Abstract
Campylobacter jejuni is the etiologic agent of human bacterial gastroenteritis worldwide. In contrast, despite heavy colonization, C. jejuni maintains a commensal mode of existence in chickens. The consumption of contaminated chicken products is thought to be the principal mode of C. jejuni transmission to the human population. C. jejuni harbors a system for N-linked protein glycosylation that has been well characterized and modifies more than 60 periplasmic and membrane-bound proteins. However, the precise role of this modification in the biology of C. jejuni remains unexplored. We hypothesized that the N-glycans protect C. jejuni surface proteins from the action of gut proteases. The C. jejuni pglB mutant, deficient in the expression of the oligosaccharyltransferase, exhibited reduced growth in medium supplemented with chicken cecal contents (CCC) compared with that of wild-type (WT) cells. Inactivation of the cecal proteases by heat treatment or with protease inhibitors completely restored bacterial viability and partially rescued bacterial growth. Physiological concentrations of trypsin, but not chymotrypsin, also reduced C. jejuni pglB mutant CFU. Live or dead staining indicated that CCC preferentially influenced C. jejuni growth as opposed to bacterial viability. We identified multiple chicken cecal proteases by mass fingerprinting. The use of protease inhibitors that target specific classes indicated that both metalloproteases and serine proteases were involved in the attenuated growth of the oligosaccharyltransferase mutant. In conclusion, protein N-linked glycosylation of surface proteins may enhance C. jejuni fitness by protecting bacterial proteins from cleavage due to gut proteases.
INTRODUCTION
The zoonotic pathogen Campylobacter jejuni is a major causative agent of human bacterial diarrhea worldwide. Persons with antecedent C. jejuni infections may also develop the severe paralytic neuropathy Guillain-Barré syndrome (1). Despite being pathogenic in humans, these bacteria are typically commensals in other animals, such as chickens. In poultry, C. jejuni displays a propensity for the deep cecal crypts, where it colonizes mucus with avidity and in high numbers (2). The gut, which contains digestive proteases and other innate immune mechanisms against invading pathogens (3), is a harsh environment for a fastidious organism such as C. jejuni. However, these bacteria have evolved mechanisms to evade host defenses and to reproduce (4, 5) and survive in the gastrointestinal tract.
C. jejuni modifies its flagellar proteins with O-linked glycans and modifies numerous periplasmic and membrane proteins with N-linked glycans (6). N-linked protein glycosylation has been demonstrated to occur in all 3 domains of life: in eukaryotes, in bacteria, and in archaea (7). In bacteria, the N-glycosylation pathway of C. jejuni has been extensively characterized (2, 8, 9). This pathway involves the transfer en bloc of a conserved heptasaccharide onto asparagine residues of the extended sequon D/E-X1-N-X2-S/T (where X1 and X2 are any amino acid except proline) on nascent proteins in the periplasmic side of the inner membrane by the oligosaccharyltransferase (OST) PglB (2, 8, 10). More than 60 C. jejuni N-glycoproteins have been identified, with even more predicted to be modified (11).
Protein N-glycosylation influences the pathogenesis of C. jejuni. Mutants with mutations in this pathway display reduced chicken (12) and mouse (13) colonization and a diminished ability to adhere to and invade intestinal epithelial cells in vitro (13). In addition, N-glycosylation-deficient mutants show altered immunoreactivity (9), and the C. jejuni N-glycan is immunomodulatory (14). Furthermore, loss of N-glycosylation results in decreased DNA uptake by the type IV secretion system (11) and also affects iron transport (15). Given the abundance of C. jejuni N-glycoproteins, it is thought that the observed effects on bacterial pathogenicity may be a consequence of pleiotropic effects due to the number of proteins involved. Therefore, the precise role of this modification in the pathophysiology of C. jejuni remains unknown.
In this study, we tested the hypothesis that C. jejuni may have evolved an N-glycosylation pathway in part to enhance bacterial survival and fitness in the gut. We demonstrate the surface localization of some C. jejuni N-glycans. We show that gut proteases attenuate the growth of the C. jejuni oligosaccharyltransferase mutant (pglB) compared with that of the wild type (WT) and that complementation of the pglB mutant with WT pglB fully restored bacterial numbers. Furthermore, we identify chicken cecal proteases and demonstrate that both metalloproteases and serine proteases synergistically mediate the observed reduction in bacterial growth, indicating that C. jejuni N-linked glycosylation may promote bacterial fitness in the gut.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Campylobacter jejuni 11168 is a reference strain that has been sequenced and extensively characterized (16). The C. jejuni 11168 pglB (oligosaccharyltransferase) mutant and the pglB mutant complemented with wild-type pglB have recently been described (17). Bacterial strains were grown for 16 to 18 h under microaerobic conditions on Mueller-Hinton (MH) agar plates. For growth of the pglB mutant, MH agar plates were supplemented with 10 μg/ml kanamycin. The pglB mutant complemented with wild-type pglB was grown on MH agar containing 10 μg/ml kanamycin and 10 μg/ml chloramphenicol.
Cell surface labeling and fluorescence microscopy.
Campylobacter cells were harvested from MH agar with 2 ml MH broth, cooled on ice for 10 min, and centrifuged for 5 min at 6,000 × g. Cells were kept on ice for all further labeling and washing steps using precooled (4°C) solutions. Cells were washed twice with 2 ml washing buffer (50 mM potassium phosphate, 100 mM NaCl) and blocked with 1% skim milk in washing buffer for 30 min. C. jejuni N-glycan-specific (hR6) antiserum (8) served as the primary antibody (1:1,000 dilution in washing buffer with 0.5% skim milk) and was applied for 30 min. Cells were washed 3 times with 2 ml washing buffer before the fluorescently labeled secondary antibody (anti-rabbit IgG–Alexa Fluor 546, diluted 1:100 in washing buffer with 0.5% skim milk) was applied for 30 min. After 4 washing steps with washing buffer, cell surface labeling was monitored using a Leica DMRXA upright microscope at 100-fold magnification equipped with an Optronics MacroFire digital camera (LM-MFCCD). Each image was taken under identical software settings.
Collection of CCC.
Chickens were obtained from the Poultry Research Facility, Department of Agriculture, Food and Nutritional Science, University of Alberta. Chicken cecal contents (CCC) were harvested from 1- and 5-week-old leghorn chickens, which were confirmed to be free of C. jejuni, as described by Waseh et al. (18). Briefly, chicken ceca were excised, and the contents were collected in Eppendorf tubes from euthanized birds. Cecal contents were diluted 1:1 in phosphate-buffered saline (PBS) supplemented with 10% glycerol and stored at −80°C until further use.
Viability and growth of C. jejuni in medium supplemented with CCC.
Wild-type (WT) C. jejuni and its isogenic pglB mutant were harvested from MH plates and diluted to an optical density at 600 nm (OD600) of ∼0.1 in MH broth. MH broth containing bacteria (4 ml) was transferred into 25-cm2 tissue culture flasks and supplemented with ∼1.5 mg of 1-week CCC. CCC proteases were inactivated by heat treatment (95°C for 45 min) or by the addition of Proteoblocker (Fermentas) protease inhibitor cocktail (PIC) (10 μl/100 μl CCC) at 37°C for 30 min prior to supplementation of MH broth containing either WT C. jejuni or the pglB mutant. MH broth containing bacteria but without CCC was used as a control. After 2 h of incubation at 37°C, 10-fold serial dilutions of aliquots in PBS were made prior to plating on Karmali (Oxoid) agar containing Karmali supplement, a Campylobacter selective medium. Bacterial CFU were enumerated after 48 to 72 h.
