Abstract
Life cycle alternation between arthropod and mammals forces the Lyme disease spirochete, Borrelia burgdorferi, to adapt to different host milieus by utilizing diverse carbohydrates. Glycerol and chitobiose are abundantly present in the Ixodes tick. B. burgdorferi can utilize glycerol as a carbohydrate source for glycolysis and chitobiose to produce N-acetylglucosamine (GlcNAc), a key component of the bacterial cell wall. A recent study reported that Rrp1, a response regulator that synthesizes cyclic diguanylate (c-di-GMP), governs glycerol utilization in B. burgdorferi. In this report, we found that the rrp1 mutant had growth defects and formed membrane blebs that led to cell lysis when GlcNAc was replaced by chitobiose in the growth medium. The gene chbC encodes a key chitobiose transporter of B. burgdorferi. We found that the expression level of chbC was significantly repressed in the mutant and that constitutive expression of chbC in the mutant successfully rescued the growth defect, indicating a regulatory role of Rrp1 in chitobiose uptake. Immunoblotting and transcriptional studies revealed that Rrp1 is required for the activation of bosR and rpoS and that its impact on chbC is most likely mediated by the BosR-RpoS regulatory pathway. Tick-mouse infection studies showed that although the rrp1 mutant failed to establish infection in mice via tick bite, exogenous supplementation of GlcNAc into unfed ticks partially rescued the infection. The finding reported here provides us with new insight into the regulatory role of Rrp1 in carbohydrate utilization and virulence of B. burgdorferi.
INTRODUCTION
The spirochete Borrelia burgdorferi is the causative agent of Lyme borreliosis, the most frequently reported vector-borne disease in the United States and many parts of the globe (1). In its natural environment, B. burgdorferi cycles between the Ixodes tick vector and small mammals, such as the white-footed mouse (2, 3). During the transmission process, B. burgdorferi is challenged with rigorous changes in the surrounding milieus, such as shifts in temperature as well as in the availability of different carbon sources for growth. The necessity to utilize different carbohydrates is further supported by the presence of several carbon metabolism pathways and transporters in the genome of this spirochete (4). For instance, in addition to the glycolysis pathway utilizing glucose as the main carbon source, B. burgdorferi also encodes genes essential for metabolizing glycerol, chitobiose, and N-acetylglucosamine (GlcNAc) (4–8).
During the mammalian phase of the enzootic life cycle of spirochetes, glucose is abundantly present in the host blood which can be utilized by B. burgdorferi for metabolism and growth (9, 10). On the other hand, during the tick phase, alternative carbohydrates, such as glycerol and chitin, become available. Glycerol is produced by the Ixodes tick as one of the cryoprotective agents, and chitin is the major component of cuticle of arthropod, including ticks (11–13). In vitro growth analysis has shown that B. burgdorferi is able to utilize glycerol as its carbon source in the absence of glucose (5, 7, 14). Also, mutants defective in glycerol metabolism are attenuated in overall survival within the tick vector. These mutants exhibit delayed replication after a blood meal or are unable to persist effectively in feeding ticks, emphasizing the requirement of glycerol for maximal fitness of B. burgdorferi during the tick phase of the life cycle (5, 14).
The genome of B. burgdorferi encodes several genes that constitute the phosphotransferase system (PTS) required for chitobiose uptake and utilization (4, 6). Among the three putative chitobiose transporter genes, chbA (BBB05), chbB (BBB06), and chbC (BBB04), only chbC is essential for chitobiose utilization in vitro (6, 15). The expression of chbC was shown to be regulated by both RpoD and RpoS (16), and its expression is highly elevated in ticks (5). In vitro growth analysis showed that the chbC mutant is unable to utilize chitobiose as a carbon source when GlcNAc availability is limited. Membrane blebs formed on the surface of the chbC mutant cells when GlcNAc was replaced with chitobiose in the growth medium, indicating a defect in utilizing chitobiose as a source of GlcNAc for cell wall synthesis (6). However, despite its having the complete set of genes for chitobiose metabolism, in vivo studies indicated that the chbC mutant is able to survive in the tick and complete the enzootic life cycle, suggesting that the chitobiose metabolism system is not essential for the survival of B. burgdorferi (17).
During the enzootic cycle, B. burgdorferi differentially regulates its tick-phase and mammal-phase genes, primarily via the RpoN-RpoS regulon and two important two-component systems (TCS), the Hk2-Rrp2 and the Hk1-Rrp1 signaling pathways (for reviews, see references 18 and 19). The Rrp2-RpoN-RpoS regulatory system has been well investigated and is known for its essential role during the transition from tick to mammal as well as in the mammalian phase of the life cycle (20–24). Several important regulatory elements of this signaling pathway, such as DsrABb, acetylphosphate, and BosR, have also been identified (for reviews, see references 18 and 19). However, little is known about the role of the second TCS. A recent breakthrough in investigating the function of Hk1-Rrp1 highlighted the significance of this TCS in the tick phase of the enzootic cycle (14, 25–27). Rrp1, a diguanylate cyclase, is the sole protein that produces cyclic diguanylate (c-di-GMP) in the genome of B. burgdorferi (28). Disruption of c-di-GMP production has been shown to result in global alteration of gene expression, as revealed by microarray analysis of an rrp1 mutant (14, 27). In vitro transcriptional analysis indicated that rrp1 is strongly induced upon tick feeding and that the induction is independent of changes in temperature (27). An rrp1-defective mutant is infectious but attenuated in overall virulence in mice via needle inoculation, and it failed to be transmitted to naïve ticks upon feeding (26). In addition, a mutant defective in rrp1 has reduced fitness in unfed ticks and is quickly killed upon feeding (14). These observations imply that Rrp1 is required for the basic survival of B. burgdorferi in the arthropod vector as well as for protection from host factors/immunity, further emphasizing the importance of Rrp1 during the tick phase of the life cycle.
Based on transcriptional analysis, Rrp1 has been shown to regulate genes involved in the metabolism of GlcNAc (26). This is required for in vitro growth of Borrelia (10), presumably both for energy production through glycolysis and for synthesizing the precursor for cell wall construction (4). Live-imaging analysis from Dunham-Ems et al. showed that B. burgdorferi replicates extensively in the tick gut at the onset of feeding (29). In order to multiply, B. burgdorferi requires a large supply of GlcNAc for cell wall synthesis. The one possible source of GlcNAc available upon feeding comes from chitin/chitobiose, which is shed from the peritrophic matrix upon feeding (30). In this study, we sought to investigate the role of Rrp1 in the regulation of chitobiose metabolism by the Lyme disease spirochete. Using a mutant defective in rrp1, we showed that Rrp1 is essential for chitobiose utilization in vitro and that supplementation of GlcNAc in ticks successfully rescued the mutant transmission to mice via tick bite. Immunoblotting and quantitative reverse transcription-PCR (qRT-PCR) analyses showed that Rrp1 affects chbC, the chitobiose transporter gene, by regulating the level of RpoS, probably through BosR, an important modulator of RpoS (31, 32). A model for Rrp1 regulation on chitobiose utilization is also proposed in this report.
MATERIALS AND METHODS
Ethics statement.
All animal experimentation was conducted following the NIH guidelines for housing and care of laboratory animals. The protocol of using mice and ticks was approved by the Committee on the Ethics of Animal Experiments and the Institutional Animal Care and Use Committee of the University of Maryland (permit number R-12-33).
Bacterial strains and growth conditions.
Infectious clone A3-68 derived from the B. burgdorferi sensu stricto B31A3 strain was used in this study (33). Cells were maintained at 34°C in Barbour-Stoenner-Kelly (BSK-II) medium supplemented with 6% rabbit serum in the presence of 3.4% carbon dioxide. The strains were grown in the following appropriate antibiotic(s) for selection pressure as needed: kanamycin (300 μg/ml) and/or gentamicin (40 μg/ml). Escherichia coli TOP10 strain (Invitrogen, Carlsbad, CA) was used for DNA cloning and cultured in lysogeny broth (LB) supplemented with appropriate concentrations of antibiotics.
Construction and complementation of B. burgdorferi rrp1mut.
The rrp1::kan plasmid was constructed for the targeted mutagenesis of rrp1 (bb0419) and the plasmid Rrp1/cis com for the complementation of the mutant. To construct the rrp1::kan plasmid, the entire open reading frames (ORF) of the bb0419 gene and the kanamycin resistance gene (kan) were PCR amplified using primer pair P1 and P2 and primer pair P3 and P4, respectively (see Table S1 in the supplemental material). The resulting PCR products were cloned into pGEM-T-Easy vector (Promega, Madison, WI). A 151-bp DNA fragment was deleted from bb0419 using the restriction cut sites XbaI and BalI. Then, the kan cassette was inserted into the obtained fragment at XbaI and BalI cut sites, generating the rrp1::kan plasmid (Fig. 1a).
Fig 1.
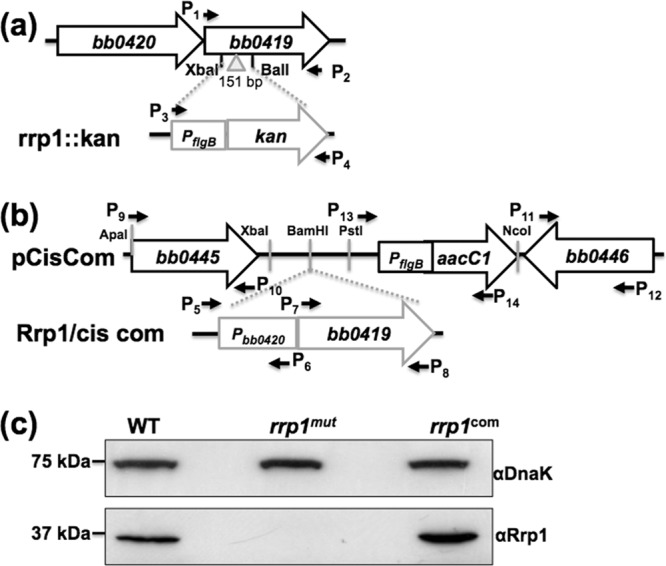
Construction of the rrp1mut mutant and its complemented rrp1com strain. (a) To construct the rrp1::kan plasmid, the entire ORF of bb0419 (rrp1) was PCR amplified. A kan cassette was inserted into the XbaI and BalI cut sites that are present within the rrp1 ORF, resulting in a 151-bp deletion, generating the rrp1::kan plasmid. (b) Pbb0420, 218 bp from the upstream region of bb0420 that contains the native promoter of the bb0419-bb0420 operon, was PCR amplified and fused to the 5′ end of bb0419. The obtained fragment was then inserted into the pCisCom construct at the BamHI cut site, yielding Rrp1/cis com. (c) Immunoblot analysis of the whole-cell lysate of the wild-type (WT), rrp1mut, and rrp1com strains probed with a specific antibody against Rrp1. DnaK was used as an internal control, as previously described (42). α, anti-.
