Abstract
A panel of commensal bacteria was screened for the ability to interact with galectin-3. Two strains of Bifidobacterium longum subsp. infantis interacted to a greater extent than did the pathogenic positive control, Escherichia coli NCTC 12900. Further validation of the interaction was achieved by using agglutination and solid-phase binding assays.
TEXT
Galectins are a family of evolutionarily conserved proteins found across a variety of species ranging from lower invertebrates to mammals (1). These lectins are involved in a wide range of biological processes, including tumor cell adhesion and progression, inflammation, wound healing, development, and immunity (2–4). A number of natural ligands of galectins have been identified. These include galactose, lactose, polylactosamine, and N-acetyllactosamine (LacNAc) (1). Recently, galectins have also been shown to bind to blood group antigens expressed on the surface of bacterial cells (5).
Galectin-3 (gal-3) is a 31-kDa chimera-type lectin containing a C-terminal carbohydrate recognition domain (CRD) possessing an affinity for β-galactoside residues (6). Gal-3 is one of the most ubiquitously expressed members of the galectin family, demonstrating elevated expression levels in the epithelial cells of the digestive tract (7, 8). Within the intestinal tract, gal-3 is detected predominantly in the villus tips (9) and interacts with MUC2, a secreted mucin found in the intestine (10). This multifunctional molecule exhibits a range of functions, at times opposing one another, depending on cell localization (11). Among these functions, gal-3 is known to interact with pathogenic microorganisms. Gal-3 was first demonstrated to associate with the lipopolysaccharide (LPS) of Klebsiella pneumoniae through the binding of β-galactoside glycans in the outer core. In addition, interactions with Escherichia coli (LPS) (12), Pseudomonas aeruginosa (outer core of LPS) (13), Neisseria gonorrhoeae (lipooligosaccharides) (14), and Helicobacter pylori (O antigen of LPS) (15) have been described. In a recent paper, Fermino et al. (12) suggested that gal-3 may have a broader specificity than LPS. Indeed, gal-3 has been shown to bind mycolic acid, the major constituent of the Mycobacterium tuberculosis cell envelope (16). Additionally, Salmonella minnesota LPS is devoid of β-galactosides but can bind to a site within the N-terminal domain of gal-3 through its lipid A portion (17). The ability of gal-3 to bind two hydrophobic ligands, such as mycolic acid and lipid A, opens the possibility that gal-3 may also interact with lipoteichoic acid, a major constituent of the cell wall of Gram-positive bacteria (18).
Many negative effects of the gal-3–pathogen interaction have been identified. Pathogenic interaction with gal-3 may result in suppressed or exaggerated states of endotoxic shock (12, 19) or increased adhesion to host tissues (20). These studies highlight the ability of pathogens to capitalize on the presence of gal-3 to augment their capacity to colonize and survive within the host environment. Despite numerous examples of pathogen interactions with gal-3, there is very little evidence to suggest that commensal bacteria interact with and influence the activity of this protein.
In this study, we demonstrated, by using an indirect surface plasmon resonance (SPR)-based approach, the ability of commensal bacteria to interact with gal-3. The resulting interaction was further validated by using agglutination and solid-phase binding assays. Of six commensal strains examined, two strains of bifidobacteria were identified that interacted strongly with gal-3. The ability of commensal strains to interact with gal-3 may indicate an ability to exclude and displace pathogens, which has major implications for gut health.
