Abstract
Phagocytosis plays a pivotal role in nutrient acquisition and evasion from the host defense systems in Entamoeba histolytica, the intestinal protozoan parasite that causes amoebiasis. We previously reported that E. histolytica possesses a unique class of a hydrolase receptor family, designated the cysteine protease-binding protein family (CPBF), that is involved in trafficking of hydrolases to lysosomes and phagosomes, and we have also reported that CPBF1 and CPBF8 bind to cysteine proteases or β-hexosaminidase α-subunit and lysozymes, respectively. In this study, we showed by immunoprecipitation that CPBF6, one of the most highly expressed CPBF proteins, specifically binds to α-amylase and γ-amylase. We also found that CPBF6 is localized in lysosomes, based on immunofluorescence imaging. Immunoblot and proteome analyses of the isolated phagosomes showed that CPBF6 mediates transport of amylases to phagosomes. We also demonstrated that the carboxyl-terminal cytosolic region of CPBF6 is engaged in the regulation of the trafficking of CPBF6 to phagosomes. Our proteome analysis of phagosomes also revealed new potential phagosomal proteins.
INTRODUCTION
Phagocytosis and phagosome biogenesis play indispensable and pivotal roles in the pathogenesis of Entamoeba histolytica. E. histolytica trophozoites ingest and digest microorganisms in the large intestine, for the acquisition of nutrients (1), and also host cells (2) during tissue invasion, for the creation of a parasitic niche. Phagocytosis also plays a role in the evasion from host immune systems (3). It has been demonstrated that the levels of in vitro and in vivo virulence of clinical and laboratory strains correlate well with the ability for phagocytosis (4–7). In addition, phagosomes contain a panel of proteins that have been implicated in pathogenesis, such as cysteine proteases (CPs) (8), amoeba pores (9), and galactose-/N-acetylgalactosamine-specific lectin (10, 11). Therefore, an understanding of the molecular mechanisms of phagosome biogenesis and trafficking, together with the characterization of “uncharacterized hypothetical” proteins in phagosomes, should further uncover the links between phagocytosis and pathogenicity.
Recently, the molecular mechanisms of phagocytosis have started to be unveiled. For example, an unconventional myosin, myosin IB, was shown to be involved in cytoskeleton rearrangement during phagocytosis (12). It was also shown that phosphatidylinositol signaling is involved in phagocytosis. The time- and position-specific accumulations of phosphatidylinositol-3-phosphate (PI3P) and phosphatidylinositol (3,4,5)-trisphosphate (PIP3) (13–15) on the phagosomal membrane were demonstrated. Surface Ca2+-binding kinase (C2PK) was shown to be involved in the initiation of phagocytosis (16). C2PK localizes in close proximity to the plasma membrane through phosphatidylserine in the presence of Ca2+ and thereby recruits E. histolytica CaBP1 (EhCaBP1) and actin to the phagocytic cup during erythrophagocytosis. Conditional suppression of C2PK expression and overexpression of a mutant form demonstrated its role in the initiation of the formation of the phagocytic cup. Surface transmembrane kinase (TMK96) and p21-activated kinase (PAK) also have been shown to play important roles in phagocytosis of human erythrocytes (17, 18). TMK96 was identified in the proteome of isolated phagosomes and plays an important role in the clearance of dead host cells. PAK was shown to be highly concentrated in the nascent pseudopod in motile trophozoites and appears to be a regulatory element controlling pseudopod extension and cell polarity. Overproduction of the carboxyl-terminal kinase domain of PAK causes multiple pseudopod formation and enhanced erythrophagocytosis.
Although the mechanisms of initiation of phagocytosis have been vigorously investigated, it is only recently that scientists have started to understand the fundamental mechanisms of how hydrolases are selectively transported to phagosomes, leading to organelle maturation. Transport and activation of hydrolytic enzymes are tightly regulated. Otherwise, they are deleterious to the cell. In yeast and mammals, trafficking of major hydrolytic enzymes is mediated by the specific cargo receptors mannose-6-phosphate receptor (M6PR) and Vps10/sortilin. M6PR regulates the trafficking of M6P-modified lysosomal hydrolases, including cathepsin L and β-hexosaminidase, while Vps10p/sortilin regulates the trafficking of carboxypeptidase Y in yeast or cathepsin D in mammals. E. histolytica appears to lack a Vps10/sortilin homolog. Furthermore, immuno-affinity pulldown of a putative M6PR (EHI_096320) failed to coprecipitate any cargo protein, suggesting that putative M6PRs are not likely receptors/carriers of lysosomal hydrolases (K. Nakada-Tsukui, unpublished data). We recently discovered a novel class of a single-transmembrane carrier/receptor family, designated the cysteine protease-binding protein family (CPBF), that specifically binds to various lysosomal hydrolytic enzymes and regulates their trafficking in E. histolytica (19, 20). CPBF consists of 11 proteins (CPBF1 to -11) with significant mutual identity and structural conservation: the signal peptide at the amino terminus, a single transmembrane domain close to the carboxyl terminus, and the YxxΦ motif at the carboxyl terminus. CPBF1 was initially discovered as a receptor/carrier for one of three major cysteine proteases, EhCP-A5 (20). We further demonstrated that CPBF1 is essential for the processing and lysosomal transport of EhCP-A5. On the other hand, CPBF8 binds to β-hexosaminidase α-subunit and lysozymes and transports them to phagosomes (19). We further showed that repression of CPBF8 by gene silencing decreased degradation of the Gram-positive bacillus Clostridium perfringens and in vitro cytopathic activity against mammalian cells.
In the present study, we have characterized another member of the most highly expressed CPBF genes among the family, CPBF6. We show that CPBF6 is distributed to lysosomes in steady state and transported to phagosomes upon phagocytosis. We have further identified cargo proteins, by affinity immunoprecipitation of CPBF6, followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis, to be α-amylase and γ-amylase. We also confirmed the role of CPBF6 in phagosomal transport of α-amylase and γ-amylase by repression of CPBF6, based on Western blotting and proteome analyses. In addition, this proteome analysis shows a new potential for phagosomal proteins. Finally, we show that the carboxyl-terminal region of CPBF6 plays a crucial role in phagosomal localization.
MATERIALS AND METHODS
Microorganisms and cultivation.
Trophozoites of E. histolytica HM-1:IMSS Cl-6 and G3 strains (21, 22) were cultured axenically at 35°C in 13- by 100-mm screw-cap Pyrex glass tubes or 25-cm2 plastic culture flasks in BI-S-33 medium as previously described (23, 24). Chinese hamster ovary (CHO) cells were grown in F-12 medium (Invitrogen, CA) supplemented with 10% fetal bovine serum on a 10-cm-diameter tissue culture dish (Iwaki, Tokyo, Japan) under 5% CO2 at 37°C.
Plasmid construction and production of E. histolytica transformants.