Complementation of the rrp1mut mutant was performed using a previously described approach (34). To construct the suicide vector pCisCom for cis complementation, the 5′ and 3′ arms for homologous recombination were PCR amplified using primer pair P9 and P10 and primer pair P11 and P12, and the flgBp-aacC1 cassette (35) was amplified from pBSV2G vector (36) using primer pair P13 and P14. The resulting fragments were cloned into pGEM-T vector (Promega). The aacC1 cassette was inserted downstream of the 5′ arm at the engineered XbaI and NcoI restriction cut sites. The 5′ arm and the aacC1 cassette were then placed upstream of the 3′ arm using ApaI and NcoI cut sites, yielding the pCisCom plasmid. To construct the Rrp1/cis com plasmid, the full length of bb0419 and 218 bp from the upstream region of bb0420 containing the native promoter of the bb0419-bb0420 operon were amplified using primer pair P5 and P6 and primer pair P7 and P8 (see Table S1 in the supplemental material). The two resulting PCR fragments were then PCR ligated using primer pair P5 and P8. The resultant Pbb0420-bb0419 fragment was ligated to the pGEM-T-vector (Promega). BamHI was then used to digest the resulting plasmid, and the obtained BamHI-cut Pbb0420-bb0419 fragment was inserted into suicide vector pCisCom to generate Rrp1/cis com (Fig. 1b).
Measuring growth rates of B. burgdorferi.
To measure the growth rates of the wild-type (WT) A3-68, rrp1mut, rrp1com, and rrp1mut chbC-positive (chbC+) strains, 5 μl of the stationary-phase cultures (1 × 108 cells/ml) was inoculated into 5 ml of normal BSK-II medium or BSK-II medium without added exogenous GlcNAc but supplemented with 20 μM chitobiose, in the form of N,N-diacetylchitobiose (designated BSK-II(−GlcNAc)+chitobiose), and incubated at 23°C and pH 7.6 (unfed tick condition) and at 34°C and pH 7.6 as described before (37). The bacterial concentrations of the cultures were measured every 24 to 48 h for up to 24 days using a Petroff-Hausser counting chamber (38). Counts were repeated in triplicate with at least two independent samples, and the results are expressed as the means ± standard errors of the means (SEM).
chbC rescues growth of the rrp1mut mutant.
To rescue the growth of the rrp1mut mutant in the BSK-II(−GlcNAc)+chitobiose medium, a plasmid that overexpresses chbC (bbb04) was constructed. The flgBp and the ORF of chbC were PCR amplified using primer pair P15 and P16 and primer pair P17 and P18, respectively. The resulting fragments were cloned into pGEM-T vector (Promega). The chbC fragment was then digested with NdeI and SphI restriction enzymes and ligated to the pGEM-T-flgBp plasmid. The entire flgBp-chbC fragment was then digested with SphI and ligated to the shuttle vector, pBBE22G (39). The resultant vector was transformed into the rrp1mut competent cells via electroporation.
SDS-PAGE and immunoblotting.
B. burgdorferi cells were cultivated at 34°C and pH 7.6 until the stationary phase (∼108 cells/ml) was reached. Cells were harvested for immunoblot analysis at the early stationary phase (day 1 [D1]) and at day 2 and day 3 (late stationary phase [D2 and D3, respectively]). Equal amounts (10 to 50 μg) of whole-cell lysates were separated on SDS-PAGE gels and transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad Laboratories, Hercules, CA). The immunoblots were probed with specific antibodies against various proteins, including BosR, DbpA, DbpB, OspA, OspC, Pta, Rrp1, Rrp2, RpoS, and DnaK (an internal control). These antibodies were kindly provided by F. T. Liang (Louisiana State University), X. F. Yang (Indiana University), M. J. Caimano (University of Connecticut Health Center), J. Skare (Texas A&M University), and J. Seshu (University of Texas at San Antonio). Immunoblots were developed using horseradish peroxidase secondary antibody with an enhanced chemiluminescence (ECL) luminol assay, as previously described (40).
RNA preparation and qRT-PCR.
RNA isolation was performed as previously described (41, 42). Briefly, B. burgdorferi strains were cultivated at 34°C, and 50 ml (∼ 108 cells/ml) of the stationary-phase cultures was harvested for RNA preparation. Total RNA was extracted using TRI reagent (Sigma-Aldrich, St. Louis, MO), following the manufacturer's instructions. The resultant samples were treated with Turbo DNase (Ambion, Austin, TX) at 37°C for 2 h to eliminate genomic DNA contamination. The resultant RNA samples were reextracted using acid phenol-chloroform (Ambion), precipitated in isopropanol, and washed with 70% ethanol. The RNA pellets were resuspended in RNase-free water. cDNA was generated from the purified RNA using an iScript cDNA synthesis kit (Bio-Rad). Quantitative reverse transcription-PCR (qRT-PCR) was performed using iQ SYBR green Supermix and a MyiQ thermal cycler (Bio-Rad). The flaB (bb0147) transcript was amplified and used as an internal control to normalize the qRT-PCR data as described before (43, 44). The enolase gene (eno; bb0337) was included as a positive-control housekeeping gene as described previously (41). The results were expressed as the expression (2−ΔΔCT) of the rrp1mut transcripts relative to that of the wild type. The primers used for qRT-PCR are listed in Table S1 in the supplemental material.
Microinjection and analysis of ticks for the assessment of spirochete persistence and transmission from ticks to mice.
C3H mice (Jackson Laboratory, Bar Harbor, ME) at 4 to 6 weeks of age were used in this study. Ixodes scapularis nymphal ticks (Oklahoma State University, Stillwater, OK) were microinjected with wild-type or genetically manipulated spirochetes as described earlier (37, 45, 46). Cultures of B. burgdorferi were grown in BSK-II medium at a density of 107 cells/ml and centrifuged at 4,000 rpm for 20 min. The resulting pellet was then washed 3 times with phosphate-buffered saline (PBS) and finally resuspended either in GlcNAc-free BSK-II medium or in the medium supplemented with 10 mM GlcNAc at a cell density of 109 cells/ml. Equal volumes of the resultant cell suspensions were injected into the rectal aperture of unfed nymph ticks using a femtojet microinjector system (Eppendorf AG, Hamburg, Germany) as previously described (45). After injection, ticks were allowed to remain in the incubator for 24 h for recovery and were subsequently placed on mice (3 animals/group) and allowed to engorge. Postfed ticks were collected and analyzed for the presence of bacteria using qRT-PCR as described previously (37). Parallel groups of microinjected unfed ticks were kept in the incubator for 5 days and used to assess the persistence of spirochetes using qRT-PCR. For assessment of pathogen transmission from ticks to the host, mice were sacrificed following 2 weeks of tick engorgement. The heart, bladder, skin, and joints were collected and subjected to qRT-PCR analysis for measuring B. burgdorferi burdens as previously detailed (37). Spleen tissues were collected from mice and assessed for the presence of viable spirochetes using culture analysis.
Sample preparation for quantification of intracellular N-acetylglucosamine.
To measure the intracellular concentration of GlcNAc, B. burgdorferi cells were cultured in BSK-II or BSK-II(−GlcNAc)+chitobiose medium and harvested by centrifugation at 5,000 rpm. Cells were washed twice and resuspended in sterilized PBS. Cell densities were adjusted to 109cells/200 μl, and cells were sonicated for 20 s. Lysates were subjected to high-speed centrifugation at 15,000 rpm and 4°C for 20 min. The resulting supernatants were transferred to clean microcentrifuge tubes and analyzed by liquid chromatography tandem mass spectrometry (LC-MS).
LC-MS.
All samples were analyzed on the LC-MS/MS system consisting of a Shimadzu Prominence high-performance LC (HPLC) system (Kyoto, Japan) and an API 3000 Turbo Ionspray ionization triple-quadrupole mass spectrometer (Applied Biosystems, Foster City, CA) controlled by Analyst 1.4.2 software (Applied Biosystems). A simple and rapid protein precipitation method was used for sample preparation. Two hundred microliters of acetonitrile containing 50 μM N-(2,4,5-trihydroxy-6-hydroxymethyl-tetrahydropyran-3-yl)-formamide (internal standard [IS]) was added to 100 μl of sample to precipitate protein. After mixing and centrifugation at 14,000 rpm for 10 min, 5 μl of supernatant was injected for LC-MS/MS analysis. Separation of analytes was achieved using a 3-μm-pore-size Thermo Aquasil C18 column (4.6 mm by 50 mm) maintained at 25°C and an isocratic elution with 80% methanol and 20% distilled water at a flow rate of 0.2 ml/min in which both contained 0.1% formic acid. The total run time was 5 min for each injection. The mass spectrometer was operated in the turbo ion spray mode with negative ion detection. The detection and quantitation of GlcNAc, N-valeryl-glucosamine, and the IS were accomplished by multiple-reaction monitoring (MRM) with the transitions m/z 220.2→118.9 for GlcNAc, 262.2→118.9 for N-valerylglucosamine, and 206.1→118.9 for the IS. The instrumental parameters were tuned to maximize the MRM signals. An online motorized six-port divert valve was used to introduce the LC eluent to the mass spectrometer over a period of 1.0 to 5.0 min for data acquisition, whereas eluent from 0 to 1.0 min was diverted to the waste.