To investigate the growth of WT C. jejuni and the pglB mutant in medium supplemented with CCC, 100 μl of bacteria at an OD600 of ∼0.1 in MH broth was inoculated into 5 ml fresh MH broth. The bacterium-containing medium (2 ml) was supplemented with 1.5 to 2.5 mg CCC and incubated as described above, and CFU were enumerated at 0, 6, 24, and 30 h postinoculation. To verify whether complementing the C. jejuni 11168 pglB mutant with WT pglB would restore bacterial CFU when coincubated with CCC, separate experiments were carried out with the C. jejuni WT 11168, the pglB mutant, and the complemented pglB mutant as described above for a 24-h period at the higher level of CCC (2.5 mg). To confirm that the observed influence of CCC on C. jejuni was not strain specific, CFU of C. jejuni 81-176 (19) and its pglB mutant (20) were quantified after incubation in MH broth containing 1-week-old bird CCC after plating on Karmali agar containing Karmali supplement. Experiments were also performed with CCC obtained from 5-week-old birds, as described above, over a 24-h period. To investigate a potential influence of the chicken microbiota on the growth of the wild type and the pglB mutant, 5-week CCC in PBS were filtered through a 0.22-μm filter prior to incubation with bacteria for 24 h. Total protein concentrations of CCC in all experiments were determined using the Bradford assay (Bio-Rad).
Estimation of the percentage of live bacteria following treatment with CCC.
The Live/Dead BacLight bacterial viability kit (Molecular Probes) was used to quantify the percentage of live bacteria following treatment with CCC according to the manufacturer's protocol. Live/dead staining of C. jejuni has been previously described (21). C. jejuni 11168 and its pglB mutant were incubated with CCC as described above, with Karmali supplement included in the broth to prevent growth of cecal flora. After the incubation period (2, 6, 24, and 30 h), bacterial CFU were determined as described above and cells from each flask were pelleted by centrifugation at 4,100 × g for 10 min. The pellets were washed twice and resuspended in 5 ml of 0.9% NaCl. Equal proportions of a 2× working solution (100 μl each) of the Live/Dead BacLight staining reagent and the bacterial suspensions were mixed in a 24-well plate and incubated in the dark for 15 min. The fluorescence of live bacteria (green) was measured at excitation and emission wavelengths of 470 nm and 530 nm, respectively, while the fluorescence of dead bacteria (red) was measured with the same excitation wavelength but with an emission wavelength of 650 nm. The blank (2× reagent) readings were subtracted from the fluorescence measurements. Each treatment had 5 samples measured, and the average of both live and dead fluorescence was determined. The percentage of live bacteria in each treatment was calculated using the following formula: % live bacteria = (average live [green] bacterial fluorescence at 530 nm/total live [green] and dead [red] bacterial fluorescence) × 100. To verify the accuracy of the live/dead staining kit, controls were carried out using live bacteria, killed bacteria (treated with 70% isopropanol as recommended by the kit instructions), and a 1:1 mixture of live and dead bacteria. Staining of control bacteria with the live/dead stain and estimation of the percentage of live bacteria showed the expected trend (data not shown).
Effect of trypsin and chymotrypsin on C. jejuni WT and pglB mutant CFU.
Physiological concentrations of trypsin (∼0.9 mg/ml; Bioshop) (22) and chymotrypsin (∼0.5 mg/ml; Bioshop) (23) were incubated with C. jejuni 11168 and its pglB mutant in 40 mM ammonium bicarbonate for 1 h, after which bacterial CFU were determined. As controls, trypsin and chymotrypsin was heat inactivated at 95°C for 10 min.
Degradation of CmeA by CCC.
The model glycoprotein CmeA, which is a component of the C. jejuni CmeABC multidrug efflux pump, was His tagged, heterologously expressed in Escherichia coli, and purified as described previously (24). Unglycosylated CmeA (20 μl) containing 10 μg of CmeA protein was incubated with 1.25 μg total CCC for 1 h at 37°C. The reaction was stopped by the addition of 4 μl of 6× protein loading buffer, and the mixture was incubated at 95°C for 5 min prior to SDS-PAGE analysis. Untreated CmeA served as a control. The proteins were separated on 10% SDS-polyacrylamide gels which were either Coomassie blue stained or electroblotted onto a polyvinylidene difluoride (PVDF) membrane. CmeA proteins were detected using a 1:1,000 dilution of mouse monoclonal anti-His antibody (Santa Cruz) followed by a 1:2,000 dilution of goat anti-mouse secondary antibody conjugated to alkaline phosphatase. All antibody treatments were for 1 h. Membranes were washed between antibody treatments with 1× PBS–Tween 20. Bound antibody was detected with 5-bromo-4-chloro-3-indolyl phosphate–nitroblue tetrazolium (NBT/BCIP) (Bio-Rad) (17).
Identification of chicken cecal proteases by MS.
The chicken cecal protein content was identified by tandem mass spectrometry (MS/MS) data obtained from a mass spectrometer coupled with a nano-ultra-performance liquid chromatography (UPLC) system (25). One- and 5-week-old chicken CCC (200 μl and ∼13 mg/ml total protein) were precipitated with 25% (final concentration) trichloroacetic acid (TCA) for 10 min at 4°C, pelleted by centrifugation, and washed twice with 200 μl ice-cold acetone. The pellet was dried by vacuum centrifugation and resuspended with 200 μl 40 mM ammonium bicarbonate containing 6 M urea and 2 M thiourea. Proteins were reduced with a final concentration of 10 mM dithiothreitol (DTT) and then carbamidomethylated with 25 mM iodoacetamide (in the dark) sequentially for 1 h at room temperature. Reduced and carbamidomethylated proteins were diluted with 1 ml of 40 mM ammonium bicarbonate and then incubated with 20 μg tosyl phenylalanyl chloromethyl ketone (TPCK) trypsin (Bioshop) overnight at 30°C. The sample was acidified with a final concentration of 2% formic acid and pelleted by vacuum centrifugation. Cleanup was carried out with a 2-ml-total-capacity C18 column (Waters) according to the manufacturer's protocol. The peptides were eluted with 60% acetonitrile in 0.1% trifluoroacetic acid (TFA), dried by vacuum centrifugation, reconstituted with 6 μl of 0.25% formic acid, and diluted to 500 μl with 0.1% formic acid. The resultant peptides were subjected to LC-MS/MS analysis on a UPLC system (nanoAcquity UPLC; Waters, Milford, MA) coupled with a q-Tof premier mass spectrometer (Waters, Milford, MA). Five microliters was loaded onto a nanoAcquity UPLC system with a peptide trap (180 µm by 20 mm) (Symmetry C18 nanoAcquity column; Waters, Milford, MA) and an analytical column (75 µm × 150 mm) (Atlantis dC18 nanoAcquity column; Waters, Milford, MA). Desalting on the peptide trap was achieved by flushing the trap with 2% acetonitrile and 0.1% formic acid at a flow rate of 10 µl/min for 3 min. Peptides were separated with a gradient of 2 to 90% solvent B (acetonitrile-0.1% formic acid) over 120 min at a flow rate of 350 nl/min. The obtained MS/MS data were analyzed with the proteomic software Mascot (version 2.2; Matrix Science). Settings for the database search were as follows: parent ion and MS/MS tolerances were set to 0.1 Da and 0.2 Da, respectively; trypsin as enzyme was specified and up to 2 missed cleavages were selected; carbamidomethylation on cysteine was selected as fixed modifications; and oxidation on methionine was selected as variable modification. The confidence of positive protein identification was judged by high protein and peptide scores in the search results. Manual inspection of the original MS/MS spectra was performed to ensure that major peaks in the MS/MS spectra were matched and explained.
