Abstract
Decomposition of plant residues is largely mediated by soil-dwelling microorganisms whose activities are influenced by both climate conditions and properties of the soil. However, a comprehensive understanding of their relative importance remains elusive, mainly because traditional methods, such as soil incubation and environmental surveys, have a limited ability to differentiate between the combined effects of climate and soil. Here, we performed a large-scale reciprocal soil transplantation experiment, whereby microbial communities associated with straw decomposition were examined in three initially identical soils placed in parallel in three climate regions of China (red soil, Chao soil, and black soil, located in midsubtropical, warm-temperate, and cold-temperate zones). Maize straws buried in mesh bags were sampled at 0.5, 1, and 2 years after the burial and subjected to chemical, physical, and microbiological analyses, e.g., phospholipid fatty acid analysis for microbial abundance, community-level physiological profiling, and 16S rRNA gene denaturing gradient gel electrophoresis, respectively, for functional and phylogenic diversity. Results of aggregated boosted tree analysis show that location rather soil is the primary determining factor for the rate of straw decomposition and structures of the associated microbial communities. Principal component analysis indicates that the straw communities are primarily grouped by location at any of the three time points. In contrast, microbial communities in bulk soil remained closely related to one another for each soil. Together, our data suggest that climate (specifically, geographic location) has stronger effects than soil on straw decomposition; moreover, the successive process of microbial communities in soils is slower than those in straw residues in response to climate changes.
INTRODUCTION
Saprophytic microorganisms play an essential role in nutrient recycling of an ecosystem. In terrestrial agricultural systems, plant residues returned to the fields are the major source of soil organic carbon (1). About 3.4 billion tons of crop residues are produced annually worldwide, with 0.47 billion tons being estimated for maize (2). Decomposition of plant residues is largely mediated by microorganisms such as bacteria and fungi in the soil (3); the resultant nutritional carbon substrates can either support the growth of crops or be partially stored in the form of soil humus. Given the importance of decomposition in soil carbon sequestration, there has been continued interest in elucidating the dynamic changes of microbial communities during the course of residue decomposition (4–7).
Like many other complex microbial processes in nature, the rate of straw decomposition in agricultural soils is determined by a combination of various environmental factors, which include climate conditions (e.g., temperature and precipitation), biotic and abiotic properties of the soil (e.g., pH and contents of water, minerals, and nutrients) as well as tillage (4, 8, 9). Despite abundant evidence in the literature detailing the effects of these environmental factors on the structure and function of microbial communities, the relative importance of climate (or geographic location) versus soil has yet not been assessed. Given that different types of soils are commonly found in areas of similar climate and the same types of soils also exist across different climate regions, knowledge with regard to the relative strength of the effects (geographic location versus soil type) will help for the selection of suitable crops based on their decomposition qualities (2, 10, 11), with the aim of increasing the amount of carbon sequestered in the soil and mitigating global climate change (12). Significantly, such an assessment is also related to the current debate in microbial biogeography with regard to the power of geographic factors relative to local environments in driving microbial diversity, i.e., climate regimen versus soil type in the case of residue decomposition (13–15).
Climate, specifically temperature, seemingly has greater impacts than soil on straw-decomposing microbial communities, according to current ecological theories highlighting the role of temperature in the determination of biodiversity (16, 17). The enzymatic reactions catalyzing the chemical breakdown of plant residues, as well as the growth kinetics of microorganisms secreting those digestive enzymes, all will be accelerated by an increase of temperature. Consistent with this prediction is the fact that plant residue decomposition occurs faster in warm climate regions and slower in cold climate regions (18). Growth at higher temperatures will lead to higher levels of diversity of the microbial community, arguably because of elevated mutation rates (19). A strong impact of temperature on the decomposition rates of soil organic matter has been observed by many studies under both laboratory and field conditions (reviewed in references 20 and 21). However, it has been noted that temperature sensitivity varies depending on straw type or chemical composition of the organic matter. In general, slower processes of decomposition are more sensitive to changes of temperature (20, 22).
Bacteria are single-celled organisms that are very sensitive to changes in their immediate environments, such as soils (23). Recent work has implicated a primary role of soil characteristics (notably soil pH and C/N ratios) in shaping bacterial community composition (24–26). Soil pH is one of the most influential chemical factors affecting the soil microbial community. Rousk et al. (27) recently examined the relative abundance of bacterial and fungal decomposers in soils across a pH gradient from pH 4.0 to 8.3 using phospholipid fatty acid (PLFA) analysis, and their results showed that pH variation produces contrasting effects on the growth of bacteria and fungi in soil; thus, it is likely the most important determining factor for bacterium/fungus ratios in soil (26).
Methodologically, questions pertaining to the relative impacts of climate and soil can be most powerfully addressed through studies of reciprocal soil transplantation, whereby different types of soils are reciprocally transferred to different climate regions and subjected to parallel testing over a short or long period of time (28–30). To this end, we performed the first large-scale soil transplantation experiment under open field conditions. It involved a reciprocal transfer of three agricultural soils from three geographic locations in China (see details in Fig. S1 in the supplemental material). In this work, maize straws were placed into mesh bags and buried in the same three soils located in three geographic regions, and the decomposition process was chemically, physically, and microbiologically monitored at 0.5, 1.0, and 2.0 years after the burial. Subsequent statistic analysis of the obtained data across 3 different locations and 3 different soils provides a direct assessment of the relative importance of climate conditions versus soil on the rate of straw decomposition and structure of the associated microbial communities.
MATERIALS AND METHODS
Experimental setup of soil transplantation and straw decomposition.
To dissect the impacts of climate and soil on agricultural productivity and sustainability, a large-scale soil transplantation experiment was set up in October 2005 which involved three Agroecological Experimental Stations (Hailun, Fengqiu, and Yingtan) administered by the Chinese Ecological Research Network (CERN) (see Fig. S1 in the supplemental material). Soil transplantation was performed with three types of agricultural soils from the local area of each of the three experimental stations: neutral black soil (Phaeozem) in Hailun, alkaline Chao soil (Cambisol) in Fengqiu, and acidic red soil (Acrisol) in Yingtan (see Table S1).
An overview of one of the three experimental plots is provided in Fig. S1B in the supplemental material. The experimental plot was composed of 21 cell units that were organized in three rows: one row for local soil and two rows for soils transferred from the other two experimental stations. Each unit was built with concrete walls (20 cm in thickness) with a final volume of 1.68 m3 (1.2 m width, 1.4 m length, and 1 m depth); the bottom of the unit was paved with quartz sand; the inner walls of the cell unit were covered with one layer of waterproof membrane to prevent infiltration. The units were designed to be 30 cm above the ground and allow free passage of drainage water at the bottom. To set up the soil transplantation, in each of the three locations, soil samples were taken from a field of the same size as the final experimental plot shown in Fig. S1. It was divided into 21 grids of 1.2 by 1.4 m, and for each grid, soils were sampled at a depth of 1 m and taken out by layers every 20 cm (for a total of 5 layers). Soils from each layer of a unit were mixed and packed into 10 plastic bags, which were then distributed to the three experimental sites and placed into the cell units according to their original sequences. The work thus involved the transportation of ∼60 tons of soils by truck to the other two sites.