Blank media were spiked with the standard working solution and then serially diluted with blank media to generate the following concentrations for both analytes: 10, 1.0, 0.5, 0.1, 0.01, 0.005, and 0.002 μM. These matrix-based calibration standards were treated the same as the samples. The calibration plots were constructed using linear regressions of the peak area ratio of analyte over IS (y axis) against the corresponding nominal concentration of the analyte (x axis). The calibration linear range of GlcNAc was 0.002 to 10 μM (on column). Three independent assays were conducted, and GlcNAc concentrations were expressed as averages of μM/108 cells for intracellular GlcNAc concentrations and μM for culture media GlcNAc concentrations.
Scanning electron microscope.
Wild-type, rrp1mut, rrp1com, and rrp1mut chbC+ cells were centrifuged and washed once with PBS. Cells were then resuspended in fresh PBS. SEM determinations were performed as described before (47, 48). Briefly, 10 μl of cell suspensions was settled on poly-l-lysine-coated round coverslips (BD BioSciences, San Jose, CA). Samples were fixed statically in 0.1 M sodium cacodylate buffer (pH 7.2) containing 2.5% glutaraldehyde, 0.075% ruthenium red, and 0.075 M lysine acetate for 1 h at room temperature. Three 10-min washes were performed using 0.2 M sodium cacodylate buffer (pH 7.2) containing 0.075% ruthenium red at room temperature without agitation. Samples were then dehydrated with a graded ethanol series (30, 50, 75, 95, and 100%) at room temperature for 10 min for each incubation step. Samples were exchanged into 100% hexamethyldisilazane and allowed to air dry before analysis was performed using a Hitachi SU-70 scanning electron microscope at an acceleration voltage of 10.0 kV.
Statistical analysis.
The significance of differences between different experimental groups was evaluated with two-way analysis of variance (ANOVA) (P values of <0.05 were considered significant).
RESULTS
Isolation of the rrp1mut mutant and its rrp1com complemented strain.
Inactivation of the rrp1 gene (bb0419) was performed using the rrp1::kan vector (Fig. 1a) by first linearizing it with SphI and then transformation into infectious A3-68 (33) competent cells by electroporation (49). After 14 days of incubation, a previously described PCR method (50) was used to screen for clones with the desired targeted mutagenesis. One clone that contained the full plasmid profile of its parental strain (see Fig. S1a in the supplemental material) was selected for further analysis and was named rrp1mut. Immunoblotting analysis using antiserum against Rrp1 confirmed that the production of the cognate gene product was abolished in the mutant (Fig. 1c). For the complementation of the rrp1mut mutant, a cis complementation approach was performed by inserting the bb0419 gene together with its native promoter into the intergenic region between bb0445 and bb0446 (34) at the BamHI-cut site as shown in Fig. 1b. The Rrp1/cis com vector was linearized with ApaI and electrotransformed into rrp1mut competent cells. Immunoblotting was used to screen for complementation. One clone containing the same plasmid profile as its parental strain (see Fig. S1b in the supplemental material) was selected for further analysis and named rrp1com. As shown in Fig. 1c, immunoblotting analysis confirmed that the expression of Rrp1 was restored in the rrp1com strain.
Rrp1 affects the ability of B. burgdorferi to utilize chitobiose for its growth.
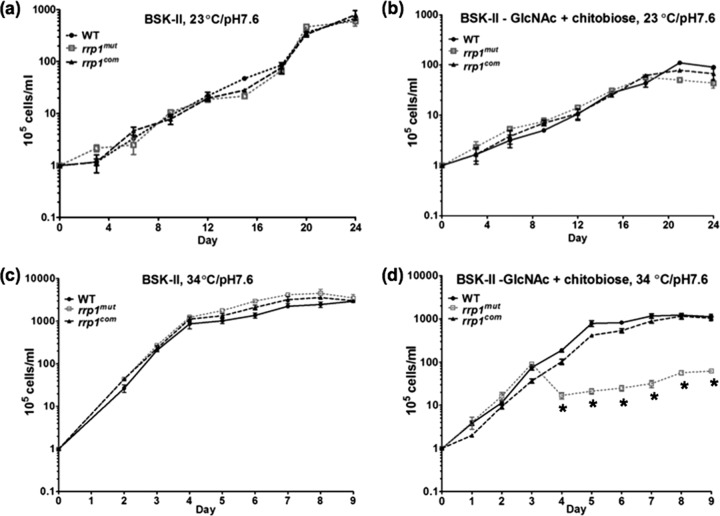
Previous studies by Kostick et al. and He et al. have established the importance of Rrp1 for the survival of B. burgdorferi in the tick after a blood meal (14, 26). During the feeding process, chitin or chitobiose is shed from the peritrophic membrane, which can be utilized as a carbon source for growth (15, 30, 51). In vitro growth analyses have shown that B. burgdorferi is still able to grow when GlcNAc is replaced with chitobiose in the BSK-II medium (6, 15, 16). To investigate if the failure of the rrp1mut mutant to survive in feeding tick was due to its inability to utilize chitobiose, which becomes available after a blood meal, we compared the growth of the rrp1mut strain to that of the wild-type and rrp1com strains in BSK-II medium as well as in BSK-II(−GlcNAc)+chitobiose medium in which GlcNAc was replaced with 20 μM chitobiose. The reason of choosing this concentration was that a previous study showed that addition of 18 μM is sufficient to restore normal growth of B. burgdorferi in vitro when GlcNAc is omitted from the BSK-II medium (6). Two conditions were included in the comparison to mimic the life cycle of the spirochete, the unfed-tick condition (23°C and pH 7.6) and the routine laboratory culture condition (34°C and pH 7.6), as described previously (37). In normal BSK-II medium, the rrp1mut mutant exhibited a growth pattern similar to that exhibited by the wild type under both culture conditions (Fig. 2a and c), consistent with the observations made by He et al. (14). In BSK-II(−GlcNAc)+chitobiose medium, while the mutant had a growth pattern similar to that of the wild type at 23°C and pH 7.6 (Fig. 2b), it exhibited a severe growth defect at 34°C and pH 7.6 (Fig. 2d). Under this culture condition, the number of mutant cells rapidly declined after the level of growth reached the mid-log phase (∼107 cell/ml). At the stationary phase, the mutant cell density was approximately 100-fold less than that of the wild type. The observed growth defect was fully restored in the rrp1com strain (Fig. 2d). Of note, the same experiment with independent cultures was repeated at least three times, and a similar growth pattern was observed. These results indicate that inactivation of rrp1 somehow impairs the ability of B. burgdorferi to utilize chitobiose for in vitro growth.
Fig 2.
The growth curves of WT, rrp1mut, and rrp1com strains. Growth curves were measured under the following conditions: (a) cells were grown in BSK-II medium at 23°C and pH 7.6; (b) cells were grown in BSK-II(−GlcNAc)+chitobiose medium at 23°C and pH 7.6; (c) cells were grown in BSK-II medium at 34°C and pH 7.6; and (d) cells were grown in BSK-II(−GlcNAc)+chitobiose medium at 34°C and pH 7.6. Asterisks indicate that the difference in cell densities between the rrp1mut mutant and the WT was statistically significant at a P value of <0.001. Cell counting was repeated in triplicate with at least two independent samples, and the results are expressed as means ± SEM.
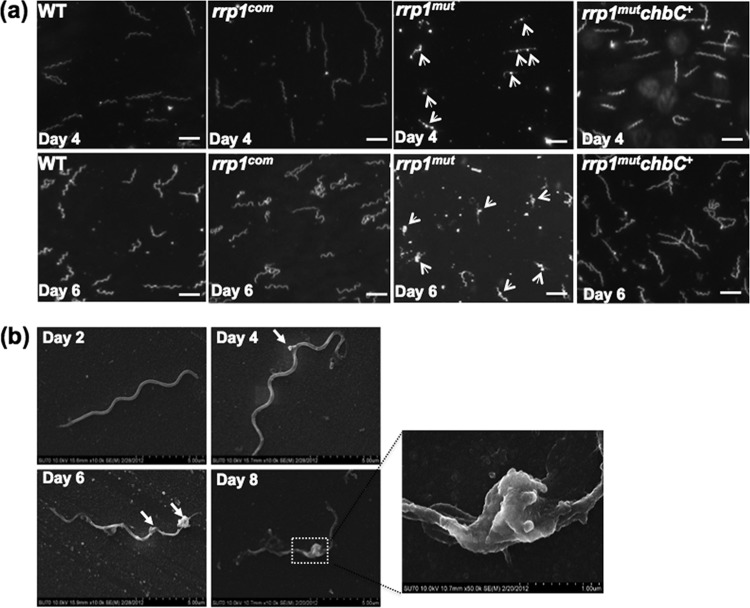
The rrp1mut mutant forms membrane blebs.
As illustrated in Fig. 2d, after day 4 (D4), the number of rrp1mut cells remained steady. However, dark-field microscopic analysis revealed that the mutant appeared to enter a “lysis phase” (Fig. 3a). During this phase, membrane blebs were progressively appearing on the cell surface. At D4, membrane blebs were seen forming on the cell surfaces in roughly 60% of the rrp1mut mutant population (Fig. 3a). At day 6 (D6), more than 80% of the mutant population had visible membrane blebs whereas both the wild-type and rrp1com strains remained normal (Fig. 3a). A scanning electron microscope was used to further visualize the membrane bleb formation throughout growth (from D2 to D8) in BSK-II(−GlcNAc)+chitobiose medium. At the time of initial growth (D2), the rrp1mut cells appeared normal (Fig. 3b). When cells entered the lysis phase (D4 to D8), blebs began to form and multiply in size and number, which eventually led to cell lysis (Fig. 3b). The formation of membrane blebs was abolished upon complementation in rrp1com cells (Fig. 3a; see also Fig. S2 in the supplemental material). These results indicate that inactivation of rrp1 induces membrane bleb formation, which further leads to cell lysis of the mutant. B. burgdorferi needs exogenous GlcNAc to grow. Failure to utilize chitobiose as a source of GlcNAc for cell wall synthesis may explain the growth and morphological defects observed in rrp1mut when GlcNAc is replaced by chitobiose in the growth medium.
Fig 3.