Bacterial interaction with gal-3 was assessed indirectly by assaying various commensal strains for their respective abilities to inhibit the well-defined interaction between gal-3 and asialofetuin (ASF) by using an SPR-based biosensor system as outlined by Maljaars et al. (21) (Biacore X100 system, solutions, and reagents; GE Healthcare UK Ltd., Buckinghamshire, United Kingdom). The experimental design is summarized in Fig. 1. SPR is a microfluidics-based methodology used to investigate the real-time interaction between two or more unlabeled molecules under flow conditions. Interactions between two molecules are measured as a change in mass on the chip surface, detected as a change in the reflected pathway of a laser beam directed at the gold chip surface. An indirect approach was used because of the functional nature of gal-3. As oligomerization is often required for full activity, immobilization would prevent a representative biological response. A carboxymethyl dextran-coated (CM5) chip was prepared in accordance with the manufacturer's instructions. Briefly, flow cell 1 was prepared by the injection of a 1:1 mixture of 0.05 M N-hydroxysuccinimide (NHS) and 0.2 M 1-ethyl-3-(3-dimethylpropyl)-carbodiimide (EDC) at a flow rate of 10 μl/min, and ASF was immobilized (10 μl, 0.2 mg/ml in 10 mM sodium acetate buffer, pH 4.5; flow rate, 5 μl/min) to a level of ∼11,000 resonance units (RU). The remaining NHS esters were blocked by the injection of a 1 M ethanolamine hydrochloride solution (70 μl, pH 8.5; flow rate, 10 μl/min). Flow cell 2 was activated with NHS-EDC and then blocked with ethanolamine to create the reference flow cell to control for nonspecific binding to the carboxymethyl dextran-coated surface. A stock concentration of 0.22 mg/ml of human recombinant gal-3 in HBS-EP buffer (0.01 M HEPES [pH 7.4], 0.15 M NaCl, 3 mM EDTA, 0.005% [vol/vol] Surfactant P20) was used for all injections following preliminary optimization experiments. Gal-3 stock solution (10 μl) was incubated with 30 μl (optical density at 600 nm of 1.0) of a bacterial suspension in HBS-EP buffer for 3 min prior to each injection. The mixture was injected over the sensor chip surface for 2 min. Results were calculated on the basis of the difference between the number of RU immediately before injection and that following the postinjection washing. Five measurements were taken, commencing with completion of the postinjection washing and every 20 s thereafter for 80 s. The measurements were averaged to calculate the response value for each injection. The surfaces were then regenerated with 0.1 M lactose (a well-characterized ligand of gal-3) in HBS-EP (10 μl) for 2 min. All measurements were performed at a flow rate of 5 μl/min. Each injection was repeated in triplicate for each bacterial strain tested, and the average of the three results is reported. Mean responses were compared against those obtained with the positive control, E. coli NCTC 12900, by using Student's t test, and significance was defined as P < 0.05.
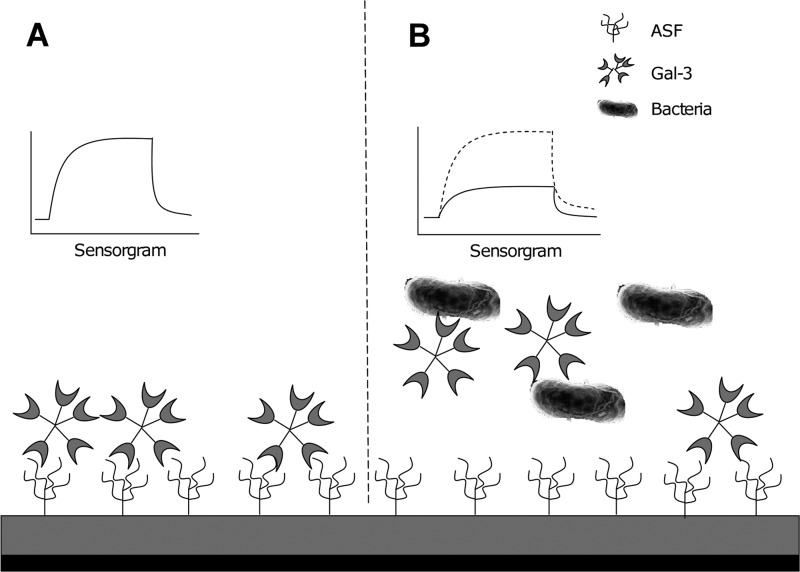
Fig 1.
Experimental overview of SPR screening study. (A) Gal-3 interacts with ASF on the chip surface. (B) Preincubation of gal-3 with bacteria decreases the gal-3 interaction with ASF, as indicated by a reduced resonance response.