The protein-coding region of CPBF6 was amplified by PCR from cDNA using the sense and antisense oligonucleotides 5′-GCGAGATCTATGTCAATGAGTTGTCTGACTT-3′ and 5′-CAGCGAGATCTAGAAAGGTCATGATAACCA-3′ (BglII restriction sites are underlined). The amplified PCR product was digested with BglII and ligated into BglII-digested pEhExHA (25), to produce pEhEx-CPBF6-HA. The plasmid to produce the mutant form of CPBF6 that lacks a 43-amino-acid (aa)-long serine-rich region (SRR, aa 815 to 857; SSDSSSSSSPSASSTQPSTSSSAPNPSGESSNNTEDNNGTKIG; pEhEx-CPBF6ΔSRR-HA) was constructed as follows. A DNA fragment was amplified by PCR using pEhEx-CPBF6-HA as a template and the sense and antisense oligonucleotides 5′-TGGATTATTTTTGGTGTTGTTCTCTTCTTG-3′ and 5′-TGGTATTTTTTCAAGTTTCATCAATATTTC-3′. The PCR product was treated with the BKL kit (TaKaRa, Shiga, Japan) and self-ligated to produce pEhEx-CPBF6ΔSRR-HA. The resulting plasmid carries a mutant CPBF6 protein that lacks the SRR. pEhEx-CPBF6Δcytosol-HA, which lacks a 15-aa-long cytosolic carboxyl-terminal region after the transmembrane region, and CPBF6ΔNNGYHDLS-HA, which lacks an 8-aa-long cytosolic carboxyl-terminal region, were constructed as follows. The protein-coding region of CPBF6Δcytosol and CPBF6ΔNNGYHDLS were amplified by PCR by using the oligonucleotides 5′-GCGAGATCTATGTCAATGAGTTGTCTGACT-3′ and 5′-GCGAGATCTTATGAAACCAATAATAAAGGC-3′ (for CPBF6Δcytosol) and 5′-GGAAGATCTATGTCAATGAGTTGTCTGACT-3′ and 5′-GGAAGATCTCTTTTTACGATATATTATGAA-3′ (for CPBF6ΔNNGYHDLS) (BglII restriction sites are underlined). respectively. The amplified PCR products were digested with BglII and ligated into BglII-digested pEhExHA to produce pEhEx-CPBF6Δcytosol-HA and CPBF6ΔNNGYHDLS-HA, respectively. The transformants that expressed full-length or truncated forms of CPBF6-HA were established by transfection of the wild-type HM1:IMSS Cl6 strain with pEhEx-CPBF6-HA, pEhEx-CPBF6ΔSRR-HA, pEhEx-CPBF6Δcytosol-HA, or CPBF6ΔNNGYHDLS-HA by liposome-mediated transfection, as previously described (26). For gene silencing of CPBF6, the 420-bp-long 5′ end of the CPBF6 protein-coding region was amplified by PCR from cDNA using the oligonucleotides 5′-CGCAGGCCTATGTCAATGAGTTGTCTGACT-3′ and 5′-GCAGAGCTCATAAGCACCAGGGGCAGAATA-3′ (StuI and SacI restriction sites are underlined). The PCR-amplified DNA fragment was digested with StuI and SacI and ligated into StuI- and SacI-digested pSAP2-Gunma, to produce pSAP2-CPBF6. The CPBF6 gene-silenced strain was established by transfection of the G3 strain with pSAP2-CPBF6 as described above. We also established a control strain by transfection of the G3 strain with pSAP2-Gunma.
Antibodies.
Anti-α-amylase and anti-γ-amylase antibodies were raised against a mixture of the peptides DIPLEEFDRLKSKG (aa 45 to 58) and GAHFVENHDENRAV (aa 319 to 332) for α-amylase and RIKKHLNKIRSDLT (aa 547 to 560) and LADEIDGHKSLTAN (aa 607 to 620) for γ-amylase. Pyridine nucleotide transhydrogenase (EhPNT) antibody was previously described (27). Anti-hemagglutinin (HA) 11MO mouse monoclonal antibody was purchased from Berkeley Antibody (Berkeley, CA). Alexa Fluor anti-mouse and anti-rabbit IgG and horseradish peroxidase-conjugated goat anti-mouse and anti-rabbit IgG were purchased from Invitrogen.
Immunofluorescene assay.
For the staining of lysosomes, amoebae were incubated in the BI-S-33 medium containing LysoTracker Red DND-99 (Invitrogen; diluted 1:500) at 35°C. CHO cells grown in F-12 medium supplemented with 10% fetal bovine serum were incubated with 10 μM CellTracker Blue (Invitrogen) at 37°C for 1 h. After washing with BI-S-33 medium three times, CellTracker-labeled CHO cells were added to the 8-mm-diameter wells of an 8-well glass slide (Thermo Scientific, Rockford, IL) containing 5 × 104 E. histolytica trophozoites, and the mixture was further incubated at 35°C in the BI-S-33 medium. After incubation, cells were fixed with 3.7% paraformaldehyde–phosphate-buffered saline (PBS) for 10 min and permeabilized with 0.2% saponin–PBS for 10 min at ambient temperature. The cells were then allowed to react with anti-HA 11MO mouse monoclonal antibody (diluted 1:1,000) and Alexa Fluor-488 anti-mouse secondary antibody (1:1,000). The samples were examined on a Carl-Zeiss (Thornwood, NY) LSM510 confocal laser-scanning microscope. Images were further analyzed using LSM510 software. We defined CPBF6-positive vacuoles based on criteria described elsewhere (19).
Immunoprecipitation.
Approximately 3 × 106 trophozoites of strains expressing CPBF6-HA, CPBF6ΔSRR, CPBF6Δcytosol, or CPBF6ΔNNGYHDLS were lysed in 2 ml of lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100 [Tokyo Kasei, Tokyo, Japan], 0.5 mg/ml E-64 [Sigma-Aldrich, St. Louis, MO], and Complete mini protease inhibitor cocktail [Roche, Basel, Switzerland]). Approximately 10 mg of the lysate was incubated with protein G-Sepharose beads (50 μl of an 80% slurry; Amersham Biosciences, Uppsala, Sweden) in 2 ml of lysis buffer at 4°C for 90 min, and centrifuged at 800 × g at 4°C for 3 min to remove proteins that were nonspecifically bound to the protein G-Sepharose beads. The precleared lysate was mixed with 90 μl of anti-HA antibody conjugated to agarose (50% slurry; Sigma-Aldrich) and incubated at 4°C for 3.5 h. The agarose beads were collected by centrifugation at 800 × g at 4°C for 3 min and washed four times with wash buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100). The agarose beads were then incubated with 180 μl of HA peptide (20 μg/ml) at 4°C overnight to dissociate proteins from the beads. The eluate was applied to SDS-PAGE gels, and silver stained, immunoblotting, activity assays, and protein sequencing were performed.
Protein digestion, LC-MS/MS), and database search for protein identification.
For identification of phagosomal proteins, we carried out two different digestion methods for LC-MS/MS analysis. Briefly, for the solution method, a 50-μg equivalent of the protein solution was extracted with 300 μl methanol–75 μl chloroform–225 μl water. After the upper aqueous layer was discarded, 325 μl of methanol was added to the organic phase. The sample was vitiated and centrifuged for 5 min. The pellet was air dried and resuspended in ammonium bicarbonate, reduced, alkylated, and digested with 1 μg trypsin (Promega, Madison, WI) overnight at room temperature. The sample was acidified to 5% with acetic acid and then subjected to LC-MS/MS. For the in-gel digestion method, a 10-μg equivalent of the solution was subjected to SDS-PAGE until the dye front migrated ∼1 cm. An approximately 1-cm2 piece was excised from the gel, transferred to a siliconized tube, and destained in 200 μl of 50% methanol overnight. The gel pieces were dehydrated in acetonitrile, rehydrated in 30 μl of 10 mM dithiothreitol in 0.1 M ammonium bicarbonate, and reduced with dithiothreitol at room temperature for 0.5 h. The dithiothreitol solution was removed and the sample alkylated in 30 μl 50 mM iodoacetamide in 0.1 M ammonium bicarbonate at room temperature for 0.5 h. The reagent was removed, and the gel pieces were dehydrated in 100 μl acetonitrile. The acetonitrile was removed, and the gel pieces were rehydrated in 100 μl 0.1 M ammonium bicarbonate. The pieces were dehydrated in 100 μl acetonitrile, the acetonitrile was removed, and the pieces were completely dried by vacuum centrifugation. The gel pieces were rehydrated in 20 ng/μl trypsin in 50 mM ammonium bicarbonate on ice for 10 min. Any excess enzyme solution was removed, and 20 μl of 50 mM ammonium bicarbonate was added. The sample was digested overnight at 37°C, and the peptides that formed were extracted from the polyacrylamide in two 30-μl aliquots of 50% acetonitrile–5% formic acid. These extracts were combined and evaporated to 15 μl for MS analysis.
The LC-MS/MS system consisted of a Thermo Electron Orbitrap Velos ETD mass spectrometer system with a Protana nanospray ion source interfaced with a self-packed 8-cm by 75-μm (inner diameter) Phenomenex Jupiter 10-μm C18 reversed-phase capillary column. Approximately 2-μg equivalents of each sample were injected, and the peptides were eluted from the column with an acetonitrile–0.1 M acetic acid gradient at a flow rate of 0.5 μl/min over 2 h. The nanospray ion source was operated at 2.5 kV. The digest was analyzed using the double-play capability of the instrument, acquiring full scan mass spectra to determine peptide molecular weights and product ion spectra to identify amino acid sequences in sequential scans. This mode of analysis produces approximately 30,000 CAD spectra of ions that range in abundance over several orders of magnitude.