The relative abundances of proteins in 1- and 5-week-old chicken CCC (with normalized total protein concentrations) were estimated using the exponentially modified protein abundance index (emPAI), which offers approximate, label-free, relative quantitation of proteins in a mixture based on protein coverage by the peptide matches in a Mascot database search result (26, 27).
Specific-class protease inhibitors and C. jejuni WT and pglB mutant CFU in medium containing chicken CCC.
To determine the class of proteases involved in the attenuated CFU of the C. jejuni 11168 pglB mutant following incubation in MH broth containing 1-week CCC, 1 mM phenylmethylsulfonyl fluoride (PMSF) and 2 mM EDTA were used to inactivate serine proteases and metalloproteases, respectively, in 2.5 mg CCC prior to supplementation of bacterium-containing medium. Protease-inactivated CCC were also used as controls. All samples were incubated with bacteria in MH broth for 24 h, and bacterial CFU were quantified as described above.
Statistics.
Unless otherwise indicated, results are expressed as the mean ± standard deviation (error bars) from three biological replicate experiments. The unpaired Student t test was used to estimate statistical significance. A P value of <0.05 was considered significant.
RESULTS
Some C. jejuni N-linked glycans are surface localized.
Given that in the chicken gut, proteases directly encounter bacterial whole cells, WT C. jejuni and its isogenic pglB mutant were investigated for the presence of surface N-glycans as described above. N-glycans were detected on the surface of C. jejuni WT cells but not pglB mutant cells by fluorescence microscopy (Fig. 1B and D). The absence of fluorescence on the pglB mutant cells was not due to the lack of bacteria but rather was due to the absence of surface N-glycans, as the same fraction of the slide that lacked fluorescence was shown to contain cells by phase-contrast microscopy (Fig. 1A and C). These results indicate that some N-glycans are located on the surface of C. jejuni cells.
Fig 1.
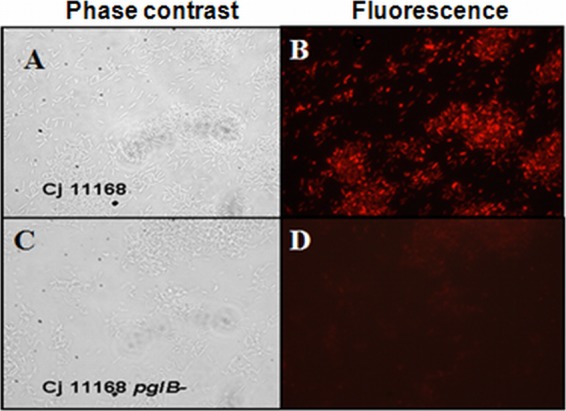
Surface localization of some C. jejuni N-glycans. WT C. jejuni and its isogenic pglB mutant were fixed onto microscope slides and probed with anti-N-glycan antibodies (hR6) (8). Secondary antibodies conjugated to Alexa Fluor 546 (red) were used to reveal bound antibodies prior to fluorescence microscopy. N-linked glycans were detected on the surface of WT C. jejuni (B) but not the pglB mutant (D). Phase-contrast microscopy (A and C) showed the presence of bacterial cells. Magnification, ×100.
N-glycosylation promotes C. jejuni survival in CCC.
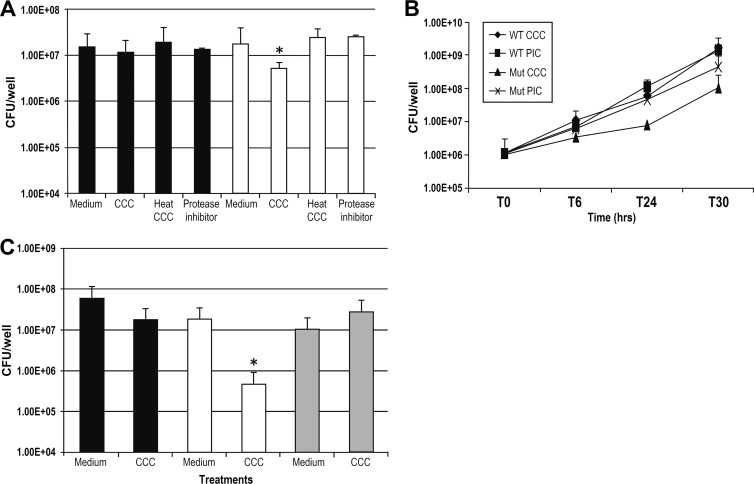
We speculated that one of the reasons why C. jejuni N-glycosylates its surface proteins may be to protect itself from the harsh environment of the gut, which consists of several digestive proteases. C. jejuni resides in the ceca of chickens in high numbers (up to 1010 CFU per gram intestinal contents) (28). Thus, we excised chicken ceca and collected the chicken cecal contents (CCC). The viability and growth of WT C. jejuni and its pglB mutant were investigated in growth medium supplemented with CCC. Following an incubation period of 2 h in CCC, there was an approximately 5-fold reduction in the recovered CFU of the pglB mutant compared with that of the WT (Fig. 2A). This reduction was completely abolished by inactivation of chicken cecal proteases either by heat treatment or with protease inhibitors (Fig. 2A), indicating that the observed effect was due to proteases. Likewise, there was a 10- to 100-fold increase in survival of WT C. jejuni compared with the pglB mutant in medium supplemented with CCC by 24 h of incubation, depending on the CCC concentration used (Fig. 2; see Fig. 6C). This effect was partially rescued by inactivation of proteases with protease inhibitors. Growth of the C. jejuni WT and that of the pglB mutant have previously been shown to be similar in MH broth (20). Complementation of the C. jejuni 11168 pglB mutant with WT pglB restored bacterial CFU, demonstrating that the observed reduced CFU in the presence of CCC by the pglB mutant was due to the lack of protein N-linked glycosylation (Fig. 2C). Taken together, these results indicate that N-glycosylation is beneficial for C. jejuni in the presence of cecal contents, and that this enhancement in growth is at least partially due to protection against chicken cecal gut proteases.
Fig 2.