Bifidobacterium longum subsp. infantis ATCC 15702 and ATCC 15697 were found to be capable of significantly inhibiting the interaction between gal-3 and ASF (Fig. 2). Indeed, these strains inhibited the interaction to a greater extent than did the positive control, E. coli O157:H7 NCTC 12900. B. longum subsp. infantis ATCC 15702 and ATCC 15697 demonstrated 93.1% (P < 0.0001) and 73.1% (P < 0.0001) inhibition of the ASF–gal-3 interaction, respectively. The remaining commensal strains displayed intermediate inhibition of the gal-3 interaction with ASF, ranging between 36 and 44%. The positive pathogen control used in the study exhibited 54% inhibition (Fig. 2). To rule out the possibility of bacterial interference with the chip surface, the SPR experiments were replicated with the two strains of bifidobacteria, as performed previously, followed by a centrifugation step (8,000 × g for 5 min) to remove bacteria prior to injection. The results indicate that the bacteria did not actively or passively interfere with the gal-3–ASF interaction at the chip surface (Fig. 3).
Fig 2.
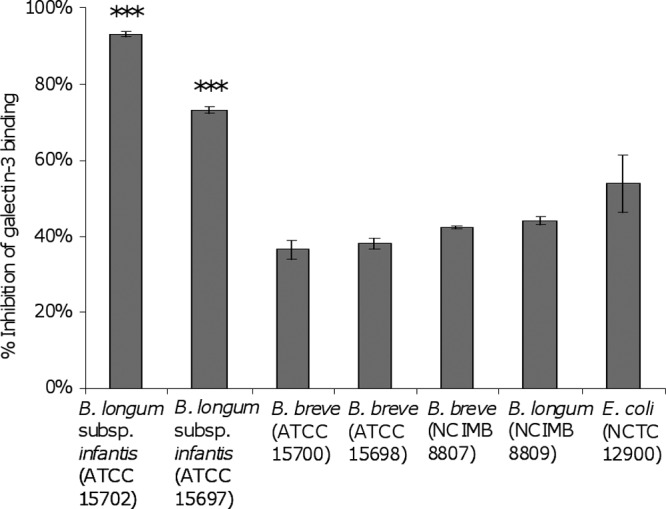
Inhibition of gal-3 interaction with immobilized ASF by a panel of commensal bacteria by an SPR biosensor approach. E. coli NCTC 12900 was included as a positive control. Inhibition is defined as the ability of bacteria to interact with gal-3, thereby reducing its ability to generate a surface plasmon response through interaction with the ASF chip surface. Gal-3 injected over the ASF surface in the absence of bacteria was used to define the 0% inhibition SPR response. Experiments were carried out in triplicate (n = 3), and the results are presented as mean inhibition ± the standard deviation. ***, P < 0.0001.
Fig 3.
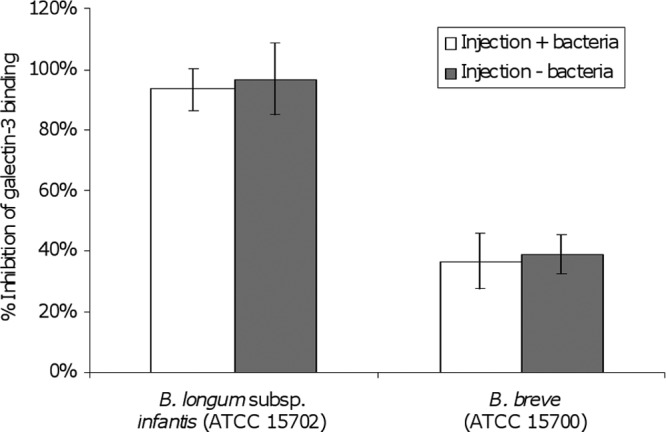
SPR analysis of bacterium-free injections of gal-3 over an immobilized ASF surface. Inhibition is defined as the ability of bacteria to interact with gal-3, thereby reducing its ability to generate a surface plasmon response through interaction with the ASF chip surface. Gal-3 injected over the ASF surface in the absence of bacteria was used to define the 0% inhibition SPR response. The effects of injections with and without bacteria are compared, and data are presented as mean inhibition ± the standard deviation. Experiments were carried out in triplicate (n = 3).