The data were analyzed by database searches using the Sequest search algorithm against the E. histolytica genome database at the J. Craig Venter Institute (http://www.jcvi.org/). The identified proteins were considered phagosomal when two unique peptides were detected.
Transcriptomic analysis.
Expression analysis was performed by using a custom-made E. histolytica array from Affymetrix, Inc. (Santa Clara, CA), as previously described (28–30). Labeled cRNA for hybridization was prepared from 5 μg of total RNA, according to the protocol of the manufacturer. Hybridization and scanning were also performed according to the protocols of the manufacturer. Raw probe intensities were generated by using the GeneChip operating software (GCOS) and a GeneTitan instrument. The averages of triplicate raw probe intensities are reported.
Phagosome purification.
Phagosome purification was performed as previously described (19, 31, 32). Briefly, amoebae were incubated with carboxylated latex beads for 1 h, and after brief centrifugation, the cell pellet was resuspended and mechanically homogenized by using a Dounce homogenizer. The phagosomes were isolated by ultracentrifugation on a sucrose gradient.
Enzymatic assay.
The amylase assay was performed by using the EnzChek amylase assay kit (Invitrogen). Briefly, the amoeba lysate, culture supernatant, or phagosomal fraction was mixed with 200 μg/ml substrate solution containing BODIPY FL-conjugated DQ starch in 100 μl of 0.1 M morpholinepropanesulfonic acid (MOPS; pH 6.9). The fluorescence was measured at excitation and emission wavelengths of 485 and 530 nm, respectively, at 25°C for 60 min.
RESULTS
CPBF6 binds to α-amylase and γ-amylase.
In order to identify cargo proteins that bind to CPBF6, we immunoprecipitated CPBF6 from the lysates of the transformant in which HA-tagged CPBF6 was ectopically expressed, by using an anti-HA antibody (Fig. 1A). SDS-PAGE analysis followed by silver staining revealed two major bands of 75 and 50 kDa (bands B and C) that were exclusively found in the immunoprecipitated sample from the CPBF6-HA strain, but not from CPBF8-HA or the HA control strain. These bands, together with a smeary region of about >120 kDa (band A), were excised and subjected to LC-MS/MS analysis (Table 1; see also Table S1 in the supplemental material). Bands B and C were identified as γ-amylase (XM_647289; EHI_044370) and α-amylase (XM_650544, EHI_023360), with 7.6% and 11.4% coverage, respectively. α-Amylase was previously demonstrated in phagosomes in our previous proteome analysis (31, 32). γ-Amylase was also confirmed to be present in phagosomes (see below). Band A was identified as CPBF6, and its apparent molecular mass based on SDS-PAGE (>120 kDa) was larger than the predicted size (99.3 kDa), suggesting that CPBF6 is posttranslationally modified, like CPBF8 (19). Western blotting with anti-α-amylase and anti-γ-amylase antibodies indicated that these amylases specifically bind to CPBF6 but not CPBF8 or HA (Fig. 1B).
Fig 1.
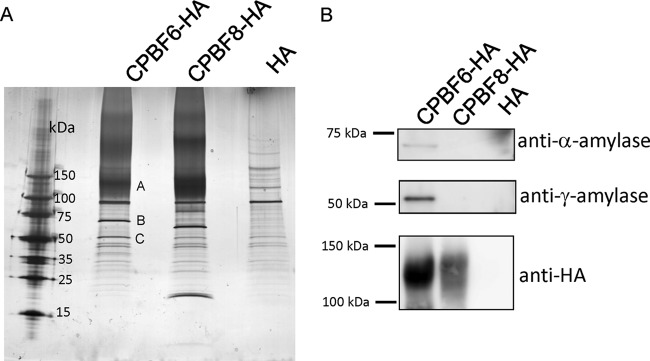
Isolation and identification of CPBF6-binding proteins. Lysates of CPBF6-HA, CPBF8-HA, and control (HA) transformants were mixed with anti-HA-antibody-conjugated agarose, washed, and eluted with HA peptide. The eluates were separated on SDS-PAGE and silver stained (A) or subjected to Western blotting with anti-α-amylase, anti-γ-amylase, and anti-HA antibody (B). Apparent molecular masses of standards are indicated on the left. Three bands excised for protein identification are labeled (A to C).
Table 1.
CPBF6-binding proteins identified by LC-MS/MS analysis
| Excised band, mass (kDa) | AmoebaDB gene ID (GenBank accession no.) | Coverage (%)a | No. of detected peptides | Annotation | Predicted mass (kDa) |
|---|---|---|---|---|---|
| A, 100–150 | EHI_178470 (XP_653036) | 30.9 | 10 | CPBF6 | 97.8 |
| B, 75 | EHI_044370 (XP_652381) | 11.5 | 4 | γ-Amylase | 77.6 |
| C, 50 | EHI_023360 (XP_655636) | 7.6 | 3 | α-Amylase | 56.7 |
Coverage was determined based on the peptides with over 95% probability.
CPBF6 is localized to phagosomes and lysosomes.
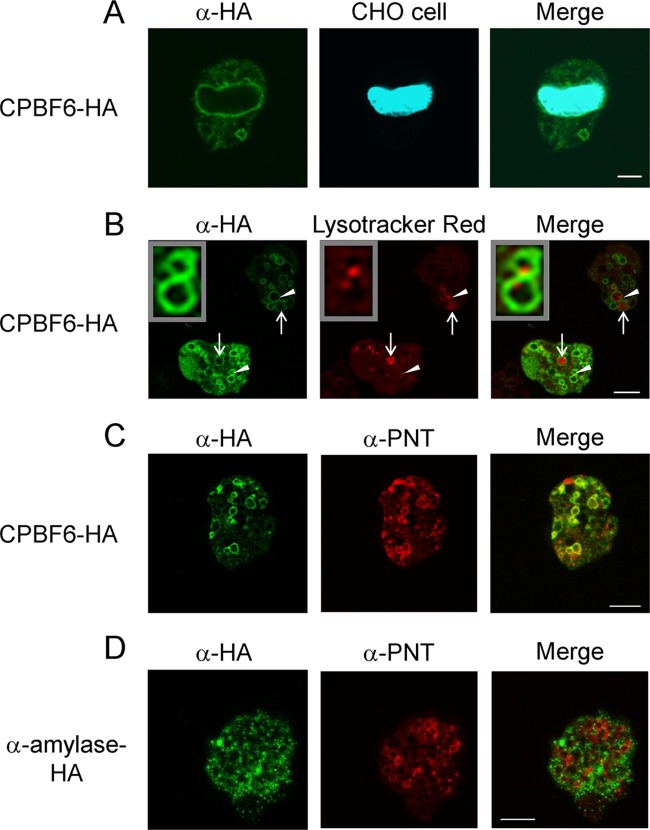
As demonstrated above, CPBF6 bound to α- and γ-amylases, the former of which and CPBF6 were previously demonstrated in our phagosome proteome analysis (32). We examined localization of CPBF6 during phagocytosis of CHO cells. Trophozoites of the CPBF6-HA-expressing strain were incubated with CellTracker-loaded CHO cells for 60 min to allow ingestion of CHO cells. An immunofluorescence assay using anti-HA antibody showed that CPBF6 localized to phagosomes containing CHO cells (Fig. 2A). We estimated that approximately 81% of CHO-containing phagosomes had colocalized with CPBF6 (n = 20). As CPBF6 was also distributed to a large number of vesicles and vacuoles under quiescent (i.e., nonphagocytic) conditions, we next examined the nature of these compartments. CPBF6-HA associated with both small (360- to 720-nm diameter) and large (1.75- to 2.5-μm diameter) acidic organelles labeled with the membrane-diffusible LysoTracker dye under steady-state conditions (83% of LysoTracker positive vesicles/vacuoles were positive for CPBF6 [n = 20]) (Fig. 2B). The large CPBF6-associated vacuoles appeared to be multivesicular bodies (33) that contained vesicles, including LysoTracker-positive vesicles (Fig. 2B, arrowheads and inset). CPBF6 also nicely colocalized with pyridine nucleotide transhydrogenase (EhPNT) (27), which converts NADH to NADPH by direct transfer of a hydride ion via the proton gradient (Fig. 2C). Since antibodies against α- and γ-amylases were not usable in the immunofluorescence assay (data not shown), we attempted to examine localization of α- and γ-amylases by using E. histolytica lines that expressed carboxyl-terminal HA-tagged α- and γ-amylases. The fluorescent signal of HA-tagged α-amylase rarely overlapped that of PNT (Fig. 2D), which coincided with CPBF6 (Fig. 2C). This indicated that CPBF6 and α-amylase likely interact only transiently. Since HA-tagged γ-amylase wasn't detected in an E. histolytica transformant, as verified by immunoblot analysis, the localization of γ-amylase remains unknown.