The straw decomposition experiment was conducted in three cell units per site, and each contained a unique type of soil. First, fresh maize straws were cut into 5-cm lengths. After being dried at 60°C for 24 h, 100 g was placed into a double-layer nylon mesh bag (15 cm in length and 10 cm in width). The mesh size of the bags was 0.074 mm, which allows free access of microorganisms (i.e., bacteria and fungi) from the soil. Bags were separately buried in a vertical position at an average depth of 12 cm. Three bags, which served as three replicates for each treatment, were taken out 0.5, 1, and 2 years after the burial, with a total of 81 bags being analyzed. The residual samples were weighed, and dry weights were later calculated according to the measured water content. Soil properties at the beginning and end of the experiment are shown in Table S1 and S2 in the supplemental material, respectively, with a comparison of pH values between maize straw and soil being shown in Table S3. The climate conditions during the experiment are listed in Table S4.
Chemical and microbiological analysis of straw residues.
Total carbon (TC) was measured using the standard method of dichromate oxidation (31). Total nitrogen (TN) was determined using the Kjeldahl method after the straw residues were wet digested with a mixture of HClO4-H2O2 (32).
A modified Bligh-Dyer procedure was used to extract the microbial phospholipid fatty acids (PLFAs) from 1 g of wet straw (33). Briefly, raw lipids were extracted with a mixed solution of methanol, chloroform, and citric acid (2:1:0.8). The glycolipid and neutral lipid fractions then were removed via passage through silicic acid-bonded solid-phase extraction columns (Waters). The resulting phospholipids were saponified and methylated to fatty acid methyl esters (FAME), which were subsequently analyzed using the MIDI Sherlock microbial identification system (MIDI, Newark, DE) with FAME 19:0 as the internal standard. A portion of each PLFA sample was used for the principal component analysis (PCA) (33).
The Biolog EcoPlate system (Biolog Inc., Hayward, CA) was used to estimate the functional diversity as previously described (34). Mashed straw residues (0.1 g dry weight) were resuspended into 49.9 ml sterile physiological saline. After shaking at 150 rpm for 30 min, the supernatant was diluted 10 times, and 150 μl was inoculated into each well of the Biolog EcoPlates. The plates were then incubated at 25°C for 6 days, and absorbance at 590 nm was measured every 24 h. Data at 72 h showed the largest difference among treatments and thus were used for the calculation of average well color development (AWCD) and diversity indices (35).
Analysis of 16S rRNA genes.
Straw residues (0.1 g per sample) were homogenized under sterile conditions, and total DNAs were extracted using a FastDNA spin kit from MP Biomedicals (Santa Ana, CA) by following the manufacturer's recommendation. For 16S rRNA gene denaturing gradient gel electrophoresis (DGGE) analysis, general primers PRBA338F (with the GC clamp) and PRUN518R were used to amplify the variable V3 region of bacterial 16S rRNA genes (36, 37); the resultant PCR products were subjected to polyacrylamide gel electrophoresis with a denaturing gradient from 40 to 60%. A typical gel photo is shown in Fig. S5 in the supplemental material. Band position and intensity, normalized using a low-molecular-weight DNA ladder (N3233; New England BioLabs) with the assistance of Quantity One software (from Bio-Rad), were used to identify ribosomal genotypes and their relative abundances, respectively (38).
The full-length 16S rRNA genes were amplified using a pair of universal primers, 27f and 1492r (39), and the PCR products were cloned into a TA cloning vector of pMD18-T (Biocolors, Shanghai, China). About 120 transformants were randomly selected for each straw sample in black soil and subjected to restriction fragment length polymorphism (RFLP) analysis (40). To do this, the cloned 16S rRNA gene fragments were PCR amplified using two primers located in the vector, RV-M (5′-GAGCGGATAATTTCACACAGG-3′) and M13-47 (5′-CGCCAGGGTTTTCCCAGTCACGA-3′), and PCR products were digested by two enzymes, Sau3AI and HhaI. Clones with unique RFLP profiles were then subjected to full-length 16S rRNA gene sequencing at Majorbio (Shanghai, China).
Data analysis.
The commonly used Shannon's (H) and Simpson's (D) indices were employed in this work to assess bacterial diversity, and they were calculated using the equations H = −[summ]Pi lnPi and D = [summ][ni(ni − 1)]/[N(N − 1)]. For analysis based on DGGE data, ni is the height of a peak, N is the sum of heights for all peaks in the densitometric curve, and Pi is the probability of importance for all bands in a track. For the analysis of Biolog EcoPlate data, Pi is the proportional color development of the ith well (ni) relative to the total color development (N) of all wells.
Treatment effects were assessed by either three-way or two-way crossed analysis of variance (ANOVA) (SPSS 16.0) with climate (cold temperate, warm temperate, and midsubtropic), soil type (black soil, Chao soil, red soil), or decomposition time (0.5, 1, and 2 years) as the fixed factor. Length gradient analysis showed that the unconstrained ordination linear model was appropriate for estimating the relatedness of the microbial communities. PCA was performed using the correlation matrix method (SPSS 16.0). Aggregated boosted tree (ABT) analysis, performed with the gbmplus package in R, version 2.7.1, was used to evaluate the relative influence of environmental variables (location, soil, and time) on microbial community (41). It was selected because of the involvement of both categorical environmental variables and numerical response variables (PLFA, community-level physiological profile [CLPP], and DGGE). The analysis was based on the assumption that the observed variation can be fully explained by the combined effects of the three environmental variables, and the contribution of each can then be quantitatively estimated.
Nucleotide sequence accession numbers.
The sequences of the 137 clones determined in the course of this work were deposited in the GenBank database under accession numbers JQ798397 to JQ798534.
RESULTS
Maize straws buried in three types of soils (i.e., black soil, Chao soil, and red soil) in three geographic locations in China (Hailun, Fengqiu, and Yingtan, listed in order from high to low latitude in the northern hemisphere), and sampled at three time points (0.5, 1.0, and 2.0 years after the burial), giving rise to a total of 27 treatments, were subjected to the chemical, physical, and microbiological analyses described below.
Chemical and physical analysis of straw decomposition.
The extent of straw decomposition was estimated by measuring the weights of residual straws and their chemical compositions in terms of total C and N. As expected, straw decomposition resulted in a decrease of total C but an increase of total N; thus, a significant decrease of C/N ratio over time was observed for all treatments (F2,81 = 72.68; P < 0.0001). The rate of decomposition was then calculated as the annual decrease of C/N ratio, or loss of mass relative to their original values, over a period of 0.5, 1, or 2 years (Fig. 1).
