypoxia-mediated changes in eNOS expression (see Fig. S7B and C), threonine phosphorylation (see Fig. S7B and D), or hnRNP E1 subcellular localization (see Fig. S7E and F). Thus, in hypoxic ECs, serine phosphorylation (including serine 43) and nuclear localization of hnRNP E1 are increased in an Akt-dependent manner. These two events significantly inhibit the ability of hnRNP E1 to bind and stabilize eNOS mRNAs. Together with the increased cytoplasmic expression of the negative posttranscriptional regulator sONE, these mechanisms account, at least in part, for the decrease of eNOS mRNA stability and overall expression in hypoxic ECs.
DISCUSSION
eNOS mRNA is an extremely stable species whose expression is highly restricted to ECs. However, little has been known about the underlying molecular mechanisms behind eNOS mRNA stabilization, and more importantly, why eNOS mRNAs need to be stabilized. Previous studies have elucidated several possible rationales for maintaining stable transcripts in other cell types: reticulocytes express highly stable mRNAs, such as α2-globin, because these cells become transcriptionally quiescent and enucleated in later stages of development (48); neurons express highly stable mRNAs that have to survive the transport from the nucleus to the termini of long axons (58), and oocytes express maternal mRNAs that are highly stable since DNA replication and cell cycle progression preoccupy the chromatin template immediately following fertilization (59, 60). In addition, it is also intuitive that genes that exhibit low levels of transcription would stabilize their mRNAs so as to increase overall gene expression. However, none of the explanations above are compelling with respect to the stability of the eNOS mRNA in the vascular endothelium. Why does eNOS need to be stable? We have previously reported the characterization of an overlapping cis-antisense transcript to eNOS, sONE, that is involved in the posttranscriptional downregulation of eNOS (17). Furthermore, we have also demonstrated that in models of disease, such as hypoxia, decreases in eNOS expression can be attributed, at least in part, to sONE-mediated posttranscriptional downregulation (11). In this study, we report the identification of multiple evolutionarily conserved pyrimidine (C and CU)-rich elements in the eNOS mRNA 3′-UTR that contribute to the exceptional stability of the eNOS transcript through the formation of stabilizing hnRNP E1-containing RNP complexes.
hnRNP E1 is a member of the hnRNP E protein family that is known to regulate several highly stable mRNAs (48, 61–64). The three best-studied isoforms, hnRNP E1, E2, and E2-KL, interact with 3′-UTR pyrimidine-rich elements (e.g., α2-globin) to form stabilizing α-complexes (36). hnRNP E proteins are also implicated in the regulation of mRNA stability for collagen α1(I) (65), tyrosine hydroxylase (66), and erythropoietin (67) and are involved in the translational regulation of 15-LOX (68, 69) via interactions with 3′-UTR pyrimidine-rich elements (48). However, a role for these proteins in the cardiovascular system has not been identified until our current work. Employing multiple independent experimental approaches, we show here that hnRNP E1 is a major component of the eNOS-stabilizing RNP complex that forms on the conserved pyrimidine-rich elements in the eNOS mRNA 3′-UTR. Indeed, our evidence strongly supports the model that hnRNP E1 actively stabilizes and protects eNOS mRNA from inhibitory factors, such as antisense RNAs, microRNAs, and siRNAs, that target the eNOS 3′ UTR. To our knowledge, this represents the first time that the interaction of hnRNP E1 with mRNA 3′-UTR regulatory cis elements has been implicated in the protection of an mRNA from antisense RNA- and microRNA-mediated downregulation. Future detailed studies are required to identify potential additional factors that interact with hnRNP E1 as part of the eNOS-stabilizing 3′-UTR RNP complex, as well as whether hnRNP E1 interacts directly or indirectly with eNOS mRNA.
Importantly, we are the first to report an eNOS-targeting microRNA, i.e., hsa-miR-765. This microRNA targets the eNOS 3′ UTR at the conserved pyrimidine-rich element P1. Interestingly, the P1 element is the site of both stabilization (by hnRNP E1) and destabilization (by sONE and hsa-miR-765). Consequently, the P1 cis element mutation abrogates both of these opposing effects, which might explain why eNOS mRNA stability remained unchanged when the P1 element was mutated in isolation.
