Abstract
Apical membrane antigen 1 (AMA1) is a leading vaccine candidate, but the allelic polymorphism is a stumbling block for vaccine development. We previously showed that a global set of AMA1 haplotypes could be grouped into six genetic populations. Using this information, six recombinant AMA1 proteins representing each population were produced. Rabbits were immunized with either a single recombinant AMA1 protein or mixtures of recombinant AMA1 proteins (mixtures of 4, 5, or 6 AMA1 proteins). Antibody levels were measured by enzyme-linked immunosorbent assay (ELISA), and purified IgG from each rabbit was used for growth inhibition assay (GIA) with 12 different clones of parasites (a total of 108 immunogen-parasite combinations). Levels of antibodies to all six AMA1 proteins were similar when the antibodies were tested against homologous antigens. When the percent inhibitions in GIA were plotted against the number of ELISA units measured with homologous AMA1, all data points followed a sigmoid curve, regardless of the immunogen. In homologous combinations, there were no differences in the percent inhibition between the single-allele and allele mixture groups. However, all allele mixture groups showed significantly higher percent inhibition than the single-allele groups in heterologous combinations. The 5-allele-mixture group showed significantly higher inhibition to heterologous parasites than the 4-allele-mixture group. On the other hand, there was no difference between the 5- and 6-allele-mixture groups. These data indicate that mixtures with a limited number of alleles may cover a majority of the parasite population. In addition, using the data from 72 immunogen-parasite combinations, we mathematically identified 13 amino acid polymorphic sites which significantly impact GIA activities. These results could be a foundation for the rational design of a future AMA1 vaccine.
INTRODUCTION
Malaria is still one of the major global health problems, and there were an estimated 225 million cases in 2009 (1), despite the decades of efforts made to reduce the malaria burden. Although vaccination against malaria is considered to be the most cost-effective control method, once applied, only one vaccine candidate, the RTS,S vaccine, has been shown to have the ability to offer partial clinical protection in several phase 2 trials in Africa, and a large phase 3 trial is under way (2). The RTS,S vaccine is targeted against preerythrocytic stages of malaria. None of the vaccine candidates against blood stages of the malaria parasite, which is responsible for all pathological manifestations of this disease, have been shown to provide significant clinical protection to date. A passive transfer study conducted in the1960s has shown that gamma globulin is a critical factor for protection in the blood stage of Plasmodium falciparum malaria (3). However, the target antigens and the mechanisms of protection have not yet been completely elucidated.
Apical membrane antigen 1 (AMA1) is one of the best-studied blood-stage antigens. AMA1 contains 8 disulfide bonds and consists of three subdomains (4, 5). While a study by Triglia et al. has shown that AMA1 is an essential protein for erythrocyte invasion (6), the amino acid polymorphism in the AMA1 protein has hampered AMA1 vaccine development. Indeed, there is evidence of balancing selection in domains I and III in field parasites from Nigeria (7), Papua New Guinea (8), Thailand (9), and Kenya (10). We and other investigators have conducted multiple phase 1 trials of AMA1 using AMA1-3D7, AMA1-FVO, or a mixture of both (AMA1-C1), and the antibodies induced by the vaccines showed strain-specific functional activities, as judged by the in vitro growth inhibition assay (GIA; also referred to as the invasion inhibition assay [IIA]) (11–14). Only two phase 2 field trials with AMA1 vaccines have been conducted, and neither of them showed significant clinical protection in a target population of African children (15, 16). However, the latter trial indicated that the AMA1-3D7-based vaccine might be able to induce strain-specific protection if the 8 polymorphic sites in the cluster 1 loop (C1L) of domain I were used to categorize the parasite AMA1 genotypes (16). A study was conducted where rabbits were immunized with chimeric AMA1 proteins in which certain portions (either whole subdomains or a part of domain I) of AMA1 were selectively switched between FVO and 3D7 and antibodies against the FVO and 3D7 parasites were tested (17). The study showed that five polymorphic sites in C1L are the most important for determination of invasion-blocking activity. A recent follow-up study of the phase 2 field trial suggested that amino acid position 197 in C1L is the most critical polymorphic site for determination of allele-specific protection (18). However, it is still unclear whether C1L is the only important area when different sequences of AMA1 are used for immunization. Indeed, another recent study in rabbits indicated that other sites, in addition to the C1L region, could also determine the functional activity of the anti-AMA1 antibody (19).
We have previously shown that the global set of AMA1 haplotypes could be grouped into six genetic populations, assuming that all amino acid polymorphic sites are equally important (20). We hypothesized that immunization with a single allelic form of AMA1 could cover other AMA1 alleles in the same population and that immunization with a mixture of proteins from the six AMA1 populations could induce cross-reactive antibodies against all parasites in the field. To test the hypothesis, we produced and characterized six allelic forms of AMA1 in this study. Rabbits were immunized with either a single recombinant AMA1 protein or a mixture of them (4, 5, or 6 different AMA1 proteins in each mixture), and the purified IgG from each rabbit was used for GIA with 12 different clones of parasites (a total of 108 immunogen-parasite combinations). The data indicate that mixtures with a limited number of alleles may cover the various genetic types of AMA1. In addition, we mathematically identified 13 key amino acid polymorphic loci which significantly impact GIA activity.
MATERIALS AND METHODS
Cloning, expression, refolding, and purification of recombinant AMA1.
The AMA1-3D7 (21) and AMA1-L32 recombinant proteins were produced in Pichia pastoris. In brief, synthetic genes encoding amino acids (aa) 25 to 546 in which the N-glycosylation sites had been mutated were made on the basis of the native sequences (GenBank accession numbers U65407 and ABN12123, respectively) using codons optimized for expression in P. pastoris. The synthetic genes were cloned into pPIC9K (Invitrogen) such that they contained heterologous YV and TSHHHHHH amino acids at the amino and carboxyl termini, respectively. The P. pastoris AMA1 (PpAMA1) expression clones were fermented in 5-liter bioreactors essentially as described previously (21) and purified using a series of column chromatography steps, including Ni Sepharose HP (GE Healthcare) capture, hydrophobic interaction chromatography (Butyl 650 M; Tosoh Bioscience), anion-exchange chromatography (Q Sepharose HP; GE Healthcare), and size-exclusion chromatography (SEC; Superdex 200; GE Healthcare). Each column step used a series of phosphate-buffered solutions (PBS) with pHs ranging from 6 to 8 and the conductivity being between 5 mS and 80 mS, obtained using NaCl. The SEC buffer was PBS, pH 7.4.
The Escherichia coli-expressed proteins AMA1-CAMP (GenBank accession number ACB87777.1), HP47 (png9_1; GenBank accession number ACB87894.1), M24 (GenBank accession number ACB87844.1), and HP22 (jan_s305; GenBank accession number ACB87812.1) were produced essentially as described previously (22). In brief, synthetic genes codon optimized for expression in E. coli were cloned into the NdeI and XhoI sites of pET-24a(+) (Novagen Inc., Madison, WI) downstream of the T7 promoter. The vectors were then transformed into the E. coli BL21(DE3) expression line (Novagen) for recombinant expression of AMA1 proteins. All of the E. coli-produced recombinant AMA1 proteins contained an additional LEHHHHHH sequence at the C terminus. The panel of recombinant E. coli-expressed AMA1 proteins was purified essentially as previously described (22), with the exception that the E. coli AMA1 (EcAMA1)-enriched solubilized inclusion bodies were refolded by dilution at 100 μg/ml for 18 h, concentrated, and buffer exchanged by tangential flow filtration (10-kDa molecular weight cutoff) and sequentially loaded on Ni Sepharose 6 FF and Q Sepharose HP columns (GE Healthcare, NJ).