Fig 2.
Localization of CPBF6 in E. histolytica. (A) Phagosome localization of CPBF6. Amoebae of the CPBF6-HA transformant were incubated with CellTracker Blue-stained CHO cells (blue) for 60 min, fixed, and reacted with anti-HA antibody and Alexa Fluor-488-conjugated anti-mouse IgG secondary antibody (green). Bar, 5 μm. (B) Lysosome localization of CPBF6. Amoebae of the CPBF6-HA transformant were labeled with LysoTracker Red (red) and subjected to an immunofluorescence assay with anti-HA antibody and Alexa Fluor-488-conjugated anti-mouse IgG secondary antibody (green). Bar, 10 μm. Arrows indicate representative large acidic vacuoles. Arrowheads indicate representative a small acidic vesicles of multivesicular bodies (MVB). The inset shows an enlarged image of the MVB. (C) Colocalization of CPBF6 and EhPNT. The cells of the CPBF6-HA transformant were fixed and reacted with anti-EhPNT antibody, Alexa Fluor-568-conjugated anti-rabbit IgG secondary antibody (red), anti-HA antibody, and Alexa Fluor-488-conjugated anti-mouse IgG secondary antibody (green). Bar, 10 μm. (D) Localization of α-amylase and EhPNT. The cells of the α-amylase-HA transformant were fixed and reacted with anti-EhPNT antibod and Alexa Fluor-568-conjugated anti-rabbit IgG secondary antibody (red) or anti-HA antibody and Alexa Fluor-488-conjugated anti-mouse IgG secondary antibody (green). Bar, 10 μm.
Repression of CPBF6 by gene silencing hampers trafficking of α-amylase and γ-amylase to phagosomes.
To demonstrate the role of CPBF6, we created a CPBF6 gene-silenced strain (CPBF6gs) with the genetic background of the G3 strain and in which amoebapore genes had already been repressed (21, 22). DNA microarray analysis verified that CPBF6 transcript was reduced by 2,419-fold while expression of other CPBF genes remained unchanged (Table 2).
Table 2.
Specific repression of CPBF6 by gene silencinga
| Gene | CPBF6gs strain | Control strain | Fold change |
|---|---|---|---|
| CPBF1 | 3,110 (±1,010) | 2,780 (±268) | 1.12 |
| CPBF2 | 135 (±15.4) | 141 (±16.7) | 0.95 |
| CPBF3 | 638 (±45.0) | 482 (±18.4) | 1.32 |
| CPBF4 | 1,120 (±347) | 1,040 (±269) | 1.08 |
| CPBF5 | 127 (±44.0) | 136 (±38.9) | 0.93 |
| CPBF6 | 1.58 (±0.75) | 3,820 (±87.8) | 4.13 × 10−4 |
| CPBF7 | 1,730 (±282) | 1,540 (±190) | 1.13 |
| CPBF8 | 5,620 (±556) | 5,630 (±454) | 1.00 |
| CPBF9 | 311 (±24.9) | 248 (±30.7) | 1.26 |
| CPBF10 | 123 (±90.2) | 94.5 (±26.8) | 1.30 |
| CPBF11 | 96.3 (±14.6) | 77.0 (±10.7) | 1.25 |
Total RNA was extracted from the CPBF6gs and control strains, and expression analysis was performed using a custom-made E. histolytica DNA microarray. Average (± standard deviation) normalized signal intensities from DNA microarrays were determined in triplicate and indicate relative levels of the transcripts, in arbitrary units, of CPBF genes (CPBF1 to CPBF11) in the CPBF6gs and control strains. The fold change indicates the relative change in signal intensity in the CPBF6gs strain relative to that in the control strain.
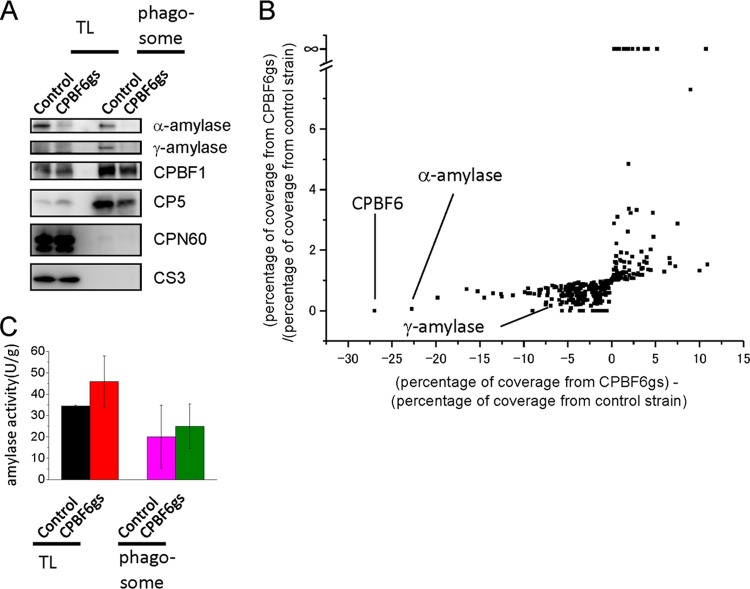
We examined the protein amounts of α-amylase and γ-amylase in the whole-cell lysates and purified phagosomes from CPBF6gs and control strains by immunoblot analysis (Fig. 3A). The amounts of α-amylase in both the whole-cell lysate and, particularly, phagosomes from CPBF6gs were greatly reduced compared to the control strain. The protein amount of γ-amylase in phagosomes was also largely reduced, whereas γ-amylase in the whole-cell lysates could not be detected, due to either the scarcity of γ-amylase or the low sensitivity of the anti-γ-amylase antibody. Despite our efforts, including truncation at both the amino and carboxyl termini of CPBF6, we were unable to produce either full-length or truncated forms of CPBF6 in order to raise anti-CPBF6 antibody (data not shown). We also carried out the proteome analysis of isolated phagosomes from CPBF6gs and control strains. The percent coverage levels of the peptides corresponding to α-amylase and γ-amylase detected from phagosomes derived from the CPBF6gs strain were significantly lower than that detected from phagosomes of the control strain (Fig. 3B; see also Table S2 in the supplemental material). In contrast, the amount of CP5 (CP-A5), which was shown to be transported via CPBF1, in the whole-cell lysate and phagosomes remained unaffected; neither chaperonin 60 (Cpn60), the mitosomal control, nor cysteine synthase 3 (CS3), the cytoplasmic control, was detected in the phagosomes (Fig. 3A). Taken together, the repression of CPBF6 hampered, if not abolished, trafficking of α-amylase and γ-amylase to phagosomes. The amount of other proteins, namely, CP2 (EAL45256), hypothetical proteins EAL48580, EAL49001, and EAL46996, and the WD domain containing protein (EAL45537), detected in CPBF6gs strain, were also significantly lower than in control strain. Since these proteins were not detected in silver-stained SDS-PAGE gels of the CPBF-coimmunoprecipitated sample, repression of CPBF6 may have indirectly caused mistargeting of these proteins. We next measured amylase activities in the whole-cell lysates and the phagosome fractions from CPBF6gs and control strains (Fig. 3C). Unexpectedly, amylase activities toward the starch in both the whole-cell lysate and the isolated phagosomes were comparable between CPBF6gs and control strains.