N-glycosylation promotes increased C. jejuni CFU and bacterial growth in medium supplemented with chicken cecal contents (CCC). (A) The CFU of WT C. jejuni and its pglB mutant were quantified in MH broth supplemented with CCC after 2 h of incubation. CCC were inactivated by heat treatment or with protease inhibitors. *, statistical difference compared with WT CCC (P < 0.005). Black bars, WT C. jejuni; white bars, C. jejuni pglB mutant. (B) Growth of WT C. jejuni and its pglB mutant (Mut) in MH broth containing CCC. A protease inhibitor cocktail (PIC) was used to inactivate chicken gut proteases. (C) The C. jejuni pglB mutant complemented with WT pglB shows CFU similar to those of the WT when incubated with medium comprising CCC. *, statistical difference compared with WT CCC (P < 0.005). Black bars, WT C. jejuni; white bars, C. jejuni pglB mutant; gray bars, C. jejuni pglB mutant complemented with WT pglB.
Fig 6.
Influence of adult CCC and inhibition of specific classes of proteases in CCC on C. jejuni WT and pglB mutant CFU. Asterisks represent statistically significant differences: *, P < 0.05; **, P < 0.0005. (A) Effect of 5-week CCC on C. jejuni WT and pglB mutant CFU. Unlike 1-week CCC, 5-week CCC influenced C. jejuni WT CFU. Means ± standard deviations (error bars) from two independent experiments are shown. **, P < 0.0005 for both groups versus WT only and WT CCC. (B) Filtration of 5-week CCC recovered C. jejuni WT CFU and showed less activity against the C. jejuni pglB mutant than 1-week CCC. *, P < 0.05 compared with WT CCC. Means ± standard deviations (error bars) from two independent experiments are shown. (C) Serine proteases and metalloproteases are involved in the reduced CFU displayed by the pglB mutant in medium containing CCC. Serine proteases and metalloproteases in CCC were inactivated with PMSF and EDTA, respectively, prior to 24 h of incubation in medium containing bacteria. CCC was also either treated with a general protease inhibitor or heat inactivated. **, P < 0.0005 for WT CCC versus pglB mutant CCC. *, P < 0.05 for all other groups compared to WT CCC.
N-glycosylation influences C. jejuni growth compared with bacterial viability.
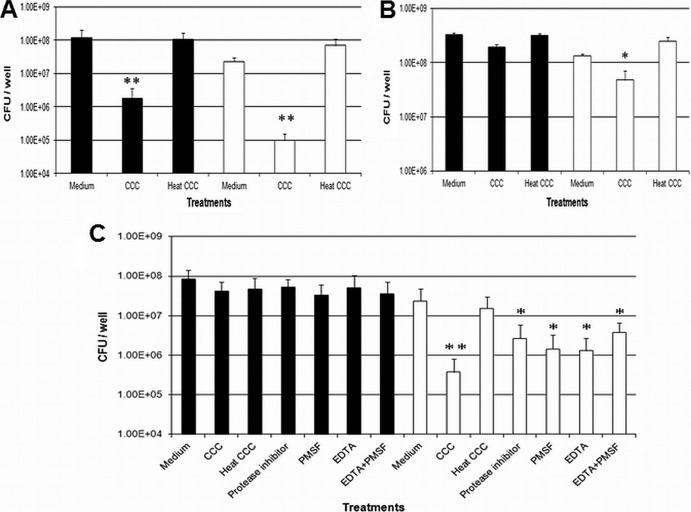
Given the observed reduction in the C. jejuni pglB mutant CFU due to CCC, we wanted to verify whether CCC preferentially influenced bacterial viability or growth. Live/dead staining was employed to estimate the percentage of live bacteria in the growth medium supplemented with CCC over time. Following incubation of WT C. jejuni and its pglB mutant in MH broth with CCC for 24 h, the percentages of live bacteria remained similar, at ∼90% (Fig. 3A), in the presence or absence of CCC in medium containing both WT and pglB mutant cells, despite a significant drop in the CFU of the pglB mutant (Fig. 3B). A similar result was obtained when the percentage of live bacteria was estimated after incubation of bacteria in medium supplemented with CCC for 2, 6, and 30 h (data not shown). These results indicate that bacterial growth or fitness was preferentially influenced by CCC as opposed to bacterial viability and that CCC appear to render pglB mutant cells viable but nonculturable (VBNC) (21).
Fig 3.
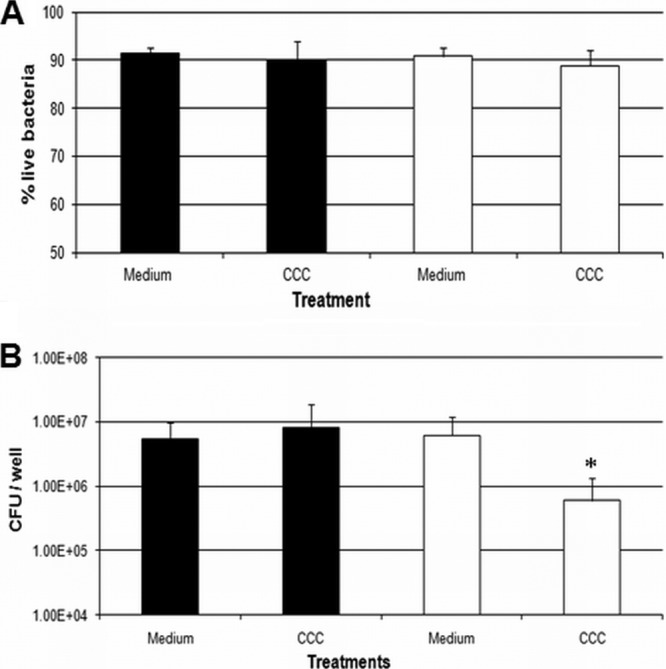
The percentage of live bacteria (A) and bacterial CFU (B) in growth medium supplemented with CCC for 24 h were determined using live/dead staining. *, statistical difference compared with WT CCC (P < 0.05). Black bars, WT C. jejuni; white bars, C. jejuni pglB mutant.
Physiological concentrations of trypsin attenuate C. jejuni 11168 pglB mutant CFU.
We sought to determine whether N-linked glycosylation would be beneficial to C. jejuni when incubated with two common commercially available human gut proteases, trypsin and chymotrypsin. Physiological concentrations of trypsin (23) reduced C. jejuni pglB mutant CFU by approximately 5-fold compared with that of the WT (Fig. 4). Heat inactivation of trypsin partially recovered pglB mutant CFU, highlighting a trypsin-mediated effect. However, physiological concentrations of chymotrypsin (23) did not affect bacterial CFU (data not shown). Bacteria showed diminished survivability in ammonium bicarbonate (data not shown), thus limiting the incubation period with proteases to 1 h. These data indicate that C. jejuni N-linked glycosylation may provide a more general protective effect against some proteases in the niche.
Fig 4.
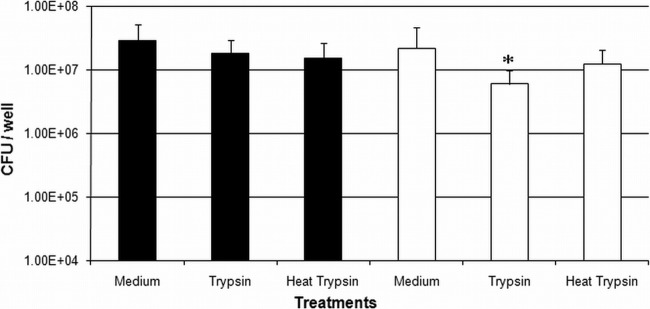
Physiological concentrations of trypsin attenuate C. jejuni pglB mutant CFU after 1 h of incubation. *, a significant difference compared with WT trypsin (P < 0.05). Black bars, WT C. jejuni; white bars, C. jejuni pglB mutant.