The recombinant AMA1-3D7 and L32 proteins correspond to native amino acids Q25 to K546, while the CAMP, HP47, M24, and HP22 proteins correspond to native amino acids G24 to K546.
Biochemical characterization of recombinant AMA1.
The panel of recombinant AMA1 proteins was characterized essentially as described by Plassmeyer et al. (23) and Uchime et al. (24). In brief, the following analytical procedures were included here: Edman degradation, reversed-phase high-performance liquid chromatography (HPLC), analytical SEC with in-line multiangle light scattering (MALS)-HPLC, matrix-assisted laser desorption ionization (MALDI) mass spectrometry, endotoxin concentration determination, immunoblotting with rat monoclonal antibody (MAb) 4G2dc1 (25) and anti-His6 (Qiagen, Inc.), and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Rabbit immunization.
The animal study was done in compliance with National Institutes of Health (NIH) guidelines and under the auspices of an Animal Care and Use Committee-approved protocol (protocol MVDB119). New Zealand White rabbits (n = 6 per group) were immunized with either a single AMA1 protein (20 μg) or a mixture of AMA1 proteins (total of 80, 100, or 120 μg of AMA1 protein for the 4-allele-mixture, 5-allele-mixture, and 6-allele-mixture groups, respectively) formulated with the Montanide ISA720 adjuvant (SEPPIC Inc., Fairfield, NJ) on days 0, 28, and 56. The 4-allele mixture contained AMA1-3D7, CAMP, HP47, and M24 proteins; the 5-allele mixture contained the proteins in the 4-allele mixture plus L32; and the 6-allele mixture contained the proteins in the 4-allele mixture plus L32 and HP22. The serum samples were collected on day 70. As a control, a group of 3 rabbits was immunized with Montanide ISA720 adjuvant alone with the same schedule.
ELISA and GIA.
The standardized methodology for performing the enzyme-linked immunosorbent assay (ELISA) has been described previously (26). The absorbance of each test sample was converted into ELISA units using a standard curve generated by serially diluting the standard in the same plate.
The standard methodology for the GIA has been described previously (27). In brief, total IgG was purified using a protein G affinity column and then buffer exchanged to RPMI 1640. The assay was performed with the purified IgGs at a final concentration of 2.5 mg/ml of total IgG against 12 different clones of P. falciparum parasites (3D7, C2A, GB4, CAMP, MT/S1, 102_1, HB3, L32, Dd2, M24, FVO, and 425). The AMA1 sequence of each clone of the parasite and the absence of mycoplasma contamination were checked by PCR (Mycotrace PCR detection kit; PAA Laboratories, Morningside, Australia) before GIA analysis. The origins of the parasite clones were Africa, Thailand, Ghana, Malaysia, Thailand, Sudan, Honduras, Liberia, Laos, Kenya, Vietnam, Gambia, Papua New Guinea, and Africa for 3D7, C2A, GB4, CAMP, MT/S1, 102_1, HB3, L32, Dd2 (Indochina III/CDC, W2-mef), M24, FVO, 425, HP47, and HP22, respectively.
Structural modeling.
Structural alignment of the PfAMA1 (1Z40) (28) and Plasmodium vivax AMA1 (PvAMA1) (1W81) (5) was done on the PAN domains. The root-mean-square differences between the two structures (domain I and II) were 0.416, suggesting that the backbones of the two structures overlap very well, and the differences were mostly seen in the flexible loop regions (see Fig. S1 in the supplemental material). On the basis of the sequence and the structural alignments, we ascertained the three-dimensional position of the polymorphic sites of PfAMA1 on the PvAMA1 structure, because the structure of PfAMA1 domain III has not been solved. All the structural figures were generated using the PyMOL molecular graphics system (www.pymol.org).
Statistical analysis.
The Mann-Whitney test was utilized to compare data for two groups. A Kruskal-Wallis test followed by a Dunn's multiple-comparison test was used to compare data for four groups. The correlation between number of ELISA units and the percent inhibition in GIA was evaluated using a Spearman rank test.
The key amino acid polymorphic sites which significantly impact GIA activity were determined as follows using GIA data with IgGs from rabbits immunized with a single AMA1. Since PfAMA1-1 is proteolytically cleaved between Ser96 and Ile97 when AMA1 translocates onto the merozoite surface (29), the polymorphic sites prior to Ser96 were excluded from this analysis. We selected the key amino acid sites by a two-step procedure. In the first step, we measured the impact of each individual amino acid site (a total of 50 sites) on GIA activity using univariate regression analysis. In each set of univariate regressions, the amino acid sites with P values of less than 0.001 (0.05/50) were considered significant. To minimize the bias from data generated from a particular parasite clone, the univariate analyses were carried out 12 times by removing data for one parasite each time (e.g., in the first analysis we removed 3D7 parasite data, and then we removed C2A parasite data for the second analysis). The amino acid sites commonly selected in all 12 univariate analyses were chosen, and a total of 22 sites were selected. In the second step, we performed a multivariate regression analysis involving the 22 amino acid sites to find jointly important amino acid sites. The multivariate regression identified 9 amino acid sites with coefficients near 0 (i.e., they had no significant impact on GIA activity), and a total of 13 amino acid sites were determined to be the key amino acid sites.
To select the best mixtures of AMA1 haplotypes, we focused only on the 13 key amino acid sites. Each of the 13 sites was weighted using the coefficient of the multivariate regression analysis. We used two types of coverage: (i) minimum coverage, which was the coverage of a haplotype with the lowest coverage in all AMA1 haplotypes which were not included in a proposed AMA1 vaccine, and (ii) average coverage, which was the average coverage of all nonincluded AMA1 haplotypes. The first component was selected from all 107 haplotypes by searching for a single haplotype with the highest coverage over the rest of the 106 haplotypes. From the remaining 106 haplotypes, the second component was selected so that the mixture of the first and second components showed the highest coverage over the rest of the 105 haplotypes. In a similar fashion, the third, fourth, and fifth components were selected.
All statistical tests were performed by R (version 2.14.0) or Prism (version 5; GraphPad Software Inc., La Jolla, CA) software, and probability values of less than 0.05 were considered significant.
RESULTS
Characterization of recombinant AMA1 proteins.