The rrp1mut mutant forms membrane blebs in chitobiose-supplemented medium. (a) Dark-field microscopic analysis of B. burgdorferi strains grown in BSK-II(−GlcNAc)+chitobiose medium at 34°C and pH 7.6. The cultures were collected at day 4 and day 6 and B. burgdorferi cells observed under dark-field illumination at ×200 magnification using a Zeiss Axiostar Plus microscope. Scale bars represent 10 μm. (b) Scanning electron microscope analysis of the rrp1mut membrane bleb formation. A total of 105 cells/ml of the rrp1mut mutant were inoculated into 5 ml BSK-II(−GlcNAc)+chitobiose medium, and cells were collected for scanning electron microscope analysis at the different time points indicated to observe the formation of membrane blebs. Arrowheads point to membrane blebs observed.
Constitutive expression of chbC rescues the growth defects of the rrp1mut strain.
The chbC gene encodes a key chitobiose transporter of B. burgdorferi. Previous studies showed that chbC mutants are defective in growth when GlcNAc is replaced by chitobiose (6, 15, 16), which is similar to the phenotype of the rrp1mut strain (Fig. 2d). Upon analysis via qRT-PCR, we found that the transcript level of chbC in the rrp1mut strain was decreased by approximately 70% compared to that of the wild type and was restored in the rrp1com strain (Fig. 4a). On the basis of this observation, we hypothesized that the repression of chbC expression in the rrp1mut strain accounts for the growth defect and membrane bleb formation. To test this hypothesis, the rrp1mut chbC+ strain, a mutant that constitutively expresses chbC, was constructed and experiments similar to those described above were repeated. Our results showed that constitutive expression of chbC fully restored the ability of the rrp1mut strain to grow in BSK-II(−GlcNAc)+chitobiose medium whereas the empty pBBE22G vector alone failed to do so (Fig. 4b). In addition, dark-field and scanning electron microscopic analyses did not show significant membrane blebbing in rrp1mut chbC+ cells during the growth in BSK-II(−GlcNAc)+chitobiose medium (Fig. 3a; see also Fig. S2 in the supplemental material). Collectively, these results suggest that the observed growth defects of the rrp1mut strain in BSK-II(−GlcNAc)+chitobiose medium was most likely due to the repression of chbC expression in the mutant.
Fig 4.
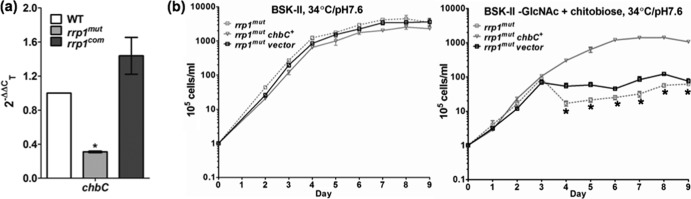
Constitutive expression of chbC restored the growth defects of the rrp1mut mutant. (a) The rrp1 mutant has decreased chbC transcript. The WT rrp1mut, and rrp1com strains were cultured at 34°C and pH 7.6 and harvested at the early stationary phase (∼108 cells/ml). Total RNA was extracted for qRT-PCR. Data are expressed as expression of the rrp1mut or rrp1com transcripts relative to the wild-type level. Asterisks indicate that the difference between WT and rrp1mut transcript levels was statistically significant at a P value of <0.01 (two-way ANOVA). (b) Constitutive expression of chbC rescues the growth defects of the rrp1mut mutant. Growth curve experiments similar to those described for Fig. 2 were repeated using BSK-II and BSK-II(−GlcNAc)+chitobiose media at 34°C and pH 7.6. Cell counting was repeated in triplicate with at least two independent samples, and the results are expressed as means ± SEM. Asterisks indicate that the difference between the rrp1mut and rrp1mut chbC+ strains in cell densities was statistically significant at a P value of <0.001.
B. burgdorferi can utilize chitobiose to obtain GlcNAc.
We reasoned that the growth defects of the rrp1mut strain in BSK-II(−GlcNAc)+chitobiose medium may have been due to its inability of utilizing chitobiose. To test this hypothesis, we developed an LC-MS method (sensitivity reached the nanomole level) to directly measure the intracellular levels of GlcNAc in B. burgdorferi cells under different culture conditions. In normal BSK-II medium at 34°C and pH 7.6, the intracellular concentration of GlcNAc in the rrp1mut cells (1.35 ± 0.23 μM, n = 3 independent samples) in the late stationary phase was similar to that in the wild-type (1.52 ± 0.027 μM, n = 3) and rrp1com (1.34 ± 0.32 μM, n = 3) cells (Fig. 5a), suggesting that inactivation of rrp1 has no impact on GlcNAc uptake and/or metabolism. Under the same culture conditions in BSK-II(−GlcNAc)+chitobiose medium, the levels of GlcNAc detected in the wild-type (1.42 ± 0.57 μM, n = 3) and rrp1com (0.96 ± 0.42 μM, n = 3) cells were similar to that in normal BSK-II medium. Of note, the level of GlcNAc in the rrp1com cells was slightly less than that in normal BSK-II medium, but the results were not statistically significant (P > 0.05). We also found that the intracellular levels of GlcNAc in the wild-type and rrp1com cells were augmented during the transition from the early log phase to the stationary phase (Fig. 5a). In contrast to the wild-type and rrp1com strains, no evident GlcNAc peak was detected by LC-MS in the rrp1mut cells after the early log phase in BSK-II(−GlcNAc)+chitobiose medium (Fig. 5a). These results indicate that the intracellular level of GlcNAc in the rrp1mut cells is too low to be detected when GlcNAc is replaced by chitobiose in the growth medium.
Fig 5.
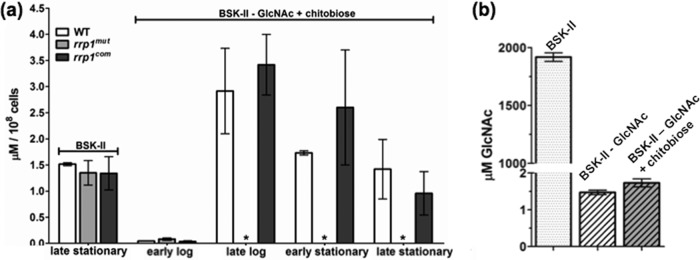
Detecting N-acetylglucosamine (GlcNAc) in B. burgdorferi culture media and cells. The intracellular level of GlcNAc and the level of GlcNAc in culture media were measured using LC-MS/MS. (a) The intracellular concentration of GlcNAc was determined at various stages of the growth curve for BSK-II(−GlcNAc)+chitobiose medium as indicated and at the late stationary phase for cultures in BSK-II medium. Samples were measured in triplicate, and data are represented as means ± SEM in μM/108 cells. (b) Concentration of GlcNAc in normal BSK-II, GlcNAc-depleted BSK-II BSK-II(−GlcNAc) and BSK-II(−GlcNAc)+chitobiose. Asterisks indicate no GlcNAc peak detected.
We also measured the basal level of GlcNAc in BSK-II medium without supplementing exogenous GlcNAc (BSK-II(−GlcNAc). The detected GlcNAc concentration was approximately 1.47 ± 0.06 μM (Fig. 5b). Addition of 20 μM chitobiose did not contribute significantly to the measurable amount of GlcNAc in BSK-II(−GlcNAc)+chitobiose medium (1.73 ± 0.11 μM). The detected GlcNAc levels were much lower than that in normal BSK-II medium (1.92 ± 0.037 mM) or the minimal concentration of GlcNAc (180 μM) required to support high-cell-density growth (6). B. burgdorferi lacks a de novo biosynthesis pathway of GlcNAc, and there is no other significant source of GlcNAc in BSK-II(−GlcNAc)+chitobiose medium. Conceivably, the GlcNAc detected in the wild-type and rrp1com cells is converted from chitobiose in the medium and this conversion is severely impaired in the rrp1mut mutant.
The rrp1mut mutant has decreased RpoS expression.
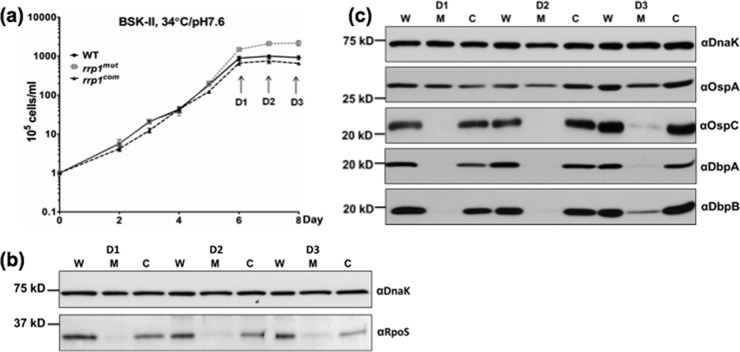
A previous study by Rhodes et al. showed that RpoS regulates chbC expression (16). Mutation of rpoS led to a delay in chitobiose utilization, and the mutant formed membrane blebs in a chitobiose-supplemented growth medium, similar to the phenotype of the rrp1mut mutant (Fig. 2d and 3; see also Fig. S2 in the supplemental material). Based on these facts, we hypothesized that inactivation of rrp1 affects the level of RpoS, which in turn leads to the repression of chbC expression in the rrp1mut mutant. To test this hypothesis, the levels of RpoS in the wild-type, rrp1mut, and rrp1com strains were evaluated by immunoblot analyses. We found that the levels of RpoS were substantially repressed in the rrp1mut mutant (Fig. 6b). At the early (D1) and middle (D2) stationary phase, RpoS was almost undetectable in the mutant; only a trace amount of RpoS was detected when the mutant entered the late stationary phase (D3) (Fig. 6a and b). To confirm this result, we examined the levels of expression of other known RpoS-regulated genes (e.g., ospC, dbpA, and dbpB) (20, 24, 52), as well as that of ospA, a gene that is not activated by RpoS (53, 54). Immunoblotting analyses revealed that the expression levels of OspC, DbpA, and DbpB were substantially decreased in the rrp1mut mutant and were restored to the wild-type levels in the rrp1com strain. In contrast, the level of OspA was unaffected in the rrp1mut strain (Fig. 6c). Consistent with the immunoblot results, qRT-PCR analysis showed that the transcript level of rpoS in the rrp1mut mutant was less than 10% of the wild-type level (Fig. 7b). Collectively, these data support the hypothesis that the impact of Rrp1 on chbC expression is mediated via RpoS.
