Abstract
Gut bacteria play a key role in the metabolism of dietary isoflavones, thereby influencing the availability and bioactivation of these polyphenols in the intestine. The human intestinal bacterium Slackia isoflavoniconvertens converts the main soybean isoflavones daidzein and genistein to equol and 5-hydroxy-equol, respectively. Cell extracts of S. isoflavoniconvertens catalyzed the conversion of daidzein via dihydrodaidzein to equol and that of genistein to dihydrogenistein. Growth of S. isoflavoniconvertens in the presence of daidzein led to the induction of several proteins as observed by two-dimensional difference gel electrophoresis. Based on determined peptide sequences, we identified a cluster of eight genes encoding the daidzein-induced proteins. Heterologous expression of three of these genes in Escherichia coli and enzyme activity tests with the resulting cell extracts identified the corresponding gene products as a daidzein reductase (DZNR), a dihydrodaidzein reductase (DHDR), and a tetrahydrodaidzein reductase (THDR). The recombinant DZNR also converted genistein to dihydrogenistein at higher rates than were observed for the conversion of daidzein to dihydrodaidzein. Higher rates were also observed with cell extracts of S. isoflavoniconvertens. The recombinant DHDR and THDR catalyzed the reduction of dihydrodaidzein to equol, while the corresponding conversion of dihydrogenistein to 5-hydroxy-equol was not observed. The DZNR, DHDR, and THDR were expressed as Strep-tag fusion proteins and subsequently purified by affinity chromatography. The purified enzymes were further characterized with regard to their activity, stereochemistry, quaternary structure, and content of flavin cofactors.
INTRODUCTION
Intestinal bacteria play a crucial role in the conversion of dietary isoflavones and may therefore contribute to the proposed health-promoting effects of these polyphenols (1–3). Knowledge on the bacteria and their enzymes responsible for polyphenol conversion is still limited even though these bacterial activities contribute to different polyphenol-metabolizing phenotypes of the host (referred to as metabotypes) (4). The soybean isoflavone daidzein is activated to equol exclusively by intestinal bacteria, and this activation occurs only in a subset of humans (5). Equol shows biological properties superior to those of its precursor daidzein, suggesting that equol formation results in enhanced beneficial effects on human health (6, 7). Slackia isoflavoniconvertens is one of the few characterized equol-forming gut bacteria isolated from humans (8). Besides conversion of daidzein via dihydrodaidzein to equol, S. isoflavoniconvertens, moreover, catalyzes the corresponding formation of 5-hydroxy-equol from genistein via dihydrogenistein (8). Interestingly, 5-hydroxy-equol has increased antioxidant activity compared to its precursor genistein (9). Daidzein and genistein are converted by S. isoflavoniconvertens not only in vitro but also in vivo (10). Studies with S. isoflavoniconvertens and the mouse intestinal bacterium Enterorhabdus mucosicola indicated that expression of the isoflavone-converting enzymes is induced by their substrates (8, 11). For a long time, nothing was known about the bacterial enzymes involved in isoflavone conversion. The mechanism of dihydrodaidzein conversion to equol via tetrahydrodaidzein has been studied in Eggerthella sp. strain Julong 732 (12, 13), and genes involved in daidzein conversion to equol have been identified only recently in two bacterial strains from the human intestine, Lactococcus sp. strain 20-92 and Slackia sp. strain NATTS (14–17).
Here we describe the detection of proteins upregulated in S. isoflavoniconvertens in the presence of daidzein and the identification of the corresponding genes. Three of the genes were functionally expressed in Escherichia coli. They encode a daidzein reductase (DZNR), a dihydrodaidzein reductase (DHDR), and a tetrahydrodaidzein reductase (THDR). The DZNR also catalyzed the conversion of genistein to dihydrogenistein. The three reductases were subsequently purified as tagged proteins and characterized.
MATERIALS AND METHODS
Chemicals.
Daidzein was purchased from Acros Organics (Geel, Belgium). Dihydrodaidzein was obtained from Toronto Research Chemicals (Toronto, Canada) and equol from Fluka (Deisenhofen, Germany). Genistein was purchased from Roth (Karlsruhe, Germany). Dihydrogenistein and 5-hydroxy-equol were available from a previous study (11). The isoflavonoids were dissolved in dimethyl sulfoxide (DMSO) and, for supplementation of growth media, subjected to sterile filtration.
Bacterial strains, media, and culture conditions.
S. isoflavoniconvertens DSM 22006 was grown under anoxic conditions in brain heart infusion (BHI) broth (Roth, Karlsruhe, Germany) as described previously (8) or in Gifu anaerobic medium (GAM) broth (Nissui Pharmaceutical, Tokyo, Japan) supplemented with 10 g/liter arginine. Competent cells of E. coli JM109 and E. coli KRX (Promega, Germany) were used for cloning and expression experiments. Transformants were selected on Luria-Bertani (LB) agar Lennox (Roth) containing carbenicillin (50 to 100 μg/ml), isopropyl-β-d-thiogalactopyranoside (IPTG; 120 μg/ml), and 5-bromo-4-chloro-indolyl-β-d-galactopyranoside (X-Gal; 40 to 80 μg/ml). The resulting E. coli JM109 clones were grown in LB broth containing carbenicillin (100 μg/ml). E. coli KRX clones were grown in Terrific broth (TB) medium containing (per liter) 12 g of tryptone, 24 g of yeast extract, 4 ml of glycerol, and 20 ml of 4.45 M potassium phosphate buffer (pH 8.2) and supplemented with carbenicillin (50 μg/ml). For growth under anoxic conditions, the media were supplemented with cysteine hydrochloride (0.5 g/liter) and resazurin (1 mg/liter) (both from Roth). The gas phase was H2-CO2 (80:20 [vol/vol]) for S. isoflavoniconvertens and N2-CO2 (80:20 [vol/vol]) for E. coli. The incubation temperature was 37°C unless otherwise noted. Bacterial growth was monitored turbidometrically by evaluation of the optical density at 600 nm (OD600).
Preparation of cell extracts.
S. isoflavoniconvertens was grown in GAM broth supplemented with 50 μM daidzein (added from a 40 mM stock solution; 1.25 ml/liter) or with the solvent DMSO. The preparation of cell extracts was done under strictly anoxic conditions by use of an anoxic workstation (MACS anaerobic workstation; Don Whitley Scientific Ltd., Shipley, United Kingdom) containing a gas atmosphere of N2-CO2-H2 (80:10:10 [vol/vol/vol]). The cells were pelleted by centrifugation (10,000 × g, 15 min, 4°C) and washed twice with 50 mM reduced potassium phosphate buffer (pH 6.8) containing cysteine hydrochloride (0.5 g/liter) and resazurin (1 mg/liter) (PPBred). The cell pellet was resuspended in PPBred supplemented with a protease inhibitor cocktail (Complete; Roche, Mannheim, Germany) (1×), and aliquots were transferred to 2-ml screw-cap tubes with an O-ring seal containing 0.5 g of 0.1-mm-diameter zirconium/silica beads (Roth, Karlsruhe, Germany). The cells were mechanically disrupted at a speed of 6.0 m/s applied twice for 20 s each time with a 5-min rest period on ice between runs (FP120 FastPrep Cell Disrupter; Qbiogene, Carlsbad, CA). Cell extracts were subsequently obtained by centrifugation (14,000 × g, 20 min, 4°C). The protein concentration was determined by the Bio-Rad Bradford assay using bovine serum albumin as the standard.