In addition to showing the basal stabilization of eNOS mRNA by hnRNP E1-containing RNP complexes, we have also provided strong evidence here that the interaction of hnRNP E1 with eNOS mRNA is inhibited during the environmental stress of hypoxia. Importantly, we provide evidence that serine phosphorylation of hnRNP E1 and Akt phosphorylation/activation are significantly increased under hypoxic conditions. An important recent study has reported that transforming growth factor β (TGF-β)-dependent, Akt2-mediated phosphorylation of hnRNP E1 serine 43 inhibits hnRNP E1 binding to its target transcripts (56). Significantly, we show here that phosphorylation of serine 43 impairs hnRNP E1 binding to eNOS mRNA. Consistent with the multifunctional roles of hnRNP E proteins, including hnRNP E1 (57, 70), we and others have observed that hnRNP E1 exists in both the nucleus and the cytoplasm (52, 71). Importantly, we show for the first time that hypoxia leads to increased nuclear localization of hnRNP E1, presumably due, at least in part, to Akt-mediated phosphorylation events, since this phenotype can be reversed by Akt inhibition. Indeed, our findings strongly support the model wherein Akt-mediated serine phosphorylation and nuclear sequestration prevents hnRNP E1 from interacting with and protecting eNOS mRNA from the inhibitory effects of sONE in hypoxic ECs. Notably, hsa-miR-765 is downregulated while sONE is highly induced under hypoxic conditions. eNOS is regulated at the posttranscriptional level by a number of stimuli other than hypoxia, such as TNF-α (14) and entry into cell cycle (12). For example, TNF-α (14) leads to decreases in eNOS mRNA and protein and destabilizes eNOS mRNA. The relative contributions of hsa-miR-765, sONE, and hnRNP E1 to the posttranscriptional regulation of eNOS in these other models of endothelial activation remain to be studied.
In summary, we have shown that the basal stabilization of human eNOS mRNA by hnRNP E1-containing RNP complexes is a key protective mechanism against the posttranscriptional inhibitory effects of the eNOS antisense transcript sONE and functionally important microRNAs, such as hsa-miR-765. However, the interaction between hnRNP E1 and eNOS mRNA is disrupted in hypoxia due to increased Akt-mediated serine phosphorylation (including serine 43) and increased nuclear localization of hnRNP E1, thus rendering eNOS susceptible to sONE-mediated downregulation. These mechanisms contribute significantly to the decrease in eNOS mRNA stability and overall eNOS expression in hypoxic endothelial cells.
Supplementary Material
ACKNOWLEDGMENTS
We declare that there are no financial or other conflicts of interest related to this work.
J.J.D.H. is the recipient of an Ontario Graduate Scholarship award. G.B.R. is the recipient of a Canadian Institute of Health Research/Heart and Stroke Foundation of Canada Doctoral Research Award. P.A.M. is a Heart and Stroke Foundation of Canada Career Investigator and is supported by a grant from the Heart and Stroke Foundation of Canada (grant T-6777).
We thank Stephen A. Liebhaber, Howard Hughes Medical Institute, University of Pennsylvania School of Medicine, for kindly providing anti-hnRNP antibodies. We thank Philip H. Howe, Hollings Cancer Center, Medical University of South Carolina, for kindly providing FLAG-tagged wild-type and S43A mutant hnRNP E1 plasmids.
Footnotes
Published ahead of print 11 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01257-12.