Fig 6.
The rrp1mut mutant has decreased RpoS expression. A total of 105 cells/ml of the WT, rrp1mut, and rrp1com strains were inoculated into BSK-II and cultivated at 34°C and pH 7.6. (a) Cells were harvested after the stationary phase (108 cells/ml) at the indicated time points (D1 to D3). Similar amounts of whole-cell lysates were analyzed by SDS-PAGE. (b) Detection of RpoS. (c) Detection of OspA, OspC, DbpA, and DbpB. DnaK was used as an internal control as previously described (42). W = WT strain, M = rrp1mut mutant, C = rrp1com strain.
Fig 7.
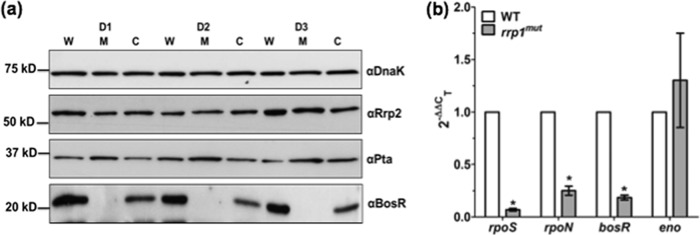
Rrp1 regulates RpoS expression through BosR. (a) Detection of BosR, Pta, and Rrp2 via immunoblot analyses was performed as described for Fig. 6. (b) qRT-PCR analysis of bosR, rpoN, rpoS, and eno transcripts was performed as described for Fig. 4a. Data are expressed as expression of the rrp1mut transcripts relative to that of the corresponding wild-type transcripts. Asterisks indicate that the differences between WT and rrp1mut transcript levels were statistically significant at a P value of <0.01 (two-way ANOVA).
The rrp1mut mutant has decreased BosR expression.
Several regulators, including Rrp2, RpoN, BosR, and Pta, modulate the transcription of the rpoS gene (for reviews, see references 18 and 19). We hypothesized that Rrp1 may regulate the expression of rpoS via one of these known regulators. To test this hypothesis, immunoblotting analyses were carried out to measure the levels of these regulators in the rrp1mut mutant. Our results showed that the levels of both Rrp2 and Pta were not significantly affected in the rrp1mut mutant (Fig. 7a). However, the level of BosR was significantly repressed in the mutant compared to the wild type. Complementation of Rrp1 restored the expression of BosR in the rrp1com strain (Fig. 7a), which indicates that Rrp1 most likely regulates RpoS via BosR, an activator of rpoS expression (31, 32). Consistent with the immunoblot results, the qRT-PCR analysis showed an approximately 80% reduction at the transcript level of bosR, suggesting that the impact of Rrp1 on bosR occurs at the transcriptional level (Fig. 7b). We also examined the transcript level of rpoN and found that it was also significantly downregulated in the rrp1mut mutant (Fig. 7b). In contrast to rpoS, rpoN, and bosR, the expression level of the enolase (eno) gene remained unchanged, indicating that the observed reduction of transcript levels in the mutant is gene specific and not due to global repression of transcriptome following the mutagenesis of rrp1. Collectively, these results suggest that Rrp1 can regulate the expression of RpoS, probably through both BosR and RpoN.
Supplementation of GlcNAc enables the rrp1mut mutant to be transmitted to mice via tick bite.
During a blood meal, chitobiose shed from the tick peritrophic membrane (30) can be potentially metabolized and provide a sufficient amount of GlcNAc for B. burgdorferi to rapidly replicate, which may facilitate the transition of the spirochetes from the tick to a mammalian host (55). Previous studies showed that a mutant defective in rrp1 expression fails to be transmitted to mice upon tick bite (14, 26). We reasoned that this failure is probably due to the defect of the rrp1 mutant in utilizing chitobiose during the transmission. If this hypothesis is correct, we would expect that external supplementation of GlcNAc should rescue, at least in part, the observed transmission defect. To test this hypothesis, naïve Ixodes scapularis nymphs were artificially infected with equal numbers of the wild type, the rrp1mut mutant, the rrp1com strain, and the rrp1mut mutant supplemented with 10 mM GlcNAc via microinjection. Infected nymphs were allowed to feed on naïve C3H mice until repletion. The ticks were then subjected to qRT-PCR analyses to determine the spirochete burden. Using flaB as a surrogate marker, we were able to detect the rrp1mut mutant in replete ticks, albeit at a much lower burden (P < 0.05) than in both the wild-type- and rrp1com strain-infected ticks. Supplementation of 10 mM GlcNAc did not increase the bacterial load of the rrp1mut mutant in replete ticks (Fig. 8a). To investigate if supplementation of GlcNAc can assist in transmission of the rrp1mut mutant from tick to mammal, C3H mice were sacrificed 14 days after tick feeding and transmission was assessed via tissue culture and qRT-PCR. Spleen tissue cultures from mice fed upon by both the wild-type- and rrp1com-infected ticks were positive 1 week after isolation but were negative for spirochetes from mice fed upon by rrp1mut mutant-infected nymphs even after 1 month of culturing (Table 1), which is consistent with previous observations (14, 26). However, this phenotype was partially rescued by providing GlcNAc to the unfed ticks prior to feeding. Two out of 3 mice fed upon by the nymphs infected with the rrp1mut mutant with supplementation of 10 mM GlcNAc were positive for spirochetes in spleen tissue culture, indicating a successful transmission from ticks to mice (Table 1). Follow-up qRT-PCR analysis on tissues from mice fed upon by the nymphs infected with the rrp1mut mutant with supplementation of 10 mM GlcNAc showed positive results for the flaB transcripts (Fig. 8b), further validating the tissue culture results. This finding supports our hypothesis that the defect in utilizing chitobiose as a source of GlcNAc contributes to the failure of the rrp1 mutant in the transmission from ticks to mice.
Fig 8.
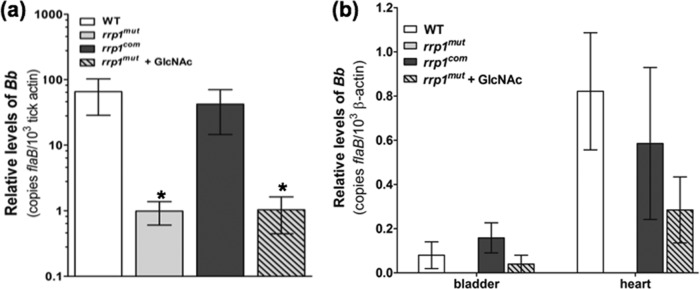
Supplementation of GlcNAc fails to rescue the rrp1mut bacterial load in fed ticks but enables transmission via tick bite. (a) RNA samples were extracted from whole fed ticks (after repletion; 5 to 7 days) and subjected to qRT-PCR analysis. The bacterial burdens in ticks were measured by the number of copies of flaB mRNA compared to the number of copies of tick-actin transcript as previously described (37). The data are presented as the means of relative levels of flaB transcript ± SEM for three groups (each group contained 4 replete ticks) for each strain (WT, rrp1mut mutant, and rrp1com strain). Asterisks indicate that the differences in bacterial load between the WT strain, the rrp1mut mutant, and the rrp1mut mutant supplemented with GlcNAc were statistically significant at a P value of <0.05. (b) Detection of spirochete burdens in C3H mice infected via tick bite using qRT-PCR. At day 14 after tick feeding, mice were sacrificed. Tissues (skin, heart, joint, and bladder) were collected and total RNA from heart and bladder tissues subjected to qRT-PCR analysis as described in Materials and Methods. Statistical analysis showed no significant difference between the WT, the rrp1com strain, and the rrp1mut mutant supplemented with GlcNAc at P = 0.05.
Table 1.
Tick-to-mouse-infection study
| Strain | No. of mice infected/total no. of micea |
|---|---|
| Wild type | 3/3 |
| rrp1mut | 0/3 |
| rrp1com | 3/3 |
| rrp1mut + GlcNAc | 2/3b |
Mouse spleen tissues were used for the reisolation.
Spirochete cells were observed 2 weeks after tissue isolation.
DISCUSSION
Owing to its complex enzootic life cycle alternating between arthropod vector and mammalian hosts, highly coordinated expression of the host-specific genes is indispensable for the survival and persistence of B. burgdorferi in nature. These extensive transcriptional adjustments are primarily governed by the Rrp2-RpoN-RpoS regulon and the Hk1-Rrp1 signaling pathway (for recent reviews, see references 18 and 19). Exhaustive investigations on the Rrp2-RpoN-RpoS regulatory pathway have provided compelling evidence that it is responsible for the differential expression of various genes required for infection and persistence in the mammalian host, including several outer surface proteins essential for the pathogenicity of B. burgdorferi such as OspC and DbpA (19–21, 23, 24, 56, 57). The second signaling pathway is comprised of a diguanylate cyclase (Rrp1) and a histidine kinase (Hk1) (25, 28). Recent studies by three independent laboratories have established the significance of both Rrp1 and Hk1 in the survival of B. burgdorferi during the blood meal (14, 25, 26). Rrp1 mutants and Hk1 mutant exhibited similar defects in feeding ticks, where the spirochetes were killed during the blood meal, indicating that the Hk1-Rrp1 TCS is essential during the transmission process (25, 26).
The role of c-di-GMP in virulence in many pathogenic bacteria has been well established (58–60). Rrp1 is the sole c-di-GMP-synthesizing protein encoded in the genome of B. burgdorferi (28), and mutagenesis of rrp1 has been shown to result in alteration of the B. burgdorferi transcriptome (14, 27). Among the genes affected by Rrp1 are carbohydrate metabolic pathways and the GlcNAc transporter systems (27). In this study, we provided evidence showing that Rrp1 is involved in the regulation of chitobiose metabolism in B. burgdorferi. In vitro growth analysis indicated that the rrp1mut mutant is unable to grow in medium containing chitobiose in place of GlcNAc as the carbon source, and restoration of Rrp1 in the mutant successfully rescued the growth defect (Fig. 2d). In addition, overexpression of a single chitobiose transporter gene, chbC, is able to salvage the growth of the rrp1mut mutant in chitobiose-supplemented medium (Fig. 4b), further supporting our hypothesis that Rrp1 can regulate the chitobiose metabolism pathway and, more specifically, the chbC gene. Furthermore, transcriptional analysis showed that the transcript level of chbC was significantly repressed in the rrp1mut mutant relative to wild-type expression, suggesting that Rrp1 is an activator of the chbC gene (Fig. 4a).