PAGE.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was carried out according to Laemmli (18) using 13% polyacrylamide gels and prestained marker proteins (PageRuler Plus prestained protein ladder; Fermentas, St. Leon-Rot, Germany). Native PAGE was performed with 4% to 15% precast linear gradient polyacrylamide gels (Ready Gel Tris-HCl; Bio-Rad, Erlangen, Germany). High-molecular-mass marker proteins (HMW electrophoresis calibration kit; GE Healthcare, Freiburg, Germany) served as standards. Gels were run in Tris/glycine buffer (24 mM Tris, 188 mM glycine, pH 8.5) at 150 V. Proteins were stained with Coomassie brilliant blue R250 (0.2% in methanol/acetic acid/water) (45:45:10 [vol/vol/vol]).
Enzyme assays.
The assays were performed at 37°C under anoxic conditions, if not indicated otherwise. In a final volume of 1 ml, the assay contained 50 mM PPBred, 2.5 μl of a 40 mM stock solution of the isoflavonoid (initially available concentration determined by high-performance liquid chromatography [HPLC] based on calibration curves), 0.6 mM NADPH or NADH or no coenzyme, and 50 μl of an enzyme source (approximately 4 mg protein/ml). For assays performed under oxic conditions, PPBred was replaced by potassium phosphate buffer (pH 6.8) and NADPH was used as a coenzyme. The activity of purified enzymes was determined in the presence of 1.2 mM NADPH. For determination of kinetic parameters (Km, Vmax), the assay contained 50 mM PPBred, a 0.5 to 40 μM concentration of the isoflavonoid (addition of 25 μl of 10 to 800 μM stock solutions), 0.6 mM NADPH, and 25 μl of an enzyme source (approximately 0.16 mg protein/ml) in a final volume of 0.5 ml. Samples were withdrawn at different time points and mixed with one volume of methanol-H2O-acetic acid (10:1:1 [vol/vol/vol]) to stop the reaction. The mixture was centrifuged (18,000 × g, 5 min), and an aliquot (20 μl) of the supernatant was analyzed by reversed-phase HPLC. Enzyme activities were determined based on the product formed within the initial 5 min of the reaction. Data are presented as the means of the results of triplicate experiments ± standard errors of the means (SEM). Kinetic parameters (Km, Vmax) were determined with GraFit data analysis software (Erithacus Software, Surrey, United Kingdom).
HPLC analyses.
Analysis was done as described previously (11) using an Ultimate 3000 system (Dionex, Idstein, Germany) equipped with a WPS-3000T SL autosampler, a LPG-3400SD pump, a DAD-3000 diode array detector, and a TCC-3000SD column oven and controlled by Chromeleon software (version 6.80; Dionex, Sunnyvale, CA). Calibration curves of the corresponding standard compounds were used for quantification. HPLC fractions containing chiral metabolites were collected manually and analyzed on a Chiralcel OJ-RH column (Daicel, Eschborn, Germany) (5 μm pore size, 150 by 4.6 mm) using the HPLC system described above. The column temperature was kept at 35°C. The mobile phase was a mixture of water acidified to pH 3 with trifluoroacetic acid and acetonitrile (60:40 [vol/vol]) maintained at a flow rate of 0.5 ml/min for 30 min. Detection was at 280 nm. UV spectra were recorded in the range of 200 to 400 nm. Enantiomers were assigned based on standard compounds and their elution behavior according to literature data (19, 20).
Two-dimensional gel electrophoresis and analysis of peptide sequences.
The cell extracts were prepared from S. isoflavoniconvertens grown overnight in 250 ml BHI broth supplemented with 300 μl of 40 mM daidzein or DMSO. The cells were centrifuged (10,000 × g, 4°C, 10 min) and washed five times in washing buffer (10 mM Tris [pH 8], 5 mM magnesium acetate, 30 mg/liter chloramphenicol) supplemented with a protease inhibitor mix (GE Healthcare) (diluted 1:100). The preparation of cell extracts was done with a cell disrupter as described previously (21) at a speed of 6.0 m/s. The protein concentration was determined as described above. Two-dimensional difference gel electrophoresis (2D-DIGE), electrophoresis of preparative gels, in-gel protein digestion, and mass spectrometric (MS) analysis employing automated data-dependent acquisition (DDA) of tryptic peptides were performed as described previously (21). Samples of bacterial extracts contained 50 μg of protein for DIGE gels and 500 μg of protein for preparative gels. For de novo sequencing performed using ProteinLynx Global Server software (version 2.3; Waters), a fragment tolerance of 0.05 Da and an estimated calibration error of 5 ppm were applied.
Amplification of gene fragments by PCR.
Genomic DNA of S. isoflavoniconvertens was isolated from cells grown in BHI broth using an RTP Bacteria DNA minikit (Invitek, Berlin, Germany). Degenerate primers based on determined peptide sequences (see Table S1 in the supplemental material) were used for amplification of specific DNA regions. The PCR mixture (25 μl) contained 1× DreamTaq Green buffer, 0.5 mM MgCl2, 0.2 mM (each) deoxynucleoside triphosphate (dNTP), 0.2 μM (each) primer, 2.5 U DreamTaq DNA polymerase (Fermentas, St. Leon-Rot, Germany), and 0.5 µl genomic DNA. The PCR program was as follows: 95°C for 3 min, 30 cycles of 95°C for 1 min, 62 to 65°C for 1 min, and 72°C for 5 min, and, finally, 72°C for 10 min. The PCR products were purified from a 1% agarose gel (innuPREP DOUBLEpure kit; Biometra, Goettingen, Germany). Ligation of the PCR products into the plasmid pGEM-T Easy and subsequent transformation of E. coli JM109 were carried out with a pGEM-T Easy Vector system (Promega, Madison, WI). Plasmid DNA was isolated from positive transformants with an innuPREP Plasmid minikit (Biometra, Goettingen, Germany) and sequenced (Eurofins MWG Operon, Ebersberg, Germany).
Inverse PCR.
The flanking regions of the amplified DNA sequence described above were obtained by inverse PCR (22) using oligonucleotide primers homologous to the known sequence but facing in opposite orientations (see Table S1 in the supplemental material). Genomic DNA of S. isoflavoniconvertens was isolated as described above and digested with SphI (FastDigest; Fermentas, St. Leon-Rot, Germany). The resulting DNA fragments were purified (innuPREP PCR Pure kit; Biometra, Goettingen, Germany), diluted to 10 ng/μl, and circularized by incubation with T4 DNA ligase (NEB, Germany) (20 U/μl) overnight at 16°C. The DNA was precipitated by adding 4 μl of 3 M sodium acetate and 100 μl of ice-cold ethanol to 40 μl of the ligation mixture and incubation at −20°C for 20 min. The precipitated DNA was collected by centrifugation (16,000 × g, 4°C, 20 min), washed with 70% ice-cold ethanol, air dried, and resuspended in 20 μl of water. The PCR mixture (25 μl) contained 1× LongAmp Taq reaction buffer, 0.3 mM (each) dNTP, 0.4 μM (each) primer, 2.5 U LongAmp Taq DNA polymerase (NEB, Germany), and 4 μl of the ligation mixture as the template. The PCR program was as follows: 94°C for 30 s, 30 cycles of 94°C for 30 s, 55°C for 1 min, and 65°C for 7 min, and, finally, 65°C for 10 min. The resulting PCR product was purified from a 1% agarose gel, cloned with a pGEM-T Easy Vector system as described above, and sequenced. Further inverse PCR experiments were performed with the restriction enzymes FspI, SalI, SapI, EcoRI (FastDigest; Fermentas, St. Leon-Rot, Germany), and BsgI (NEB, Germany) and primer sets based on the sequence information obtained before. Additional primers were used for sequencing of the obtained PCR products (see Table S1 in the supplemental material).
Sequence analysis.