The direct interaction between B. longum subsp. infantis ATCC 15702 and gal-3 was further assessed by employing an agglutination assay as outlined by Hynes et al. (22). Gal-3 effectively agglutinated the bacterial suspension of B. longum subsp. infantis ATCC 15702, as demonstrated by a carpet of aggregated cells on the bottoms of the wells (Fig. 4, column A). Alternatively, in the absence of gal-3, control wells exhibited a tight bacterial pellet, indicating a lack of agglutination (Fig. 4, column B).
Fig 4.
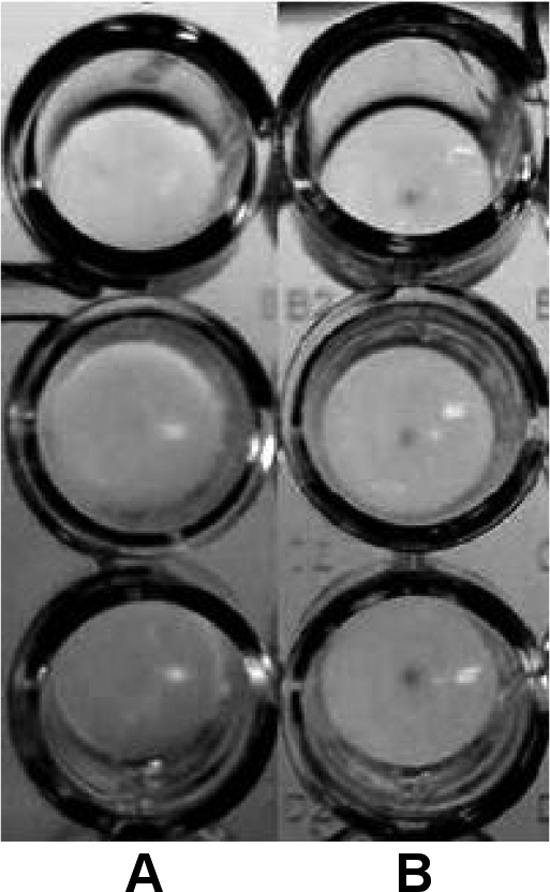
Agglutination assay for direct visualization of bacterium–gal-3 interaction. Gal-3 or PBS alone (negative control) was incubated with B. longum subsp. infantis ATCC 15702. Gal-3 exposure resulted in agglutination of the bacteria, as evidence by the absence of a dense pellet of bacteria in the bottom of the well (A). PBS alone did not agglutinate the bacteria, resulting in pellet formation at the bottom of the well (B).
In order to characterize the interaction between B. longum subsp. infantis ATCC 15702 and gal-3, a solid-phase binding platform was used. The two B. longum subsp. infantis strains and E. coli NCTC 12900 were stained with TAMRA (carboxytetramethylrhodamine; Invitrogen) (intracellular localization) as outlined by Alemka et al. (23) and exposed to gal-3-coated wells (InnoCyte ECM cell adhesion assay kit; Calbiochem [Merck, Darmstadt, Germany]) under anaerobic conditions (Anaerocult A gas pack; Merck, Darmstadt, Germany) for 1 h at 37°C. The wells were washed three times with phosphate-buffered saline (PBS), and the fluorescence in each well was recorded at an excitation wavelength of 530 nm with detection at 590 nm with a Synergy HT plate reader (BIO-TEK, Bedfordshire, United Kingdom; sensitivity setting of 45). B. longum subsp. infantis ATCC 15702 displayed a significantly higher level of interaction with immobilized gal-3 than did the other two bacterial strains assayed (Fig. 5A). While B. longum subsp. infantis ATCC 15702 and ATCC 15697 both demonstrate a comparatively high level of gal-3 inhibition in the SPR experiments, ATCC 15697 does not interact as strongly under the solid-phase experimental conditions used, likely because of the difference in incubation times with gal-3 (1 h versus 3 min), as well as the differing means of detection. Resuspension of