Abstract
A broadly reactive and highly sensitive reverse transcription-quantitative PCR assay to detect salivirus/klassevirus was developed. By means of the developed assay, salivirus/klassevirus was detected in 13 (93%) raw sewage, 4 (29%) secondary-treated sewage, and 9 (16%) river water samples, with a maximum concentration of 9.7 × 106 copies/liter.
TEXT
Salivirus is a novel member of the family Picornaviridae that was recently discovered in the feces of children with nonpolio acute flaccid paralysis (AFP) in Nigeria (1). Another picornavirus, klassevirus, has also been found in the feces of children with gastroenteritis in the United States and Australia (2, 3). Salivirus is genetically indistinguishable from klassevirus (1–3), and both salivirus and klassevirus are currently classified as the novel genus Salivirus according to the Picornaviridae Study Group of the International Committee on Taxonomy of Viruses (ICTV) (http://www.picornastudygroup.com/).
Several reverse transcription-PCR (RT-PCR) and RT-nested PCR assays have been developed based on the conserved nucleotide sequences in the 5′ untranslated region (UTR) and 2C, 3D, and viral protein 0 (VP0)/VP3 regions (1–5). These assays have succeeded in detecting salivirus/klassevirus in the feces of patients with gastroenteritis and nonpolio AFP in several countries worldwide, indicating the pathogenicity of this virus to humans (1–6).
Human enteric viruses are known to be abundant in wastewater and receiving water bodies (7). Monitoring of wastewater samples from a wastewater treatment plant (WWTP) is considered the most promising approach for understanding the true incidence of viruses that are shed from both symptomatic and asymptomatic patients in the WWTP service area; this approach is especially effective for rare and/or emerging viruses (8–11).
The first detection of salivirus/klassevirus in water was reported in 2009: the virus was detected in a raw sewage sample from a WWTP in Spain (3). Subsequently, in our previous study, salivirus/klassevirus was detected in 50% (7/14) of raw sewage samples and 29% (4/14) of secondary-treated sewage samples from a Japanese WWTP (12). Metagenomic analysis revealed the presence of salivirus/klassevirus in untreated sewage in Nepal and Thailand (13). These results indicate the possibility that salivirus/klassevirus is ubiquitous in wastewater worldwide. To further develop understanding of the prevalence of salivirus/klassevirus in water, further studies should be conducted extensively from the quantitative viewpoint.
Design of primers and probe.
This study aimed to develop an RT-quantitative PCR (RT-qPCR) assay for the highly sensitive and specific detection of salivirus/klassevirus. The 5′ UTR was chosen as a target to design oligonucleotide primers and probe that specifically reacted with salivirus/klassevirus because it has been widely used for the primer/probe design for many picornaviruses (14–17). The 5′ UTR nucleotide sequences of 13 salivirus/klassevirus isolates were obtained from the GenBank database (accession numbers GU245894, GQ184145, GQ179640, JN379039, and GU376738 to GU376746). Because these isolates originated from a limited number of countries (1, 2, 5), three 5′ UTR sequences identified in Japanese wastewater samples in this study (GenBank accession numbers AB775007 to AB775009) by the conventional RT-nested PCR (5) were also included to develop a broadly reactive assay. All 16 salivirus/klassevirus sequences were aligned using Jalview software version 2.8 (18), with 9 non-salivirus/klassevirus sequences of the family Picornaviridae, such as Aichi virus (GenBank accession number AB040749), poliovirus (GenBank accession number NC_002058), and hepatitis A virus (GenBank accession number M59810).