The Vector NTI Suite 9 software package (Invitrogen, Carlsbad, CA) was employed to process and assemble the sequenced DNA fragments. Sequence similarity searching was done with the Basic Local Alignment Search Tool (BLAST; http://www.ncbi.nlm.nih.gov/blast). Searching for putative transcription promoter and terminator sequences was done using the Web-based programs BPROM (Softberry, Mount Kisco, NY) and ARNold (http://rna.igmors.u-psud.fr/toolbox/arnold), respectively. For prediction of the protein localization and the membrane protein topology, the Web-based program PSORT (http://psort.hgc.jp) was applied.
Heterologous gene expression.
For the construction of expression plasmids, DNA regions of S. isoflavoniconvertens comprising the individual genes were amplified by PCR. The conditions were as described for inverse PCR using appropriate primers (see Table S1 in the supplemental material) and 1 μl genomic DNA as the template. After ligation of the purified PCR products into pGEM-T Easy, E. coli JM109 or E. coli KRX was transformed with the resulting plasmids. E. coli cultures were grown either aerobically or anaerobically. The expression of heterologous genes in E. coli JM109 transformants was induced with 0.4 mM IPTG 3 h after inoculation, and cultivation was continued overnight. Heterologous gene expression in E. coli KRX clones was induced via T7 RNA polymerase expression with 0.1% (wt/vol) rhamnose 3 h after inoculation, and cultivation was continued overnight at 25°C. Cell extracts of E. coli clones were prepared under oxic or anoxic conditions as described above for S. isoflavoniconvertens.
Expression and purification of tagged enzymes.
For purification, Strep-tag II was genetically fused to the C-terminal end of each protein by use of a StarGate cloning system (IBA, Goettingen, Germany). Appropriate primers were designed with StartPrimer D'Signer software for gene amplification by PCR using the expression plasmids (5 ng) as the template (see Table S1 in the supplemental material). The general composition of the PCR mixture was as described for inverse PCR. The PCR program was as follows: 94°C for 30 s, 20 cycles of 94°C for 30 s, 56°C for 1 min, and 65°C for 5 min, and, finally, 65°C for 10 min. The PCR products were ligated using the donor vector pENTRY-IBA51 into the acceptor vector pPSG-IBA3 providing C-terminal Strep-tag II, and E. coli KRX was transformed with the resulting plasmids. Expression of the recombinant tagged proteins was induced by rhamnose, and the preparation of cell extracts was performed as described before. Approximately 11 mg of total protein was obtained from 140 mg cell mass and used for the following one-step purification procedure. The Strep-tag II proteins were purified using a Strep-tag starter kit (IBA, Goettingen, Germany). Cell extract (1 ml; 10 to 14 mg protein) was loaded by gravity flow onto a 1-ml Strep-Tactin Sepharose column, and, after washing, the Strep-tag II proteins were eluted with the elution buffer containing 2.5 mM desthiobiotin. The concentration of the purified proteins was determined as described above. Depending on the individual enzyme, the yield ranged from 0.1 to 2.3 mg of protein per run.
Determination of flavin cofactors.
UV and visible light (UV/Vis) spectra of purified enzymes were recorded on a Tecan Infinite M200 PRO multimode reader in the range of 230 to 600 nm. The protein concentration ranged from 0.1 to 3.5 mg/ml. The flavin cofactors were extracted from the proteins by trichloroacetic-acid treatment and identified by thin-layer chromatography (TLC) on silica gel 60 (Merck) as described previously (23) using flavin adenine dinucleotide (FAD) (100 pmol; Rf = 0.17), flavin mononucleotide (FMN) (100 pmol; Rf = 0.36), and riboflavin (50 pmol, Rf = 0.61) as standards. The amount of samples applied was equivalent to approximately 30 μg of extracted protein. The bands were visualized with an UV lamp at 366 nm.
Nucleotide sequence accession numbers.
The genomic DNA sequences from S. isoflavoniconvertens have been deposited in the GenBank database under accession no. JQ358709 (ifcA, ifcB, ifcC, tdr, ddr, ifcD, and dzr) and accession no. KC143072 (ifcE).
RESULTS
Conversion of daidzein and genistein by cell extracts of S. isoflavoniconvertens.
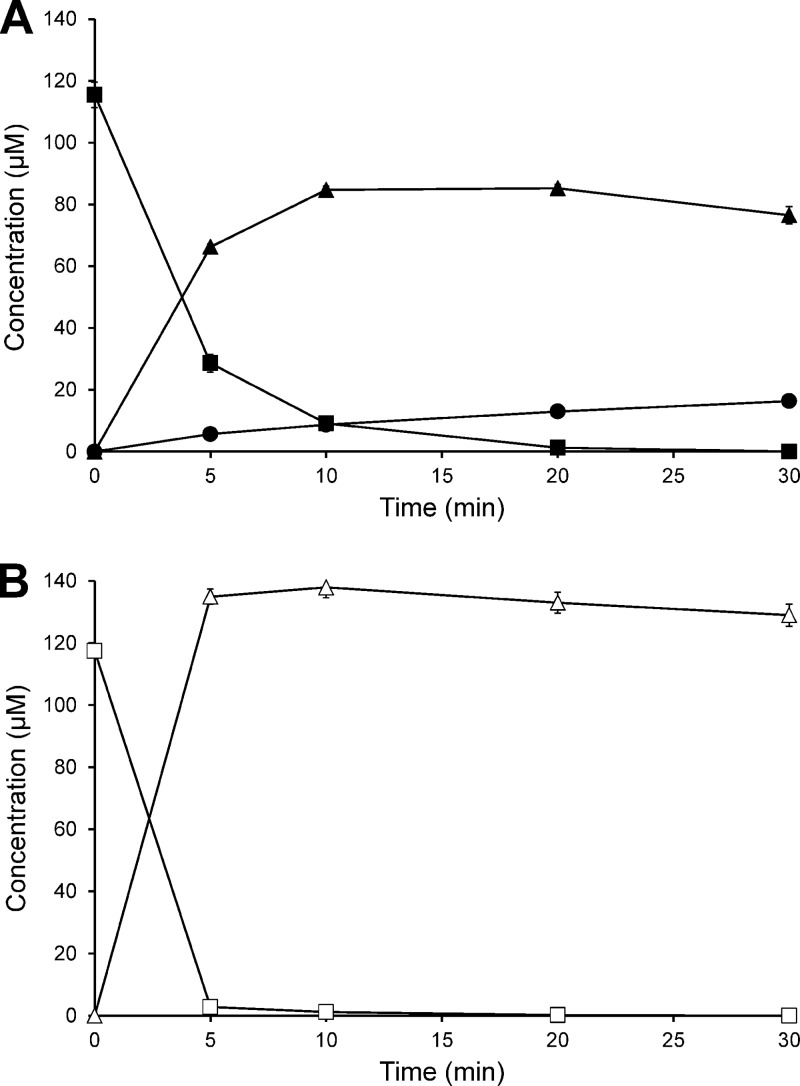
Cell extracts of S. isoflavoniconvertens grown in the presence of daidzein converted daidzein to dihydrodaidzein and small amounts of equol with NADPH as a coenzyme (Fig. 1A). No daidzein conversion was observed with cell extracts of S. isoflavoniconvertens grown in the absence of this isoflavone. Genistein was transformed in the presence of NADPH to dihydrogenistein by cell extracts of S. isoflavoniconvertens grown with daidzein, and no further conversion to 5-hydroxy-equol was observed within 30 min (Fig. 1B). Analysis of the metabolites by chiral HPLC revealed that a racemic mixture (1:1 ratio) of (S)-dihydrodaidzein (retention time [Rt] = 8.6 min) and (R)-dihydrodaidzein (Rt = 10.2 min) and, subsequently, (S)-equol (Rt = 16.1 min) was formed from daidzein. The conversion of genistein yielded a racemic mixture (1:1 ratio) of (S)-dihydrogenistein (Rt = 11.4 min) and (R)-dihydrogenistein (Rt = 13.6 min).