REFERENCES
- 1. Yetik-Anacak G, Catravas JD. 2006. Nitric oxide and the endothelium: history and impact on cardiovascular disease. Vascul. Pharmacol. 45:268–276 [DOI] [PubMed] [Google Scholar]
- 2. Huang PL, Huang Z, Mashimo H, Bloch KD, Moskowitz MA, Bevan JA, Fishman MC. 1995. Hypertension in mice lacking the gene for endothelial nitric oxide synthase. Nature 377:239–242 [DOI] [PubMed] [Google Scholar]
- 3. Shesely EG, Maeda N, Kim HS, Desai KM, Krege JH, Laubach VE, Sherman PA, Sessa WC, Smithies O. 1996. Elevated blood pressures in mice lacking endothelial nitric oxide synthase. Proc. Natl. Acad. Sci. U. S. A. 93:13176–13181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rudic RD, Shesely EG, Maeda N, Smithies O, Segal SS, Sessa WC. 1998. Direct evidence for the importance of endothelium-derived nitric oxide in vascular remodeling. J. Clin. Invest. 101:731–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wilcox JN, Subramanian RR, Sundell CL, Tracey WR, Pollock JS, Harrison DG, Marsden PA. 1997. Expression of multiple isoforms of nitric oxide synthase in normal and atherosclerotic vessels. Arterioscler. Thromb. Vasc. Biol. 17:2479–2488 [DOI] [PubMed] [Google Scholar]
- 6. Faller DV. 1999. Endothelial cell responses to hypoxic stress. Clin. Exp. Pharmacol. Physiol. 26:74–84 [DOI] [PubMed] [Google Scholar]
- 7. Ho JJ, Man HS, Marsden PA. 2012. Nitric oxide signaling in hypoxia. J. Mol. Med. 90:217–231 [DOI] [PubMed] [Google Scholar]
- 8. Tai SC, Robb GB, Marsden PA. 2004. Endothelial nitric oxide synthase: a new paradigm for gene regulation in the injured blood vessel. Arterioscler. Thromb. Vasc. Biol. 24:405–412 [DOI] [PubMed] [Google Scholar]
- 9. McQuillan LP, Leung GK, Marsden PA, Kostyk SK, Kourembanas S. 1994. Hypoxia inhibits expression of eNOS via transcriptional and posttranscriptional mechanisms. Am. J. Physiol. 267:H1921–H1927 [DOI] [PubMed] [Google Scholar]
- 10. Ziesche R, Petkov V, Williams J, Zakeri SM, Mosgoller W, Knofler M, Block LH. 1996. Lipopolysaccharide and interleukin 1 augment the effects of hypoxia and inflammation in human pulmonary arterial tissue. Proc. Natl. Acad. Sci. U. S. A. 93:12478–12483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fish JE, Matouk CC, Yeboah E, Bevan SC, Khan M, Patil K, Ohh M, Marsden PA. 2007. Hypoxia-inducible expression of a natural cis-antisense transcript inhibits endothelial nitric-oxide synthase. J. Biol. Chem. 282:15652–15666 [DOI] [PubMed] [Google Scholar]
- 12. Flowers MA, Wang Y, Stewart RJ, Patel B, Marsden PA. 1995. Reciprocal regulation of endothelin-1 and endothelial constitutive NOS in proliferating endothelial cells. Am. J. Physiol. 269:H1988–H1997 [DOI] [PubMed] [Google Scholar]
- 13. Yoshizumi M, Perrella MA, Burnett JC, Jr, Lee ME. 1993. Tumor necrosis factor downregulates an endothelial nitric oxide synthase mRNA by shortening its half-life. Circ. Res. 73:205–209 [DOI] [PubMed] [Google Scholar]
- 14. Yan G, You B, Chen SP, Liao JK, Sun J. 2008. Tumor necrosis factor-alpha downregulates endothelial nitric oxide synthase mRNA stability via translation elongation factor 1-alpha 1. Circ. Res. 103:591–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lu JL, Schmiege LM, III, Kuo L, Liao JC. 1996. Downregulation of endothelial constitutive nitric oxide synthase expression by lipopolysaccharide. Biochem. Biophys. Res. Commun. 225:1–5 [DOI] [PubMed] [Google Scholar]
- 16. Liao JK, Shin WS, Lee WY, Clark SL. 1995. Oxidized low-density lipoprotein decreases the expression of endothelial nitric oxide synthase. J. Biol. Chem. 270:319–324 [DOI] [PubMed] [Google Scholar]
- 17. Robb GB, Carson AR, Tai SC, Fish JE, Singh S, Yamada T, Scherer SW, Nakabayashi K, Marsden PA. 2004. Post-transcriptional regulation of endothelial nitric-oxide synthase by an overlapping antisense mRNA transcript. J. Biol. Chem. 279:37982–37996 [DOI] [PubMed] [Google Scholar]