Proteolytic activity and identification of chicken cecal proteases.
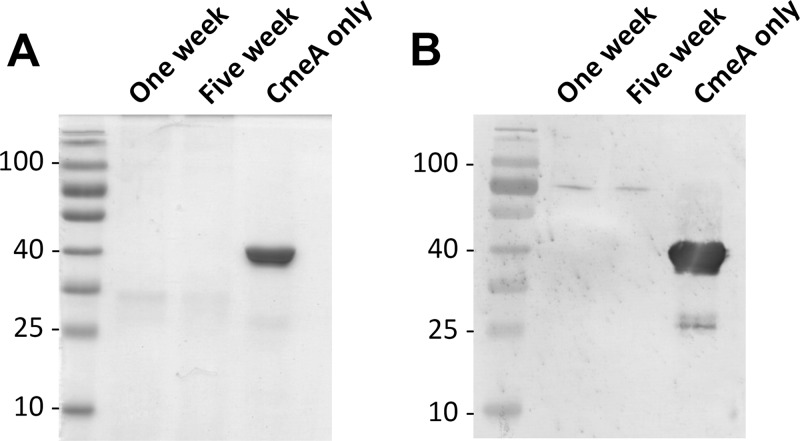
To verify proteolytic activity in the CCC, we incubated CCC with a C. jejuni model glycoprotein, CmeA (29, 30). Immunoblotting demonstrated complete degradation of CmeA by both 1-week and 5-week CCC within 1 h (Fig. 5). We then went on to identify the proteases in the CCC using a proteomics approach. A combination of protein precipitation, trypsin digestion, LC-MS/MS, and Mascot database searches led to the identification of numerous proteins in 1- and 5-week-old CCC, with the increased presence of microorganisms in the adult CCC making up for the increase in protein hits in 5-week CCC (Table 1; see Tables S1 and S2 in the supplemental material). Metalloproteases and serine proteases constituted the main classes of proteases in the CCC. Bacterial, viral, fungal, and parasitic protein products were also identified in the CCC (see Tables S1 and S2 in the supplemental material). These data confirm both the proteolytic activity and the presence of proteases in the CCC.
Fig 5.
Proteolytic activity of CCC. CCC collected from 1- and 5-week-old birds degrade purified C. jejuni His-tagged CmeA after a 1-h incubation period. (A) Coomassie blue staining; (B) immunoblotting with anti-His antibodies. Molecular masses are indicated on the left of each image and shown in kDa. Samples loaded in each lane are indicated. CmeA incubated in buffer without CCC was included as a control.
Table 1.
Identification of chicken cecal proteases by LC/MS/MS
| Protease hits (NCBI accession no.; protein description) | 1 wk |
5 wka |
Trend in relative abundance from 1 wk to 5 wkb | ||
|---|---|---|---|---|---|
| Mascot score | emPAI | Mascot score | emPAI | ||
| gi|118101154; predicted similar to MGC80117 protein (chymotrypsin C) (Gallus gallus)c | 2,719 | 2.81 | 1,143 | 2.37 | Decrease |
| gi|272753691; aminopeptidase N (Gallus gallus) | 2,697 | 2.44 | 1,499 | 1.46 | Decrease |
| gi|50752241; predicted similar to procarboxypeptidase B (Gallus gallus) | 1,554 | 2.11 | 1,032 | 1.67 | Decrease |
| gi|45383013; carboxypeptidase A1 preproprotein (Gallus gallus) | 966 | 1.5 | 531 | 0.84 | Decrease |
| gi|118090349; predicted similar to aminopeptidase A (Gallus gallus) | 738 | 0.55 | 121 | 0.14 | Decrease |
| gi|326911021; predicted carboxypeptidase A2-like (Meleagris gallopavo) | 527 | 0.6 | 135 | 0.24 | Decrease |
| gi|71895185; dipeptidyl peptidase 4 (Gallus gallus) | 442 | 0.52 | 165 | 0.13 | Decrease |
| gi|157817197; chymotrypsin-like elastase family member 2A (Gallus gallus)c | 347 | 1.62 | 756 | 2.75 | Increase |
| gi|25814806; trypsinogen (Gallus gallus)c | 265 | 0.3 | 765 | 1.18 | Increase |
| gi|326927553; dipeptidase 1 (Meleagris gallopavo) | 241 | 0.26 | X | X | NA |
| gi|118096208; predicted similar to chymotrypsinogen A (Gallus gallus)c | 144 | 0.65 | 63 | 0.29 | Decrease |
| gi|118124356; similar to chymotrypsinogen 2 precursor, partial (Gallus gallus)c | 102 | 0.45 | 54 | 0.45 | Equal |
| gi|118089320; predicted similar to aminopeptidase P (Gallus gallus) | 70 | 0.14 | X | X | NA |
| gi|118083765; predicted similar to enteropeptidase (Gallus gallus)c | 65 | 0.07 | X | X | NA |
| gi|119331076; Xaa-Pro dipeptidase (Gallus gallus) | 53 | 0.07 | X | X | NA |
| gi|326924266; predicted Xaa-Pro aminopeptidase 2-like (Meleagris gallopavo) | 51 | 0.05 | X | X | NA |
X, undetected.
NA, not applicable.
Serine proteases and remaining enzymes are metalloproteases.
Adult CCC are attenuated in activity compared with 1-week-old CCC.
Similar concentrations of 5-week-old CCC and 1-week-old CCC were coincubated with WT C. jejuni and its pglB mutant to verify the effect of adult CCC on bacterial CFU. Adult CCC reduced C. jejuni pglB mutant numbers by only 20-fold, compared with 100-fold when 2.5 mg of 1-week CCC was used (Fig. 2C and 6C versus 6A). In addition, there was a 10-fold reduction in the CFU of WT C. jejuni in the presence of 5-week-old CCC, which was not observed with 1-week-old CCC. This decrease in WT CFU likely correlated with the increased diversity of microbial species in the adult CCC compared with 1-week-old CCC as determined by tryptic mass fingerprinting (see Tables S1 and S2 in the supplemental material). We thus speculated that microbes in the adult CCC were responsible for the attenuated CFU of WT C. jejuni when exposed to adult CCC. Five-week CCC were filtered to remove microbes prior to incubation with WT C. jejuni and its pglB mutant. Following filtration of CCC and removal of microbial flora in the adult CCC, only the growth of the pglB mutant was affected by the CCC, and there was no significant effect on the WT (Fig. 6A and B). These results demonstrate that adult CCC microflora influenced C. jejuni WT CFU compared with 1-week CCC. In addition, CCC from younger chickens appear to be more active than CCC from older birds, mediating 100- and 4-fold reductions, respectively, in the C. jejuni pglB mutant numbers in comparison to the WT. Furthermore, despite having lower protease activity than 1-week CCC, 5-week CCC still completely degraded CmeA after a 1-h incubation period (Fig. 5).