Fig 1.
Time course of the conversion of (A) daidzein and (B) genistein by a cell extract of S. isoflavoniconvertens in the presence of NADPH under strictly anoxic conditions. ■, daidzein; ▲, dihydrodaidzein; •, equol; □, genistein; Δ, dihydrogenistein. The symbols represent the means of the results of triplicate experiments. Error bars indicate SEM.
In the presence of NADPH, dihydrodaidzein formation from daidzein occurred at an initial rate of 36.0 ± 1.1 nmol min−1 mg protein−1. NADPH-dependent dihydrogenistein formation from genistein proceeded at a 1.8-fold-higher rate (63.5 ± 1.7 nmol min−1 mg protein−1). Formation rates were considerably lower when NADH was used instead of NADPH (for dihydrodaidzein, 6.20 ± 0.07 nmol min−1 mg protein−1; for dihydrogenistein, 9.85 ± 0.08 nmol min−1 mg protein−1). The formation of dihydrodaidzein and dihydrogenistein from daidzein and genistein, respectively, occurred at considerably lower rates of 3.93 ± 0.01 nmol min−1 mg protein−1 and 6.48 ± 0.04 nmol min−1 mg protein−1, respectively, when no coenzyme was added. Further conversion to equol or 5-hydroxy-equol was observed neither with NADH nor without coenzymes within 30 min of incubation. Dihydrodaidzein alone was slowly converted to equol (0.36 ± 0.01 nmol min−1 mg protein−1) in the presence of NADPH, whereas 5-hydroxy-equol formation from dihydrogenistein was not observed.
Identification of genes encoding daidzein-induced proteins.
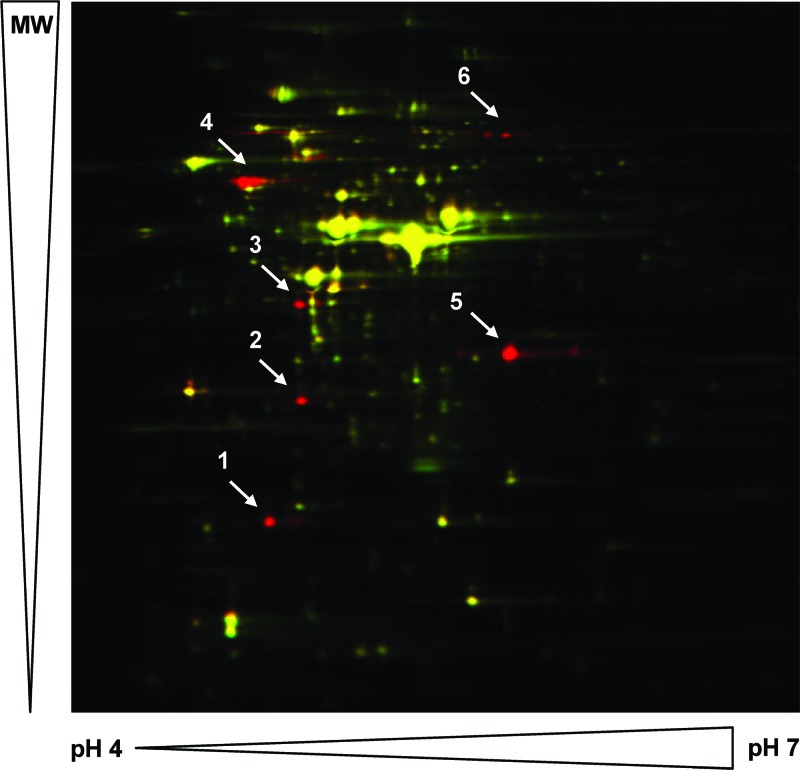
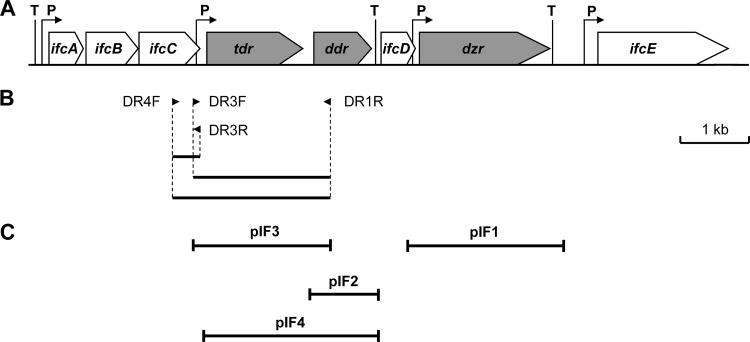
Proteins showing increased expression in S. isoflavoniconvertens cells grown in the presence of daidzein (48 μM) were detected by the 2D-DIGE technique. Six protein spots or groups of proteins were clearly upregulated 24- to 210-fold in response to the presence of daidzein (Fig. 2; see also Table S2 in the supplemental material). To identify their encoding genes, the spots of the upregulated proteins were cut out of the gel and tryptically digested and the resulting peptides subjected to de novo sequencing by MS analyses. Taking the codon usage of the closely related Slackia heliotrinireducens into consideration, degenerate primers were deduced from three peptide sequences of two of the induced proteins and used subsequently to amplify a 2.3-kb genomic DNA region of S. isoflavoniconvertens (Fig. 3). Following amplification of adjacent DNA regions by inverse PCR, a genomic DNA region of 10.4 kb was subsequently sequenced. Analysis of the sequence revealed eight open reading frames (ORFs), all oriented in the same direction (Fig. 3A). The derived amino acid sequences of six ORFs matched several peptide sequences of the daidzein-induced proteins determined by de novo sequencing (see Table S2 in the supplemental material), which confirmed the successful identification of their encoding genes. The calculated molecular masses and isoelectric points of the gene products (Table 1) were in good agreement with the positions of the corresponding proteins in the 2D-DIGE gel (Fig. 2). The gene products of dzr and ifcD could not be identified as induced proteins by DIGE analysis. For the putative IfcD protein, this may be explained by its high isoelectric point, which is outside the range of the 2D gel. However, the dzr gene product should have been detectable on the gel according to its size and isoelectric point. Thus, this protein was possibly lost during the experimental procedure.
Fig 2.
2D-DIGE gel image overlay of proteins extracted from S. isoflavoniconvertens grown in the presence of daidzein (red spots) and in its absence (green spots). Spot 1, IfcA; spot 2, IfcB; spot 3, IfcC; spot 4, THDR; spot 5, DHDR; spot 6, IfcE. MW, molecular weight.
Fig 3.
(A) Schematic representation of gene organization of daidzein-induced proteins from S. isoflavoniconvertens. (B) PCR products obtained by the use of peptide-based degenerate primers. (C) Insertions of plasmids used for heterologous gene expression. Genes with clarified function are marked in gray. P, putative promoter; T, putative transcriptional terminator.
Table 1.
Selected characteristics of the daidzein-induced genes and their corresponding gene products
| Gene | Length (bp) | Gene product | No. of amino acids | Molecular mass (kDa) | pI |
|---|---|---|---|---|---|
| ifcA | 471 | IfcA | 157 | 17 | 4.8 |
| ifcB | 720 | IfcB | 240 | 26 | 4.9 |
| ifcC | 906 | IfcC | 302 | 33 | 4.8 |
| tdr | 1458 | THDR | 486 | 55 | 4.7 |
| ddr | 855 | DHDR | 285 | 29 | 5.4 |
| ifcD | 483 | IfcD | 161 | 18 | 8.3 |
| dzr | 1932 | DZNR | 644 | 70 | 6.0 |
| ifcE | 1836 | IfcE | 612 | 66 | 5.3 |
Analysis of the identified genes and their organization.