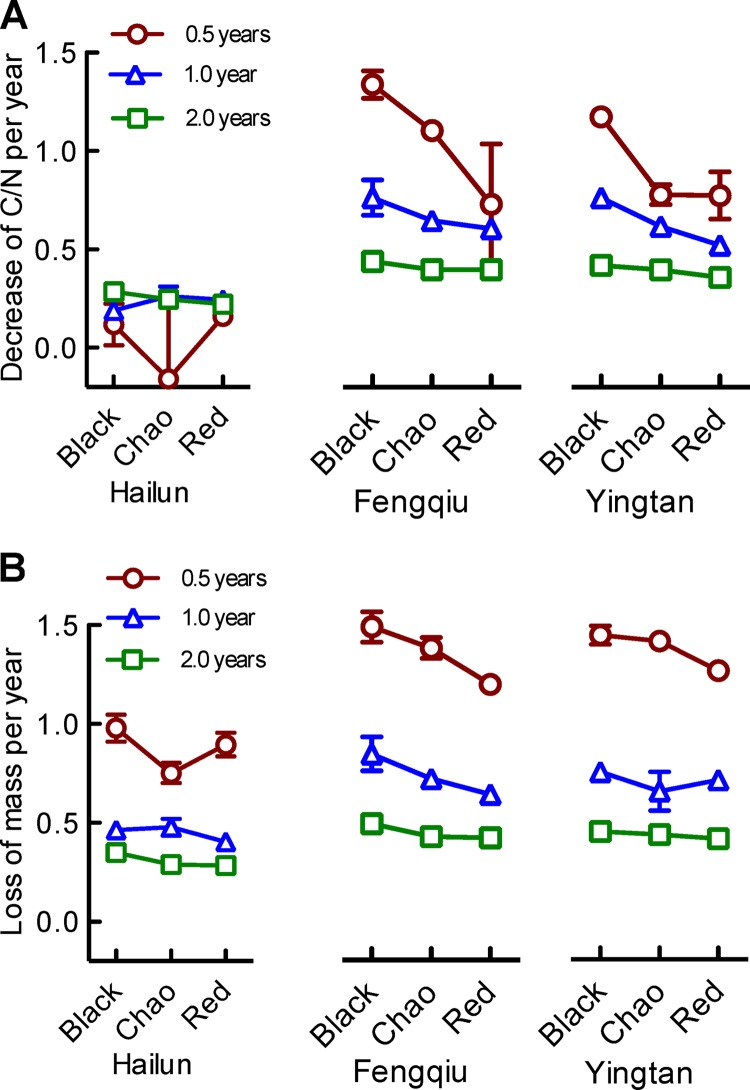
FIG 1.
Rates of straw decomposition as measured by the decrease of C/N ratio (A) and loss of residual mass (B) per year within a period of 0.5, 1.0, and 2.0 years. Data are means and standard errors from 3 replicates.
A cursory comparison of the C/N and biomass data, presented in Fig. 1A and B, respectively, shows the similar effects of location, soil, and time. Indeed, multivariate analysis revealed that the effects of location, soil, and time are highly significant (see Table S5 in the supplemental material). Decomposition appears to occur more rapidly in warm-climate regions (i.e., Fengqiu and Yingtan) than in Hailun, which is located in the cold-temperate zone. The effects of soil type are significant but relatively small compared to the effects of location, with a general trend, in order from a high to low rate of decomposition, of black soil > Chao soil > red soil under the same climate conditions. Of particular note is a seeming delay between C/N ratio reduction and mass reduction. For example, after half a year of decomposition, straws buried in Hailun halved their weight but their C/N ratios remained unchanged (data not shown); as a result, the decomposition rate was detectable in terms of the loss of mass (Fig. 1B) but not the reduction of C/N ratios (Fig. 1A). Finally, the rate data were subjected to aggregated boosted tree (ABT) analysis. Results shown in Table 1 indicate that location had larger effects than soil, and the effect of site location was even greater than time over a period of 2 years of decomposition.
TABLE 1.
Percent contribution of the three environmental variables to the rates of straw decomposition as revealed by ABT analysis
| Variable | Contribution (%) to the rate of straw decomposition as measured by: |
|
|---|---|---|
| Relative decrease of C/N ratio (per yr) | Relative loss of residual mass (per yr) | |
| Time | 36.53 | 82.08 |
| Location | 46.12 | 12.84 |
| Soil | 17.35 | 5.08 |
PLFA analysis of microorganisms associated with straw decomposition.
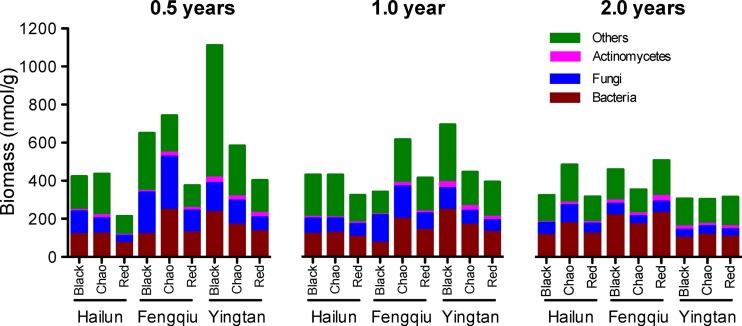
Phospholipid fatty acid (PLFA) profiles were employed to estimate the microbial biomass, and the results are shown in Fig. 2. Multifactor analysis of variance showed significant differences in microbial biomass according to location, soil type, and time (see Table S5 in the supplemental material). It is predicted that microbial biomass is high during the early stages of straw decomposition and then declines along with the decrease of available carbon sources. Such a trend was observed in some instances; for example, the total biomass for black soil in Yingtan was the highest at year 0.5, dropped to 62.7% at year 1, and further dropped to 26.8% at year 2 (Fig. 2). The scores of sum biomass are highly variable among the nine treatments at year 0.5 and year 1 and become less variable at year 2 (coefficient of variation between treatment means was calculated as 48.1, 26.9, and 22.4% for year 0.5, 1, and 2, respectively).
FIG 2.
Microbial biomass as measured by PLFA analysis. Data are means from three replicates for each of the three groups of microorganisms (bacteria, fungi, and actinomycetes). “Others” refers to PLFAs that are not strictly associated with bacteria, fungi, and actinomycetes.
Soil produced a clear impact on microbial biomass, particularly for straws decomposed in the midsubtropical area (Yingtan). It showed an abundance trend of black > Chao > red soils at year 0.5 (Fig. 2). This trend was also observed for the three soils at year 1 but not at year 2. At year 2, straw residues for all three soils in Yingtan possessed similar levels of microbial biomass (Fig. 2) despite slight differences in C/N ratios (Fig. 1).
Warm climates support high levels of microbial abundance under the same conditions of other environmental factors (i.e., soil type and straw material) (42). As expected, the year 0.5 data consistently showed that biomass in Hailun (an experimental site in the cold-temperate zone) is the lowest for each of the three soil types; for black soil, in particular, the biomass varied, in order from lowest to highest scores, in Hailun < Fengqiu < Yingtan (Fig. 2). The effects of location (and soil type) on microbial biomass weakened as straw decomposition progressed, and no significant effects were found at year 1 and year 2 at a level of P < 0.05 (see Table S6 in the supplemental material).
CLPP analysis of microbial communities associated with maize straws.
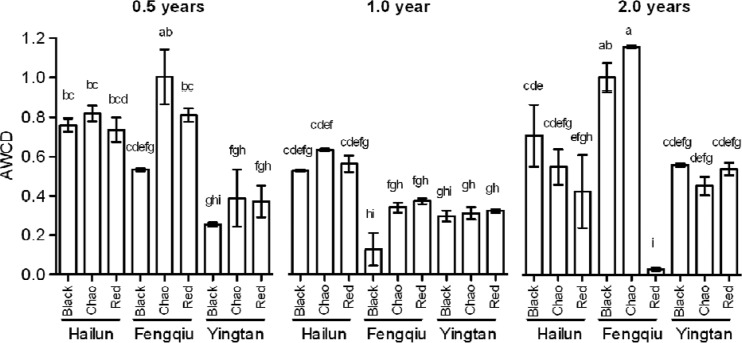
Metabolic activity of the straw-associated microorganisms was examined using the Biolog EcoPlate system, a technique known as the community-level physiological profile (CLPP) analysis (34). The ability to utilize 31 carbon sources was assessed in triplicate, and the data were calculated as AWCD (average well color development). Multivariate analysis showed significant effects of time, climate, and soil (see Table S7 in the supplemental material).