B. longum subsp. infantis ATCC 15702 in PBS containing 50 mM lactose (a known ligand for gal-3) prior to interaction with immobilized gal-3 resulted in a 90% reduction of bacterial adherence (P < 0.05) (Fig. 5B), indicating the involvement of a carbohydrate ligand in the bacterium–gal-3 interaction. To investigate the interaction further, studies were carried out with full-length soluble gal-3 protein and the gal-3 CRD. Resuspension of B. longum subsp. infantis ATCC 15702 in PBS containing a 20-fold molar excess (3.85 μM, in relation to that immobilized on the well surface) of full-length gal-3 prior to well exposure resulted in a 44% reduction of bacterial binding (P < 0.01; Fig. 5C), while the presence of the gal-3 CRD (3.85 μM) did not result in a significant level of inhibition. These results highlight the importance of the full-length gal-3 protein for maximal binding. Negative-control experiments with bacteria that are known not to interact with gal-3 were difficult to design. Such controls are not available, given that galactose and other moieties recognized by gal-3 are commonly found on the surface of most bacteria (12, 17). Further work is required to fully characterize the interaction between gal-3 and B. longum subsp. infantis ATCC 15702 or ATCC 15697 and to determine its implications for host health and the integrity of the intestinal microflora.
Fig 5.
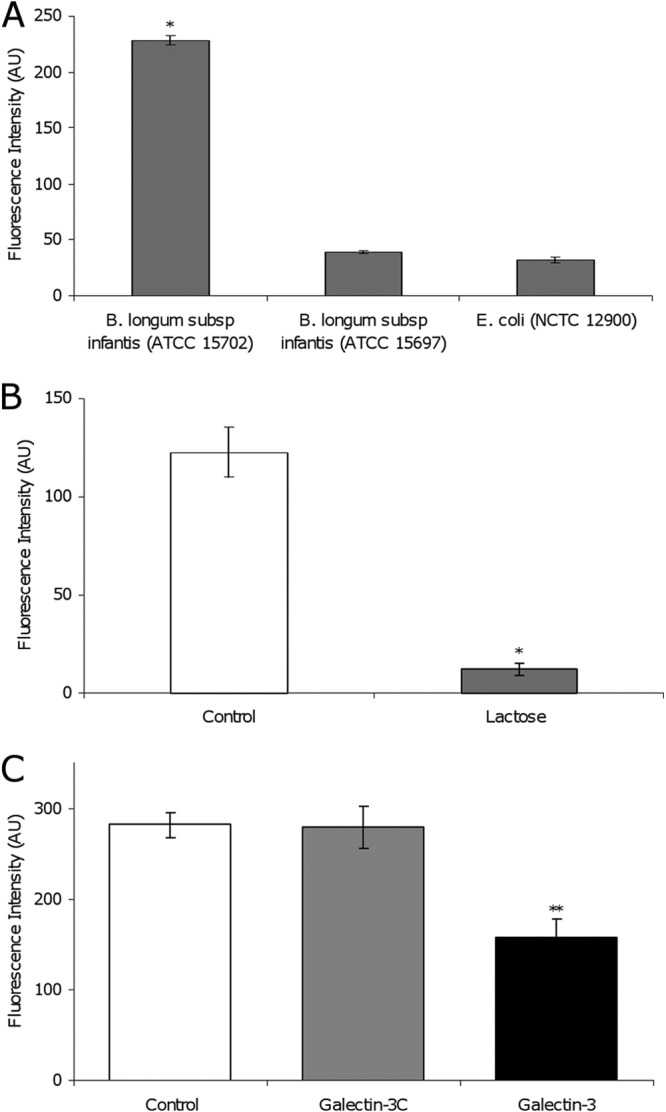
(A) Binding of TAMRA-labeled bacteria to recombinant gal-3 in a solid-phase binding assay. B. longum subsp. infantis ATCC 15702 and ATCC 15697 and E. coli O157:H7 NCTC 12900 were incubated in gal-3-coated wells for 1 h. (B) Binding of TAMRA-labeled B. longum subsp. infantis ATCC 15702 to recombinant gal-3 in a solid-phase binding assay. TAMRA-labeled bacteria were incubated in gal-3-coated wells (white bars) or in gal-3-coated wells in the presence of lactose (gray bars). (C) Binding of TAMRA-labeled B. longum subsp. infantis ATCC 15702 to recombinant gal-3 in a solid-phase binding assay. Bacteria were incubated in gal-3-coated wells (white) or in gal-3-coated wells in the presence of the soluble gal-3 CRD (gal-3C; gray) or full-length soluble gal-3 (black). All experiments were carried out in triplicate (n = 3). The data are presented as means ± the standard deviations. *, P < 0.05; **, P < 0.01; AU, arbitrary units.