The amino acid sequences of three of the ORFs (tdr, ddr, and dzr) showed high similarity to those of the tetrahydrodaidzein reductase (THDR), the dihydrodaidzein reductase (DHDR), and the daidzein reductase (DZNR) of other human gut bacteria (14, 15, 17, 24) (Table 2). The amino acid sequence of the protein encoded by tdr (AFV15450) comprises an N-terminal dinucleotide-binding motif (GXGX2G; positions 14 to 19) and a central glycyl radical enzyme motif (RVXG; positions 174 to 177). The product of tdr shows 90% sequence identity with the THDR of Slackia sp. strain NATTS (BAL46928) and 87% to the THDR of Lactococcus sp. strain 20-92 (BAJ72747). Based on its length of 285 amino acids and the cofactor and active site sequence motifs, the product of ddr (AFV15451) belongs to the “classical” short-chain dehydrogenases/reductases (SDR) (25). The motif YX3K (positions 90 to 94) is highly conserved in SDR and part of the catalytic tetrad. The amino acid sequence of the product encoded by ddr is 93% identical to that of the DHDR of Slackia sp. strain NATTS (BAL46929) and 86% identical to that of Lactococcus sp. strain 20-92 (BAJ72748). The product of dzr (AFV15453) is characterized by a 4Fe-4S cluster-binding motif (CX2CX3CX12C; positions 343 to 363) and an extended dinucleotide-binding motif (hhhXGXGX2GXE, where h is a hydrophobic residue; positions 386 to 397). It shows 95% identity with the DZNR of Slackia sp. strain NATTS (BAL46930) and 42% identity with the DZNR of Lactococcus sp. strain 20-92 (BAJ72750).
Table 2.
Amino acid sequence identities of THDR, DHDR, and DZNR from S. isoflavoniconvertens with the corresponding enzymes from the other equol-forming human gut bacteria and the GC content of the complete genomes and the encoding genes
| Bacterial strain [% GC content] | % identity to indicated S. isoflavoniconvertensa enzyme (accession no.) [% GC content] |
||
|---|---|---|---|
| THDR (AFV15450) [59.8] | DHDR (AFV15451) [61.4] | DZNR (AFV15453) [59.5] | |
| Slackia sp. strain NATTS [NRb] | 90 (BAL46928) [59.5] | 93 (BAL46929) [61.7] | 95 (BAL46930) [60.2] |
| Lactococcus sp. strain 20-92 [39.0] | 87 (BAJ72747) [63.7] | 86 (BAJ72748) [67.4] | 42 (BAJ72750) [64.5] |
| Eggerthella sp. strain YY7918 [56.2] | 87 (BAK44716)c [63.7] | 86 (BAK44715)c [67.5] | 42 (BAK44713)c [64.8] |
% GC content for the complete genome, 58.5.
NR, not reported.
Hypothetical protein deduced from complete genome sequencing.
The product of ifcA (AFV15447) includes a dinucleotide-binding motif (GXGX2G; positions 73 to 78). IfcA shows 81% amino acid sequence identity with the dihydrodaidzein racemase of Lactococcus sp. strain 20-92 (BAM25050) (16), which comprises a central dinucleotide-binding motif also. The predicted amino acid sequences of ifcB (AFV15448) and ifcC (AFV15449) are similar to those of the β-subunit and the α-subunit, respectively, of electron-transferring flavoproteins. The sequence of IfcC contains three glycyl radical enzyme motifs (RVXG; positions 41 to 44, 187 to 190, and 232 to 235). The amino acid sequence of the ifcC product shows the highest (61%) identity with those of putative proteins of Lactococcus sp. strain 20-92 (BAJ72746) and Eggerthella sp. strain YY7918 (BAK44717), while the product of ifcB is most similar (65% identity) to the adjacently encoded putative protein from Eggerthella sp. strain YY7918 (BAK44718). As observed for S. isoflavoniconvertens, all these putative genes are also located upstream of the corresponding tdr gene. The product of ifcD (AFV15452) exhibits no conserved domains. However, the putative IfcD shows 63% identity with the hypothetical protein encoded by the ORF located between the genes encoding DHDR and DZNR of Lactococcus sp. strain 20-92 (BAJ72749). The protein sequence encoded by ifcE includes a 4Fe-4S cluster-binding motif (CX2CX2CX2C; positions 49 to 58) and a dinucleotide-binding motif (GXGX2G; positions 233 to 238). The gene product is similar to putative bacterial NADPH-dependent glutamate synthase β chain–like oxidoreductases, including that of S. heliotrinireducens with 67% identity (ACV22673). Moreover, 65% amino acid identity with Lactococcus sp. strain 20-92 (BAJ72752) and Eggerthella sp. strain YY918 (BAK44710) putative proteins, both of which are encoded by an ORF located downstream of the DZNR-encoding gene, was observed.
Based on analysis of their predicted amino acid sequences, DZNR, DHDR, THDR, and IfcA are most likely localized in the cytoplasm, whereas one potential transmembrane region is present in IfcB (positions 116 to 132), IfcC (positions 93 to 109), and IfcE (positions 455 to 471). For IfcD, a cleavable N-terminal signal sequence (positions 1 to 33) and three transmembrane helices (positions 39 to 55, 87 to 103, and 125 to 141) are predicted.
The GC content of tdr, ddr, and dzr (Table 2) and the other five identified genes ranges from 56.6% to 63.3% and matches well with that of the complete genome of S. isoflavoniconvertens, which is 58.5% (8). Eighteen transcriptional sigma70 promoters were predicted for the sequenced DNA region of S. isoflavoniconvertens. Four of these promoters may be functional based on the high similarity of their sequences to the consensus sequences of the −10 and −35 boxes and/or their location upstream of ifcA, tdr, dzr, and ifcE (positions of transcription start site, 189, 2454, 5578, and 8103) (Fig. 3A). The two promoters upstream of each tdr and ifcE include the −16 region TRTG motif, which is typical of Gram-positive bacteria and known to enhance transcription (26). Sequences of predicted Rho-independent transcription terminators are located upstream of ifcA and downstream of ddr and dzr (Fig. 3A).
Heterologous expression of dzr, ddr, and tdr in E. coli.
To identify their function, the putative genes encoding DZNR, DHDR, and THDR in S. isoflavoniconvertens were subcloned (Fig. 3C) and expressed in E. coli under the control of the lac or the T7 promoter. Each of the three genes was expressed individually; ddr and tdr were additionally expressed in combination. SDS-PAGE analysis of the cell extracts of E. coli clones carrying dzr [E. coli KRX(pIF1)], ddr [E. coli JM109(pIF2)], and tdr [E. coli JM109(pIF3)] revealed predominant protein bands of approximately 70 kDa, 30 kDa, and 55 kDa, respectively, which were not detected in the cell extract from the corresponding control clone carrying the empty vector [E. coli JM109(pGEM-T Easy) or E. coli KRX(pGEM-T Easy)] (data not shown). The observed molecular masses are in agreement with those deduced from the amino acid sequence of the products of individual genes (Table 1). DHDR appeared to be expressed at a much lower level than DZNR and THDR. The combined expression of ddr and tdr by E. coli KRX(pIF4) resulted, as expected, in two additional bands of 30 kDa and 55 kDa compared to the control. The protein band of DHDR was much weaker than that of THDR, as was observed for the proteins expressed separately.