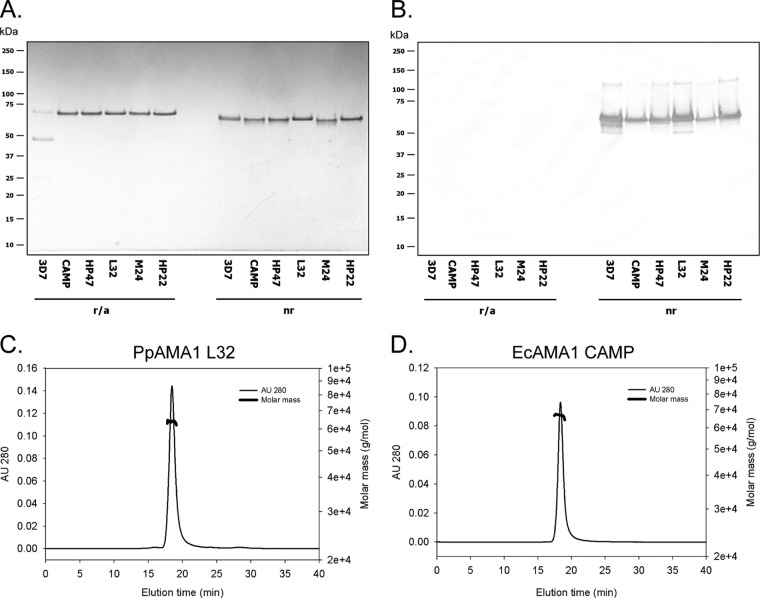
We have previously shown that the global set of AMA1 haplotypes could be grouped into 6 genetic populations (20), assuming that all polymorphic sites were equally important. Using this information, 6 AMA1 proteins representing each population (AMA1-3D7, CAMP, HP47, L32, M24, and HP22) were identified (Fig. 1), and all amino acid polymorphic sites are shown in Fig. 2. Recombinant proteins of all six forms of AMA1 were produced using either a P. pastoris (AMA1-3D7 and AMA1-L32) or an E. coli (CAMP, HP47, M24, and HP22) expression system. Each protein was characterized to confirm the identity of the amino terminus, and the result was as expected compared to the theoretical result (data not shown). The endotoxin levels were all below 20 endotoxin units per mg (data not shown). The integrity was assessed by reversed-phase HPLC, and the primary elution peak comprised greater than 90% of the total elution product (data not shown). Analysis of the integrity by SDS-PAGE showed that each purified recombinant protein migrated essentially as a single band which had a demonstrable migration shift upon reduction of disulfide bonds (Fig. 3A). An assessment of the proteins by immunoblotting showed that the conformation-dependent functional rat MAb 4G2 (25) reacted with all recombinant forms of AMA1 only under nonreduced conditions (Fig. 3B), which is consistent for a P. pastoris-expressed protein as well as a refolded E. coli protein. Immunoblotting with an anti-His6 MAb showed reactions with both the reduced and nonreduced panels of proteins (data not shown). Finally, an analysis of the AMA1 panel of proteins in solution by SEC-MALS demonstrated that each recombinant protein appeared to be monomeric with an appropriate average molar mass. Examples for P. pastoris- and E. coli-expressed proteins are shown in Fig. 3C (AMA1-L32) and D (AMA1-CAMP), respectively.
Fig 1.

AMA1 haplotypes selected in this study. The global set of AMA1 haplotypes could be grouped into 6 genetic populations. One of the AMA1 haplotypes was selected from each population for the rabbit immunization study, and 1 to 3 parasites per population were selected for GIA.
Fig 2.

AMA1 amino acid polymorphic sites in the 14 parasite clones. The AMA1 amino acid polymorphic sites of 14 parasite clones used in this study are shown (the 3D7 sequence was used as a reference). For each parasite, the AMA1 population number (Pop 1 to 6) and whether it was used for immunization (Immu) are shown. The arrow indicates the cleavage site from the 83-kDa form of AMA1 to the 66-kDa form.
Fig 3.
Characterizations of recombinant AMA1 proteins. Characterizations of a panel of recombinant AMA1 proteins were determined by Coomassie blue-stained SDS-polyacrylamide gel analysis under reduced/alkylated conditions (r/a) and nonreduced conditions (nr) (A), by immunoblotting with MAb 4G2dc1 (B), and by analytical SEC-MALS-HPLC (C and D). Examples for P. pastoris- and E. coli-expressed proteins are shown in panels C (AMA1-L32) and D (AMA1-CAMP), respectively. AU 280, absorbance units at 280 nm.
Immunogenicity of AMA1 recombinant proteins.
Rabbits (n = 6 per group) were immunized with either a single AMA1 protein (20 μg) or a mixture of them (total of 80, 100, or 120 μg of AMA1 protein for the 4-allele-mixture, 5-allele-mixture, and 6-allele-mixture groups, respectively), and the levels of antibody against each of the 6 allelic forms of AMA1 in each rabbit were determined by ELISA. As a control, 3 rabbits were immunized with adjuvant alone and showed less than 450 ELISA units for antibodies to all of the AMA1 proteins (data not shown). In general, homologous antisera and antisera from rabbits immunized with mixtures of AMA1 showed larger numbers of ELISA units than heterologous antisera (Fig. 4). To test the effect of homologous/heterologous combinations of the immunogen-ELISA antigen and the effect of single-allele and allele mixture immunization on antibody levels, the ELISA data sets were grouped into 4 categories. The four categories were homologous combinations with immunization with a single allele (homol-single; e.g., anti-AMA1-3D7 antisera were tested using AMA1-3D7-coated ELISA plates), homologous combinations with immunization with allele mixtures (homol-mix; e.g., antisera to the 4-allele mix, which contained AMA1-3D7, CAMP, HP47, and M24, were tested using AMA1-3D7-coated ELISA plates), heterologous combinations with immunization with a single allele (hetero-single; e.g., anti-3D7 antisera were tested using AMA1-CAMP-coated ELISA plates), and heterologous combinations with immunization with allele mixtures (hetero-mix; e.g., antisera of the 4-allele mix were tested using AMA1-L32-coated ELISA plates). When the 4 sets of ELISA data were compared, only the hetero-single group showed significantly lower antibody levels than the other 3 groups, and no significant differences were detected among the other 3 groups (Fig. 5). The data indicate that the immunizations with single allele AMA1 induced allele-specific antibodies, as judged by ELISA.
Fig 4.
Antibody levels are higher against homologous than heterologous AMA1. Rabbits (n = 6 per group) were immunized with either a single AMA1 protein (20 μg) or mixtures of proteins (total of 80, 100, or 120 μg of AMA1 protein for the 4-allele-mixture, 5-allele-mixture, and 6-allele-mixture groups, respectively). Different symbols represent different immunization groups. Each individual rabbit serum sample was tested using AMA1-3D7 (A), AMA1-CAMP (B), AMA1-HP47 (C), AMA1-L32 (D), AMA1-M24 (E), or AMA1-HP22 (F) ELISA plates. Bars represent the geometric mean of the group. The homologous combinations of immunogen-ELISA antigen (rabbits were immunized with a single homologous immunogen or with a mixture containing the homologous antigen) are highlighted by gray boxes.
Fig 5.
Single-allele AMA1 immunization showed a lower level of antibody to heterologous antigens. The ELISA data presented in Fig. 4 are grouped on the basis of whether the immunogen-ELISA combinations are homologous (homol) or heterologous (hetero) and whether rabbits were immunized with a single AMA1 protein (single) or a mixture (mix) of AMA1 proteins. Box-and-whiskers plots are shown. The number under each box represents the total number of immunogen-ELISA combinations in each set of grouped data.
Growth-inhibitory activity of purified IgGs against various parasites.