We hypothesized that differences in the composition and relative abundance of proteases in 1-week and 5-week CCC may account for the diminished proteolytic activity in the adult CCC. We used Mascot-generated emPAI values, previously described and used by Ishihama et al. (26, 27), to estimate the relative protein abundances in 1-week and 5-week CCC (Table 1). With the exception of a few proteases such as trypsinogen and chymotrypsin-like elastase, emPAI values were generally higher in 1-week CCC than in adult CCC, suggesting a general reduction in the abundance of proteases in the 5-week CCC. Additionally, chymotrypsinogen 2 precursor appeared to be equally abundant in young and adult CCC, with an emPAI score of 0.45 each. There was also an increased diversity of proteases in 1-week compared with 5-week CCC, as evidenced by proteases such as aminopeptidase P, enteropeptidase, and Xaa-Pro dipeptidase, which were undetected in older CCC (Table 1). These data suggest a differential protease composition between 1-week and adult CCC, since protein concentrations of 1-week and 5-week CCC were normalized prior to mass spectrometry. Therefore, differences in protease composition and abundance may at least partially account for the reduced activity of adult CCC on the C. jejuni pglB mutant in comparison to 1-week CCC.
Serine proteases and metalloproteases in 1-week CCC are involved in the reduction of the C. jejuni pglB mutant CFU.
We determined that the main classes of proteases found in the CCC are metalloproteases and serine proteases (Table 1). Serine proteases and metalloproteases in the CCC were inactivated with PMSF and EDTA, respectively, and protease-inactivated CCC were coincubated with WT C. jejuni and its pglB mutant in growth medium. A 100-fold reduction in the pglB mutant CFU in medium supplemented with CCC compared with that of the WT was fully or partially restored by heat treatment or general protease inhibitor cocktail (PIC), respectively (Fig. 6C), consistent with results shown in Fig. 2A and B. Both PMSF and EDTA partially rescued the pglB mutant CFU at levels that were approximately half the CFU numbers (∼1.5 × 106 CFU/well) in samples treated with general PIC. Intriguingly, pretreatment of the CCC with a combination of both PMSF and EDTA recovered pglB mutant CFU at levels (∼3 × 106 CFU/well) similar to those for the general PIC-treated samples (Fig. 6C). Taken together, these data indicate that both serine proteases and metalloproteases in the CCC synergistically influence the C. jejuni pglB mutant CFU compared with the WT.
DISCUSSION
The general role for N-glycosylation in C. jejuni is not known and has been the subject of research since the discovery of this pathway in bacteria (9). The vast array of proteins modified (11) and the inability of C. jejuni N-glycosylation mutants to colonize chickens led us to the hypothesis that this modification may be important for bacterial survival and persistence in the niche. Thus, we explored the possibility that among other functions of the C. jejuni N-glycosylation pathway, surface N-glycans may also provide C. jejuni with a distinctive advantage by protecting it from the harsh environment of the gut. In this study, we demonstrated that N-glycosylation enhances C. jejuni growth. We showed proteolytic activity in CCC and identified several cecal serine proteases and metalloproteases by mass spectrometry. In addition, both classes of proteases are involved in the attenuated numbers of the C. jejuni pglB mutant compared with the WT when grown in the presence of CCC.
Intriguingly, despite a reduction in the C. jejuni pglB mutant CFU compared with WT cells recovered postincubation with CCC, quantification of the percentage of live bacteria indicated that similar amounts of live WT and pglB mutant cells were present in the growth medium from 2 to 30 h following incubation with CCC. Indeed, it has been shown that C. jejuni can switch into a “viable but nonculturable” (VBNC) state in response to stress (21, 31). In recent studies (21, 32, 33), C. jejuni has been demonstrated to alter its metabolism in response to stress, leading to reduced culturability. In one such study (32), the C. jejuni genes fdhT and fdhU (encoding the proteins FdhT and FdhU of unknown function) were identified as crucial for the ability to recover these bacteria from standard growth media. Clearly, then, C. jejuni alters its metabolism under different environmental conditions to sustain its pathogenic and commensal modes of existence (32, 33). It is possible that cecal proteases may have triggered bacterial switch to a VBNC state, a possible explanation for the reduced pglB mutant CFU recovered despite similar levels of live WT and pglB mutant cells. Therefore, in this study, CCC may have preferentially influenced bacterial growth.
An effect comparable to that observed with CCC on the C. jejuni pglB mutant CFU was also observed with commercially available trypsin but not chymotrypsin. Physiologically relevant concentrations of trypsin normally found in the human gut (22) significantly reduced pglB mutant CFU after a short incubation period (Fig. 4). Even though this effect was specific to trypsin but not chymotrypsin, combined with the activity elicited by diverse chicken cecal proteases on the C. jejuni pglB mutant, it is possible that N-glycosylation may more generally protect C. jejuni surface proteins from the action of proteases in the niche.
The protein content and identity of proteases in the chicken cecum remain uncharacterized. Trypsin-like, chymotrypsin-like, and aminopeptidase activities have been demonstrated in the chicken gut (34, 35); however, chicken cecal proteases are largely unknown. Increased knowledge of the cecal protein content has a potential to improve our understanding of how both pathogenic and commensal microbes interact within this niche and can lead to intervention strategies aimed at lowering the burden of human diseases transmitted through the food chain. We used LC-MS/MS to identify chicken cecal proteases among the chicken and microbial protein products. These proteases were active, since they completely degraded purified C. jejuni CmeA within 1 h. Interestingly, 1-week-old chicken ceca appear to harbor less diversity in microbial products but more individual proteases than 5-week-old birds. Metalloproteases and serine proteases constitute the main classes of proteases in CCC. We observed that specific inhibitors of proteases EDTA and PMSF partially rescued the pglB mutant CFU, and a combination of the two inhibitors recovered pglB mutant numbers to levels comparable to those with the more general protease inhibitor cocktail, strongly suggesting that both serine proteases and metalloproteases are involved in the attenuated CFU of the C. jejuni pglB mutant compared with the WT in the presence of CCC.
Our results are consistent with a recent study that identified the proteases aminopeptidase, procarboxipeptidase, and trypsin in chicken ceca (36). In addition, nonprotease components of the chicken cecum, such as superoxide dismutase, observed in this study (see Table S1 in the supplemental material) were also identified by Gielda and Dirita (36). Our data further corroborate another recent study that showed dipeptidyl peptidase 4 and aminopeptidase A at the transcript level in chicken cecal samples (37). Interestingly, in the latter study, there was increased gene expression of the identified proteases in the ceca of birds resistant to C. jejuni colonization compared to chickens that were susceptible to bacterial colonization. Furthermore, the data presented here are consistent with the severely attenuated levels of colonization displayed by the C. jejuni pglB mutant and other C. jejuni N-glycosylation-deficient mutants in chickens (38, 39). These results point to the fact that chicken cecal proteases may influence C. jejuni colonization.