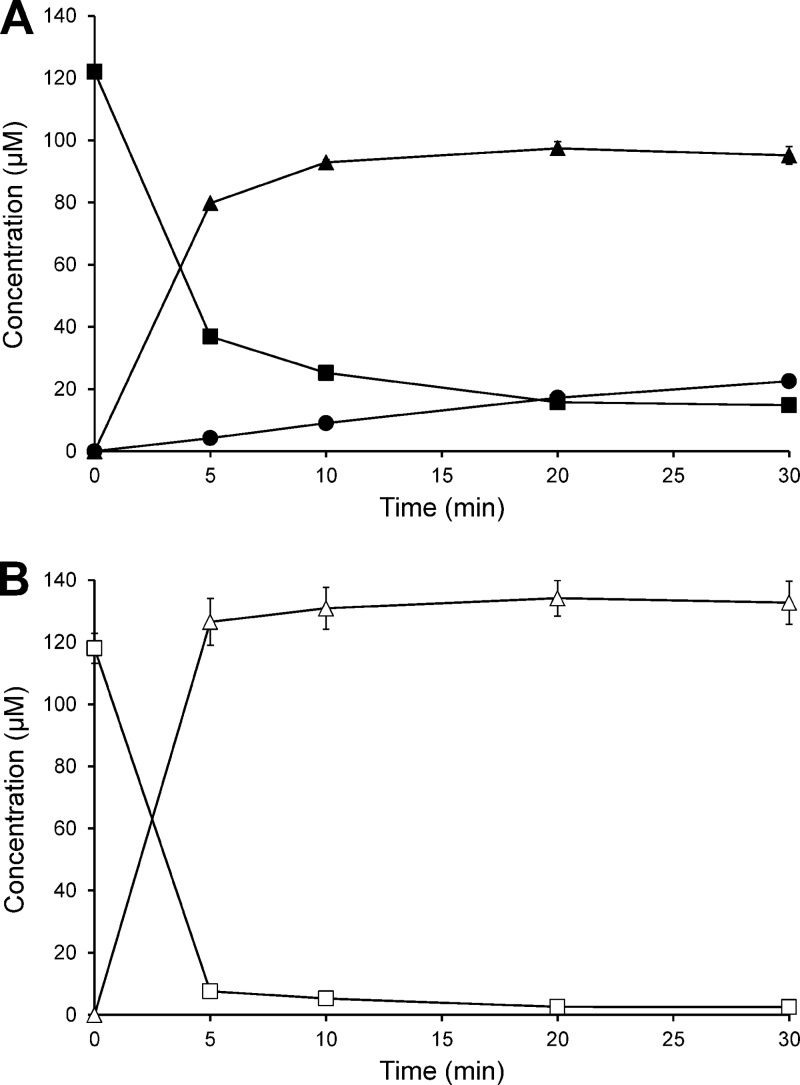
For testing the activity of the recombinant enzymes, cultivation of the E. coli clones, preparation of cell extracts, and enzyme assays were performed under anoxic conditions. The complete conversion of daidzein via dihydrodaidzein to equol was achieved by combining the cell extract of the DZNR-expressing E. coli KRX(pIF1) with that of E. coli KRX(pIF4) expressing both DHDR and THDR (Fig. 4A). The incubation of genistein with the combined extracts resulted in formation of dihydrogenistein, but 5-hydroxy-equol was not detected (Fig. 4B). Dihydrodaidzein was formed from daidzein in the presence of NADPH at an initial rate of 27.1 ± 0.4 nmol min−1 mg protein−1, while the formation of dihydrogenistein from genistein proceeded under identical conditions at a 3.3-fold-higher rate (89.9 ± 3.8 nmol min−1 mg protein−1). The replacement of NADPH by NADH led to only minor formation of dihydrodaidzein (0.35 ± 0.11 nmol min−1 mg protein−1) and dihydrogenistein (0.02 ± 0.01 nmol min−1 mg protein−1). No products were observed in the absence of NADPH or NADH. In accordance with the results reported above, cell extracts containing both DHDR and THDR catalyzed the conversion of dihydrodaidzein to equol but not the corresponding reduction of dihydrogenistein to 5-hydroxy-equol. The rate of equol formation was higher in the presence of NADPH (0.33 ± 0.01 nmol min−1 mg protein−1) than with NADH (0.18 ± 0.00 nmol min−1 mg protein−1). Cell extracts of E. coli KRX(pIF1) expressing only DZNR catalyzed the NAD(P)H-dependent conversion of daidzein and genistein to dihydrodaidzein and dihydrogenistein, respectively.
Fig 4.
Time course of the conversion of (A) daidzein and (B) genistein by combined cell extracts of E. coli expressing the DZNR, DHDR, and THDR from S. isoflavoniconvertens. The assays were performed in the presence of NADPH under strictly anoxic conditions. ■, daidzein; ▲, dihydrodaidzein; •, equol; □, genistein; Δ, dihydrogenistein. The symbols represent the means of the results of triplicate experiments. Error bars indicate SEM.
During incubation of daidzein with the combined extracts containing DZNR, DHDR, and THDR, only (R)-dihydrodaidzein was initially formed. After 20 min of incubation, small amounts of (S)-dihydrodaidzein were observed and amounted to 6% of the total dihydrodaidzein after 30 min. Equol was exclusively present as (S)-enantiomer. Analogously, the incubation of genistein with the recombinant enzymes led to the formation of mainly (R)-dihydrogenistein. However, (S)-dihydrogenistein was already observed after 5 min and represented 25% of the total dihydrogenistein after 30 min.
Expression and purification of tagged recombinant DZNR, DHDR, and THDR.
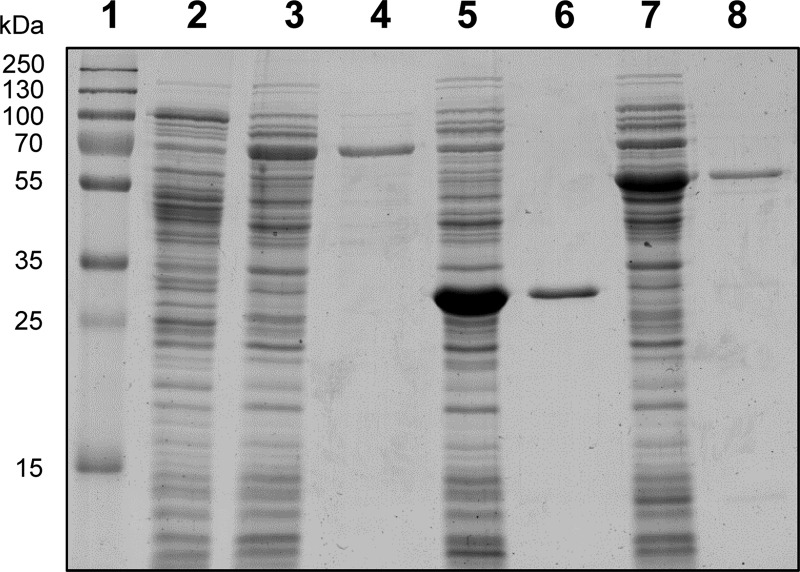
To facilitate their purification, the three enzymes were expressed individually as fusion proteins with a C-terminal Strep-tag in anaerobically grown E. coli. Cultures of the generated E. coli KRX(pST1) exhibited moderate expression of DZNR, whereas DHDR and THDR were highly expressed by E. coli KRX(pST2) and E. coli KRX(pST3), respectively (Fig. 5). The tagged proteins were anoxically purified from the corresponding E. coli cell extracts by a one-step affinity chromatographic procedure (Fig. 5). Based on the amount of protein applied on the column, the yields were up to 32% for DHDR, 25% for THDR, and 1% for DZNR. The low yield of DZNR is ascribed to its relatively low expression and poor accessibility of the tag. Analysis of the wash fractions revealed only minor binding of DZNR to the purification matrix.