Purified IgG from each individual rabbit was tested at 2.5 mg/ml against 6 P. falciparum clones (Fig. 6). For the HP47 and HP22 haplotypes, we used clones 102_1 (9 aa differences from HP47) and 425 (15 aa differences from HP22), respectively, instead of homologous parasites, since culture-adapted parasites with the particular AMA1 haplotypes were not available. The averages of the percent inhibition of control IgGs from rabbits immunized with adjuvant alone were −7, −15, 23, −10, 12, and 13 for the 3D7, CAMP, 102_1, L32, M24, and 425 parasites, respectively. The percent inhibitions in GIA were plotted against the number of ELISA units measured with homologous AMA1 (e.g., antibody levels of all samples were measured using AMA1-3D7-coated ELISA plates, and samples were tested by GIA with 3D7 parasites; Fig. 6). All data points followed a sigmoid curve, regardless of the immunogen (in the cases of GIA with parasites 102_1 and 425, numbers of anti-HP47 and anti-HP22 ELISA units, respectively, were used on the x axes in Fig. 6). When tested statistically, there were significant correlations between the number of ELISA units and percent inhibition (P < 0.001 by Spearman rank test for all parasites).
Fig 6.
Strong correlation between numbers of ELISA units and functional activity. Each purified rabbit IgG was tested at 2.5 mg/ml against the 3D7 (A), CAMP (B), 102_1 (C), L32 (D), M24 (E), or 425 (F) clone of P. falciparum parasites by GIA. The numbers of homologous anti-AMA1 ELISA units in the GIA well (x axis) are plotted against percent inhibition (y axis). For GIA with the 102_1 clone of parasites (C), the numbers of anti-AMA1-HP47 ELISA units were used, and the numbers of anti-AMA1-HP22 ELISA units were used for clone 425 (F). The line represents the best fit of the data from all 9 groups, and the Spearman correlation coefficient (rs) is shown.
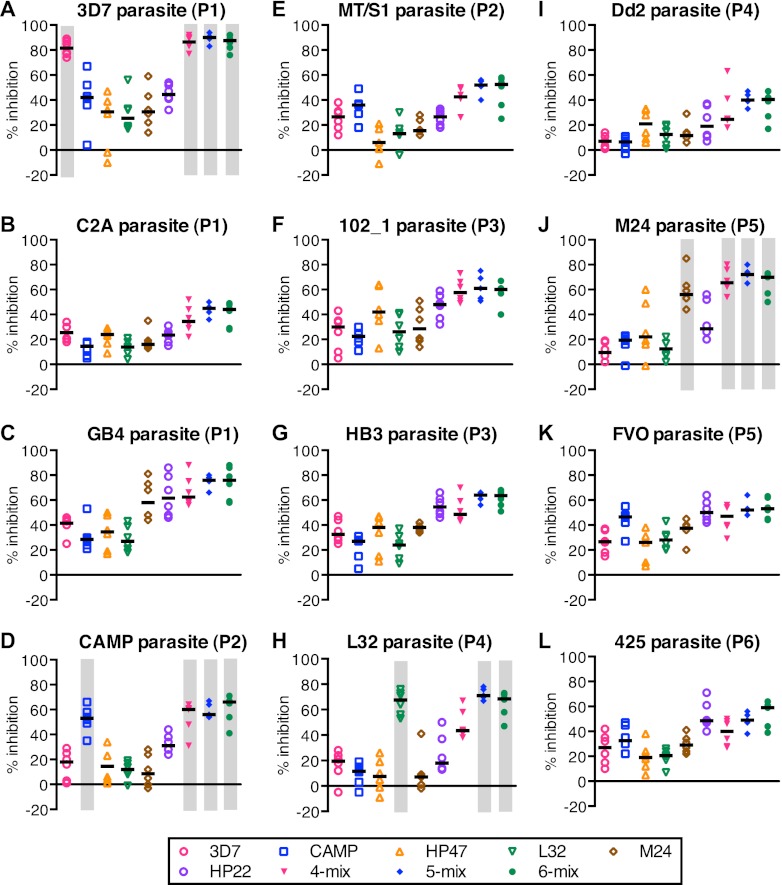
We then tested the IgGs with an additional 6 parasites. The adjuvant control IgGs showed between 1 and 14% inhibition, on average, against those 6 parasites (data not shown). Similar to the ELISA data, IgGs from the groups immunized with single alleles and allele mixtures showed higher levels of percent inhibition when tested with homologous parasites (Fig. 7A, D, H, and J). However, the grouping of AMA1 haplotypes into 6 genetic populations did not necessarily explain the GIA results. For example, while the GB4 clone (Fig. 7C) belonged to population 1, the anti-M24 (population 5) and anti-HP22 (population 6) antibodies showed higher inhibition than the anti-3D7 (population 1) antibody. Similar mismatches were also observed in other clones, such as HB3 (Fig. 7G) and Dd2 (Fig. 7I). The data suggest that it was inadequate to assume that all the polymorphic sites in the AMA1 molecule are equally important in terms of GIA.
Fig 7.
Higher inhibition in homologous combinations. Each purified IgG was tested at 2.5 mg/ml against 3D7 (A), C2A (B), GB4 (C), CAMP (D), MT/S1 (E), 102_1 (F), HB3 (G), L32 (H), Dd2 (I), M24 (J), FVO (K), or 425 (L) clones of P. falciparum parasites. Bars represent the median of the group. The homologous combinations of immunogen-parasite (rabbits were immunized with a single homologous immunogen or one component of the mixture was homologous) are highlighted by gray boxes. 3D7, C2A, and GB4 belong to population 1 (P1); CAMP and MT/S1 belong to population 2 (P2); HP47 (orange triangles), 102_1, and HB3 belong to population 3 (P3); L32 and Dd2 belong to population 4 (P4); M24 and FVO belong to population 5 (P5); and HP22 (purple circles) and 425 belong to population 6 (P6).
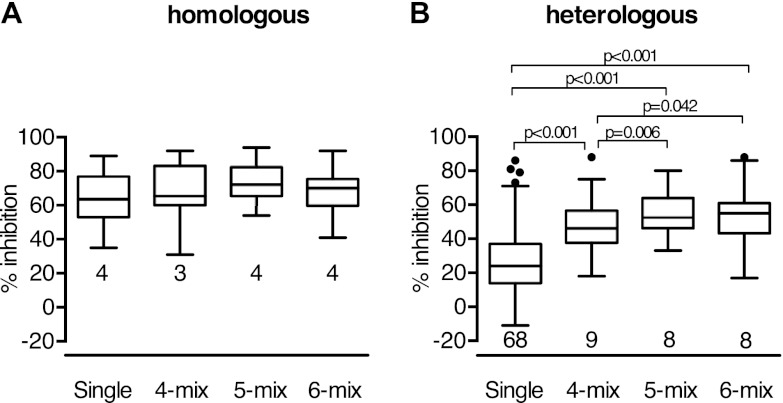
The data from 108 immunogen-parasite combinations were grouped into 4 categories. The four categories were homologous combinations with single-allele immunization (homol-single; e.g., anti-AMA1-3D7 IgGs were tested against 3D7 parasites), homologous combinations with allele mixture immunization (homol-mix), heterologous combinations with single-allele immunization (hetero-single), and heterologous combinations with allele mixture immunization (hetero-mix). There was no significant difference between the homol-single and homol-mix groups (P = 0.160 by Dunn's multiple-comparison test). On the other hand, there was a significant difference between the hetero-single and hetero-mix groups (P < 0.001). The effect of an allele mixture vaccine on heterologous parasites was further analyzed (Fig. 8). The single-allele group showed significantly lower inhibition than any of the allele mixture groups (P < 0.001 for all comparisons). When 4-, 5-, and 6-allele-mix groups were compared, the 4-allele-mix group showed significantly lower inhibition than the 5- and 6-allele-mix groups (P = 0.006 and 0.042, respectively). However, inhibition for the 6-allele-mix group was not significantly better than that for the 5-allele-mix group (P = 0.56). The data suggest that the mixtures of AMA1 induced higher percent inhibition against heterologous parasites and that the 5-allele mix of AMA1 was as good as the 6-allele-mix vaccine in terms of the coverage of heterologous parasites.