A previous report established that RpoS is involved in the transcription of the chbC gene (16), which prompted us to investigate the linkage between Rrp1 and RpoS. Using immunoblotting analyses, we showed that the rrp1mut mutant has reduced levels of RpoS protein and of several proteins encoded by RpoS-dependent genes, including OspC, DbpA, and DbpB (Fig. 6b and c). Upon analysis of several known regulatory elements of rpoS, we found that the protein level of BosR was also downregulated in the rrp1mut mutant (Fig. 7a), which indicates that Rrp1 regulates RpoS, probably by affecting the expression of BosR. The oxidative stress response regulator BosR is an important activator of the RpoS pathway (31, 32, 61). The expression of bosR is induced upon tick feeding and reaches the maximal level in the murine host (62). As rrp1 is also highly expressed in the feeding tick (27), it may assist the induction of bosR following a blood meal. Thus, a decrease in bosR expression in the rrp1mut strain leads to a reduction in rpoS expression, which subsequently results in reduced transcription of the RpoS-dependent chbC gene.
Cultivation of the chbC mutant in chitobiose-supplemented medium has been shown to lead to the formation of membrane-attached masses (6). In addition, when the rpoS mutant is cultured in a chitobiose-supplemented medium, membrane blebs similar to those in the chbC mutant were observed (16). Our scanning electron microscope analysis showed that similar round membrane blebs were seen forming on the rrp1mut cells when cultured in BSK-II(−GlcNAc)+chitobiose medium and that the formation of the blebs progressed throughout growth and, eventually, cell lysis (Fig. 3). The formation of membrane blebs was reversible upon overexpression of the chbC gene in the rrp1mut background (Fig. 3a and 4b; see also Fig. S2 in the supplemental material), suggesting that the mutant has a defect in chitobiose utilization and that the membrane blebs were most likely formed due to failure to convert chitobiose to GlcNAc for cell wall synthesis. Dunham-Ems et al. recently showed that the rpoS mutant, when present in fed ticks, formed an unusual morphotype, which they termed “round bodies” (63). The observation of formation of such “round bodies” may be related to the blebbings observed in the chbC and rpoS mutants as well as in our rrp1mut mutant (6, 16). Under unfavorable environmental conditions, initial membrane blebbing occurs, which then progresses to cell lysis. Part of the cell may convert to a cystic form or “round bodies,” which, upon encountering a suitable environment, can be quickly reverted to a live spirochete (64–66). The initial failure to utilize chitobiose after a blood meal could cause an unfavorable environment for growth, which subsequently may induce the formation of “round bodies” as a mechanism of protection against hostile conditions.
How does regulation of chitobiose metabolism integrate into the life cycle of B. burgdorferi? Previous reports by Tilly et al. indicated that a chbC mutant can complete the enzootic cycle and that chitobiose metabolism is likely unimportant for B. burgdorferi growth in vivo (17). In their study, the chbC mutant was able to infect mice via needle inoculation, be acquired by naïve ticks, survive the molting process, and complete the transmission to naïve mice (17), implying that chitobiose metabolism is not essential for the life cycle of this spirochete. The genome of B. burgdorferi includes several copies of chitobiose transporter genes that are predicted to form the chitobiose PTS, which includes chbA (bbb05), chbB (bbb06), and chbC (bbb04) (4, 6). Although in vitro growth analysis indicated that both chbA and chbB do not play a role in chitobiose metabolism, they may be able to compensate for the loss of chbC during the in vivo growth in the arthropod vector, which could potentially rescue the chbC mutant during the enzootic cycle and subsequently enable transmission. An in vivo study using various chb mutants (i.e., double or triple chb mutants) would help to clarify the exact role of these chitobiose transporter genes in the infectious cycle of B. burgdorferi. In addition to the chitobiose transporter system, the genome of B. burgdorferi also encodes several other phospotransferase components for carbohydrate uptake (4). The import of GlcNAc into the B. burgdorferi cell is believed to be performed through the glucose PTS transporters (7), similar to the case in other bacteria such as E. coli (8). Hence, it is possible that an alternative uncharacterized PTS with a loose specificity for carbohydrate may be able to transport chitobiose, albeit at a lower capacity than the characterized chitobiose transporters. Identifying an additional PTS(s) for chitobiose uptake will definitely help increase understanding of the significance of chitobiose in the life cycle of this spirochete.
During the tick phase of the enzootic cycle, B. burgdorferi encounters different carbohydrates for metabolism, one of which is glycerol, the natural antifreeze agent produced by the Ixodes tick (12, 13). Studies have shown that the rrp1 mutant is defective in glycerol metabolism and fails to survive after tick feeding (14). A different group also reported that mutagenesis of a glycerol utilization gene, glycerol 3-phosophate dehydrogenase (glpD), led to a reduced spirochete burden in unfed ticks and impaired replication in feeding ticks, emphasizing the importance of glycerol metabolism during the tick phase of the life cycle (5). Microarray analysis indicated that the expression of several glycerol metabolism genes is regulated by Rrp1 (14). Constitutive expression of the glycerol metabolic operon in the rrp1 mutant was shown to partially rescue the mutant in ticks after a blood meal. However, the mutant remained incapable of transmission to mice, implying that another Rrp1-regulated factor(s) may be at play (14). It is reasonable to suggest that the chitobiose metabolic pathway (in addition to the glycerol metabolism) may be one of the factors that is required to fully rescue the rrp1 mutant in ticks. In the unfed tick, glycerol is the major carbon source available to the spirochete. Glycerol can be channeled into the glycolysis pathway for energy production, and thus a mutant defective in glycerol metabolism would have reduced metabolic fitness. Upon feeding, chitobiose becomes available and can be converted into GlcNAc for cell wall synthesis. The rrp1 mutant, which has a preexisting reduced fitness, encounters a second defect in chitobiose utilization, further compromising the condition of the spirochetes. Together, these defects eventually result in clearance of the spirochetes from the vector. This may explain why overexpressing the glycerol utilization pathway alone is unable to salvage the rrp1 mutant in the feeding tick. In addition, our in vitro growth analysis showed that the rrp1mut strain can grow in the chitobiose-supplemented medium under unfed tick conditions but not at elevated temperature (Fig. 2). As chbC is dually regulated by both RpoD and RpoS (16), under unfed tick conditions where RpoS is not expressed, chbC is solely regulated by RpoD, which can explain the lack of growth defect in the rrp1mut mutant (Fig. 2b). Upon transition to an elevated temperature where RpoS is induced (56), a low level of RpoS in the rrp1mut mutant leads to decreased transcription of chbC, which subsequently results in failure to utilize chitobiose for growth. Both in vitro growth analysis (Fig. 2) and transcriptional (Fig. 4a and 7b) and translational (Fig. 6) analyses seemed to support this proposition. Along with this proposition, a study from Rhodes et al. showed that at elevated temperature (33°C), the rpoS mutant showed reduced transcription of chbC as well as a defect in utilizing chitobiose for growth (16), emphasizing the importance of RpoS in the regulation of chitobiose metabolism.
In our tick infection study, when 10 mM GlcNAc was supplemented, we were able to partially rescue the rrp1mut mutant in fed ticks to allow transmission to mice (Table 1). Although the bacterial burden (as measured by qRT-PCR) in fed ticks remained low in the GlcNAc-supplemented rrp1mut mutant group, this could be explained by the additional defect in the glycerol metabolic pathway in the rrp1 mutant as mentioned earlier. Addition of GlcNAc may provide only enough of a utilizable carbon source to rescue a small percentage of the spirochetes to multiply and complete the transmission process following a blood meal (Fig. 8). In order to fully rescue the mutant phenotype, both glycerol and chitobiose metabolism pathways may be necessary, as the former is necessary for glycolysis while the latter is needed for GlcNAc production for cell wall synthesis (4). In our tick study, we did not include the rrp1mut chbC+ strain as a rescue strategy due to several concerns. First, we observed a significantly higher expression of RpoS in this strain when we cultivated it under routine laboratory culture conditions (data not shown). Second, the rrp1mut chbC+ strain exhibited poor growth in normal BSK-II medium under conditions mimicking the unfed-tick conditions (data not shown). We reasoned that the untimely and high induction of RpoS may have been responsible for the poor growth pattern observed in this strain. We currently do not have an explanation as to how overexpression of the chbC gene can affect RpoS expression. Perhaps constitutive expression of the chitobiose transporter forces a switch in the carbohydrate metabolism preferences and stimulates the spirochete to induce fed-tick or mammal-phase genes, such as RpoS. The expression of RpoS as well as other important surface lipoproteins such as OspC is necessary to establish an infection in mammals (see reviews in references 18 and 19). In vitro immunoblot results indicated that the rrp1mut mutant has minimal expression of RpoS and RpoS-regulated proteins (Fig. 6b and c). Reduced expression of these important virulence factors may explain the partial restoration of transmission to mice (Table 1) as well as the late emergence of spirochetes from the tissue infected by the rrp1mut mutant plus GlcNAc even though qRT-PCR analysis showed no significant difference between the WT and the rrp1mut mutant plus GlcNAc in the spirochete burden of infected mice tissues (Fig. 8b).
A study by Caimano et al. showed that RpoS acts as a repressor of the glycerol metabolic pathway (20). In addition, RpoS has been shown to be involved in the transcription of the chbC gene (16). Based on this information together with the data in this study, we propose a regulatory model for Rrp1 during the feeding-tick stage of the B. burgdorferi enzootic cycle (Fig. 9). During a blood meal, host factors or chitobiose shed from the peritrophic membrane or both serve as a signal(s) to activate the Hk1 histidine kinase, leading to the phosphorylation of the response regulator, Rrp1. Activated Rrp1 induces BosR and RpoN expression, which, together with other factors (such as the Rrp2-RpoN signaling and DsrABb), leads to the expression of RpoS. Induction of RpoS expression results in repression of the glycerol metabolism genes and activation of the chitobiose transporter chbC gene. Activation of the chitobiose utilization pathway allows a switch in carbohydrate metabolism, changing from glycerol to chitobiose as the main carbon source for replication and subsequent transmission to a mammalian host.