- 18. Yamada T, Carson AR, Caniggia I, Umebayashi K, Yoshimori T, Nakabayashi K, Scherer SW. 2005. Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J. Biol. Chem. 280:18283–18290 [DOI] [PubMed] [Google Scholar]
- 19. Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- 20. Bartel DP. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carthew RW, Sontheimer EJ. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Friedman RC, Farh KK, Burge CB, Bartel DP. 2009. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19:92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chaudhury A, Chander P, Howe PH. 2010. Heterogeneous nuclear ribonucleoproteins (hnRNPs) in cellular processes: focus on hnRNP E1's multifunctional regulatory roles. RNA 16:1449–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kourembanas S, Marsden PA, McQuillan LP, Faller DV. 1991. Hypoxia induces endothelin gene expression and secretion in cultured human endothelium. J. Clin. Invest. 88:1054–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dumont DJ, Gradwohl GJ, Fong GH, Auerbach R, Breitman ML. 1993. The endothelial-specific receptor tyrosine kinase, tek, is a member of a new subfamily of receptors. Oncogene 8:1293–1301 [PubMed] [Google Scholar]
- 26. Marsden PA, Sultan P, Cybulsky M, Gimbrone MA, Jr, Brenner BM, Collins T. 1992. Nucleotide sequence of endothelin-1 cDNA from rabbit endothelial cells. Biochim. Biophys. Acta 1129:249–250 [DOI] [PubMed] [Google Scholar]
- 27. Marsden PA, Schappert KT, Chen HS, Flowers M, Sundell CL, Wilcox JN, Lamas S, Michel T. 1992. Molecular cloning and characterization of human endothelial nitric oxide synthase. FEBS Lett. 307:287–293 [DOI] [PubMed] [Google Scholar]
- 28. Marsden PA, Heng HH, Scherer SW, Stewart RJ, Hall AV, Shi XM, Tsui LC, Schappert KT. 1993. Structure and chromosomal localization of the human constitutive endothelial nitric oxide synthase gene. J. Biol. Chem. 268:17478–17488 [PubMed] [Google Scholar]
- 29. Gustin KE, Burk RD. 1993. A rapid method for generating linker scanning mutants utilizing PCR. Biotechniques 14:22, 24 [PubMed] [Google Scholar]
- 30. Teichert AM, Miller TL, Tai SC, Wang Y, Bei X, Robb GB, Phillips MJ, Marsden PA. 2000. In vivo expression profile of an endothelial nitric oxide synthase promoter-reporter transgene. Am. J. Physiol. Heart Circ. Physiol. 278:H1352–H1361 [DOI] [PubMed] [Google Scholar]
- 31. Chomczynski P, Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156–159 [DOI] [PubMed] [Google Scholar]
- 32. Loflin PT, Chen CY, Xu N, Shyu AB. 1999. Transcriptional pulsing approaches for analysis of mRNA turnover in mammalian cells. Methods 17:11–20 [DOI] [PubMed] [Google Scholar]
- 33. Ross J. 1995. mRNA stability in mammalian cells. Microbiol. Rev. 59:423–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kiledjian M, Day N, Trifillis P. 1999. Purification and RNA binding properties of the polycytidylate-binding proteins alphaCP1 and alphaCP2. Methods 17:84–91 [DOI] [PubMed] [Google Scholar]
- 35. Dandekar T, Stripecke R, Gray NK, Goossen B, Constable A, Johansson HE, Hentze MW. 1991. Identification of a novel iron-responsive element in murine and human erythroid delta-aminolevulinic acid synthase mRNA. EMBO J. 10:1903–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chkheidze AN, Lyakhov DL, Makeyev AV, Morales J, Kong J, Liebhaber SA. 1999. Assembly of the alpha-globin mRNA stability complex reflects binary interaction between the pyrimidine-rich 3′ untranslated region determinant and poly(C) binding protein alphaCP. Mol. Cell. Biol. 19:4572–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ho