In conclusion, the results presented here, to the best of our knowledge, demonstrate for the first time an interaction between gal-3 and commensal intestinal bacteria. The results identify a higher affinity of gal-3 for two specific strains of bifidobacteria, with gal-3 requiring the full-length protein for enhanced activity, similar to findings reported by Fermino et al. (12). This novel interaction may potentially play a role in colonization resistance and host infection by limiting the availability of gal-3 to pathogens.
ACKNOWLEDGMENTS
Devon Kavanaugh is in receipt of a Teagasc Walsh Fellowship. This work was supported by Science Foundation Ireland under grant 08/SRC/B1393 and the Alimentary Glycoscience Research Cluster (AGRC).
We thank John O'Callaghan for contributions to content discussion and editing of the text.
Footnotes
Published ahead of print 22 March 2013
REFERENCES
- 1. Gunning AP, Bongaerts RJ, Morris VJ. 2009. Recognition of galactan components of pectin by galectin-3. FASEB J. 23: 415–424 [DOI] [PubMed] [Google Scholar]
- 2. Cao Z, Said N, Amin S, Wu HK, Bruce A, Garate M, Hsu DK, Kuwabara I, Liu FT, Panjwani N. 2002. Galectins-3 and -7, but not galectin-1, play a role in re-epithelialization of wounds. J. Biol. Chem. 277: 42299–42305 [DOI] [PubMed] [Google Scholar]
- 3. Dumic J, Dabelic S, Flogel M. 2006. Galectin-3: an open-ended story. Biochim. Biophys. Acta 1760: 616–635 [DOI] [PubMed] [Google Scholar]
- 4. Leffler H, Carlsson S, Hedlund M, Qian Y, Poirier F. 2004. Introduction to galectins. Glycoconj. J. 19: 433–440 [DOI] [PubMed] [Google Scholar]
- 5. Stowell SR, Arthur CM, Dias-Baruffi M, Rodrigues LC, Gourdine JP, Heimburg-Molinaro J, Ju T, Molinaro RJ, Rivera-Marrero C, Xia B, Smith DF, Cummings RD. 2010. Innate immune lectins kill bacteria expressing blood group antigen. Nat. Med. 16: 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barondes SH, Castronovo V, Cooper D, Cummings RD, Drickamer K, Feizi T, Gitt MA, Hirabayashi J, Hughes C, Kasai K. 1994. Galectins: a family of animal beta-galactoside-binding lectins. Cell 76: 597–598 [DOI] [PubMed] [Google Scholar]
- 7. Huflejt ME, Jordan ET, Gitt MA, Barondes SH, Leffler H. 1997. Strikingly different localization of galectin-3 and galectin-4 in human colon adenocarcinoma T84 cells. J. Biol. Chem. 272: 14294–14303 [DOI] [PubMed] [Google Scholar]
- 8. Stillman BN, Hsu DK, Pang M, Brewer CF, Johnson P, Liu F-T, Baum LG. 2006. Galectin-3 and galectin-1 bind distinct cell surface glycoprotein receptors to induce T cell death. J. Immunol. 176: 778–789 [DOI] [PubMed] [Google Scholar]
- 9. Nio-Kobayashi J, Takahashi-Iwanaga H, Iwanaga T. 2009. Immunohistochemical localization of six galectin subtypes in the mouse digestive tract. J. Histochem. Cytochem. 57: 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bresalier RS, Byrd JC, Wang L, Raz A. 1996. Colon cancer mucin: a new ligand for the β-galactoside-binding protein galectin-3. Cancer Res. 56: 4354–4357 [PubMed] [Google Scholar]