As illustrated in Fig. 1, a forward primer (KLA-F; 5′-TCTGCTTGGTGCCAACCTC-3′), a reverse primer (KLA-R; 5′-CCARGCACACACATGAGRGGATAC-3′), and a TaqMan MGB probe (KLA-TP; 5′-FAM-TGCGGGAGTGCTCT-MGB-NFQ-3′) were designed from sequences that were highly conserved only among the salivirus/klassevirus isolates by using Primer Express software version 3.0 (Applied Biosystems, Foster City, CA). The number of nucleotide mismatches and gaps between the designed primers/probe and the non-salivirus/klassevirus isolates was 11 to 20 nucleotides (nt) for the forward primer, 7 to 23 nt for the reverse primer, and 3 to 14 nt for the probe. A nucleotide BLAST search of each primer and probe yielded no significant homology to non-salivirus/klassevirus sequences. These results confirm the specificity of the designed primers and probe for salivirus/klassevirus detection.
Fig 1.
Alignment of 16 nucleotide sequences of the 5′ UTR of salivirus/klassevirus, including 3 sequences identified in this study (GenBank accession numbers AB775009). The nucleotide sequences corresponding to nucleotide positions 270 to 374 of the 5′ UTR of the salivirus/klassevirus isolate SH1 (GU245894) are shown. The accession number is indicated on the left side of the sequence. Nucleotide sequences corresponding to the primers (KLA-F and KLA-R) and the TaqMan MGB probe (KLA-TP) are boxed by solid lines. Arrows and double lines show the locations of the primers and TaqMan MGB probe, respectively. The amplicon length is 87 bp.
Performance of qPCR.
Double-stranded DNA containing the 486-bp 5′ UTR sequence of the salivirus/klassevirus isolate SH1 (GU245894) that was amplified by outer primers for the nested PCR (SAL-L1 and SAL-R1) (5) was artificially synthesized to be used as a template for plasmid DNA construction. The double-stranded DNA was cloned into the T-Vector pMD20 (TaKaRa Bio, Otsu, Japan), and the concentration of the resulting purified plasmid DNA was determined by measuring the optical density at 260 nm.
The serially diluted standard samples of salivirus/klassevirus plasmid DNA (1.0 × 106 to 1.0 × 100 copies/reaction) were subjected to the qPCR run using the designed primers and probe to determine the sensitivity of the assay. Briefly, a 5-μl aliquot of each standard sample was mixed with 20 μl of a PCR mixture containing 12.5 μl of Premix Ex Taq (probe qPCR) (TaKaRa Bio), 400 nM (each) forward and reverse primers, and 200 nM TaqMan MGB probe. PCR tubes containing the mixtures were placed into the Thermal Cycler Dice real-time system TP800 (TaKaRa Bio) and incubated at 95°C for 30 s, followed by 45 cycles at 95°C for 5 s and 58°C for 30 s. Amplification data were collected and analyzed with the Thermal Cycler Dice real-time system software version 3.00D (TaKaRa Bio).
As summarized in Table 1, the developed qPCR assay was consistently able to detect as few as 2.0 × 101 copies/reaction and sometimes even 1.0 × 100 copies/reaction. A fairly linear correlation (correlation coefficient r = −0.997) was observed between the log10 initial plasmid concentrations and the threshold cycle (CT) values at which the fluorescence intensity exceeded the threshold value over a wide range of 6 log10 between 1.0 × 106 and 1.0 × 100 copies/reaction. The slope (S) of the linear regression curve was −3.302, and the efficiency (E) of the PCR amplification was calculated to be 100.8% according to the following formula: E = (10−1/S − 1) × 100. Compared with the conventional nested PCR assay (5), the developed qPCR assay showed a similar or higher sensitivity for plasmid DNA amplification: both methods were able to detect as little as 1 copy of the plasmid DNA (Table 1). Based on these experimental data, we concluded that a highly sensitive qPCR assay had been successfully developed.
Table 1.