As shown in Fig. 3 (year 0.5), AWCD scores for a given soil type varied against the thermal gradient according to Hailun ≥ Fengqiu > Yingtan. Notably, this trend did not hold when straw decomposition progressed to year 1 and year 2. The effects of soil type on microbial activities were small and complex; in most cases, the three soils in the same geographic location showed similar levels of AWCD.
FIG 3.
Metabolic activities of bacterial communities associated with straw decomposition as estimated using the Biolog EcoPlates analysis. Average well color development (AWCD) data are means and standard errors from 3 replicates. Bars that are not connected by the same letter (shown above each bar) are significantly different (P < 0.05) by Tukey's honestly significant difference test.
When the functional diversity was calculated in the form of Simpson's (D) and Shannon's (H) indices, multivariate analysis revealed significant effects of location, soil type, and time of decomposition (see Table S7 in the supplemental material). The data of Simpson's index are more variable than those of Shannon's index (see Fig. S2), and no general trends were observed regarding the impacts of location or soil. However, there is a decrease of functional diversity over time (at least in red soil): the observed reduction of species abundance (H, 0.5 > 1 > 2 years) is coupled with an increase of frequency of the dominant types (D, 0.5 < 1 ≤ 2 years) as the straw decomposition progresses (see Fig. S2).
DGGE analysis of straw-decomposing microbial communities.
To determine the phylogenetic structure of the straw-decomposing bacterial community, total DNA was prepared from each of the residual samples and used for PCR amplification of 16S rRNA genes and subsequent DGGE analysis. Diversity was calculated as the Simpson's (D) and Shannon's (H) indices. Multivariate analysis showed significant effects of location but no significant effect of soil type at the level of P < 0.05 (see Table S8 in the supplemental material).
The data were then plotted to show the detailed effects of location (see Fig. S3 in the supplemental material), and a general trend was observed only in year 2 for black and Chao soils; for the Shannon index (H), the trend was Hailun > Fengqiu > Yingtan (see Fig. S3B); the opposite trend was observed for the Simpson's index (D), Hailun < Fengqiu < Yingtan (see Fig. S3A). The negative correlation between H and D is consistent with the CLPP results described above, suggesting that the dominant genotypes are those that are metabolically active in straw decomposition.
Overall assessment of the dynamic changes of microbial communities during maize straw decomposition.
To assess the relative effects of location, soil, and time on microbial community structure, the PLFA, CLPP, and DGGE data presented above were subjected to ABT analysis. A total of eight parameters were analyzed, and the results are summarized in Table 2. Interestingly, the ABT data consistently showed that geographic location has larger effects than soil type on microbial communities; the only exception is total biomass, which was affected by location and soil type at similar levels of ∼30% (Table 2). Excluding total biomass, soil type exerts a smaller effect on microbial communities than location or time of decomposition. The effect of time is moderate, and only in two measures (total biomass and the Simpson's index of CLPP) did the time of decomposition produce the largest effects. A detailed analysis of location, soil, and time on each of the eight parameters is provided in Fig. S4 in the supplemental material. Taken together, we conclude that the structure of microbial communities associated with maize straw decomposition was predominantly determined by the factor of geographic location rather than soil type.
TABLE 2.
Percent contribution of the three environmental variables to variation of microbial communities as revealed by ABT analysis
| Variable | % variation in microbial abundance (PLFA) |
% variation in metabolic diversity (CLPP) |
% variation in phylogenetic diversity (DGGE) |
|||||
|---|---|---|---|---|---|---|---|---|
| Fungus/bacterium ratio | G+/G−a | Total biomass | AWCD | Shannon index | Simpson index | Shannon index | Simpson index | |
| Time | 36.71 | 35.13 | 37.95 | 36.05 | 33.09 | 60.98 | 29.85 | 22.61 |
| Location | 38.06 | 47.78 | 28.24 | 42.00 | 40.40 | 21.47 | 45.33 | 57.41 |
| Soil | 25.23 | 17.09 | 33.81 | 21.95 | 26.51 | 17.55 | 24.82 | 19.98 |
G+/G−, ratio of Gram-positive bacteria to Gram-negative bacteria.
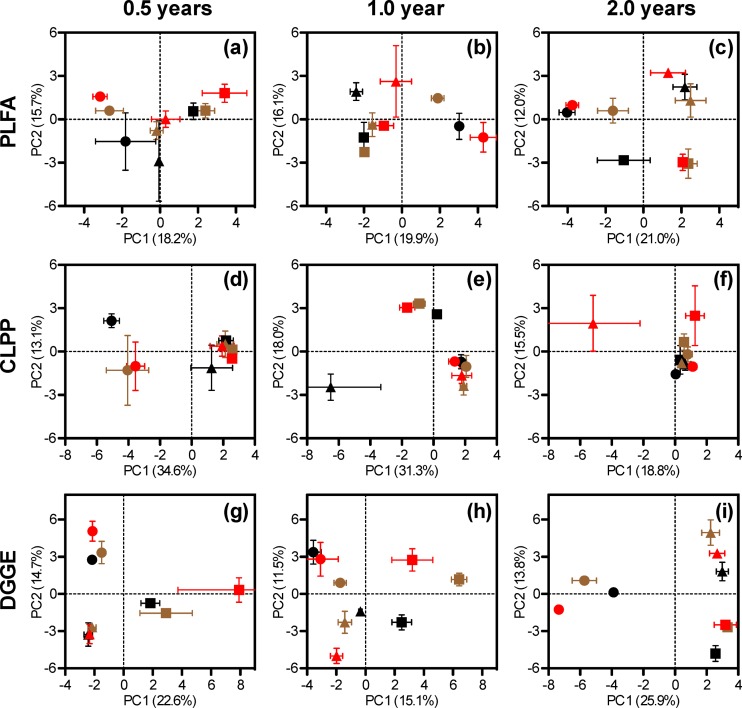
Furthermore, PCA was used to compare the effects of location and soil type over time. The results are presented in Fig. 4, with the three locations (or climates) being represented by three different shapes and the three soil types by three different colors. The analysis revealed two significant findings. First, for all 9 data sets (Fig. 4a to i), where the microbial communities can be separated from each other, they are primarily grouped by shape, not by color; for example, see the PLFA data in year 2 (Fig. 4c). Therefore, the results of PCA support the general conclusion drawn from the ABT analysis that the effects of location were larger than those of soil type on decomposing microbial communities.
FIG 4.
Principal component analysis of straw-decomposing microbial communities based on the results of PLFA, CLPP, and DGGE. Average loading scores along the first and second principal components were shown with standard errors from three repeats. Residue samples from different geographic sites and soils are differentiated by the shape and color of the symbols: Hailun (squares), Fengqiu (triangles), and Yingtan (circles), as well as black soil (black), Chao soil (brown), and red soil (red).
Second, the CLPP data in year 2 (Fig. 4f) show that the microbial communities are very similar (with the only exception being red soil in Fengqiu); however, both the PLFA and DGGE analysis (Fig. 4c and i, respectively) indicate that these communities are still very different from each other. Thus, the data indicate that after 2 years of decomposition under different soil and climate conditions, the resulting microbial communities differ in species abundance and composition but remain metabolically similar.
DNA sequencing analysis of microbial communities associated with straw decomposition.