- 11. Vasta GR. 2012. Galectins as pattern recognition receptors: structure, function, and evolution. Adv. Exp. Med. Biol. 946: 21–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fermino ML, Polli CD, Toledo KA, Liu FT, Hsu DK, Roque-Barreira MC, Pereira-da-Silva G, Bernardes ES, Halbwachs-Mecarelli L. 2011. LPS-induced galectin-3 oligomerization results in enhancement of neutrophil activation. PLoS One 6: e26004 doi:10.1371/journal.pone.0026004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gupta SK, Masinick S, Garrett M, Hazlett LD. 1997. Pseudomonas aeruginosa lipopolysaccharide binds galectin-3 and other human corneal epithelial proteins. Infect. Immun. 65: 2747–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. John CM, Jarvis GA, Swanson KV, Leffler H, Cooper MD, Huflejt ME, Griffiss JM. 2002. Galectin-3 binds lactosaminylated lipooligosaccharides from Neisseria gonorrhoeae and is selectively expressed by mucosal epithelial cells that are infected. Cell. Microbiol. 4: 649–662 [DOI] [PubMed] [Google Scholar]
- 15. Fowler M, Thomas RJ, Atherton J, Roberts IS, High NJ. 2006. Galectin-3 binds to Helicobacter pylori O-antigen: it is upregulated and rapidly secreted by gastric epithelial cells in response to H. pylori adhesion. Cell. Microbiol. 8: 44–54 [DOI] [PubMed] [Google Scholar]
- 16. Barboni E, Coade S, Fiori A. 2005. The binding of mycolic acids to galectin-3: a novel interaction between a host soluble lectin and trafficking mycobacterial lipids? FEBS Lett. 579: 6749–6755 [DOI] [PubMed] [Google Scholar]
- 17. Mey A, Leffler H, Hmama Z, Normier G, Revillard JP. 1996. The animal lectin galectin-3 interacts with bacterial lipopolysaccharides via two independent sites. J. Immunol. 156: 1572–1577 [PubMed] [Google Scholar]
- 18. Yi ZJ, Fu YR, Li M, Gao KS, Zhang XG. 2009. Effect of LTA isolated from bifidobacteria on d-galactose-induced aging. Exp. Gerontol. 44: 760–765 [DOI] [PubMed] [Google Scholar]
- 19. Li Y, Komai-Koma M, Gilchrist DS, Hsu DK, Liu FT, Springall T, Xu D. 2008. Galectin-3 is a negative regulator of lipopolysaccharide-mediated inflammation. J. Immunol. 181: 2781–2789 [DOI] [PubMed] [Google Scholar]
- 20. Kleshchenko YY, Moody TN, Furtak VA, Ochieng J, Lima MF, Villalta F. 2004. Human galectin-3 promotes Trypanosoma cruzi adhesion to human coronary artery smooth muscle cells. Infect. Immun. 72: 6717–6721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maljaars CE, Andre S, Halkes KM, Gabius HJ, Kamerling JP. 2008. Assessing the inhibitory potency of galectin ligands identified from combinatorial (glyco)peptide libraries using surface plasmon resonance spectroscopy. Anal. Biochem. 378: 190–196 [DOI] [PubMed] [Google Scholar]
- 22. Hynes SO, Hirmo S, Wadstrom T, Moran AP. 1999. Differentiation of Helicobacter pylori isolates based on lectin binding of cell extracts in an agglutination assay. J. Clin. Microbiol. 37: 1994–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alemka A, Clyne M, Shanahan F, Tompkins T, Corcionivoschi N, Bourke B. 2010. Probiotic colonization of the adherent mucus layer of HT29MTXE12 cells attenuates Campylobacter jejuni virulence properties. Infect. Immun. 78: 2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]