Detection of salivirus/klassevirus plasmid DNA by qPCR and nested PCR
| Initial concn of plasmid DNA (copies/reaction) | Detection by: |
||
|---|---|---|---|
| qPCR |
Nested PCR (no. of positive tubes/no. of tested tubes) | ||
| No. of positive tubes/no. of tested tubes | CT value (mean ± SD)a | ||
| 1.0 × 106 | 3/3 | 21.8 ± 0.1 | 1/1 |
| 1.0 × 105 | 3/3 | 24.9 ± 0.1 | 1/1 |
| 1.0 × 104 | 3/3 | 28.1 ± 0.1 | 1/1 |
| 1.0 × 103 | 3/3 | 31.2 ± 0.2 | 1/1 |
| 1.0 × 102 | 3/3 | 34.5 ± 0.1 | 1/1 |
| 5.0 × 101 | 3/3 | 36.1 ± 0.5 | 1/1 |
| 2.0 × 101 | 5/5 | 37.5 ± 1.9 | 3/3 |
| 1.0 × 101 | 4/5 | 38.7 ± 1.6 | 3/3 |
| 5.0 × 100 | 4/5 | 37.9 ± 3.1 | 1/3 |
| 1.0 × 100 | 1/5 | 41.8 | 1/3 |
| 0 | 0/3 | Not determined | 0/2 |
Mean and standard deviation (SD) values were calculated using the results of positive tubes.
Detection of salivirus/klassevirus in wastewater and river water.
Between January 2011 and February 2012, a total of 84 water samples (2 liters each) were collected monthly from 2 Japanese river basins: raw sewage samples (n = 14) and secondary-treated sewage samples before chlorination (n = 14) collected from a WWTP and river water samples of the Fujikawa River (site FRW; n = 14) and the Tamagawa River (sites TRW1, TRW2, and TRW3, from the upstream side; n = 14 each). These water samples were collected as grab samples.
Viruses in the water samples (100 ml of raw sewage and 2 liters each of secondary-treated sewage and river water) were concentrated by the electronegative membrane-vortex (EMV) method (19) as described previously (12). Viral RNA was extracted from 140 μl of the virus-concentrated sample using the QIAamp viral RNA minikit (Qiagen, Hilden, Germany), followed by RT using the High Capacity cDNA reverse transcription kit (Applied Biosystems). Then a 5-μl aliquot of cDNA was used as a template for the qPCR assay by following the above-described experimental procedure. All the water and standard samples and negative controls were analyzed in duplicate. According to the results of the experiments using murine norovirus as an internal control, no RT-qPCR inhibitory effect was observed in any of the water samples tested in this study (data not shown).
Salivirus/klassevirus was detected in 93% (13/14) of the raw sewage samples, 29% (4/14) of the secondary-treated sewage samples, and 16% (9/56) of the river water samples, showing concentrations of 3.7 × 105 to 9.7 × 106, 1.6 × 104 to 7.1 × 104, and 1.7 × 104 to 2.7 × 105 copies/liter, respectively (Fig. 2). The virus was always detected with a relatively constant concentration in the raw sewage except in one sample collected in March 2011. Although no reports have been published regarding the detection of salivirus/klassevirus in fecal specimens from Japanese patients, the results of this study show that salivirus/klassevirus infection occurs in Japan almost throughout the year.
Fig 2.
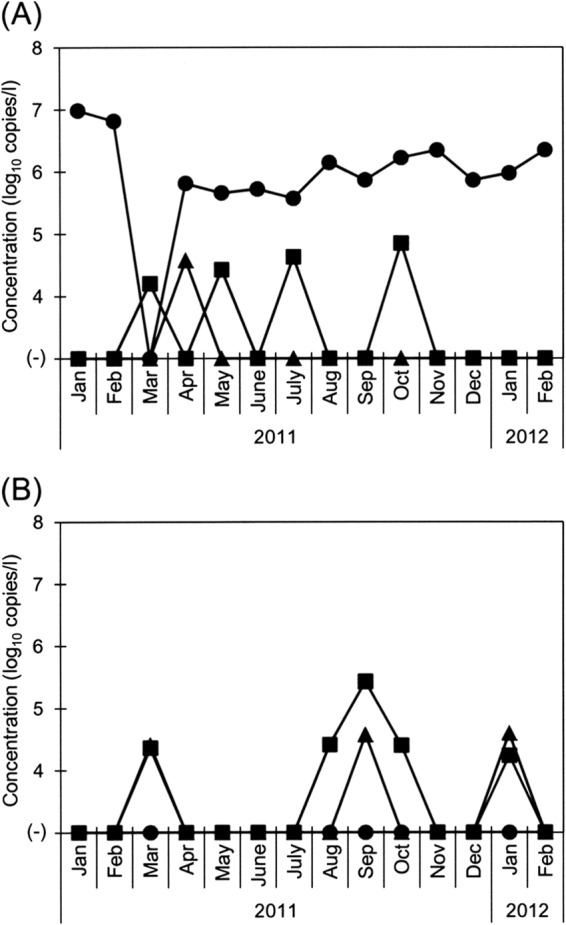
Concentrations of salivirus/klassevirus in wastewater and river water samples, determined by the developed RT-qPCR assay. (A) Raw sewage (filled circles) and secondary-treated sewage (filled squares) of a WWTP and river water from the Fujikawa River (filled triangles). (B) River water collected from 3 sites along the Tamagawa River (filled circles, site TRW1; filled squares, site TRW2; filled triangles, site TRW3).