Fig 3.
Phenotypic changes of CPBF6 gene silencing. (A) Immunoblot analysis of the whole-cell lysate and the phagosome fraction. Approximately 20 μg of the whole-cell lysate and 2 μg of the phagosome fraction were electrophoresed by SDS-PAGE and subjected to immunoblot analysis using anti-α-amylase, anti-γ-amylase, CPBF1, CP5, Cpn60, and CS3 antibodies. CPBF1 and CP5 are phagosomal proteins, while CPN60 is a mitosomal marker and CS3 is a cytosolic marker. Faint smeary bands in the whole-cell lysate of the control and CPBF6gs strains, detected with anti-γ-amylase antibody, were considered background because of their low intensities compared to those in the phagosome fraction. (B) Proteomic analysis of isolated phagosomes from CPBF6gs and control strains. Each dot represents a protein identified from phagosomes. The x axis indicates the percentage of coverage from CPBF6gs minus the percentage of coverage from the control strain), and it is theoretically close to 0 when the repression of CPBF6 does not affect the trafficking of a protein. Similarly, y axis indicates the percentage of coverage from CPBF6gs divided by the percentage of coverage from the control strain, and it is close to 1 when the repression of CPBF6 does not affect the trafficking of a protein. Individual data are shown in Table S2 in the supplemental material. (C) Enzymatic activities of amylase in whole-cell lysates and phagosomes from CPBF6gs and control strains. The amoeba lysate, culture supernatant, or phagosomal fraction was mixed with 200 μg/ml of substrate solution containing BODIPY FL-conjugated DQ starch in 100 μl of 0.1 M MOPS (pH 6.9). The fluorescence was measured at excitation and emission wavelengths of 485 and 530 nm, respectively, at 25°C for 60 min. Data shown are the means ± standard deviations of three independent experiments.
We compared our newly created phagosome proteome from the G3 strain with our previous findings with the HM-1 strain (32). In the current study, we detected 345 proteins from phagosomes that were isolated after 60 min of coincubation of the trophozoites with beads (see Table S2). We compared phagosomal proteins detected in the present and previous studies at the same time point (60 min). Sixty-nine proteins were commonly detected in both studies, while 276 or 31 proteins were identified only in the present or previous study, respectively (at 60 min in reference 32) (see Tables S4 and S3 in the supplemental material). We categorized 67 common phagosomal proteins, based on protein functions (see Table S5). All data for phagosome proteomics can be found in Tables S2 to S5 of the supplemental material.
The serine-rich region of CPBF6 is important for cargo binding and phagosomal transport.
CPBF6 contains, like CPBF8 (19) and CPBF7, an SRR located in the middle of the luminal portion of proteins. We previously demonstrated that the SRR of CPBF8 is involved in binding with the β-hexosaminidase α-subunit and lysozymes (19). We investigated whether the SRR of CPBF6 also plays a role in binding with its cargos. We created a transformant that expressed an HA-tagged mutant form of CPBF6 lacking the 43-aa-long SRR (CPBF6ΔSRR-HA). We immunoprecipitated CPBF6-HA and CPBF6ΔSRR-HA by using anti-HA antibody from lysates of the corresponding strains. The precipitated proteins were subjected to immunoblot analysis using anti-α-amylase and anti-γ-amylase antibodies (Fig. 4A). The amount of immunoprecipitated α-amylase and γ-amylase from the CPBF6ΔSRR-HA lysate was lower than that from the CPBF6-HA lysate. In addition, the immunofluorescence assay showed that CPBF6ΔSRR-HA was not localized to phagosomes (Fig. 4B). Only approximately 26% of phagosomes colocalized with CPBF6ΔSRR-HA, compared to >80% with full-length CPBF6-HA (n = 20). These data are consistent with the notion that the SRR in CPBF6 is important for both the binding to α-amylase and γ-amylase and for their phagosomal transport. Immunoblot analysis showed that CPBF6-HA was detected as a broad smeary band of 110 to 150 kDa (Fig. 1B and 4A), whereas CPBF6ΔSRR-HA was detected as an apparently single 100-kDa band. These data are consistent with the premise that the SRR contains posttranslational modifications.
Fig 4.
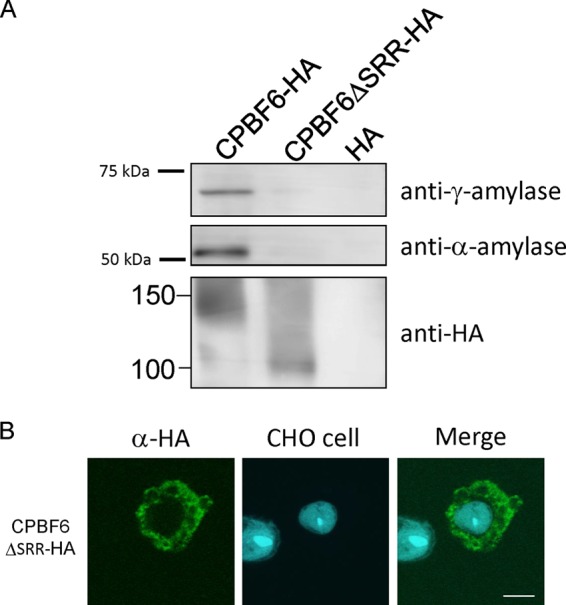
The SRR of CPBF6 is structurally important. (A) CPBF6-HA and CPBF6ΔSRR-HA were immunoprecipitated from corresponding expressing strains by using anti-HA-antibody-conjugated agarose, eluted with HA peptides, and subjected to SDS-PAGE and immunoblot analyses with anti-HA, anti-α-amylase, and anti-γ-amylase antibodies and appropriate secondary antibodies. (B) Localization of CPBF6ΔSRR-HA during phagocytosis. Amoebae were incubated with CellTracker Blue-stained CHO cells (blue) for 60 min, fixed, and reacted with anti-HA antibody and Alexa Fluor-488-conjugated anti-mouse IgG secondary antibody (green). Bar, 10 μm.
The cytosolic domain is involved in the phagosomal trafficking of CPBF6.
We speculated that the cytosolic domain containing the YxxΦ motif of CPBF proteins may also be involved in binding with accessory molecules and, therefore, plays an important role in the determination of localization. However, our previously study showed that the localization of CPBF1 was not altered by mutation of the YxxΦ motif, suggesting that the motif does not primarily determine its localization of CPBF1 (20). We asked if the cytoplasmic region containing the YxxΦ motif is involved in the phagosomal trafficking of CPBF6. We created two deletion mutants of CPBF6, CPBF6Δcytosol and CPBF6ΔNNGYHDLS, which lacked the entire 15-aa-long cytosolic portion after the transmembrane domain or the 8-aa carboxyl-terminal portion containing the YxxΦ motif. We examined the localization of these proteins in an immunofluorescence assay (Fig. 5A and B). CPBF6Δcytosol showed localization dissimilar from that of CPBF6. Only 4% (n = 20) of phagosomes were positive with the anti-HA antibody in the CPBF6Δcytosol-HA transformant (Fig. 5C). In contrast, the percentage of phagosomes that localized with CPBF6ΔNNGYHDLS-HA was 69% (n = 20), comparable to that of wild-type CPBF6-HA (81%; n = 20). These results suggest that the cytosolic region, but not the YxxL motif itself, determines the phagosomal localization of CPBF6.
Fig 5.