JJ, Metcalf JL, Yan MS, Turgeon PJ, Wang JJ, Chalsev M, Petruzziello-Pellegrini TN, Tsui AK, He JZ, Dhamko H, Man HS, Robb GB, Teh BT, Ohh M, Marsden PA. 2012. Functional importance of dicer protein in the adaptive cellular response to hypoxia. J. Biol. Chem. 287:29003–29020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fish JE, Yan MS, Matouk CC, St Bernard R, Ho JJ, Gavryushova A, Srivastava D, Marsden PA. 2010. Hypoxic repression of endothelial nitric-oxide synthase transcription is coupled with eviction of promoter histones. J. Biol. Chem. 285:810–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liebhaber SA. 1989. Alpha thalassemia. Hemoglobin 13:685–731 [DOI] [PubMed] [Google Scholar]
- 40. Wang X, Kiledjian M, Weiss IM, Liebhaber SA. 1995. Detection and characterization of a 3′ untranslated region ribonucleoprotein complex associated with human alpha-globin mRNA stability. Mol. Cell. Biol. 15:1769–1777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morales J, Russell JE, Liebhaber SA. 1997. Destabilization of human alpha-globin mRNA by translation anti-termination is controlled during erythroid differentiation and is paralleled by phased shortening of the poly(A) tail. J. Biol. Chem. 272:6607–6613 [DOI] [PubMed] [Google Scholar]
- 42. Zhang W, Wagner BJ, Ehrenman K, Schaefer AW, DeMaria CT, Crater D, DeHaven K, Long L, Brewer G. 1993. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 13:7652–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lagnado CA, Brown CY, Goodall GJ. 1994. AUUUA is not sufficient to promote poly(A) shortening and degradation of an mRNA: the functional sequence within AU-rich elements may be UUAUUUA(U/A)(U/A). Mol. Cell. Biol. 14:7984–7995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zubiaga AM, Belasco JG, Greenberg ME. 1995. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol. Cell. Biol. 15:2219–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lewis T, Gueydan C, Huez G, Toulme JJ, Kruys V. 1998. Mapping of a minimal AU-rich sequence required for lipopolysaccharide-induced binding of a 55-kDa protein on tumor necrosis factor-alpha mRNA. J. Biol. Chem. 273:13781–13786 [DOI] [PubMed] [Google Scholar]
- 46. Chen CY, Shyu AB. 1995. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 20:465–470 [DOI] [PubMed] [Google Scholar]
- 47. Wilson GM, Brewer G. 1999. The search for trans-acting factors controlling messenger RNA decay. Prog. Nucleic Acid Res. Mol. Biol. 62:257–291 [DOI] [PubMed] [Google Scholar]
- 48. Holcik M, Liebhaber SA. 1997. Four highly stable eukaryotic mRNAs assemble 3′ untranslated region RNA-protein complexes sharing cis and trans components. Proc. Natl. Acad. Sci. U. S. A. 94:2410–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kiledjian M, Wang X, Liebhaber SA. 1995. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. EMBO J. 14:4357–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang X, Liebhaber SA. 1996. Complementary change in cis determinants and trans factors in the evolution of an mRNP stability complex. EMBO J. 15:5040–5051 [PMC free article] [PubMed] [Google Scholar]
- 51. Habelhah H, Shah K, Huang L, Ostareck-Lederer A, Burlingame AL, Shokat KM, Hentze MW, Ronai Z. 2001. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat. Cell Biol. 3:325–330 [DOI] [PubMed] [Google Scholar]
- 52. Chkheidze AN, Liebhaber SA. 2003. A novel set of nuclear localization signals determine distributions of the alphaCP RNA-binding proteins. Mol. Cell. Biol. 23:8405–8415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ji X, Kong J, Liebhaber SA. 2003. In vivo association of the stability control protein alphaCP with actively translating mRNAs. Mol. Cell. Biol. 23:899–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ostrowski J, Bomsztyk K. 2003. Nuclear shift of hnRNP K protein in neoplasms and other states of enhanced cell proliferation. Br. J. Cancer 89:1493–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. 2007. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ. Res. 100:1164–1173 [DOI] [PubMed] [Google Scholar]