The range of the concentrations of salivirus/klassevirus in raw sewage was quite comparable to that of other well-known human enteric viruses, such as human adenoviruses and noroviruses (E. Haramoto and M. Otagiri, unpublished data). On the other hand, salivirus/klassevirus was less abundant than other viruses in secondary-treated sewage, resulting in a higher virus removal ratio of >3 log10 in many of the tested months. Accordingly, river water samples were less contaminated with salivirus/klassevirus, although the virus-positive ratio was relatively high at the downstream sites TRW2 (36%) and TRW3 (21%).
In addition to 3 wastewater-derived sequences that had been used to design the primers and probe, one additional sequence was obtained from the site TRW2 (GenBank accession number AB775010) by the RT-nested PCR (5) followed by direct nucleotide sequencing. When this sequence was aligned with the sequences of the designed primers and probe, a 1-nt mismatch was found in the nucleotide region at which the reverse primer bound (data not shown). Although the successful generation of the specific RT-qPCR product was confirmed by electrophoresis for this sample, further information about the diversity of the nucleotide sequences of salivirus/klassevirus isolates worldwide will be very beneficial to develop a more broadly reactive assay.
In conclusion, this study established a broadly reactive and highly sensitive RT-qPCR assay for salivirus/klassevirus and subsequently quantitatively determined the prevalence of this virus in wastewater and river water in Japan. The results from raw sewage samples show that the salivirus/klassevirus infection occurs in the studied area almost throughout the year. To our knowledge, this is the first study to quantitatively demonstrate the prevalence of salivirus/klassevirus in water. Future studies are required to more deeply understand the prevalence of this emerging virus in various types of environmental water samples worldwide. The developed assay will be further applied to fecal specimens to clarify the incidence of this virus in human society.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in the GenBank database under accession numbers AB775007 to AB775010.
ACKNOWLEDGMENTS
This study was partially supported by a grant-in-aid for the Global COE Program from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by the research fund from the Tokyu Foundation for Better Environment, Japan.
We thank the workers of the WWTP for their kind cooperation in providing wastewater samples.