Fig 9.
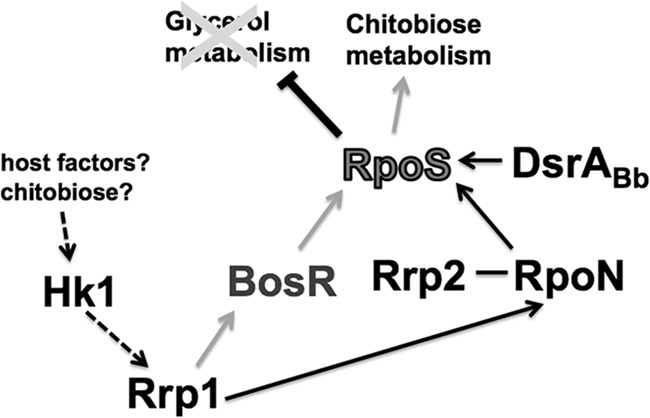
Model for Rrp1 regulation of chitobiose metabolism in B. burgdorferi. During the feeding process, certain host factors or chitobiose or both serve as a ligand(s) to activate the histidine kinase, Hk1, leading to phosphorylation of the response regulator, Rrp1. Phosphorylated Rrp1 activates BosR (the oxidative stress regulator) and RpoN, which then activate the transcription of rpoS. The expression of RpoS is further enhanced via the signaling by Rrp2-RpoN as well as other regulators such as DsrABb. An increase in the level of RpoS leads to repression of glycerol metabolism genes and activation of the components of the chitobiose metabolic pathway, especially the chitobiose transporter gene, chbC. Expression of the chitobiose utilization pathway allows B. burgdorferi to utilize chitobiose shed from the peritrophic membrane which is formed after a blood meal as a source for GlcNAc, a precursor for cell wall synthesis. A switch from glycerol to chitobiose for utilization as the main carbon source allows the spirochete to multiply in numbers and completes the transmission cycle.
The regulation by c-di-GMP can occur at various stages, including both transcriptional and posttranscriptional stages (58, 67–69). C-di-GMP affects a cellular function by binding to its receptor proteins, such as PilZ domain-containing proteins (69). Our transcriptional analysis indicated a reduction in transcripts of both the activator of the rpoS gene, bosR, and rpoN (Fig. 7b), suggesting that transcriptional regulation is involved. Currently, little is known about the regulation by c-di-GMP in B. burgdorferi. A c-di-GMP effector protein, PlzA, which contains a PilZ domain, has been identified in B. burgdorferi (70). Whether PlzA or an additional unidentified effector protein(s) is involved in the regulation by Rrp1 on bosR/rpoN requires further investigation. In summary, our findings on Rrp1 in this study show an interaction between the two important signaling pathways in B. burgdorferi, Hk1-Rrp1 and Rrp2-RpoN-RpoS. By acting as an activator of RpoS, Rrp1 serves as a carbohydrate switch in B. burgdorferi during the tick phase, allows glycerol metabolism in the unfed tick stage, and switches the carbon metabolism to chitobiose upon a blood meal by activating RpoS expression via BosR. Studies to elucidate the underlying molecular mechanism as well as the effector protein(s) involved are necessary to fully understand the function of Rrp1 in the enzootic cycle.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Rosa for providing the A3-68 strain and chbC mutant and M. Caimano, F. T. Liang, J. Seshu, J. Skare, and X. F. Yang for providing various antibodies used in this study.
This research was supported by Public Health Service grants (AI073354 and AI078958) to C.L.
Footnotes
Published ahead of print 11 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00050-13.
REFERENCES
- 1. Bacon RM, Kugeler KJ, Mead PS; Centers for Disease Control and Prevention (CDC) 2008. Surveillance for Lyme disease--United States, 1992–2006. MMWR Surveill. Summ. 57:1–9 [PubMed] [Google Scholar]
- 2. Tsao JI. 2009. Reviewing molecular adaptations of Lyme borreliosis spirochetes in the context of reproductive fitness in natural transmission cycles. Vet. Res. 40:36 doi:10.1051/vetres/2009019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Levine JF, Wilson ML, Spielman A. 1985. Mice as reservoirs of the Lyme disease spirochete. Am. J. Trop. Med. Hyg. 34:355–360 [DOI] [PubMed] [Google Scholar]
- 4. Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fuji C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580–586 [DOI] [PubMed] [Google Scholar]
- 5. Pappas CJ, Iyer R, Petzke MM, Caimano MJ, Radolf JD, Schwartz I. 2011. Borrelia burgdorferi requires glycerol for maximum fitness during the tick phase of the enzootic cycle. PLoS Pathog. 7:e1002102 doi:10.1371/journal.ppat.1002102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tilly K, Elias AF, Errett J, Fischer E, Iyer R, Schwartz I, Bono JL, Rosa P. 2001. Genetics and regulation of chitobiose utilization in Borrelia burgdorferi. J. Bacteriol. 183:5544–5553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. von Lackum K, Stevenson B. 2005. Carbohydrate utilization by the Lyme borreliosis spirochete, Borrelia burgdorferi. FEMS Microbiol. Lett. 243:173–179 [DOI] [PubMed] [Google Scholar]
- 8. Hoon-Hanks LL, Morton EA, Lybecker MC, Battisti JM, Samuels DS, Drecktrah D. 2012. Borrelia burgdorferi malQ mutants utilize disaccharides and traverse the enzootic cycle. FEMS Immunol. Med. Microbiol. 66:157–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Young DS, Harris EK, Cotlove E. 1971. Biological and analytic components of variation in long-term studies of serum constituents in normal subjects. IV. Results of a study designed to eliminate long-term analytic deviations. Clin. Chem. 17:403–410 [PubMed] [Google Scholar]
- 10. Barbour AG. 1984. Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57:521–525 [PMC free article] [PubMed] [Google Scholar]
- 11. Hackman RH. 1982. Structure and function in tick cuticle. Annu. Rev. Entomol. 27:75–95 [DOI] [PubMed] [Google Scholar]
- 12. Lee RJ, Jr, Baust JG. 1987. Cold-hardiness in the Antarctic tick, Ixodes uriae. Physiol. Zool. 60:499–506 [Google Scholar]
- 13. Burks CS, Stewart RL, Jr, Needham GR, Lee RE., Jr 1996. Cold hardiness in the Ixodid ticks (Ixodidae), p 85–87 In Mitchell R, Horn DJ, Needham GR, Welbourn WC. (ed), Acarology IX: vol. 1, proceedings. Ohio Biological Survey, Columbus, OH [Google Scholar]
- 14. He M, Ouyang Z, Troxell B, Xu H, Moh A, Piesman J, Norgard MV, Gomelsky M, Yang XF. 2011. Cyclic di-GMP is essential for the survival of the Lyme disease spirochete in ticks. PLoS Pathog. 7:e1002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rhodes RG, Atoyan JA, Nelson DR. 2010. The chitobiose transporter, chbC, is required for chitin utilization in Borrelia burgdorferi. BMC Microbiol. 10:21 doi:10.1186/1471-2180-10-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rhodes RG, Coy W, Nelson DR. 2009. Chitobiose utilization in Borrelia burgdorferi is dually regulated by RpoD and RpoS. BMC Microbiol. 9:108 doi:10.1186/1471-2180-9-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tilly K, Grimm D, Bueschel DM, Krum JG, Rosa P. 2004. Infectious cycle analysis of a Borrelia burgdorferi mutant defective in transport of chitobiose, a tick cuticle component. Vector Borne Zoonotic Dis. 4:159–168 [DOI] [PubMed] [Google Scholar]
- 18. Radolf JD, Caimano MJ, Stevenson B, Hu LT. 2012. Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat. Rev. Microbiol. 10:87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Samuels DS. 2011. Gene regulation in Borrelia burgdorferi. Annu. Rev. Microbiol. 65:479–499 [DOI] [PubMed] [Google Scholar]
- 20. Caimano MJ, Iyer R, Eggers CH, Gonzalez C, Morton EA, Gilbert MA, Schwartz I, Radolf JD. 2007. Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol. Microbiol. 65:1193–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fisher MA, Grimm D, Henion AK, Elias AF, Stewart PE, Rosa PA, Gherardini FC. 2005. Borrelia burgdorferi σ54 is required for mammalian infection and vector transmission but not for tick colonization. Proc. Natl. Acad. Sci. U. S. A. 102:5162–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ouyang Z, Blevins JS, Norgard MV. 2008. Transcriptional interplay among the regulators Rrp2, RpoN and RpoS in Borrelia burgdorferi. Microbiology 154:2641–2658 [DOI] [PubMed] [Google Scholar]
- 23. Ouyang Z, Narasimhan S, Neelakanta G, Kumar M, Pal U, Fikrig E, Norgard MV. 2012. Activation of the RpoN-RpoS regulatory pathway during the enzootic life cycle of Borrelia burgdorferi. BMC Microbiol. 12:44 doi:10.1186/1471-2180-12-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hübner A, Yang X, Nolen DM, Popova TG, Cabello FC, Norgard MV. 2001. Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl. Acad. Sci. U. S. A. 98:12724–12729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Caimano MJ, Kenedy MR, Kairu T, Desrosiers DC, Harman M, Dunham-Ems S, Akins DR, Pal U, Radolf JD. 2011. The hybrid histidine kinase Hk1 is part of a two-component system that is essential for survival of Borrelia burgdorferi in feeding Ixodes scapularis ticks. Infect. Immun. 79:3117–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kostick JL, Szkotnicki LT, Rogers EA, Bocci P, Raffaelli N, Marconi RT. 2011. The diguanylate cyclase, Rrp1, regulates critical steps in the enzootic cycle of the Lyme disease spirochetes. Mol. Microbiol. 81:219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rogers EA, Terekhova D, Zhang HM, Hovis KM, Schwartz I, Marconi RT. 2009. Rrp1, a cyclic-di-GMP-producing response regulator, is an important regulator of Borrelia burgdorferi core cellular functions. Mol. Microbiol. 71:1551–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dunham-Ems SM, Caimano MJ, Pal U, Wolgemuth CW, Eggers CH, Balic A, Radolf JD. 2009. Live imaging reveals a biphasic mode of dissemination of Borrelia burgdorferi within ticks. J. Clin. Invest. 119:3652–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shao L, Devenport M, Jacobs-Lorena M. 2001. The peritrophic matrix of hematophagous insects. Arch. Insect Biochem. Physiol. 47:119–125 [DOI] [PubMed] [Google Scholar]