- 56. Chaudhury A, Hussey GS, Ray PS, Jin G, Fox PL, Howe PH. 2010. TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI. Nat. Cell Biol. 12:286–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Meng Q, Rayala SK, Gururaj AE, Talukder AH, O'Malley BW, Kumar R. 2007. Signaling-dependent and coordinated regulation of transcription, splicing, and translation resides in a single coregulator, PCBP1. Proc. Natl. Acad. Sci. U. S. A. 104:5866–5871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mohr E, Richter D. 2001. Messenger RNA on the move: implications for cell polarity. Int. J. Biochem. Cell Biol. 33:669–679 [DOI] [PubMed] [Google Scholar]
- 59. Wormington M, Searfoss AM, Hurney CA. 1996. Overexpression of poly(A) binding protein prevents maturation-specific deadenylation and translational inactivation in Xenopus oocytes. EMBO J. 15:900–909 [PMC free article] [PubMed] [Google Scholar]
- 60. Aoki K, Matsumoto K, Tsujimoto M. 2003. Xenopus cold-inducible RNA-binding protein 2 interacts with ElrA, the Xenopus homolog of HuR, and inhibits deadenylation of specific mRNAs. J. Biol. Chem. 278:48491–48497 [DOI] [PubMed] [Google Scholar]
- 61. Makeyev AV, Chkheidze AN, Liebhaber SA. 1999. A set of highly conserved RNA-binding proteins, alphaCP-1 and alphaCP-2, implicated in mRNA stabilization, are coexpressed from an intronless gene and its intron-containing paralog. J. Biol. Chem. 274:24849–24857 [DOI] [PubMed] [Google Scholar]
- 62. Makeyev AV, Liebhaber SA. 2000. Identification of two novel mammalian genes establishes a subfamily of KH-domain RNA-binding proteins. Genomics 67:301–316 [DOI] [PubMed] [Google Scholar]
- 63. Waggoner SA, Liebhaber SA. 2003. Identification of mRNAs associated with alphaCP2-containing RNP complexes. Mol. Cell. Biol. 23:7055–7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Waggoner SA, Liebhaber SA. 2003. Regulation of alpha-globin mRNA stability. Exp. Biol. Med. 228:387–395 [DOI] [PubMed] [Google Scholar]
- 65. Stefanovic B, Hellerbrand C, Holcik M, Briendl M, Aliebhaber S, Brenner DA. 1997. Posttranscriptional regulation of collagen alpha1(I) mRNA in hepatic stellate cells. Mol. Cell. Biol. 17:5201–5209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Czyzyk-Krzeska MF, Beresh JE. 1996. Characterization of the hypoxia-inducible protein binding site within the pyrimidine-rich tract in the 3′-untranslated region of the tyrosine hydroxylase mRNA. J. Biol. Chem. 271:3293–3299 [DOI] [PubMed] [Google Scholar]
- 67. Czyzyk-Krzeska MF, Bendixen AC. 1999. Identification of the poly(C) binding protein in the complex associated with the 3′ untranslated region of erythropoietin messenger RNA. Blood 93:2111–2120 [PubMed] [Google Scholar]
- 68. Ostareck-Lederer A, Ostareck DH, Standart N, Thiele BJ. 1994. Translation of 15-lipoxygenase mRNA is inhibited by a protein that binds to a repeated sequence in the 3′ untranslated region. EMBO J. 13:1476–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ostareck DH, Ostareck-Lederer A, Wilm M, Thiele BJ, Mann M, Hentze MW. 1997. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell 89:597–606 [DOI] [PubMed] [Google Scholar]
- 70. Siomi H, Dreyfuss G. 1995. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129:551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gamarnik AV, Andino R. 1997. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3:882–892 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.