We found that adult cecal microbiota may also affect C. jejuni WT CFU, consistent with recent reports showing antagonistic effects of adult chicken cecal bacteria on C. jejuni colonization (40–42). Elimination of microbial flora in 5-week CCC by filtration completely restored WT bacterial numbers. This effect was absent in 1-week CCC and correlated with increased diversity of microbial species in 5-week CCC compared with 1-week CCC as demonstrated by LC-MS/MS. Additionally, while ceca from 1-week old chickens reduced C. jejuni pglB mutant CFU by two log units, adult CCC were less active, eliciting only a 4-fold drop. Furthermore, despite a diminished effect of 5-week CCC versus 1-week CCC on C. jejuni pglB mutant CFU, adult CCC retained the ability to fully degrade C. jejuni purified CmeA, possibly due to the high concentration of CCC used in the degradation experiments.
Due to the attenuated effect of adult CCC as opposed to 1-week CCC on C. jejuni pglB mutant numbers, we compared the relative abundance of normalized 1-week and 5-week CCC using previously described Mascot emPAI scores (26, 27). In agreement with the observed diminished activity of 5-week CCC on C. jejuni pglB mutant CFU, with the exception of trypsinogen and elastase showing higher emPAI scores in adult CCC, there was a general decrease in the relative abundance of 5-week cecal proteases compared with 1-week proteases (Table 1). Earlier studies also indicate that the specific activity of chicken intestinal enzymes generally decreases with increased age (35), and this trend has also been described in turkeys (43) and rats (44). Therefore, despite the differences in composition and relative abundance between 1-week and 5-week CCC shown in this study, it is also possible that the attenuated effect of adult CCC compared with CCC from younger birds on the C. jejuni pglB mutant CFU may be due to a reduced activity of 5-week proteases (35, 43, 44).
In previous studies, O-linked glycosylation has been shown to protect proteins and peptides from proteolytic cleavage (45, 46). Gross et al. developed an in vitro protease protection assay as a strategy to discover inhibitors of the O-linked glycosylation pathway, based on the fact that O-linked glycosylation protected peptides from proteolytic cleavage by masking protease cleavage sites (45). Furthermore, recent data from our laboratory demonstrate that N-glycosylation of C. jejuni CmeA in E. coli protects this model glycoprotein from proteolysis by the E. coli protease DegP compared with unglycosylated CmeA (R. Dubb et al., unpublished data). Thus, it is likely that C. jejuni surface N-glycans act in a similar manner, protecting proteins from the action of gut proteases by limiting the accessibility to proteolytic cleavage sites. We speculate that differentially cleaved C. jejuni proteins may be involved in bacterial stress survival and/or nutrient acquisition or other physiological process affecting bacterial fitness in the niche.
In summary, we have investigated the role of surface N-glycans in the growth and survival of C. jejuni ex vivo. N-glycosylation is beneficial for C. jejuni growth and culturability in the chicken gut. This enhancement in bacterial fitness is at least partially due to protection against proteases. We have identified chicken cecal proteases and showed that serine proteases and metalloproteases in the CCC act jointly to diminish the CFU of an N-linked-glycosylation-deficient mutant. C. jejuni may maintain an N-glycosylation pathway in part to protect itself from the harsh environment that it inhabits.
Supplementary Material
ACKNOWLEDGMENTS
We thank Markus Aebi for hR6 antibodies. We thank Yasmin Barre for CmeA expression and purification.
This work was funded by an Alberta Innovates Technology Futures scholar award to C.M.S.
Footnotes
Published ahead of print 4 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01370-12.
REFERENCES
- 1. Janssen R, Krogfelt KA, Cawthraw SA, van Pelt W, Wagenaar JA, Owen RJ. 2008. Host-pathogen interactions in Campylobacter infections: the host perspective. Clin. Microbiol. Rev. 21:505–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Young KT, Davis LM, Dirita VJ. 2007. Campylobacter jejuni: molecular biology and pathogenesis. Nat. Rev. Microbiol. 5:665–679 [DOI] [PubMed] [Google Scholar]
- 3. Linden SK, Sutton P, Karlsson NG, Korolik V, McGuckin MA. 2008. Mucins in the mucosal barrier to infection. Mucosal Immunol. 1:183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alemka A, Clyne M, Shanahan F, Tompkins T, Corcionivoschi N, Bourke B. 2010. Probiotic colonization of the adherent mucus layer of HT29MTXE12 cells attenuates Campylobacter jejuni virulence properties. Infect. Immun. 78:2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Van Deun K, Pasmans F, Ducatelle R, Flahou B, Vissenberg K, Martel A, Van den Broeck W, Van Immerseel F, Haesebrouck F. 2008. Colonization strategy of Campylobacter jejuni results in persistent infection of the chicken gut. Vet. Microbiol. 130:285–297 [DOI] [PubMed] [Google Scholar]
- 6. Szymanski CM, Wren BW. 2005. Protein glycosylation in bacterial mucosal pathogens. Nat. Rev. Microbiol. 3:225–237 [DOI] [PubMed] [Google Scholar]
- 7. Nothaft H, Szymanski CM. 2011. Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8:765–778 [DOI] [PubMed] [Google Scholar]
- 8. Schwarz F, Lizak C, Fan YY, Fleurkens S, Kowarik M, Aebi M. 2011. Relaxed acceptor site specificity of bacterial oligosaccharyltransferase in vivo. Glycobiology 21:45–54 [DOI] [PubMed] [Google Scholar]
- 9. Szymanski CM, Yao R, Ewing CP, Trust TJ, Guerry P. 1999. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32:1022–1030 [DOI] [PubMed] [Google Scholar]
- 10. Kelly J, Jarrell H, Millar L, Tessier L, Fiori LM, Lau PC, Allan B, Szymanski CM. 2006. Biosynthesis of the N-linked glycan in Campylobacter jejuni and addition onto protein through block transfer. J. Bacteriol. 188:2427–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scott NE, Parker BL, Connolly AM, Paulech J, Edwards AV, Crossett B, Falconer L, Kolarich D, Djordjevic SP, Hojrup P, Packer NH, Larsen MR, Cordwell SJ. 2011. Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell Proteomics 10:M000031–MCP000201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hendrixson DR, DiRita VJ. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52:471–484 [DOI] [PubMed] [Google Scholar]
- 13. Szymanski CM, Burr DH, Guerry P. 2002. Campylobacter protein glycosylation affects host cell interactions. Infect. Immun. 70:2242–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Sorge NM, Bleumink NM, van Vliet SJ, Saeland E, van der Pol WL, van Kooyk Y, van Putten JP. 2009. N-glycosylated proteins and distinct lipooligosaccharide glycoforms of Campylobacter jejuni target the human C-type lectin receptor MGL. Cell. Microbiol. 11:1768–1781 [DOI] [PubMed] [Google Scholar]
- 15. Palyada K, Threadgill D, Stintzi A. 2004. Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186:4714–4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, Barrell BG. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668 [DOI] [PubMed] [Google Scholar]