Fig 5.
SDS-PAGE analysis of cell extracts of E. coli expressing Strep-tag-fused DZNR, DHDR, or THDR from S. isoflavoniconvertens (10 μg protein each) and the corresponding enzymes purified by affinity chromatography (1 μg protein each). Lane 1, molecular mass standards; lane 2, E. coli KRX(pGEM-T Easy); lane 3, E. coli KRX(pST1); lane 4, tagged DZNR; lane 5, E. coli KRX(pST2); lane 6, tagged DHDR; lane 7, E. coli KRX(pST3); lane 8, tagged THDR.
To test the influence of atmospheric oxygen on the heterologously expressed enzymes, cell extracts were prepared under oxic conditions from the aerobically grown recombinant E. coli clones. The combined extracts expressing the tagged DZNR, DHDR, and THDR, respectively, converted neither daidzein nor genistein in the presence of oxygen. The combined extracts expressing the tagged DHDR and THDR, likewise, did not reduce dihydrodaidzein to equol under these conditions.
Characterization of DZNR, DHDR, and THDR.
The purified DZNR catalyzed the formation of dihydrodaidzein from daidzein and dihydrogenistein from genistein, with Vmax values of 0.62 ± 0.04 and 2.14 ± 0.33 μmol min−1 mg protein−1, respectively. The Km values were 1.44 ± 0.43 μM for daidzein and 26.7 ± 8.3 μM for genistein. Chiral analysis of the products revealed that (R)-dihydrodaidzein and (R)-dihydrogenistein, respectively, were exclusively formed. The apparent molecular mass of DZNR under nondenaturing conditions was estimated by native PAGE to be 90 kDa. Based on the molecular mass of one subunit of 70 kDa, this suggests that the native DZNR is a monomer. Its UV/Vis spectrum with absorption maxima around 360 and 450 nm was indicative of a flavin-containing protein. Thus, the DZNR was subjected to acid extraction. Subsequent TLC analysis of the obtained supernatant revealed the presence of the flavin cofactors FAD (Rf = 0.17) and, in a small amount, FMN (Rf = 0.32).
The purified and combined DHDR and THDR catalyzed the conversion of dihydrodaidzein at a rate of 26.3 ± 0.8 nmol min−1 mg protein−1 and the formation of equol at 1.68 ± 0.05 nmol min−1 mg protein−1. Analysis of the ratio of dihydrodaidzein enantiomers in the course of the conversion indicated that (S)-dihydrodaidzein was the preferred substrate. The final product was (S)-equol. Native PAGE of DHDR showed an apparent molecular mass of 119 kDa, suggesting a homotetrameric structure of the 29-kDa subunits. In addition, a diffuse band of 86 kDa was observed. The result of the analysis of DHDR for the presence of flavins by UV/Vis spectroscopy and TLC was negative. The THDR migrated as multiple bands of 230, 136, and 75 kDa in native PAGE. The predominant 136-kDa band is proposed to represent the enzyme dimer composed of two identical 55-kDa subunits, whereas the other bands are presumably the tetramer and monomer. The UV/Vis spectrum of the THDR with absorption maxima at 371 and 447 nm was characteristic of a flavoprotein. The contained flavin was subsequently identified by TLC analysis to be FAD (Rf = 0.17).
DISCUSSION
The enzymes catalyzing the conversion of daidzein and genistein to equol and 5-hydroxy-equol, respectively, in S. isoflavoniconvertens are induced by these isoflavones (8). This inducibility has also been observed for the isoflavone-converting E. mucosicola isolated from the mouse intestine (11). The induction of the enzymes was confirmed here by detecting the daidzein-converting activity in cell extracts of S. isoflavoniconvertens grown in the presence of daidzein but not in its absence. The differences in the extent of upregulation of the daidzein-induced proteins may have been due to differences in gene transcription and translation and/or protein degradation. For some proteins, the induction factors may have been biased by the presence of different isoforms known as a charge train.
Based on the detection of daidzein-induced proteins from S. isoflavoniconvertens, we identified a cluster of eight genes. The identity of the genes encoding DZNR, DHDR, and THDR was confirmed by their functional expression in E. coli. The purified DZNR exhibited a 3-fold-higher Vmax for genistein than for daidzein, while the Km was much lower for daidzein than for genistein. The enzyme converted daidzein to (R)-dihydrodaidzein, as previously reported for the DZNR from Lactococcus sp. strain 20-92 (14, 16). Only one enantiomer, (R)-dihydrogenistein, was also formed from genistein by the DZNR from S. isoflavoniconvertens. In addition to catalyzing a similar reaction, the DZNR shares several features, including the presence of FAD and FMN with bacterial 2,4-dienoyl-CoA reductase. This enzyme catalyzes the reduction of double bonds in fatty acids, which starts with a hydride transfer from NADPH to FAD followed by electron transfers via the 4Fe-4S cluster and FMN (27). The purified and combined DHDR and THDR catalyzed the conversion of dihydrodaidzein to exclusively (S)-equol, which is the only enantiomer formed by intestinal bacteria (5). The preferred use of (S)-dihydrodaidzein by the DHDR from S. isoflavoniconvertens as a substrate corresponds to the activity reported for the DHDR from Lactococcus sp. strain 20-92 (16). While equol formation was still catalyzed by cell-free systems to a limited extent, neither cell extracts of S. isoflavoniconvertens nor its recombinant enzymes converted dihydrogenistein to 5-hydroxy-equol. It may be assumed that cell rupture somehow compromises the activity of DHDR or/and THDR. Moreover, the formation of 5-hydroxy-equol appears to proceed less efficiently than that of equol, as has previously been observed with intact cells of S. isoflavoniconvertens in vitro and in vivo (8, 10).
While this work was in progress, genes encoding daidzein-converting enzymes from two other equol-forming bacterial strains were identified (14, 15, 17). The amino acid sequences of the proteins encoded by dzr, ddr, and tdr of S. isoflavoniconvertens show high identity with those of DZNR, DHDR, and THDR of Lactococcus sp. strain 20-92 (14, 15) and Slackia sp. strain NATTS (17). Moreover, similar genes are present in the completely sequenced genome of the equol-producing human intestinal Eggerthella sp. strain YY7918, but their function has not yet been demonstrated (24). The DZNR is a member of the old yellow enzyme family, and the primary sequence of the enzyme from S. isoflavoniconvertens and the other bacteria contains a 4Fe-4S cluster-binding motif and an extended dinucleotide-binding motif. The latter is characteristic of the glutathione reductase subfamily of FAD-containing proteins, which possess an NAD(P)H-binding domain in addition to the FAD-binding domain (28). A second extended dinucleotide-binding motif is present only in the DZNR of Lactococcus sp. strain 20-92 and Eggerthella sp. strain YY7918. Interestingly, the DZNR from S. isoflavoniconvertens and Slackia sp. strain NATTS shows only ca. 40% amino acid identity with those of Lactococcus sp. strain 20-92 and Eggerthella sp. strain YY7918, while the similarity of the amino acid sequences of each THDR and DHDR from the four bacterial strains is considerably higher (Table 2). The DHDR from S. isoflavoniconvertens and those of the other three bacteria belong to the “classical” enzymes within the SDR superfamily, which, among other features, is characterized by a conserved dinucleotide-binding motif. The THDR sequence also contains a conserved dinucleotide-binding motif. In addition, a typical motif of glycyl radical enzymes (29) is present in the sequences of S. isoflavoniconvertens and Slackia sp. strain NATTS. This is in accordance with a radical mechanism as postulated for the THDR from Eggerthella sp. strain Julong 732 (13). However, the glycyl radical enzyme motif is not completely conserved in the THDR from Lactococcus sp. strain 20-92 and the hypothetical protein from Eggerthella sp. strain YY7918. The dinucleotide-binding motifs are assumed to bind NAD(P)H, which is required for the reduction of daidzein via dihydrodaidzein to equol by the enzymes from S. isoflavoniconvertens and those from the other bacteria (14, 15, 17). However, data on the NAD(P)H dependency of the THDR are contradictory (12, 14).