- 31. Ouyang Z, Deka RK, Norgard MV. 2011. BosR (BB0647) controls the RpoN-RpoS regulatory pathway and virulence expression in Borrelia burgdorferi by a novel DNA-binding mechanism. PLoS Pathog. 7:e1001272 doi:10.1371/journal.ppat.1001272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hyde JA, Shaw DK, Smith R, III, Trzeciakowski JP, Skare JT. 2009. The BosR regulatory protein of Borrelia burgdorferi interfaces with the RpoS regulatory pathway and modulates both the oxidative stress response and pathogenic properties of the Lyme disease spirochete. Mol. Microbiol. 74:1344–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rego RO, Bestor A, Rosa PA. 2011. Defining the plasmid-borne restriction-modification systems of the Lyme disease spirochete Borrelia burgdorferi. J. Bacteriol. 193:1161–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li X, Pal U, Ramamoorthi N, Liu X, Desrosiers DC, Eggers CH, Anderson JF, Radolf JD, Fikrig E. 2007. The Lyme disease agent Borrelia burgdorferi requires BB0690, a Dps homologue, to persist within ticks. Mol. Microbiol. 63:694–710 [DOI] [PubMed] [Google Scholar]
- 35. Elias AF, Bono JL, Kupko JJ, III, Stewart PE, Krum JG, Rosa PA. 2003. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J. Mol. Microbiol. Biotechnol. 6:29–40 [DOI] [PubMed] [Google Scholar]
- 36. Tilly K, Bestor A, Dulebohn DP, Rosa PA. 2009. OspC-independent infection and dissemination by host-adapted Borrelia burgdorferi. Infect. Immun. 77:2672–2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sze CW, Zhang K, Kariu T, Pal U, Li C. 2012. Borrelia burgdorferi needs chemotaxis to establish infection in mammals and to accomplish its enzootic cycle. Infect. Immun. 80:2485–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bakker RG, Li C, Miller MR, Cunningham C, Charon NW. 2007. Identification of specific chemoattractants and genetic complementation of a Borrelia burgdorferi chemotaxis mutant: flow cytometry-based capillary tube chemotaxis assay. Appl. Environ. Microbiol. 73:1180–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lawrenz MB, Wooten RM, Norris SJ. 2004. Effects of vlsE complementation on the infectivity of Borrelia burgdorferi lacking the linear plasmid lp28-1. Infect. Immun. 72:6577–6585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li C, Xu H, Zhang K, Liang FT. 2010. Inactivation of a putative flagellar motor switch protein FliG1 prevents Borrelia burgdorferi from swimming in highly viscous media and blocks its infectivity. Mol. Microbiol. 75:1563–1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sal MS, Li C, Motalab MA, Shibata S, Aizawa S, Charon NW. 2008. Borrelia burgdorferi uniquely regulates its motility genes and has an intricate flagellar hook-basal body structure. J. Bacteriol. 190:1912–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sze CW, Li C. 2011. Inactivation of bb0184, which encodes carbon storage regulator A, represses the infectivity of Borrelia burgdorferi. Infect. Immun. 79:1270–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Burtnick MN, Downey JS, Brett PJ, Boylan JA, Frye JG, Hoover TR, Gherardini FC. 2007. Insights into the complex regulation of rpoS in Borrelia burgdorferi. Mol. Microbiol. 65:277–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. von Lackum K, Ollison KM, Bykowski T, Nowalk AJ, Hughes JL, Carroll JA, Zuckert WR, Stevenson B. 2007. Regulated synthesis of the Borrelia burgdorferi inner-membrane lipoprotein IpLA7 (P22, P22-A) during the Lyme disease spirochaete's mammal-tick infectious cycle. Microbiology 153:1361–1371 [DOI] [PubMed] [Google Scholar]
- 45. Kariu T, Coleman AS, Anderson JF, Pal U. 2011. Methods for rapid transfer and localization of Lyme disease pathogens within the tick gut. J. Vis Exp. 2011:2544 doi:10.3791/2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pal U, Yang X, Chen M, Bockenstedt LK, Anderson JF, Flavell RA, Norgard MV, Fikrig E. 2004. OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands. J. Clin. Invest. 113:220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brossard KA, Campagnari AA. 2012. The Acinetobacter baumannii biofilm-associated protein plays a role in adherence to human epithelial cells. Infect. Immun. 80:228–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marks LR, Clementi EA, Hakansson AP. 2012. The human milk protein-lipid complex HAMLET sensitizes bacterial pathogens to traditional antimicrobial agents. PLoS One 7:e43514 doi:10.1371/journal.pone.0043514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Samuels DS. 1995. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol. Biol. 47:253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li C, Bakker RG, Motaleb MA, Sartakova ML, Cabello FC, Charon NW. 2002. Asymmetrical flagellar rotation in Borrelia burgdorferi nonchemotactic mutants. Proc. Natl. Acad. Sci. U. S. A. 99:6169–6174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hegedus D, Erlandson M, Gillott C, Toprak U. 2009. New insights into peritrophic matrix synthesis, architecture, and function. Annu. Rev. Entomol. 54:285–302 [DOI] [PubMed] [Google Scholar]
- 52. Yang XF, Alani SM, Norgard MV. 2003. The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc. Natl. Acad. Sci. U. S. A. 100:11001–11006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Alverson J, Bundle SF, Sohaskey CD, Lybecker MC, Samuels DS. 2003. Transcriptional regulation of the ospAB and ospC promoters from Borrelia burgdorferi. Mol. Microbiol. 48:1665–1677 [DOI] [PubMed] [Google Scholar]
- 54. Sohaskey CD, Zuckert WR, Barbour AG. 1999. The extended promoters for two outer membrane lipoprotein genes of Borrelia spp. uniquely include a T-rich region. Mol. Microbiol. 33:41–51 [DOI] [PubMed] [Google Scholar]
- 55. Jutras BL, Chenail AM, Stevenson B. 2013. Changes in bacterial growth rate govern expression of the Borrelia burgdorferi OspC and Erp infection-associated surface proteins. J. Bacteriol. 195:757–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Caimano MJ, Eggers CH, Hazlett KR, Radolf JD. 2004. RpoS is not central to the general stress response in Borrelia burgdorferi but does control expression of one or more essential virulence determinants. Infect. Immun. 72:6433–6445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Smith AH, Blevins JS, Bachlani GN, Yang XF, Norgard MV. 2007. Evidence that RpoS (σS) in Borrelia burgdorferi is controlled directly by RpoN (σ54/σN). J. Bacteriol. 189:2139–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Coggan KA, Wolfgang MC. 2012. Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr. Issues Mol. Biol. 14:47–70 [PubMed] [Google Scholar]
- 59. Cotter PA, Stibitz S. 2007. c-Di-GMP-mediated regulation of virulence and biofilm formation. Curr. Opin. Microbiol. 10:17–23 [DOI] [PubMed] [Google Scholar]
- 60. Ryan RP, Dow JM. 2010. Intermolecular interactions between HD-GYP and GGDEF domain proteins mediate virulence-related signal transduction in Xanthomonas campestris. Virulence 1:404–408 [DOI] [PubMed] [Google Scholar]
- 61. Ouyang Z, Kumar M, Kariu T, Haq S, Goldberg M, Pal U, Norgard MV. 2009. BosR (BB0647) governs virulence expression in Borrelia burgdorferi. Mol. Microbiol. 74:1331–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Medrano MS, Ding Y, Wang XG, Lu P, Coburn J, Hu LT. 2007. Regulators of expression of the oligopeptide permease A proteins of Borrelia burgdorferi. J. Bacteriol. 189:2653–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dunham-Ems SM, Caimano MJ, Eggers CH, Radolf JD. 2012. Borrelia burgdorferi requires the alternative sigma factor RpoS for dissemination within the vector during tick-to-mammal transmission. PLoS Pathog. 8:e1002532 doi:10.1371/journal.ppat.1002532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Brorson O, Brorson SH. 1997. Transformation of cystic forms of Borrelia burgdorferi to normal, mobile spirochetes. Infection 25:240–246 [DOI] [PubMed] [Google Scholar]
- 65. Brorson O, Brorson SH. 1998. In vitro conversion of Borrelia burgdorferi to cystic forms in spinal fluid, and transformation to mobile spirochetes by incubation in BSK-H medium. Infection 26:144–150 [DOI] [PubMed] [Google Scholar]
- 66. Miklossy J, Kasas S, Zurn AD, McCall S, Yu S, McGeer PL. 2008. Persisting atypical and cystic forms of Borrelia burgdorferi and local inflammation in Lyme neuroborreliosis. J. Neuroinflammation 5:40 doi:10.1186/1742-2094-5-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Beyhan S, Tischler AD, Camilli A, Yildiz FH. 2006. Transcriptome and phenotypic responses of Vibrio cholerae to increased cyclic di-GMP level. J. Bacteriol. 188:3600–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Méndez-Ortiz MM, Hyodo M, Hayakawa Y, Membrillo-Hernandez J. 2006. Genome-wide transcriptional profile of Escherichia coli in response to high levels of the second messenger 3′,5′-cyclic diguanylic acid. J. Biol. Chem. 281:8090–8099 [DOI] [PubMed] [Google Scholar]
- 69. Sondermann H, Shikuma NJ, Yildiz FH. 2012. You've come a long way: c-di-GMP signaling. Curr. Opin. Microbiol. 15:140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Freedman JC, Rogers EA, Kostick JL, Zhang H, Iyer R, Schwartz I, Marconi RT. 2010. Identification and molecular characterization of a cyclic-di-GMP effector protein, PlzA (BB0733): additional evidence for the existence of a functional cyclic-di-GMP regulatory network in the Lyme disease spirochete, Borrelia burgdorferi. FEMS Immunol. Med. Microbiol. 58:285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.