Fig 8.
Higher inhibition with mixtures of AMA1 in heterologous combinations. The GIA data from Fig. 7 are grouped on the basis of whether the immunogen-parasite combinations are homologous (A) or heterologous (B) and then on the basis of the immunization strategy (rabbits were immunized with either a single protein or mixtures of 4, 5, or 6 proteins). Box-and-whiskers plots are shown. The number under each box represents the total number of immunogen-parasite combinations in each set of grouped data.
Identification of key polymorphic amino acid sites and potential AMA1 mixtures.
We then mathematically calculated the key amino acid polymorphic sites which significantly correlated with percent inhibition using the GIA data with IgGs from rabbits immunized with a single form of AMA1 (72 immunogen-parasite combinations). We identified 13 key amino acid polymorphic sites, as described in the Materials and Methods section (amino acid residues 172, 196, 197, 200, 207, 224, 242, 283, 330, 435, 439, 485, and 496), and these are shown in Fig. 9. The key polymorphic sites are spread throughout the AMA1 molecule. Focusing on the 13 determined amino acid sites, we then tried to identify the best mixture of 5 allelic forms of AMA1 from 107 haplotypes (all amino acid polymorphic sites in 107 haplotypes are shown in Fig. S2 in the supplemental material). We calculated the minimum and average coverage of each predicted best mixture and the coverage of 4-allele-mix or 5-allele-mix vaccines, which were used in this study, against unselected haplotypes. The 4-allele mix showed lower minimum coverage. The 5-allele mix used in this study and the mathematically predicted best mixtures (on the basis of either the minimum or the mean coverage) had comparable levels of coverage. The data suggest that if we select 5 divergent AMA1 haplotypes, the mixture vaccine can provide ∼84% of the minimum coverage (and ∼98% of the mean coverage) to the key amino acid polymorphic sites in any given haplotype.
Fig 9.
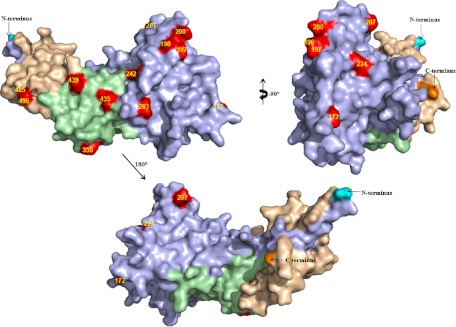
Distribution of key polymorphic residues. The key polymorphic amino acids are shown in red on the PvAMA1 structure (Protein Data Bank accession number 1W81). Domain I is light blue, domain II is green, and domain III is wheat colored. The N terminus is in cyan, and the C terminus is in orange.
DISCUSSION
In this study, we produced six recombinant AMA1 proteins, immunized rabbits with either a single protein or a mixture of proteins (mixtures of 4, 5, or 6 proteins), and tested the functional activity of each individual IgG by GIA with 12 different clones of parasites. All groups immunized with allele mixtures showed significantly higher percent inhibition than groups immunized with single alleles in the heterologous immunogen-parasite combinations, and the 5-allele-mix-immunized group was significantly better than the 4-allele-mix-immunized group in terms of GIA activities to heterologous parasites. However, there was not more improvement in the 6-allele-mix group than the 5-allele-mix group. These data indicate that a limited number of allelic mixtures may cover the majority of AMA1 populations. In addition, using the data from 72 immunogen-parasite combinations, we mathematically identified 13 key amino acid polymorphic sites for future development of an AMA1-based vaccine.
The result of a recent phase 2 trial with the AMA1-3D7 vaccine (16) raised the hope that an AMA1-based vaccine might be able to induce at least strain-specific protective immunity in a target population. If this is true, the next question is how to overcome the polymorphic nature of the AMA1 molecule. There are several pieces of encouraging data provided from previous animal studies. When rabbits were immunized with a mixture of multiple allelic forms of AMA1 and/or chimeric proteins, the vaccine could induce more cross-reactive antibodies than vaccination with a single allele of AMA1 (17, 21, 30). However, there are multiple haplotypes of AMA1 in the field (20), and from a practical point of view, it is impossible to make a mixture vaccine including all of them. We have previously shown that the global set of AMA1 haplotypes could be grouped into 6 genetic populations, assuming that all amino acid polymorphic sites are equally important (20). Therefore, if immunization with a single allelic form of AMA1 can cover other AMA1 alleles in the same genetic population, a mixture of 6 AMA1 alleles could induce cross-reactive antibodies against all parasites. The GIA with 12 clones of parasites showed that IgGs against the same population of AMA1 were not necessarily the best (Fig. 7). These results suggest that it is inadequate to assume that all the polymorphic sites in the AMA1 molecule are equally important in terms of GIA. While our original hypothesis turned out to be incorrect, the important point was that the 5-allele mix was as good as the 6-allele mix in covering heterologous parasites. The data indicate that mixtures with a limited number of alleles may cover the various genetic types of AMA1. Similarly, a recent study showed that a mixture of four anti-AMA1 antibodies (W2Mef, 3D7, HB3, and FVO) demonstrated broader coverage against 18 isolates than any of the single antibodies (19). Dutta and his colleagues also found that a mixture of 4 allelic forms of AMA1 could induce strain-transcending antibodies (S. Dutta, personal communication). An alternative approach to overcome the polymorphic nature of AMA1 has been tested (31–33). In this approach, animals were immunized with three recombinant diversity-covering (DiCo) AMA1 proteins, which were designed to cover maximal numbers of polymorphisms in 355 sequences when all of the 3 were taken together. A direct-comparison study with several antibodies (e.g., antibodies raised against a mixture of 4 or 5 allelic forms of AMA1 and the 3-DiCo-protein mixture) against multiple clones of parasites will be beneficial in selecting an optimal approach for future vaccine development.
Since the current study indicates that not all of the amino acid polymorphic sites are equally important, we tried to identify the critical sites in this study. Dutta et al. tried to answer this question before (17). In that study, they immunized rabbits with 3D7 and FVO chimeric AMA1 recombinant proteins and performed GIA with FVO and 3D7 parasites. This study has shown that the C1L cluster in domain I contains the most important residues, and the second important cluster is in domain II. A recent follow-up study of the phase 2 field trial with the AMA1-3D7 vaccine also suggested that amino acid position 197 was the most critical polymorphic site (18). However, these two studies did not address whether C1L was the only critical region for antibodies other than AMA1-3D7 and AMA1-FVO antibodies. When U.S. adults were immunized with AMA1-C1, which is a mixture of AMA1-3D7 and FVO, the purified IgGs showed inhibitory activity against 3D7 and FVO parasites, as judged by GIA, as expected, but their activity was much less than their activity against L32 and M24 parasites (11). The data suggested that there may be other critical sites which are not covered by either 3D7 or FVO. A recent study with antibodies against 4-allele forms of AMA1 tested by GIA with 18 isolates showed that the sequence similarity in the C1L region did not necessarily explain the growth inhibition of the antibodies (19). The same study also performed GIA with transgenic parasites which expressed C1L epitopes from different strains of parasites and indicated that antibodies against C1L contributed to a minor proportion of the total inhibitory antibodies, depending on the parasite-antibody combinations (19). In the present study, using the data from 72 immunogen-parasite combinations, which included 50 amino acid polymorphic sites (Fig. 2), we mathematically identified 13 key amino acid polymorphic sites (Fig. 9): amino acid residues 172, 196, 197, 200, 207, 224, 242, 283, 330, 435, 439, 485, and 496. Similar to the study by Dutta et al. (17), the polymorphic sites in the C1L region (residues 196, 197, 200, and 207) and in domain II (amino acid residues 330 and 435) were identified as the key polymorphic sites. In addition, other polymorphic sites in domain I (amino acid residues 172, 207, 224, 242, and 283) and domain III (amino acid residues 439, 485, and 496) were also found to be important. Since studies with field parasites showed a strong balancing selection in domain III and not only in domain I (7, 9, 10), it is reasonable to suggest that there are also key polymorphic sites in domain III. This study and others have made it clear that not all of the amino acid polymorphisms contribute equally to the growth-inhibitory activity. It is likely that the structure of the AMA1 molecule plays a major role (34). Therefore, we should select better combinations of AMA1 alleles by considering the AMA1 structure as well as the results from animal studies, such as this study.