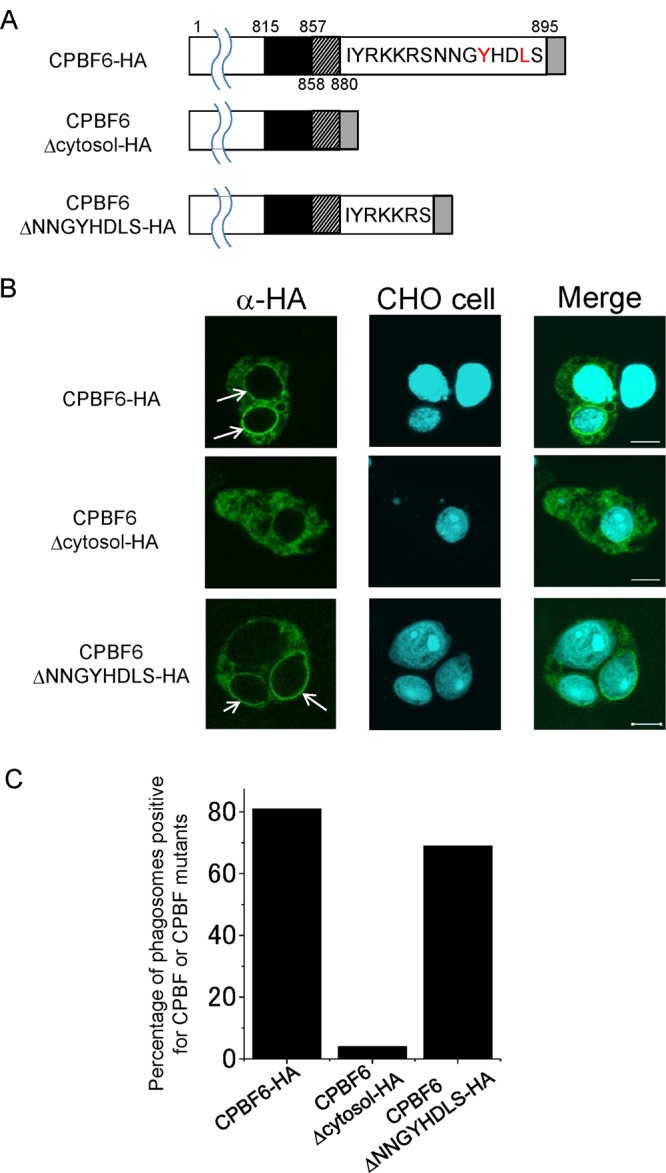
The carboxyl terminus of CPBF6 is required for the localization of CPBF6 to the phagosome. (A) Schematic representation of CPBF6-HA, CPBF6Δcytosol-HA, and CPBF6ΔNNGYHDLS-HA. Black boxes indicate the SRR. Gray boxes indicate the HA tag. Hatched boxes indicate the transmembrane region. Amino acid residues in the cytosolic region of the wild type and two CPBF6 mutants are shown, with tyrosine and leucine indicated in red. Amino acid positions are also depicted. (B) Localization of CPBF6-HA, CPBF6Δcytosol-HA, and CPBF6ΔNNGYHDLS-HA during phagocytosis. Amoebae were incubated with CellTracker Blue-stained CHO cells (blue) for 60 min, fixed, and reacted with anti-HA antibody and Alexa Fluor-488-conjugated anti-mouse IgG secondary antibody (green). Bar, 10 μm. Arrows indicate CPBF6-positive phagosomes. (C) Quantitative analysis of phagosomal localization of CPBF6-HA, CPBF6Δcytosol-HA, and CPBF6ΔNNGYHDLS-HA to phagosomes. The percentages of phagosomes with which wild-type or truncated CPBF6 colocalized in strains expressing CPBF6-HA, CPBF6Δcytosol-HA, or CPBF6ΔNNGYHDLS-HA are shown.
DISCUSSION
Cargo specificity of CPBF6.
We demonstrated in the present study that one of the three most highly expressed CPBF proteins, CPBF6, selectively binds to putative α-amylase and γ-amylase and is essential for their trafficking to phagosomes. Together with our previous reports, we have now clearly shown that three major CPBFs, CPBF1 (20), -6 (this study), and -8 (19), have remarkable differences in their affinities toward cargo proteins: CPBF1 for CP-A1 and CP-A5, CPBF8 for β-hexosaminidase α-subunit and lysozymes, and CPBF6 for α-amylase and γ-amylase. The E. histolytica genome apparently encodes six proteins annotated as α-amylase. Among them, only one α-amylase (EHI_023360) was identified in this study, by virtue of its ability to bind to CPBF6. Furthermore, phagosome proteome analysis showed that two α-amylases (EHI_023360 and EHI_152880) are transported to phagosomes in the wild-type amoeba, but CPBF6 gene silencing did not affect the transport of these amylases (EHI_023360 and EHI_152880) to phagosomes, although it abolished transport of another α-amylase (EHI_023360). Therefore, α-amylase (EHI_023360) appears to be a major, and perhaps the sole, α-amylase transported via CPBF6. In contrast, γ-amylase is encoded by a single gene in the E. histolytica genome. These findings altogether underline the strict cargo specificity of CPBF6 in trafficking of lysosomal enzymes. As we did not directly sequence the eluate of immunoprecipitation, the whole spectrum of cargos that CPBF6 binds, including ones identified in this study, remains unknown.
Differences in structural determinants for cargo binding of CPBF6.
CPBF8 has an SRR, which is essential for binding to both β-hexosaminidase α-subunit and lysozymes (19). In this study, we found that the SRR is apparently conserved in CPBF6 and is structurally important. In contrast, CPBF1 apparently lacks the SRR, suggesting that the SRR is not required for binding of CPBF1 to CPs (20). Recently, we found that another member of the family, CPBF10, binds to two isotypes of α-amylase (EHI_023360 and EHI_153100) (K. Marumo et al., unpublished data). Since CPBF10 apparently lacks the SRR and yet binds to α-amylase (EHI_023360), CPBF6 may also bind to α-amylase (EHI_023360) via a portion other than the SRR. Phylogenetic analysis of CPBFs suggests that CPBF6 belongs to the same clade as CPBF7, -8, and -10 (20). This may partially explain the overlapping cargo specificity of CPBF6 and CPBF10. These data also imply that an ancestor of CPBF6, -7, -8, and -10 may have recognized ancestral α-amylase, which is closest to the extant EHI_023360, and CPBF6 and -10 have further evolved to gain additional binding abilities while CPBF8 has lost its binding capacity to α-amylase and gained the binding capacity for the β-hexosaminidase α-subunit and lysozymes.
Physiological functions of CPBF6 and its cargos.
We have unequivocally demonstrated the cargo specificity of CPBF6. As shown by Western blotting and proteome analysis, phagosome transport of α-amylase and γ-amylase was severely affected by CPBF6 gene silencing. The amount of α-amylase and γ-amylase in the whole-cell lysate from CPBF6gs also decreased compared to the control strain. This observation was similar to that for CPBF8gs, where the amounts of β-hexosaminidase α-subunit and lysozymes in the whole-cell lysates were greatly reduced (19). Although deletion or repression of hydrolase receptors often causes missecretion of cargos (20), gene silencing of CPBF8 did not result in missecretion of the β-hexosaminidase α-subunit and lysozymes. This can be explained by swift degradation of non- or mistargeted α-amylase and γ-amylase by proteasomes in the CPBF8gs strain. Although we have clearly demonstrated that phagosomal transport of α-amylase and γ-amylase is mediated by CPBF6, enzymatic activities toward the starch were comparable between the control strain and CPBF6gs. These data indicate that another protein(s), e.g., β-amylase, compensates for the phagosomal amylase activities in the CPBF6gs strain. However, an increase of β-amylase in phagosomes from the CPBF6gs strain, compared to that from the control strain, which was detected by proteomic analysis (see Table S2 in the supplemental material), did not prove to be statistically significant. Alternatively, the current assay method used in this study did not allow measurement of the specific activities of the putative amylases transported via CPBF6. As the percent coverage of β-amylase (EAL48510) was higher in the CPBF6gs strain than in the control strain, the increased β-amylase may have compensated for the overall amylase activity in the phagosome fraction from the CPBF6gs strain. Although the amylase activity was also measured at low pH, which is favorable for γ-amylase, it was comparable between the CPBF6gs and control strains (data not shown).
α-Amylase (EHI_023360) contains the maltase (3.31 e−5) and α-phosphotrehalase (4.81 e−4) domains, based on a pfam search. γ-Amylase (EHI_044370) has similarity with glucoamylase from Thermoactiomyces vulgaris (2 e−9), which has been shown to have high hydrolytic activity toward maltooligosaccharides (34). The substrates of α-amylase and γ-amylase should be determined in the future to allow a better understanding of the biological and physiological roles of CPBF6.
Mechanisms for phagosomal localization of CPBF6.