Armed with the knowledge that geographic location has greater effects than soil type on straw decomposition, we next sought to provide further insights into the microbial community structure, with a focus on one soil (i.e., black soil) under three different climate conditions (Hailun, Fengqiu, and Yingtan) at three time points (0.5, 1, and 2 years). A total of 9 maize residue samples were subjected to construction of 16S rRNA gene libraries; from each library, 120 clones were randomly picked and analyzed first by restriction fragment length polymorphism (RFLP). An analysis with two enzymes (HhaI and Sau3AI) resulted in 137 unique restriction profiles (termed operational taxonomic units [OTUs]). The number of OTUs was plotted against the total number of clones for each of the nine libraries. Results indicate that the OTU numbers are nearly saturated for all libraries; thus, predictions of the microbial community can be made (see Fig. S6 in the supplemental material). The calculated Shannon's and Simpson's indices are consistent with the results of DGGE described above; in particular, they displayed opposite effects (data not shown). Moreover, similarities of the bacterial populations were compared using the calculated Sørensen similarity index, and results showed that population similarity decreases over time in each location (see Table S9).
We then proceeded with sequencing analysis of the full-length 16S rRNA genes that represent the 137 unique OTUs. Their phylogenetic positions were subsequently determined by a BLAST search of the 16S rRNA gene database. Results showed that the straw-decomposing bacteria belong to 10 phyla and 17 classes. The most dominant group is the Proteobacteria (62.9% in total), particularly the Rhizobiales of Alphaproteobacteria (31.0%). The next most dominant phyla are Sphingobacteria, Acidobacteria, and Planctomycetales, which take up 14.7, 11.4, and 3.8% of the total number of clones, respectively. The two most dominant clones are OTU 11 (JQ798402) and OTU 111 (JQ798528), representing 52 and 44 clones, respectively, with Variovorax sp. strain KS2D-23 (AB196432) and Granulicella sapmiensis (S002989305), respectively, as the best hits.
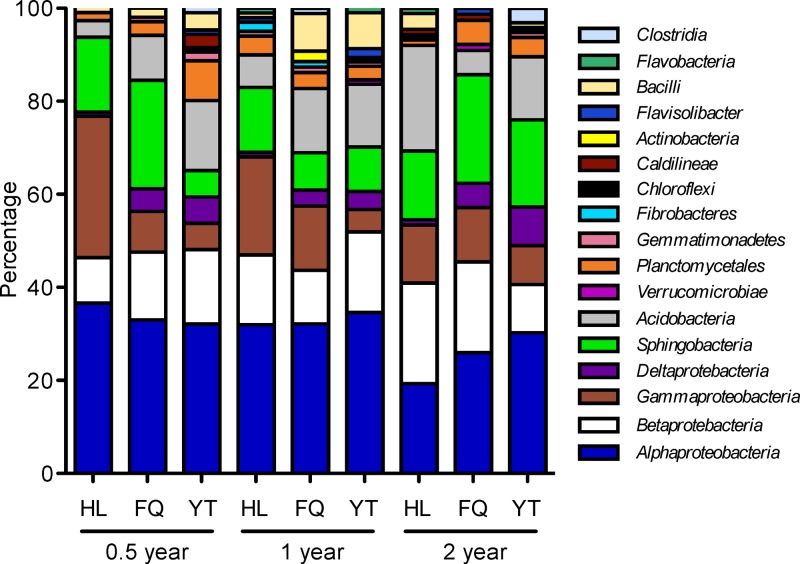
The dynamic change of microbial community at the class level is summarized in Fig. 5. Along with the progression of straw decomposition, members of Alphaproteobacteria and Gammaproteobacteria decreased in frequency, whereas Betaproteobacteria, Sphingobacteria, and Acidobacteria showed the opposite; this trend is observed more pronouncedly in the cold climate zone (i.e., Hailun). Moreover, a clear effect on relative abundance of the dominant groups of bacteria was observed by location. For example, Deltaproteobacteria showed a trend, in order from low to high abundance, in Hailun < Fengqiu < Yingtan at all three time points; the same trend was found for Alphaproteobacteria at year 2. However, the abundance of Gammaproteobacteria decreased against the thermal gradient (Hailun > Fengqiu > Yingtan).
FIG 5.
Relative abundance of different classes of bacteria associated with straw decomposition in black soil in three geographic locations of Hailun (HL), Fengqiu (FQ), and Yingtan (YT).
DGGE analysis of microbial communities in bulk soil.
The finding that straw-decomposing microbial communities was predominantly affected by location rather than soil type raises a significant question: does the same hold for microbial communities in bulk soil? To this end, we analyzed soil samples from the same sites at time zero and also at the end of the experiment (year 2) using 16S rRNA gene DGGE analysis. Of particular note are the soil microbial communities at time zero, which represent the founding microorganisms for straw decomposition. The same bunch of fresh maize straws had been subjected to heat drying at 60°C for 24 h before they were distributed to different locations for burial. Therefore, microorganisms that survived the heat drying would be negligible and consistent across all of the treatments.
Results of the PCA analysis based on the DGGE data clearly showed that at time zero (i.e., 2.5 years after the soil transplantation), the microbial communities in soils remain closely related to each other for each soil at different locations (Fig. 6A). At year 2 of the decomposition experiment (i.e., 4.5 years after transplantation), the soil microbial communities are still primarily grouped by soil type; however, cumulative succession to climate change has been observed, particularly for soils in Yingtan (Fig. 6B). Yingtan is the warmest climate region involved in this study; thus, the noticeable convergence of soil microbial communities in Yingtan suggests that microbial succession to climate change occurred more rapidly from low- to high-temperature areas compared to change from high- to low-temperature areas. Nevertheless, a comparison of the DGGE data presented in Fig. 4 and 6 revealed significant differences between microbial communities in bulk soils and those residing in straw residues. Specifically, the successive process is slower in soils than in straw residues in response to climate changes.
FIG 6.
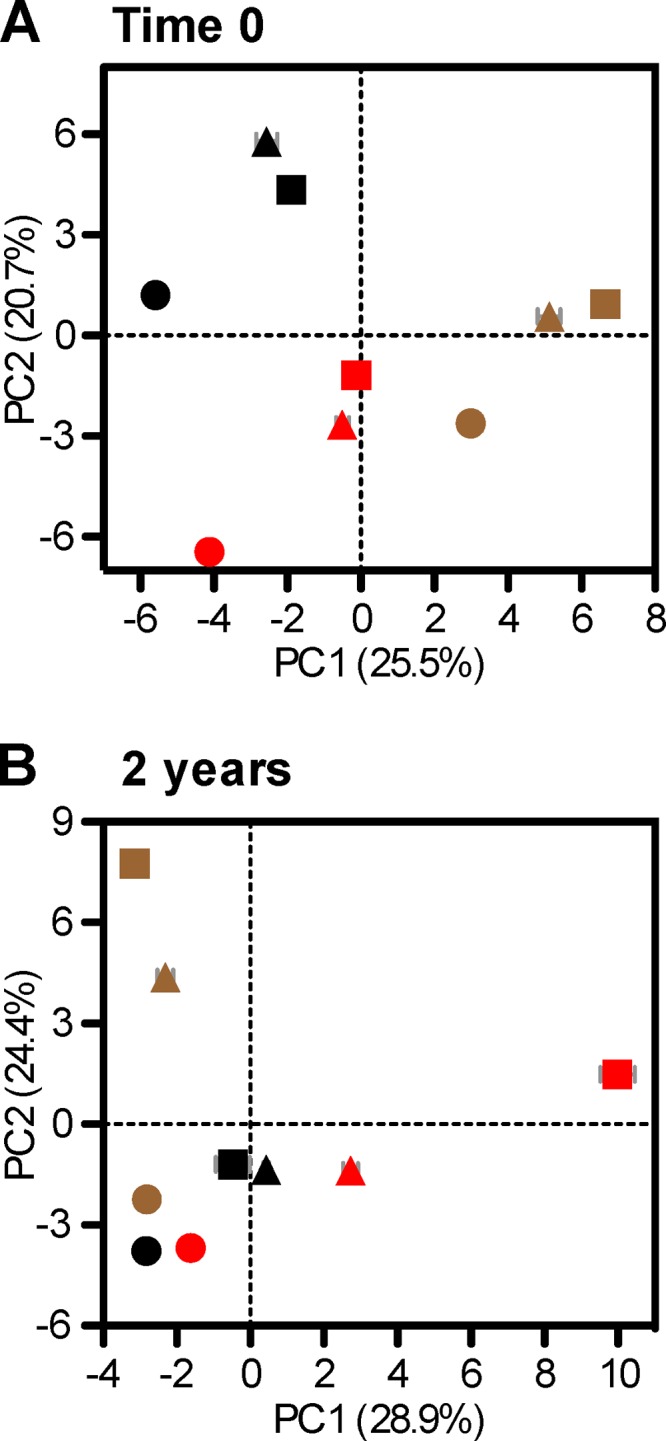
Principal component analysis of microbial communities in bulk soils based on the results of 16S rRNA gene DGGE at the beginning (A) and the end (B) of the decomposition experiment. Average loading scores along the first and second principal components were shown with standard errors from three repeats. Residue samples from different geographic sites and soils are differentiated by the shape and color of the symbols: Hailun (square), Fengqiu (triangle), and Yingtan (circle), as well as black soil (black), Chao soil (brown), and red soil (red).