Focusing on the 13 critical amino acid sites, we tried to find the best mixture of AMA1 variants. We used two strategies (i.e., minimum coverage and mean coverage) to select the best mixture, because there are no data to judge which selection strategy better shows efficacy in the field, where many different haplotypes of AMA1 exist. We also calculated the coverage of the 4-allele mix and the 5-allele mix, which were used in this study. The 6-allele mix used in this study had the same amino acid variations as those in the 5-allele mix for the 13 critical sites (Fig. 2), so we did not calculate the coverage for the 6-allele mix. As shown in Table 1, it is of note that the two mathematically predicted best mixtures and the 5-allele mix used in this study had similar coverage, while the haplotype numbers in each mixture were different. These data suggest that if we select 5 divergent allelic forms of AMA1 (using all polymorphism data in the AMA1 molecule and not only data in a certain domain of the molecule), the mixture vaccine may have a comparable level of coverage. We believe that future AMA1-based vaccines may benefit from optimizing coverage of these key polymorphic sites.
Table 1.
Best mixtures and coverage
| Mixture | Haplotype no.a | Coverage (%) |
|
|---|---|---|---|
| Minimumb | Meanc | ||
| Minimum bestd | 9, 13, 20, 31, 106 | 83.6 | 94.5 |
| Mean beste | 7, 15, 58, 87, 106 | 74.9 | 98.1 |
| 5 allelesf | 2, 6, 20, 21, 105 | 83.6 | 98.1 |
| 4 allelesg | 2, 6, 20, 105 | 74.9 | 96.5 |
Haplotype numbers are the numbers in Fig. S2 in the supplemental material.
The lowest coverage of haplotypes in unselected 103/102 haplotypes by the 4-/5-allele-mixture vaccines.
The mean coverage of the unselected 103 or 102 haplotypes by the 4- or 5-allele-mixture vaccines, respectively.
The best mixture of haplotypes was selected from 107 haplotypes of AMA1 on the basis of the minimum coverage of key amino acid sites.
The best mixture of haplotypes was selected from 107 haplotypes of AMA1 on the basis of the mean coverage of key amino acid sites.
The 5-allele-mix vaccine used in this study.
The 4-allele-mix vaccine used in this study.
There are several potential limitations in this study. The first one is that we could not evaluate the impact of amino acid polymorphisms which were not present in the 72 immunogen-parasite combinations. Since AMA1 is a highly polymorphic molecule, it is practically impossible to test all of the mixtures experimentally. However, most of the polymorphisms existing in 107 haplotypes were covered by the 13 haplotypes used in this study, and the average coverage of all polymorphisms was 99.0% (see Fig. S3 in the supplemental material). Therefore, while additional information (i.e., more amino acid haplotypes in the field, GIAs with more parasites) may improve the prediction, we believe that the majority of polymorphisms were covered in this study. The second potential limitation is whether we can predict the effect of a vaccine in the field by the in vitro GIA. It is controversial whether the GIA activity induced by natural infections correlates with clinical protection in the field (35–37). However, there are some supportive data from monkey (38) and human (39) challenge studies in the case of AMA1 immunizations, and no alternative assay is currently used to evaluate the functional activity of anti-AMA1 antibodies. Third, the AMA1 recombinant proteins were made using two different expression systems (AMA1-3D7 and L32 were produced in P. pastoris, and the others were produced in E. coli). We have previously shown that the AMA1-3D7 and -FVO recombinant proteins produced in these two expression systems were equivalent (22), and Fig. 6 clearly indicates that AMA1 recombinant proteins expressed in either of the systems induced antibodies which could equally block merozoite invasion (i.e., all data points followed a single dose-response curve). The last possible limitation is that the allele-mixture-immunized groups received 4 to 6 times more total protein than the single-allele-immunized groups. The rabbits could be immunized with the same total amount of AMA1 proteins (e.g., the 4-allele-mix group was immunized with 5 μg of each allelic form instead of 20 μg of each form), but it may be difficult to interpret the data because we need to consider both the immunogenicity effect and the allelic effect. Our preliminary data from other rabbit studies showed that immunization with an 8- to 12-μg dose of proteins sometimes elicited lower levels of antibody than immunization with 20 μg. Therefore, we decided to immunize rabbits with 20 μg of each protein in this study. There was no significant difference in antibody titers or GIA activity against homologous AMA1 protein or homologous parasite clones between the single-allele and allele mixture immunization groups. Therefore, it is reasonable to speculate that the differences in ELISA and GIA were likely caused by the allelic effect and were not a result of immunogenicity. Further studies may be required to determine whether it will be possible to reduce the dose of each allelic form of AMA1 protein if there are any practical and/or safety issues in preclinical and clinical vaccine development. Several attempts to develop multiantigen vaccines (against either the blood stage only or multiple stages) have been made, and combination of AMA1 with other antigens may be necessary to develop a clinically effective vaccine. In such a case, the dose of AMA1 proteins may be further evaluated.
Taken together, we believe that our approach and the conclusions are reasonable, despite the limitations described above. In this study, we tested 108 immunogen-parasite combinations by GIA and found that mixtures of a limited number of alleles might cover the majority of AMA1 populations. In addition, using data for 72 immunogen-parasite combinations, we mathematically identified 13 critical amino acid polymorphic sites. The results from this study can be a foundation for the rational design of an AMA1 vaccine in the future.
Supplementary Material
ACKNOWLEDGMENTS
We thank Lynn Lambert, Kelly Rausch, and their teams for the rabbit immunization study.
The study was supported by the intramural program of the National Institute of Allergy and Infectious Diseases, NIH, and GIA work was supported partially by the PATH Malaria Vaccine Initiative.
Footnotes
Published ahead of print 19 February 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01414-12.