- 17. Scott NE, Nothaft H, Edwards AV, Labbate M, Djordjevic SP, Larsen MR, Szymanski CM, Cordwell SJ. 2012. Modification of the Campylobacter jejuni N-linked glycan by EptC protein-mediated addition of phosphoethanolamine. J. Biol. Chem. 287:29384–29396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Waseh S, Hanifi-Moghaddam P, Coleman R, Masotti M, Ryan S, Foss M, MacKenzie R, Henry M, Szymanski CM, Tanha J. 2010. Orally administered P22 phage tailspike protein reduces salmonella colonization in chickens: prospects of a novel therapy against bacterial infections. PLoS One 5:e13904 doi:10.1371/journal.pone.0013904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Korlath JA, Osterholm MT, Judy LA, Forfang JC, Robinson RA. 1985. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152:592–596 [DOI] [PubMed] [Google Scholar]
- 20. Nothaft H, Liu X, McNally DJ, Li J, Szymanski CM. 2009. Study of free oligosaccharides derived from the bacterial N-glycosylation pathway. Proc. Natl. Acad. Sci. U. S. A. 106:15019–15024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Watson RO, Galan JE. 2008. Campylobacter jejuni survives within epithelial cells by avoiding delivery to lysosomes. PLoS Pathog. 4:e14 doi:10.1371/journal.ppat.0040014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lake-Bakaar G, McKavanagh S, Rubio CE, Epstein O, Summerfield JA. 1980. Measurement of trypsin in duodenal juice by radioimmunoassay. Gut 21:402–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kahns AH, Buur A, Bundgaard H. 1993. Prodrugs of peptides. 18. Synthesis and evaluation of various esters of desmopressin (dDAVP). Pharm. Res. 10:68–74 [DOI] [PubMed] [Google Scholar]
- 24. Feldman MF, Wacker M, Hernandez M, Hitchen PG, Marolda CL, Kowarik M, Morris HR, Dell A, Valvano MA, Aebi M. 2005. Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 102:3016–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schilling O, Huesgen PF, Barre O, Auf dem Keller U, Overall CM. 2011. Characterization of the prime and non-prime active site specificities of proteases by proteome-derived peptide libraries and tandem mass spectrometry. Nat. Protoc. 6:111–120 [DOI] [PubMed] [Google Scholar]
- 26. Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. 2005. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell Proteomics 4:1265–1272 [DOI] [PubMed] [Google Scholar]
- 27. Shinoda K, Tomita M, Ishihama Y. 2010. emPAI Calc—for the estimation of protein abundance from large-scale identification data by liquid chromatography-tandem mass spectrometry. Bioinformatics 26:576–577 [DOI] [PubMed] [Google Scholar]
- 28. Alemka A, Whelan S, Gough R, Clyne M, Gallagher ME, Carrington SD, Bourke B. 2010. Purified chicken intestinal mucin attenuates Campylobacter jejuni pathogenicity in vitro. J. Med. Microbiol. 59:898–903 [DOI] [PubMed] [Google Scholar]
- 29. Hogan SP, Seidu L, Blanchard C, Groschwitz K, Mishra A, Karow ML, Ahrens R, Artis D, Murphy AJ, Valenzuela DM, Yancopoulos GD, Rothenberg ME. 2006. Resistin-like molecule beta regulates innate colonic function: barrier integrity and inflammation susceptibility. J. Allergy Clin. Immunol. 118:257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wacker M, Linton D, Hitchen PG, Nita-Lazar M, Haslam SM, North SJ, Panico M, Morris HR, Dell A, Wren BW, Aebi M. 2002. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298:1790–1793 [DOI] [PubMed] [Google Scholar]
- 31. Jackson DN, Davis B, Tirado SM, Duggal M, van Frankenhuyzen JK, Deaville D, Wijesinghe MA, Tessaro M, Trevors JT. 2009. Survival mechanisms and culturability of Campylobacter jejuni under stress conditions. Antonie Van Leeuwenhoek 96:377–394 [DOI] [PubMed] [Google Scholar]
- 32. Pryjma M, Apel D, Huynh S, Parker CT, Gaynor EC. 2012. FdhTU-modulated formate dehydrogenase expression and electron donor availability enhance recovery of Campylobacter jejuni following host cell infection. J. Bacteriol. 194:3803–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu X, Gao B, Novik V, Galan JE. 2012. Quantitative proteomics of intracellular Campylobacter jejuni reveals metabolic reprogramming. PLoS Pathog. 8:e1002562 doi:10.1371/journal.ppat.1002562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kadhim K, Zuki ABZ, Noordin MM, Babjee SMA, Zamri-Saad M. 2011. Activities of amylase, trypsin and chymotrypsin of pancreas and small intestinal contents in the red jungle fowl and broiler breed. Afr. J. Biotechnol. 10:108–115 [Google Scholar]
- 35. Iji PA, Saki A, Tivey DR. 2001. Body and intestinal growth of broiler chicks on a commercial starter diet. 3. Development and characteristics of tryptophan transport. Br. Poult. Sci. 42:523–529 [DOI] [PubMed] [Google Scholar]
- 36. Gielda LM, Dirita VJ. 2012. Zinc competition among the intestinal microbiota. mBio 3(4):e00171–12 doi:10.1128/mBio.00171-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Connell S, Meade KG, Allan B, Lloyd AT, Kenny E, Cormican P, Morris DW, Bradley DG, O'Farrelly C. 2012. Avian resistance to Campylobacter jejuni colonization is associated with an intestinal immunogene expression signature identified by mRNA sequencing. PLoS One 7:e40409 doi:10.1371/journal.pone.0040409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jones MA, Marston KL, Woodall CA, Maskell DJ, Linton D, Karlyshev AV, Dorrell N, Wren BW, Barrow PA. 2004. Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infect. Immun. 72:3769–3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Karlyshev AV, Everest P, Linton D, Cawthraw S, Newell DG, Wren BW. 2004. The Campylobacter jejuni general glycosylation system is important for attachment to human epithelial cells and in the colonization of chicks. Microbiology 150:1957–1964 [DOI] [PubMed] [Google Scholar]
- 40. Ghareeb K, Awad WA, Mohnl M, Porta R, Biarnes M, Bohm J, Schatzmayr G. 2012. Evaluating the efficacy of an avian-specific probiotic to reduce the colonization of Campylobacter jejuni in broiler chickens. Poult. Sci. 91:1825–1832 [DOI] [PubMed] [Google Scholar]
- 41. Kergourlay G, Messaoudi S, Dousset X, Prevost H. 2012. Genome sequence of Lactobacillus salivarius SMXD51, a potential probiotic strain isolated from chicken cecum, showing anti-campylobacter activity. J. Bacteriol. 194:3008–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang G, Ma L, Doyle MP. 2007. Potential competitive exclusion bacteria from poultry inhibitory to Campylobacter jejuni and Salmonella. J. Food Prot. 70:867–873 [DOI] [PubMed] [Google Scholar]
- 43. Sell JL, Angel CR, Piquer FJ, Mallarino EG, al-Batshan HA. 1991. Developmental patterns of selected characteristics of the gastrointestinal tract of young turkeys. Poult. Sci. 70:1200–1205 [DOI] [PubMed] [Google Scholar]
- 44. Holt PR, Tierney AR, Kotler DP. 1985. Delayed enzyme expression: a defect of aging rat gut. Gastroenterology 89:1026–1034 [DOI] [PubMed] [Google Scholar]
- 45. Gross BJ, Swoboda JG, Walker S. 2008. A strategy to discover inhibitors of O-linked glycosylation. J. Am. Chem. Soc. 130:440–441 [DOI] [PubMed] [Google Scholar]
- 46. Han I, Kudlow JE. 1997. Reduced O glycosylation of Sp1 is associated with increased proteasome susceptibility. Mol. Cell. Biol. 17:2550–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.