DISCUSSION
Unraveling the relative importance of climate factors versus soil types in the development of soil microbial communities poses significant challenges. Data currently available are mostly based on microbiological surveys of soils under various geographic conditions or experiments of soil incubation (15, 20, 43). While these previous works have provided great insights into the effects of individual environmental factors (e.g., temperature and soil pH), the classic techniques used have a limited ability to differentiate the combined effects of climate and soil that occur in both natural and agricultural ecosystems. Thus, soil transplantation techniques have drawn increasing attention for studies of soil microbial ecology (28) and shifts of soil microbial community after transplantation have been recorded (29, 30, 44–47). To the best of our knowledge, this is the first report of a large-scale reciprocal soil transplantation experiment where the relative impacts of climate versus soil for decomposing microbial communities were assessed. The results of chemical analysis, physical analysis, and microbiological characterization consistently showed that the rate of straw decomposition (Table 1 and Fig. 1) and the community structure of decomposing microorganisms were primarily determined by the geographic locations rather than the soil types (Table 2 and Fig. 4).
The finding that a relatively smaller, albeit significant, effect was observed for soil type is very surprising. First, soil was the major (if not only) source of microorganisms involved in straw decomposition, and the initial breakdown was performed by preexisting enzymes and microbes in the soil (3). Thus, it had been our original prediction that the microbial communities should be grouped primarily by soil types, at least in the early stages of straw decomposition. In fact, the microbial communities from the same climate regions were closely related to each other, even at 0.5 years after the burial (Fig. 4). This raises the possibility that the climate-provoked successive changes of the microbial community in soil occurred rapidly within the first 2.5 years of transplantation, before the straw decomposition experiments started in 2008 (time zero). However, our subsequent analysis of the microbial communities in bulk soil indicates that this was not the case: at time zero, the initial communities for straw decomposition from the surrounding soils were similar for each type of soil in different locations (Fig. 6A).
Second, the three soils tested in this work represent three local agricultural soils from southern to northern parts of China. Their chemical properties differed significantly prior to the soils being translocated (see Table S1 in the supplemental material) and remained distinct from each other during the course of straw decomposition (see Table S2). Of particular note is the difference of soil pH for the three soils in the same location (e.g., pH 5.80 for red soil, pH 7.68 for Chao soil, and pH 6.26 for black soil in Hailun) and the more than three-times-higher levels of available nitrogen for black soil than for Chao and red soils (see Table S2). The close relatedness of straw-decomposing microbial communities for three different soils in the same climate region (Fig. 4), and also the divergence of microbial communities for the same soil in three different climate regions, strongly suggests that dynamic change of the microbial communities is driven by climate factors associated with the geographic locations; the effects of soil type are surprisingly small.
The observed strong effects of geographic location are in general agreement with many reports that highlight the importance of temperature in the decomposition of soil organic matter (20, 48, 49). As expected, maize straw decomposition occurred more slowly in the cold climate region (Hailun) than in the two warm regions, Fengqiu and Yingtan (Fig. 1). However, from our data it is unclear how the levels of bacterial diversity are determined by climate factors. For a given soil type and time point, no general trends were observed along the thermal gradient; wherever a trend can be found, it either changed over time or did not hold for another soil type. As an example of the Shannon's index of DGGE data (see Fig. S3 in the supplemental material), in year 2 there was a trend in black and Chao soils from high to low diversity (Hailun > Fengqiu > Yingtan) against the thermal gradient; however, no difference was found for red soil. It thus appears that climate change can either increase or decrease the bacterial diversity depending upon other environmental factors, such as variables of the soil. Because of the limited number of geographic sites investigated in this work, it is not possible to make a reliable assessment of the correlations between individual climate factors and the levels of bacterial diversity.
It should also be noted that the soil transplantation experiment was performed under open field conditions. Thus, local microorganisms can enter into the soil system from the top through the air, rain, or snow and from the bottom via the ground water. This mimics the natural conditions where different types of soils coexist in the same climate region. However, microorganisms from the surrounding area have been well adapted to the local climate conditions; they can potentially drive the microbial communities in the same direction (which is expected to be dominated by the local members) during the process of reestablishment of the microbial community after soil transplantation. While such an effect was undetectable for microbial communities in bulk soils by using the 16S rRNA gene DGGE analysis, it may occur for the microbial communities associated with straw decomposition. The derived nutritional substrates may disproportionately favor the growth of the invading local microorganisms, which would cause a potential overestimation of the effects of climate conditions.
Soil-dwelling bacteria can be classified into the two ecological functional categories of copiotrophs and oligotrophs (50), which are equivalent to the r- and k-strategies used to describe the ecological attributes of plants and animals (51). Copiotrophs (r-strategists) have high growth rates under nutrient-rich conditions, whereas oligotrophs (k-strategists) are slow growers that are advantageous in nutrient-poor environments. When fresh plant residues are released into the soil, the decomposing microbial community will experience a gradual shift of nutrient state from resource repletion in the early stages (as a result of a rapid degradation of labile substrates) to resource depletion in the late stages. Therefore, bacterial phyla that increase or decrease in frequency during the process of decomposition are considered copiotrophs or oligotrophs, respectively. Bastian et al. recently monitored the dynamic changes of microbial communities within the first 6 months of wheat straw decomposition (52). Their results identified Gammaproteobacteria as copiotrophs, with Actinobacteria and Deltaproteobacteria being oligotrophs.