Footnotes
Published ahead of print 29 March 2013
REFERENCES
- 1. Li L, Victoria J, Kapoor A, Blinkova O, Wang C, Babrzadeh F, Mason CJ, Pandey P, Triki H, Bahri O, Oderinde BS, Baba MM, Bukbuk DN, Besser JM, Bartkus JM, Delwart EL. 2009. A novel picornavirus associated with gastroenteritis. J. Virol. 83: 12002– 12006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Greninger AL, Runckel C, Chiu CY, Haggerty T, Parsonnet J, Ganem D, DeRisi JL. 2009. The complete genome of klassevirus—a novel picornavirus in pediatric stool. Virol. J. 6: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Holtz LR, Finkbeiner SR, Zhao G, Kirkwood CD, Girones R, Pipas JM, Wang D. 2009. Klassevirus 1, a previously undescribed member of the family Picornaviridae, is globally widespread. Virol. J. 6:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Han TH, Kim CH, Chung JY, Park SH, Hwang ES. 2010. Klassevirus infection in children, South Korea. Emerg. Infect. Dis. 16:1623– 1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shan T, Wang C, Cui L, Yu Y, Delwart E, Zhao W, Zhu C, Lan D, Dai X, Hua X. 2010. Picornavirus salivirus/klassevirus in children with diarrhea, China. Emerg. Infect. Dis. 16: 1303– 1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Greninger AL, Holtz L, Kang G, Ganem D, Wang D, DeRisi JL. 2010. Serological evidence of human klassevirus infection. Clin. Vaccine Immunol. 17: 1584– 1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fong TT, Lipp EK. 2005. Enteric viruses of humans and animals in aquatic environments: health risks, detection, and potential water quality assessment tools. Microbiol. Mol. Biol. Rev. 69: 357– 371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haramoto E, Katayama H, Phanuwan C, Ohgaki S. 2008. Quantitative detection of sapoviruses in wastewater and river water in Japan. Lett. Appl. Microbiol. 46: 408– 413 [DOI] [PubMed] [Google Scholar]
- 9. Kitajima M, Haramoto E, Phanuwan C, Katayama H. 2011. Prevalence and genetic diversity of Aichi viruses in wastewater and river water in Japan. Appl. Environ. Microbiol. 77: 2184– 2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. La Rosa G, Iaconelli M, Pourshaban M, Fratini M, Muscillo M. 2010. Molecular detection and genetic diversity of norovirus genogroup IV: a yearlong monitoring of sewage throughout Italy. Arch. Virol. 155: 589– 593 [DOI] [PubMed] [Google Scholar]
- 11. Sano D, Pérez-Sautu U, Guix S, Pintó RM, Miura T, Okabe S, Bosch A. 2011. Quantification and genotyping of human sapoviruses in the Llobregat River catchment, Spain. Appl. Environ. Microbiol. 77: 1111– 1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Haramoto E, Otagiri M. 2013. Prevalence and genetic diversity of klassevirus in wastewater in Japan. Food Environ. Virol. 5: 46– 51 [DOI] [PubMed] [Google Scholar]
- 13. Ng TFF, Marine R, Wang C, Simmonds P, Kapusinszky B, Bodhidatta L, Oderinde BS, Wommack KE, Delwart E. 2012. High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J. Virol. 86: 12161– 12175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baumgarte S, De Souza Luna LK, Grywna K, Panning M, Drexler JF, Karsten C, Huppertz HI, Drosten C. 2008. Prevalence, types, and RNA concentrations of human parechoviruses, including a sixth parechovirus type, in stool samples from patients with acute enteritis. J. Clin. Microbiol. 46: 242– 248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Corless CE, Guiver M, Borrow R, Edwards-Jones V, Fox AJ, Kaczmarski EB, Mutton KJ. 2002. Development and evaluation of a ‘real-time’ RT-PCR for the detection of enterovirus and parechovirus RNA in CSF and throat swab samples. J. Med. Virol. 67: 555– 562 [DOI] [PubMed] [Google Scholar]
- 16. Costafreda MI, Bosch A, Pintó RM. 2006. Development, evaluation, and standardization of a real-time TaqMan reverse transcription-PCR assay for quantification of hepatitis A virus in clinical and shellfish samples. Appl. Environ. Microbiol. 72: 3846– 3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Petitjean J, Vabret A, Dina J, Gouarin S, Freymuth F. 2006. Development and evaluation of a real-time RT-PCR assay on the LightCycler for the rapid detection of enterovirus in cerebrospinal fluid specimens. J. Clin. Virol. 35: 278– 284 [DOI] [PubMed] [Google Scholar]
- 18. Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189– 1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haramoto E, Katayama H, Asami M, Akiba M. 2012. Development of a novel method for simultaneous concentration of viruses and protozoa from a single water sample. J. Virol. Methods 182: 62– 69 [DOI] [PubMed] [Google Scholar]