Our mutation study showed that the YxxL motif does not determine the phagosomal localization of CPBF6. Our previous study with CPBF1 also suggested that this motif is not required for localization of CPBF1 to the endoplasmic reticulum (ER) and phagosomes (20). Thus, we concluded that the YxxL motif is not likely involved in the recruitment of this family of receptors/carriers in E. histolytica. This is surprising, as the major components of the AP complex and clathrins are conserved in E. histolytica (35, 36), and the motif is known to bind to the μ-subunit of the complex.
We further demonstrated that deletion of the cytosolic region containing a stretch of positively charged amino acids, IYRKKRS, close to the transmembrane region abolished phagosomal localization of CPBF6. In Saccharomyces cerevisiae, it has been reported that lysines and arginines in the carboxyl-terminal region are involved in the interaction with phospholipids (37) and that the higher the number of lysine/arginine residues in the region, the higher is the affinity of the protein with phospholipids. It was also demonstrated that negatively charged phosphatidylinositides such as PI3P and PIP3, accumulate in phagosomes (13–15). Thus, it is plausible that lysines and arginines in the cytosolic region of CPBF6 are involved in the recognition of a phospholipid(s) on phagosomal membranes that determines the recruitment of CPBF6 to phagosomes. CPBF1, -7, and -8 were previously found in phagosomes and have a lysine-/arginine-rich region downstream of the transmembrane region, similar to CPBF6 (19). Altogether, we tentatively propose that the lysine-/arginine-rich cytosolic region is responsible for the recruitment of these CPBF members to phagosomes.
Differential localization of CPBF1, -6, and -8 and colocalization of CPBF6 with PNT.
CPBF6 is distributed to the membrane of lysosomes and multivesicular bodies in the quiescent state and translocated to phagosomes during phagocytosis. The localization and dynamism of CPBF6 were similar to those of CPBF8 (19). In contrast, CPBF1 showed different localization from CPBF6 and CPBF8; CPBF1 mainly localizes to the ER and occasionally to phagosomes, but seldom to lysosomes (20). Phylogenetic analysis of CPBFs from three species of Entamoeba (also discussed above) clearly indicated that CPBF6, -7, 08 to -8, and -10 form monophyly (20), suggesting evolutionary kinship among them. CPBF1 appears to have diverged from the clade consisting of CPBF3, 04 to -4, and -11 and CPBF6, -7, -8, and -10 at the earlier stage of evolution. Such an evolutionary distance between CPBF6 and -8 and CPBF1 may result in the different localization of CPBF6 and -8 and CPBF1. The colocalization of both CPBF6 (Fig. 2C) and CPBF8 (19) with EhPNT was striking. PNT is distributed to both LysoTracker-stainable acidic (late endosomes, lysosomes, and phagosomes) and nonacidic vesicles and vacuoles (27), and therefore is not necessarily a lysosomal marker, despite the fact that PNT utilizes the proton gradient across the membrane. Thus, it is tempting to speculate that CPBF6/8 and PNT are cotransported and that the enzymatic reactions of putative α-amylase, γ-amylase, β-hexosaminidase α-subunit, and lysozymes transported by CPBF6/8, require NADPH produced by PNT. The mechanisms of localization of PNT to lysosomes, endosomes, and phagosomes are still unknown. We previously demonstrated that all domains, such as the amino-terminal extension, 11–13 transmembrane, and linker regions are essential for the vesicular/vacuolar distribution of PNT (27). However, in contrast to CPBF6, no apparent lysine-/arginine-rich region is present in PNT.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Nicholas Sherman, W.M. Keck Biomedical Mass Spectrometry Laboratory at the University of Virginia, for technical support on MS analyses. We are also grateful to Kazuo Ebine and Kisaburo Nagamune for helpful discussions.
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (23117001, 23117005, and 23390099), a Grant-in-Aid on Bilateral Programs of Joint Research Projects and Seminars from the Japan Society for the Promotion of Science, a Grant-in-Aid on Strategic International Research Cooperative Program (SICP) from the Japan Science and Technology Agency, a grant for research on emerging and reemerging infectious diseases from the Ministry of Health, Labor and Welfare (MHLW) of Japan (H23-Shinkosaiko-ippan-014), a grant for research to promote the development of anti-AIDS pharmaceuticals from the Japan Health Sciences Foundation (KHA1101), and by the Global COE Program (Global COE for Human Metabolomic Systems Biology) from MEXT, Japan.
Footnotes
Published ahead of print 18 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00915-12.
REFERENCES
- 1. Bracha R, Kobiler D, Mirelman D. 1982. Attachment and ingestion of bacteria by trophozoites of Entamoeba histolytica. Infect. Immun. 36:396–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tsutsumi V, Ramírez-Rosales A, Lanz-Mendoza H, Shibayama M, Chávez B, Rangel-López E, Martínez-Palomo A. 1992. Entamoeba histolytica: erythrophagocytosis, collagenolysis, and liver abscess production as virulence markers. Trans. R. Soc. Trop. Med. Hyg. 86:170–172 [DOI] [PubMed] [Google Scholar]
- 3. Guerrant RL, Brush J, Ravdin JI, Sullivan JA, Mandell GL. 1981. Interaction between Entamoeba histolytica and human polymorphonuclear neutrophils. J. Infect. Dis. 143:83–93 [DOI] [PubMed] [Google Scholar]
- 4. Bracha R, Mirelman D. 1984. Virulence of Entamoeba histolytica trophozoites. Effects of bacteria, microaerobic conditions, and metronidazole. J. Exp. Med. 160:353–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hirata KK, Que X, Melendez-Lopez SG, Debnath A, Myers S, Herdman DS, Orozco E, Bhattacharya A, McKerrow JH, Reed SL. 2007. A phagocytosis mutant of Entamoeba histolytica is less virulent due to deficient proteinase expression and release. Exp. Parasitol. 115:192–199 [DOI] [PubMed] [Google Scholar]
- 6. Katz U, Ankri S, Stolarsky T, Nuchamowitz Y, Mirelman D. 2002. Entamoeba histolytica expressing a dominant negative N-truncated light subunit of its Gal-lectin are less virulent. Mol. Biol. Cell 13:4256–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Orozco E, Guarneros G, Martinez-Palomo A, Sánchez T. 1983. Entamoeba histolytica. Phagocytosis as a virulence factor. J. Exp. Med. 158:1511–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Que X, Brinen LS, Perkins P, Herdman S, Hirata K, Torian BE, Rubin H, McKerrow JH, Reed SL. 2002. Cysteine proteinases from distinct cellular compartments are recruited to phagocytic vesicles by Entamoeba histolytica. Mol. Biochem. Parasitol. 119:23–32 [DOI] [PubMed] [Google Scholar]
- 9. Andrä J, Herbst R, Leippe M. 2003. Amoebapores, archaic effector peptides of protozoan origin, are discharged into phagosomes and kill bacteria by permeabilizing their membranes. Dev. Comp. Immunol. 27:291–304 [DOI] [PubMed] [Google Scholar]
- 10. Mann BJ. 2002. Structure and function of the Entamoeba histolytica Gal/GalNAc lectin. Int. Rev. Cytol. 216:59–80 [DOI] [PubMed] [Google Scholar]
- 11. Petri WA, Jr, Haque R, Mann BJ. 2002. The bittersweet interface of parasite and host: lectin-carbohydrate interactions during human invasion by the parasite Entamoeba histolytica. Annu. Rev. Microbiol. 56:39–64 [DOI] [PubMed] [Google Scholar]