Data presented here indicate a decrease of relative abundance over time for Gammaproteobacteria and Alphaproteobacteria (Fig. 5); moreover, there was a trend in order from high to low along the thermal gradient, particularly in year 0.5 (Hailun > Fengqiu > Yingtan). Given the lower extent of decomposition in Hailun than that in Fengqiu and Yingtan, the data consistently suggest that both Gammaproteobacteria and Alphaproteobacteria are copiotrophs. Similarly, as previously observed in wheat straw decomposition (52, 53), Deltaproteobacteria and Acidobacteria behaved like oligotrophs with higher abundance in later stages or warmer regions. Intriguingly, Sphingobacteria increased in frequency in Yingtan, as expected for an oligotroph; however, the trend did not hold at the other two sites, and in year 0.5 the proportion of Sphingobacteria was the lowest in Yingtan, which was unexpected due to its relatively higher extent of decomposition (Fig. 1 and 6). In a previous work by Fierer et al. (50), Betaproteobacteria was assigned to the copiotroph category, showing a positive relationship between its relative abundance and soil carbon availability. However, opposite dynamics were observed in this work for Betaproteobacteria, with an increase of frequency in Hailun but a decrease of frequency in Yingtan.
The inconsistency of Sphingobacteria and Betaproteobacteria levels at different sites suggests that temperature plays a very important role in determining the ecological behaviors of bacteria, and their ecological attributes may differ under different climate conditions. Responses to changes in temperature may vary between fast growers and slower growers under similar nutritional conditions. Thus, certain microbes may have adopted contrasting strategies to maximize their fitness in both warm and cold climate regions.
Taken together, our data of reciprocal soil transplantation show that climate conditions (or geographic location) played a larger role than the properties of soil in determining the rate of straw decomposition and also the structure of the associated microbial communities (bacteria in particular). Given the crucial role of plant residues in global carbon sequestration and climate change mitigation (54), the empirical data presented here provide useful information to guide the estimation or modeling of global carbon cycling in soil in the context of climate changes. Furthermore, our data suggest that microbial succession to climate change for communities in bulk soil was relatively slow (observed in this study only for soils transferred to the warmer region, Yingtan). The observed different responses between microorganisms in bulk soil and those involved in straw decomposition can be partially explained by the chemical and physical differences of the niches, pH in particular (see Table S3 in the supplemental material), and also the differential growth of a subset of microorganisms that was stimulated by the supplementation of nutritional substrates derived from the straw residues. It will be interesting to know whether or not the observations for maize straw hold for other crop residues and also in later stages of decomposition. One ongoing goal associated with this reciprocal soil transplantation project is to examine the decomposition of wheat residues.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yueyu Sui and Zhixin Dong for help with straw sampling and chemical analysis, Ryllian Zhang for critical reading of the manuscript, and the anonymous reviewers for helpful comments on the manuscript.
This work was supported by the National Basic Research Program of China (2011CB100506), National Natural Science Foundation of China (41271258), Strategic Priority Research Program of Chinese Academy of Sciences (XDA05070303), and the NZ-China Scientist Exchange Program.
Footnotes
Published ahead of print 22 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00083-13.
REFERENCES
- 1.Fageria NK. 2012. Role of soil organic matter in maintaining sustainability of cropping systems. Commun. Soil Sci. Plan. 43:2063–2113 [Google Scholar]
- 2.Lal R. 1997. Residue management, conservation tillage and soil restoration for mitigating greenhouse effect by CO2-enrichment. Soil Tillage Res. 43:81–107 [Google Scholar]
- 3.McGuire KL, Treseder KK. 2010. Microbial communities and their relevance for ecosystem models: decomposition as a case study. Soil Biol. Biochem. 42:529–535 [Google Scholar]
- 4.Baumann K, Marschner P, Smernik RJ, Baldock JA. 2009. Residue chemistry and microbial community structure during decomposition of eucalypt, wheat and vetch residues. Soil Biol. Biochem. 41:1966–1975 [Google Scholar]
- 5.Dilly O, Bloem J, Vos A, Munch JC. 2004. Bacterial diversity in agricultural soils during litter decomposition. Appl. Environ. Microbiol. 70:468–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rui J, Peng J, Lu Y. 2009. Succession of bacterial populations during plant residue decomposition in rice field soil. Appl. Environ. Microbiol. 75:4879–4886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stubbs TL, Kennedy AC, Reisenauer PE, Burns JW. 2009. Chemical composition of residue from cereal crops and cultivars in dryland ecosystems. Agron. J. 101:538–545 [Google Scholar]
- 8.Risch AC, Jurgensen MF, Frank DA. 2007. Effects of grazing and soil micro-climate on decomposition rates in a spatio-temporally heterogeneous grassland. Plant Soil 298:191–201 [Google Scholar]
- 9.Vanhala P, Karhu K, Tuomi M, Bjorklof K, Fritze H, Liski J. 2008. Temperature sensitivity of soil organic matter decomposition in southern and northern areas of the boreal forest zone. Soil Biol. Biochem. 40:1758–1764 [Google Scholar]
- 10.Christensen BT. 1985. Wheat and barley straw decomposition under field conditions: effect of soil type and plant cover on weight loss, nitrogen and potassium content. Soil Biol. Biochem. 17:691–697 [Google Scholar]
- 11.Parton W, Silver WL, Burke IC, Grassens L, Harmon ME, Currie WS, King JY, Adair EC, Brandt LA, Hart SC, Fasth B. 2007. Global-scale similarities in nitrogen release patterns during long-term decomposition. Science 315:361–364 [DOI] [PubMed] [Google Scholar]
- 12.Bardgett RD. 2011. Plant-soil interactions in a changing world. F1000 Biol. Rep. 3:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martiny JB, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, Ovreas L, Reysenbach AL, Smith VH, Staley JT. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4:102–112 [DOI] [PubMed] [Google Scholar]
- 14.de Wit R, Bouvier T. 2006. “Everything is everywhere, but, the environment selects”; what did Baas Becking and Beijerinck really say? Environ. Microbiol. 8:755–758 [DOI] [PubMed] [Google Scholar]
- 15.Maron PA, Mougel C, Ranjard L. 2011. Soil microbial diversity: methodological strategy, spatial overview and functional interest. C. R. Biol. 334:403–411 [DOI] [PubMed] [Google Scholar]
- 16.Brown JH, Gillooly JF, Allen AP, Savage VM, West GB. 2004. Toward a metabolic theory of ecology. Ecology 85:1771–1789 [Google Scholar]
- 17.Prosser JI, Bohannan BJ, Curtis TP, Ellis RJ, Firestone MK, Freckleton RP, Green JL, Green LE, Killham K, Lennon JJ, Osborn AM, Solan M, van der Gast CJ, Young JP. 2007. The role of ecological theory in microbial ecology. Nat. Rev. Microbiol. 5:384–392 [DOI] [PubMed] [Google Scholar]