REFERENCES
- 1. Cibulskis RE, Aregawi M, Williams R, Otten M, Dye C. 2011. Worldwide incidence of malaria in 2009: estimates, time trends, and a critique of methods. PLoS Med. 8:e1001142 doi:10.1371/journal.pmed.1001142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agnandji ST, Lell B, Soulanoudjingar SS, Fernandes JF, Abossolo BP, Conzelmann C, Methogo BG, Doucka Y, Flamen A, Mordmuller B, Issifou S, Kremsner PG, Sacarlal J, Aide P, Lanaspa M, Aponte JJ, Nhamuave A, Quelhas D, Bassat Q, Mandjate S, Macete E, Alonso P, Abdulla S, Salim N, Juma O, Shomari M, Shubis K, Machera F, Hamad AS, Minja R, Mtoro A, Sykes A, Ahmed S, Urassa AM, Ali AM, Mwangoka G, Tanner M, Tinto H, D'Alessandro U, Sorgho H, Valea I, Tahita MC, Kabore W, Ouedraogo S, Sandrine Y, Guiguemde RT, Ouedraogo JB, Hamel MJ, Kariuki S, Odero C, et al. 2011. First results of phase 3 trial of RTS,S/AS01 malaria vaccine in African children. N. Engl. J. Med. 365:1863–1875 [DOI] [PubMed] [Google Scholar]
- 3. Cohen S, McGregor IA, Carrington S. 1961. Gamma-globulin and acquired immunity to human malaria. Nature 192:733–737 [DOI] [PubMed] [Google Scholar]
- 4. Hodder AN, Crewther PE, Matthew ML, Reid GE, Moritz RL, Simpson RJ, Anders RF. 1996. The disulfide bond structure of Plasmodium apical membrane antigen-1. J. Biol. Chem. 271:29446–29452 [DOI] [PubMed] [Google Scholar]
- 5. Pizarro JC, Normand BV, Chesne-Seck ML, Collins CR, Withers-Martinez C, Hackett F, Blackman MJ, Faber BW, Remarque EJ, Kocken CH, Thomas AW, Bentley GA. 2005. Crystal structure of the malaria vaccine candidate apical membrane antigen 1. Science 308:408–411 [DOI] [PubMed] [Google Scholar]
- 6. Triglia T, Healer J, Caruana SR, Hodder AN, Anders RF, Crabb BS, Cowman AF. 2000. Apical membrane antigen 1 plays a central role in erythrocyte invasion by Plasmodium species. Mol. Microbiol. 38:706–718 [DOI] [PubMed] [Google Scholar]
- 7. Polley SD, Conway DJ. 2001. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics 158:1505–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cortes A, Mellombo M, Mueller I, Benet A, Reeder JC, Anders RF. 2003. Geographical structure of diversity and differences between symptomatic and asymptomatic infections for Plasmodium falciparum vaccine candidate AMA1. Infect. Immun. 71:1416–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Polley SD, Chokejindachai W, Conway DJ. 2003. Allele frequency-based analyses robustly map sequence sites under balancing selection in a malaria vaccine candidate antigen. Genetics 165:555–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Osier FH, Weedall GD, Verra F, Murungi L, Tetteh KK, Bull P, Faber BW, Remarque E, Thomas A, Marsh K, Conway DJ. 2010. Allelic diversity and naturally acquired allele-specific antibody responses to Plasmodium falciparum apical membrane antigen 1 in Kenya. Infect. Immun. 78:4625–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mullen GE, Ellis RD, Miura K, Malkin E, Nolan C, Hay M, Fay MP, Saul A, Zhu D, Rausch K, Moretz S, Zhou H, Long CA, Miller LH, Treanor J. 2008. Phase 1 trial of AMA1-C1/Alhydrogel plus CPG 7909: an asexual blood-stage vaccine for Plasmodium falciparum malaria. PLoS One 3:e2940 doi:10.1371/journal.pone.0002940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Polhemus ME, Magill AJ, Cummings JF, Kester KE, Ockenhouse CF, Lanar DE, Dutta S, Barbosa A, Soisson L, Diggs CL, Robinson SA, Haynes JD, Stewart VA, Ware LA, Brando C, Krzych U, Bowden RA, Cohen JD, Dubois MC, Ofori-Anyinam O, De-Kock E, Ballou WR, Heppner DG., Jr 2007. Phase I dose escalation safety and immunogenicity trial of Plasmodium falciparum apical membrane protein (AMA-1) FMP2.1, adjuvanted with AS02A, in malaria-naive adults at the Walter Reed Army Institute of Research. Vaccine 25:4203–4212 [DOI] [PubMed] [Google Scholar]
- 13. Remarque EJ, Roestenberg M, Younis S, Walraven V, van der Werff N, Faber BW, Leroy O, Sauerwein R, Kocken CH, Thomas AW. 2012. Humoral immune responses to a single allele PfAMA1 vaccine in healthy malaria-naive adults. PLoS One 7:e38898 doi:10.1371/journal.pone.0038898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spring MD, Cummings JF, Ockenhouse CF, Dutta S, Reidler R, Angov E, Bergmann-Leitner E, Stewart VA, Bittner S, Juompan L, Kortepeter MG, Nielsen R, Krzych U, Tierney E, Ware LA, Dowler M, Hermsen CC, Sauerwein RW, de Vlas SJ, Ofori-Anyinam O, Lanar DE, Williams JL, Kester KE, Tucker K, Shi M, Malkin E, Long C, Diggs CL, Soisson L, Dubois MC, Ballou WR, Cohen J, Heppner DG., Jr 2009. Phase 1/2a study of the malaria vaccine candidate apical membrane antigen-1 (AMA-1) administered in adjuvant system AS01B or AS02A. PLoS One 4:e5254 doi:10.1371/journal.pone.0005254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sagara I, Dicko A, Ellis RD, Fay MP, Diawara SI, Assadou MH, Sissoko MS, Kone M, Diallo AI, Saye R, Guindo MA, Kante O, Niambele MB, Miura K, Mullen GE, Pierce M, Martin LB, Dolo A, Diallo DA, Doumbo OK, Miller LH, Saul A. 2009. A randomized controlled phase 2 trial of the blood stage AMA1-C1/Alhydrogel malaria vaccine in children in Mali. Vaccine 27:3090–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Thera MA, Doumbo OK, Coulibaly D, Laurens MB, Ouattara A, Kone AK, Guindo AB, Traore K, Traore I, Kouriba B, Diallo DA, Diarra I, Daou M, Dolo A, Tolo Y, Sissoko MS, Niangaly A, Sissoko M, Takala-Harrison S, Lyke KE, Wu Y, Blackwelder WC, Godeaux O, Vekemans J, Dubois MC, Ballou WR, Cohen J, Thompson D, Dube T, Soisson L, Diggs CL, House B, Lanar DE, Dutta S, Heppner DG, Jr, Plowe CV. 2011. A field trial to assess a blood-stage malaria vaccine. N. Engl. J. Med. 365:1004–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dutta S, Lee SY, Batchelor AH, Lanar DE. 2007. Structural basis of antigenic escape of a malaria vaccine candidate. Proc. Natl. Acad. Sci. U. S. A. 104:12488–12493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ouattara A, Takala-Harrison S, Thera MA, Coulibaly D, Niangaly A, Saye R, Tolo Y, Dutta S, Heppner DG, Soisson L, Diggs CL, Vekemans J, Cohen J, Blackwelder WC, Dube T, Laurens MB, Doumbo OK, Plowe CV. 2012. Molecular basis of allele-specific efficacy of a blood-stage malaria vaccine: vaccine development implications. J. Infect. Dis. 207:511–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Drew DR, Hodder AN, Wilson DW, Foley M, Mueller I, Siba PM, Dent AE, Cowman AF, Beeson JG. 2012. Defining the antigenic diversity of Plasmodium falciparum apical membrane antigen 1 and the requirements for a multi-allele vaccine against malaria. PLoS One 7:e51023 doi:10.1371/journal.pone.0051023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Duan J, Mu J, Thera MA, Joy D, Kosakovsky Pond SL, Diemert D, Long C, Zhou H, Miura K, Ouattara A, Dolo A, Doumbo O, Su XZ, Miller L. 2008. Population structure of the genes encoding the polymorphic Plasmodium falciparum apical membrane antigen 1: implications for vaccine design. Proc. Natl. Acad. Sci. U. S. A. 105:7857–7862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kennedy MC, Wang J, Zhang Y, Miles AP, Chitsaz F, Saul A, Long CA, Miller LH, Stowers AW. 2002. In vitro studies with recombinant Plasmodium falciparum apical membrane antigen 1 (AMA1): production and activity of an AMA1 vaccine and generation of a multiallelic response. Infect. Immun. 