- 12. Voigt H, Olivo JC, Sansonetti P, Guillén N. 1999. Myosin IB from Entamoeba histolytica is involved in phagocytosis of human erythrocytes. J. Cell Sci. 112:1191–1201 [DOI] [PubMed] [Google Scholar]
- 13. Byekova YA, Powell RR, Welter BH, Temesvari LA. 2010. Localization of phosphatidylinositol (3,4,5)-trisphosphate to phagosomes in Entamoeba histolytica achieved using glutathione S-transferase- and green fluorescent protein-tagged lipid biosensors. Infect. Immun. 78:125–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ghosh SK, Samuelson J. 1997. Involvement of p21racA, phosphoinositide 3-kinase, and vacuolar ATPase in phagocytosis of bacteria and erythrocytes by Entamoeba histolytica: suggestive evidence for coincidental evolution of amebic invasiveness. Infect. Immun. 65:4243–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakada-Tsukui K, Okada H, Mitra BN, Nozaki T. 2009. Phosphatidylinositol-phosphates mediate cytoskeletal reorganization during phagocytosis via a unique modular protein consisting of RhoGEF/DH and FYVE domains in the parasitic protozoon Entamoeba histolytica. Cell. Microbiol. 11:1471–1491 [DOI] [PubMed] [Google Scholar]
- 16. Somlata Bhattacharya S, Bhattacharya A. 2011. A C2 domain protein kinase initiates phagocytosis in the protozoan parasite Entamoeba histolytica. Nat. Commun. 2:230. [DOI] [PubMed] [Google Scholar]
- 17. Boettner DR, Huston CD, Linford AS, Buss SN, Houpt E, Sherman NE, Petri WA., Jr 2008. Entamoeba histolytica phagocytosis of human erythrocytes involves PATMK, a member of the transmembrane kinase family. PLoS Pathog. 4:e8 doi:10.1371/journal.ppat.0040008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Labruyère E, Zimmer C, Galy V, Olivo-Marin Guillén J-CN. 2003. EhPAK, a member of the p21-activated kinase family, is involved in the control of Entamoeba histolytica migration and phagocytosis. J. Cell Sci. 116:61–71 [DOI] [PubMed] [Google Scholar]
- 19. Furukawa A, Nakada-Tsukui K, Nozaki T. 2012. Novel transmembrane receptor involved in phagosome transport of lysozymes and β-hexosaminidase in the enteric protozoan Entamoeba histolytica. PLoS Pathog. 8:e1002539 doi:10.1371/journal.ppat.1002539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nakada-Tsukui K, Tsuboi Furukawa K, Yamada A, Nozaki Y, T 2012. A novel class of cysteine protease receptors that mediate lysosomal transport. Cell. Microbiol. 14:1299–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bracha R, Nuchamowitz Y, Mirelman D. 2003. Transcriptional silencing of an amoebapore gene in Entamoeba histolytica: molecular analysis and effect on pathogenicity. Eukaryot. Cell 2:295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang H, Alramini H, Tran V, Singh U. 2011. Nucleus-localized antisense small RNAs with 5′-polyphosphate termini regulate long term transcriptional gene silencing in Entamoeba histolytica G3 strain. J. Biol. Chem. 286:44467–44479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Diamond LS, Mattern CF, Bartgis IL. 1972. Viruses of Entamoeba histolytica. I. Identification of transmissible virus-like agents. J. Virol. 9:326–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Diamond LS, Harlow DR, Cunnick CC. 1978. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72:431–432 [DOI] [PubMed] [Google Scholar]
- 25. Saito-Nakano Y, Mitra BN, Nakada-Tsukui K, Sato D, Nozaki T. 2007. Two Rab7 isotypes, EhRab7A and EhRab7B, play distinct roles in biogenesis of lysosomes and phagosomes in the enteric protozoan parasite Entamoeba histolytica. Cell. Microbiol. 9:1796–1808 [DOI] [PubMed] [Google Scholar]
- 26. Nozaki T, Asai T, Sanchez LB, Kobayashi S, Nakazawa M, Takeuchi T. 1999. Characterization of the gene encoding serine acetyltransferase, a regulated enzyme of cysteine biosynthesis from the protist parasites Entamoeba histolytica and Entamoeba dispar. Regulation and possible function of the cysteine biosynthetic pathway in Entamoeba. J. Biol. Chem. 274:32445–32452 [DOI] [PubMed] [Google Scholar]
- 27. Yousuf MA, Mi-Ichi F, Nakada-Tsukui K, Nozaki T. 2010. Localization and Targeting of an unusual pyridine nucleotide transhydrogenase in Entamoeba histolytica. Eukaryot. Cell 9:926–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gilchrist CA, Houpt E, Trapaidze N, Fei Z, Crasta O, Asgharpour A, Evans C, Martino-Catt S, Baba DJ, Stroup S, Hamano S, Ehrenkaufer G, Okada M, Singh U, Nozaki T, Mann BJ, Petri WA., Jr 2006. Impact of intestinal colonization and invasion on the Entamoeba histolytica transcriptome. Mol. Biochem. Parasitol. 147:163–176 [DOI] [PubMed] [Google Scholar]
- 29. Husain A, Jeelani G, Sato D, Nozaki T. 2011. Global analysis of gene expression in response to L-cysteine deprivation in the anaerobic protozoan parasite Entamoeba histolytica. BMC Genomics 12:275 doi:10.1186/1471-2164-12-275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Penuliar GM, Furukawa A, Nakada-Tsukui K, Husain A, Sato D, Nozaki T. 2012. Transcriptional and functional analysis of trifluoromethionine resistance in Entamoeba histolytica. J. Antimicrob. Chemother. 67:375–386 [DOI] [PubMed] [Google Scholar]
- 31. Okada M, Huston CD, Mann BJ, Petri WA, Kita K, Nozaki T. 2005. Proteomic analysis of phagocytosis in the enteric protozoan parasite Entamoeba histolytica. Eukaryot. Cell 4:827–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Okada M, Huston CD, Oue M, Mann BJ, Petri WA, Jr, Kita K, Nozaki T. 2006. Kinetics and strain variation of phagosome proteins of Entamoeba histolytica by proteomic analysis. Mol. Biochem. Parasitol. 145:171–183 [DOI] [PubMed] [Google Scholar]
- 33. Saito-Nakano Y, Yasuda T, Nakada-Tsukui K, Leippe M, Nozaki T. 2004. Rab5-associated vacuoles play a unique role in phagocytosis of the enteric protozoan parasite Entamoeba histolytica. J. Biol. Chem. 279:49497–49507 [DOI] [PubMed] [Google Scholar]
- 34. Uotsu-Tomita R, Tonozuka T, Sakai H, Sakano Y. 2001. Novel glucoamylase-type enzymes from Thermoactinomyces vulgaris and Methanococcus jannaschii whose genes are found in the flanking region of the a-amylase genes. Appl. Microbiol. Biotechnol. 56:465–473 [DOI] [PubMed] [Google Scholar]
- 35. Clark CG, Alsmark UCM, Tazreiter M, Saito-Nakano Y, Ali V, Marion S, Weber C, Mukherjee C, Bruchhaus I, Tannich E, Leippe M, Sicheritz-Ponten T, Foster PG, Samuelson J, Noël CJ, Hirt RP, Embley TM, Gilchrist CA, Mann BJ, Singh U, Ackers JP, Bhattacharya S, Bhattacharya A, Lohia A, Guillén N, Duchêne M, Nozaki T, Hall N. 2007. Structure and content of the Entamoeba histolytica genome. Adv. Parasitol. 65:51–190 [DOI] [PubMed] [Google Scholar]
- 36. Loftus B, Anderson I, Davies R, Alsmark UCM, Samuelson J, Amedeo P, Roncaglia P, Berriman M, Hirt RP, Mann BJ, Nozaki T, Suh B, Pop M, Duchene M, Ackers J, Tannich E, Leippe M, Hofer M, Bruchhaus I, Willhoeft U, Bhattacharya A, Chillingworth T, Churcher C, Hance Z, Harris B, Harris D, Jagels K, Moule S, Mungall K, Ormond D, Squares R, Whitehead S, Quail MA, Rabbinowitsch E, Norbertczak H, Price C, Wang Z, Guillén N, Gilchrist C, Stroup SE, Bhattacharya S, Lohia A, Foster PG, Sicheritz-Ponten T, Weber C, Singh U, Mukherjee C, El-Sayed NMWAP, Jr, Clark CG, Embley TM, Barrell B, Fraser CM, Hall N. 2005. The genome of the protist parasite Entamoeba histolytica. Nature 433:865–868 [DOI] [PubMed] [Google Scholar]
- 37. Scheglmann D, Werner K, Eiselt G, Klinger R. 2002. Role of paired basic residues of protein C-termini in phospholipid binding. Protein Eng. 15:521–527 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.