- 18.Tuomi M, Thum T, Jarvinen H, Fronzek S, Berg B, Harmon M, Trofymow JA, Sevanto S, Liski J. 2009. Leaf litter decomposition–estimates of global variability based on Yasso07 model. Ecol. Model 220:3362–3371 [Google Scholar]
- 19.Stegen JC, Enquist BJ, Ferriere R. 2009. Advancing the metabolic theory of biodiversity. Ecol. Lett. 12:1001–1015 [DOI] [PubMed] [Google Scholar]
- 20.Conant RT, Ryan MG, Agren GI, Birge HE, Davidson EA, Eliasson PE, Evans SE. 2011. Temperature and soil organic matter decomposition rates—synthesis of current knowledge and a way forward. Global Change Biol. 17:3392–3404 [Google Scholar]
- 21.Davidson EA, Janssens IA. 2006. Temperature sensitivity of soil carbon decomposition and feedbacks to climate change. Nature 440:165–173 [DOI] [PubMed] [Google Scholar]
- 22.Fierer N, Craine JM, McLauchlan K, Schimel JP. 2005. Litter quality and the temperature sensitivity of decomposition. Ecology 86:320–326 [Google Scholar]
- 23.Papke RT, Ward DM. 2004. The importance of physical isolation to microbial diversification. FEMS Microbiol. Ecol. 48:293–303 [DOI] [PubMed] [Google Scholar]
- 24.Fierer N, Strickland MS, Liptzin D, Bradford MA, Cleveland CC. 2009. Global patterns in belowground communities. Ecol. Lett. 12:1238–1249 [DOI] [PubMed] [Google Scholar]
- 25.Fierer N, Jackson RB. 2006. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U. S. A. 103:626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffiths RI, Thomson BC, James P, Bell T, Bailey M, Whiteley AS. 2011. The bacterial biogeography of British soils. Environ. Microbiol. 13:1642–1654 [DOI] [PubMed] [Google Scholar]
- 27.Rousk J, Brookes PC, Baath E. 2009. Contrasting soil pH effects on fungal and bacterial growth suggest functional redundancy in carbon mineralization. Appl. Environ. Microbiol. 75:1589–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazzaro A, Gauer A, Zeyer J. 2011. Field-scale transplantation experiment to investigate structures of soil bacterial communities at pioneering sites. Appl. Environ. Microbiol. 77:8241–8248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Waldrop MP, Firestone MK. 2006. Response of microbial community composition and function to soil climate change. Microb. Ecol. 52:716–724 [DOI] [PubMed] [Google Scholar]
- 30.Vanhala P, Karhu K, Tuomi M, Bjorklof K, Fritze H, Hyvarinen H, Liski J. 2010. Transplantation of organic surface horizons of boreal soils into warmer regions alters microbiology but not the temperature sensitivity of decomposition. Global Change Biol. 17:538–550 [Google Scholar]
- 31.Nelson DW, Sommers LE. 1996. Total carbon, organic carbon, and organic matter, p 961–1010 Sparks DL, Page AL, Helmke PA, Loeppert RH, Soltanpour PN, Tabatabai MA, Johnston CT, Sumner ME. (ed), Methods of soil analysis. Part 3–chemical methods. Soil Science Society of America, Inc., and American Society of Agronomy, Inc., Madison, WI [Google Scholar]
- 32.Novozamsky I, Houba V, Van Eck R. 1983. A novel digestion technique for multi-element plant analysis. Commun. Soil Sci. Plant Anal. 14:239–248 [Google Scholar]
- 33.Zhong WH, Gu T, Wang W, Zhang B, Lin XG, Huang QR, Shen WS. 2009. The effects of mineral fertilizer and organic manure on soil microbial community and diversity. Plant Soil 326:511–522 [Google Scholar]
- 34.Sun B, Dong ZX, Zhang XX, Li Y, Cao H, Cui ZL. 2011. Rice to vegetables: short- versus long-term impact of land-use change on the indigenous soil microbial community. Microb. Ecol. 62:474–485 [DOI] [PubMed] [Google Scholar]
- 35.Zak JC, Willig MR, Moorhead DL, Wildman HG. 1994. Functional diversity of microbial communities: a quantitative approach. Soil Biol. Biochem. 26:1101–1108 [Google Scholar]
- 36.Woese CR, Gutell R, Gupta R, Noller HF. 1983. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol. Rev. 47:621–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakatsu CH, Torsvik V, Øvreas L. 2000. Soil Community analysis using DGGE of 16S rDNA polymerase chain reaction products. Soil Sci. Soc. Am. J. 64:1382–1388 [Google Scholar]
- 38.Cahyani VR, Matsuya K, Asakawa S, Kimura M. 2003. Succession and phylogenetic composition of bacterial communities responsible for the composting process of rice straw estimated by PCR-DGGE analysis. Soil Sci. Plant Nutr. 49:619–630 [Google Scholar]
- 39.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. Wiley & Sons, Chichester, United Kingdom [Google Scholar]
- 40.Zhang XX, Kosier B, Priefer UB. 2001. Genetic diversity of indigenous Rhizobium leguminosarum bv. viciae isolates nodulating two different host plants during soil restoration with alfalfa. Mol. Ecol. 10:2297–2305 [DOI] [PubMed] [Google Scholar]
- 41.De'ath G. 2007. Boosted trees for ecological modeling and prediction. Ecology 88:243–251 [DOI] [PubMed] [Google Scholar]
- 42.Zhou J, Xue K, Xie J, Deng Y, Wu L, Cheng X, Fei S, Deng S, He Z, Nostrand V, Luo Y. 2012. Microbial mediation of carbon-cycle feedbacks to climate warming. Nat. Climate Change 2:106–110 [Google Scholar]
- 43.Fang C, Smith P, Moncrieff JB, Smith JU. 2005. Similar response of labile and resistant soil organic matter pools to changes in temperature. Nature 433:57–59 [DOI] [PubMed] [Google Scholar]
- 44.Zumsteg A, Bernasconi S, Zeyer J, Frey B. 2011. Microbial community and activity shifts after soil transplantation. Appl. Geochem. 26:S326–S329 [Google Scholar]
- 45.Boyle SA, Rich JJ, Bottomley PJ, Cromack KC, Myrold DD. 2006. Reciprocal transfer effects on denitrifying community composition and activity at forest and meadow sites in the Cascade Mountains of Oregon. Soil Biol. Biochem. 38:870–878 [Google Scholar]
- 46.Balser T, Firestone MK. 2005. Linking microbial community composition and soil processes in a California annual grassland and mixed-conifer forest. Biogeochemistry 73:395–415 [Google Scholar]
- 47.Hannam KD, Quideau SA, Kishchuk BE. 2007. The microbial communities of aspen and spruce forest floors are resistant to changes in litter inputs and microclimate. Appl. Soil Ecol. 35:635–647 [Google Scholar]
- 48.Teklay T, Shi Z, Attaeian B, Chang SX. 2010. Temperature and substrate effects on C & N mineralization and microbial community function of soils from a hybrid poplar chronosequence. Appl. Soil Ecol. 46:413–421 [Google Scholar]
- 49.Feng X, Simpson MJ. 2009. Temperature and substrate controls on microbial phospholipid fatty acid composition during incubation of grassland soils contrasting in organic matter quality. Soil Biol. Biochem. 41:804–812 [Google Scholar]
- 50.Fierer N, Bradford MA, Jackson RB. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354–1364 [DOI] [PubMed] [Google Scholar]
- 51.MacArthur R, Wilson E. 1967. The theory of island biogeography. Princeton University Press, Princeton, NJ [Google Scholar]
- 52.Bastian F, Bouziri L, Nicolardot B, Ranjard L. 2009. Impact of wheat straw decomposition on successional patterns of soil microbial community structure. Soil Biol. Biochem. 41:262–275 [Google Scholar]
- 53.Bernard L, Mougel C, Maron PA, Nowak V, Leveque J, Henault C, Haichar FZ, Berge O, Marol C, Balesdent J, Gibiat F, Lemanceau P, Ranjard L. 2007. Dynamics and identification of soil microbial populations actively assimilating carbon from 13C-labelled wheat residue as estimated by DNA- and RNA-SIP techniques. Environ. Microbiol. 9:752–764 [DOI] [PubMed] [Google Scholar]
- 54.Schimel JP, Schaeffer SM. 2012. Microbial control over carbon cycling in soil. Front. Microbiol. 3:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.