70:6948–6960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Giersing B, Miura K, Shimp R, Wang J, Zhou H, Orcutt A, Stowers A, Saul A, Miller LH, Long C, Singh S. 2005. Posttranslational modification of recombinant Plasmodium falciparum apical membrane antigen 1: impact on functional immune responses to a malaria vaccine candidate. Infect. Immun. 73:3963–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Plassmeyer ML, Reiter K, Shimp RL, Kotova S, Smith PD, Hurt DE, House B, Zou X, Zhang Y, Hickman M, Uchime O, Herrera R, Nguyen V, Glen J, Lebowitz J, Jin AJ, Miller LH, Macdonald NJ, Wu Y, Narum DL. 2009. The structure of the Plasmodium falciparum circumsporozoite protein, a leading malaria vaccine candidate. J. Biol. Chem. 284:26951–26963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Uchime O, Herrera R, Reiter K, Kotova S, Shimp RL, Jr, Miura K, Jones D, Lebowitz J, Ambroggio X, Hurt DE, Jin AJ, Long C, Miller LH, Narum DL. 2012. Analysis of the conformation and function of the Plasmodium falciparum merozoite proteins MTRAP and PTRAMP. Eukaryot. Cell 11:615–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kocken CH, van der Wel AM, Dubbeld MA, Narum DL, van de Rijke FM, van Gemert GJ, van der Linde X, Bannister LH, Janse C, Waters AP, Thomas AW. 1998. Precise timing of expression of a Plasmodium falciparum-derived transgene in Plasmodium berghei is a critical determinant of subsequent subcellular localization. J. Biol. Chem. 273:15119–15124 [DOI] [PubMed] [Google Scholar]
- 26. Miura K, Orcutt AC, Muratova OV, Miller LH, Saul A, Long CA. 2008. Development and characterization of a standardized ELISA including a reference serum on each plate to detect antibodies induced by experimental malaria vaccines. Vaccine 26:193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miura K, Zhou H, Muratova OV, Orcutt AC, Giersing B, Miller LH, Long CA. 2007. In immunization with Plasmodium falciparum apical membrane antigen 1, the specificity of antibodies depends on the species immunized. Infect. Immun. 75:5827–5836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bai T, Becker M, Gupta A, Strike P, Murphy VJ, Anders RF, Batchelor AH. 2005. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc. Natl. Acad. Sci. U. S. A. 102:12736–12741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Howell SA, Withers-Martinez C, Kocken CH, Thomas AW, Blackman MJ. 2001. Proteolytic processing and primary structure of Plasmodium falciparum apical membrane antigen-1. J. Biol. Chem. 276:31311–31320 [DOI] [PubMed] [Google Scholar]
- 30. Kusi KA, Faber BW, Thomas AW, Remarque EJ. 2009. Humoral immune response to mixed PfAMA1 alleles; multivalent PfAMA1 vaccines induce broad specificity. PLoS One 4:e8110 doi:10.1371/journal.pone.0008110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Remarque EJ, Faber BW, Kocken CH, Thomas AW. 2008. A diversity-covering approach to immunization with Plasmodium falciparum apical membrane antigen 1 induces broader allelic recognition and growth inhibition responses in rabbits. Infect. Immun. 76:2660–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kusi KA, Faber BW, Riasat V, Thomas AW, Kocken CH, Remarque EJ. 2010. Generation of humoral immune responses to multi-allele PfAMA1 vaccines; effect of adjuvant and number of component alleles on the breadth of response. PLoS One 5:e15391 doi:10.1371/journal.pone.0015391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kusi KA, Remarque EJ, Riasat V, Walraven V, Thomas AW, Faber BW, Kocken CH. 2011. Safety and immunogenicity of multi-antigen AMA1-based vaccines formulated with CoVaccine HTTM and Montanide ISA 51 in rhesus macaques. Malar. J. 10:182 doi:10.1186/1475-2875-10-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Coley AM, Gupta A, Murphy VJ, Bai T, Kim H, Anders RF, Foley M, Batchelor AH. 2007. Structure of the malaria antigen AMA1 in complex with a growth-inhibitory antibody. PLoS Pathog. 3:1308–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Crompton PD, Miura K, Traore B, Kayentao K, Ongoiba A, Weiss G, Doumbo S, Doumtabe D, Kone Y, Huang CY, Doumbo OK, Miller LH, Long CA, Pierce SK. 2010. In vitro growth-inhibitory activity and malaria risk in a cohort study in Mali. Infect. Immun. 78:737–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dent AE, Bergmann-Leitner ES, Wilson DW, Tisch DJ, Kimmel R, Vulule J, Sumba PO, Beeson JG, Angov E, Moormann AM, Kazura JW. 2008. Antibody-mediated growth inhibition of Plasmodium falciparum: relationship to age and protection from parasitemia in Kenyan children and adults. PLoS One 3:e3557 doi:10.1371/journal.pone.0003557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Perraut R, Marrama L, Diouf B, Sokhna C, Tall A, Nabeth P, Trape JF, Longacre S, Mercereau-Puijalon O. 2005. Antibodies to the conserved C-terminal domain of the Plasmodium falciparum merozoite surface protein 1 and to the merozoite extract and their relationship with in vitro inhibitory antibodies and protection against clinical malaria in a Senegalese village. J. Infect. Dis. 191:264–271 [DOI] [PubMed] [Google Scholar]
- 38. Dutta S, Sullivan JS, Grady KK, Haynes JD, Komisar J, Batchelor AH, Soisson L, Diggs CL, Heppner DG, Lanar DE, Collins WE, Barnwell JW. 2009. High antibody titer against apical membrane antigen-1 is required to protect against malaria in the Aotus model. PLoS One 4:e8138 doi:10.1371/journal.pone.0008138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Duncan CJ, Sheehy SH, Ewer KJ, Douglas AD, Collins KA, Halstead FD, Elias SC, Lillie PJ, Rausch K, Aebig J, Miura K, Edwards NJ, Poulton ID, Hunt-Cooke A, Porter DW, Thompson FM, Rowland R, Draper SJ, Gilbert SC, Fay MP, Long CA, Zhu D, Wu Y, Martin LB, Anderson CF, Lawrie AM, Hill AV, Ellis RD. 2011. Impact on malaria parasite multiplication rates in infected volunteers of the protein-in-adjuvant vaccine AMA1-C1/Alhydrogel+CPG 7909. PLoS One 6:e22271 doi:10.1371/journal.pone.0022271 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.