No further enzymes or proteins from S. isoflavoniconvertens were essential for daidzein conversion by the recombinant DZNR, DHDR, and THDR. Compared to that by S. isoflavoniconvertens cells, the rate of equol formation by cell-free systems was low. A dihydrodaidzein racemase has been described very recently in Lactococcus sp. strain 20-92 that increased equol formation, presumably by transforming (R)-dihydrodaidzein to (S)-dihydrodaidzein (16). In S. isoflavoniconvertens, this dihydrodaidzein racemase could be encoded by ifcA based on its highly similar amino acid sequence. IfcA is present in the daidzein-induced S. isoflavoniconvertens extract, as observed by 2D-DIGE analysis. The activity of an enzyme catalyzing the conversion of (R)-dihydrodaidzein and (R)-dihydrogenistein to their corresponding (S)-enantiomers is made evident by comparing the experiments with the purified DZNR and those with cell extracts of S. isoflavoniconvertens. While DZNR formed exclusively the (R)-enantiomers of dihydrodaidzein and dihydrogenistein from the parent isoflavones, racemic mixtures were observed during conversion by the cell extracts. However, the subsequent formation of equol by cell extracts was slow, indicating limited activity of DHDR and/or THDR. The products of the remaining genes of the cluster were also upregulated, which indicates their role in isoflavone conversion. Based on their dinucleotide- or Fe-S cluster-binding motifs and similarity to proteins from databases, these gene products could be involved in the electron transfer associated with isoflavonoid reduction. Genes encoding hypothetical proteins similar to the ones from S. isoflavoniconvertens are also present in Lactococcus sp. strain 20-92 (15) and Eggerthella sp. strain YY7918 (24).
The GC content of the genes from S. isoflavoniconvertens investigated here is in the range of that determined for the complete genome of this organism (Table 2). In contrast, the GC content of up to 68% of the corresponding genes from Eggerthella sp. strain YY7918 and, in particular, from Lactococcus sp. strain 20-92 greatly exceeds their genomic GC content of 56% (24) and 39% (type strain of Lactococcus garvieae) (30), respectively. The deviations in the GC content indicate that these genes were acquired by horizontal transfer from bacteria with high GC content and may confer a considerable advantage to the recipients.
The results of this study provide the basis for further elucidating the catalytic mechanisms underlying the isoflavone conversion by S. isoflavoniconvertens. The function of the daidzein-induced proteins encoded by the remaining genes of the identified cluster and the regulation of gene expression are to be clarified in future studies. The identification of conserved regions in the isoflavone-converting enzymes based on sequences from various gut bacteria enables us not only to pinpoint catalytic sites but also to detect corresponding genes in other isoflavone-activating bacteria within the microbial metagenome of the human gut. The knowledge gained on highly conserved gene sequences may subsequently be used for fecal sample analysis to characterize isoflavone metabotypes in humans.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant BR 2269/4-1 from the Deutsche Forschungsgemeinschaft.
We thank Anke Gühler for technical assistance.
Footnotes
Published ahead of print 29 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03693-12.
REFERENCES
- 1. Messina M. 2010. A brief historical overview of the past two decades of soy and isoflavone research. J. Nutr. 140: 1350S–1354S [DOI] [PubMed] [Google Scholar]
- 2. Steiner C, Arnould S, Scalbert A, Manach C. 2008. Isoflavones and the prevention of breast and prostate cancer: new perspectives opened by nutrigenomics. Br. J. Nutr. 99: ES78–E108 [DOI] [PubMed] [Google Scholar]
- 3. Barnes S. 2010. The biochemistry, chemistry and physiology of the isoflavones in soybeans and their food products. Lymphat. Res. Biol. 8: 89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bolca S, Van de Wiele T, Possemiers S. 2013. Gut metabotypes govern health effects of dietary polyphenols. Curr. Opin. Biotechnol. 24: 220–225 [DOI] [PubMed] [Google Scholar]
- 5. Setchell KD, Clerici C. 2010. Equol: history, chemistry, and formation. J. Nutr. 140: 1355S–1362S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Setchell KD, Clerici C. 2010. Equol: pharmacokinetics and biological actions. J. Nutr. 140: 1363S–1368S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jackson RL, Greiwe JS, Schwen RJ. 2011. Emerging evidence of the health benefits of S-equol, an estrogen receptor β agonist. Nutr. Rev. 69: 432–448 [DOI] [PubMed] [Google Scholar]
- 8. Matthies A, Blaut M, Braune A. 2009. Isolation of a human intestinal bacterium capable of daidzein and genistein conversion. Appl. Environ. Microbiol. 75: 1740–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arora A, Nair MG, Strasburg GM. 1998. Antioxidant activities of isoflavones and their biological metabolites in a liposomal system. Arch. Biochem. Biophys. 356: 133–141 [DOI] [PubMed] [Google Scholar]
- 10. Matthies A, Loh G, Blaut M, Braune A. 2012. Daidzein and genistein are converted to equol and 5-hydroxy-equol by human intestinal Slackia isoflavoniconvertens in gnotobiotic rats. J. Nutr. 142: 40–46 [DOI] [PubMed] [Google Scholar]
- 11. Matthies A, Clavel T, Gütschow M, Engst W, Haller D, Blaut M, Braune A. 2008. Conversion of daidzein and genistein by an anaerobic bacterium newly isolated from the mouse intestine. Appl. Environ. Microbiol. 74: 4847–4852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim M, Kim SI, Han J, Wang XL, Song DG, Kim SU. 2009. Stereospecific biotransformation of dihydrodaidzein into (3S)-equol by the human intestinal bacterium Eggerthella strain Julong 732. Appl. Environ. Microbiol. 75: 3062–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim M, Marsh EN, Kim SU, Han J. 2010. Conversion of (3S,4R)-tetrahydrodaidzein to (3S)-equol by THD reductase: proposed mechanism involving a radical intermediate. Biochemistry 49: 5582–5587 [DOI] [PubMed] [Google Scholar]
- 14. Shimada Y, Yasuda S, Takahashi M, Hayashi T, Miyazawa N, Sato I, Abiru Y, Uchiyama S, Hishigaki H. 2010. Cloning and expression of a novel NADP(H)-dependent daidzein reductase, an enzyme involved in the metabolism of daidzein, from equol-producing Lactococcus strain 20–92. Appl. Environ. Microbiol. 76: 5892–5901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shimada Y, Takahashi M, Miyazawa N, Ohtani T, Abiru Y, Uchiyama S, Hishigaki H. 2011. Identification of two novel reductases involved in equol biosynthesis in Lactococcus strain 20–92. J. Mol. Microbiol. Biotechnol. 21: 160–172 [DOI] [PubMed] [Google Scholar]
- 16. Shimada Y, Takahashi M, Miyazawa N, Abiru Y, Uchiyama S, Hishigaki H. 2012. Identification of a novel dihydrodaidzein racemase essential for equol biosynthesis from daidzein in Lactococcus strain 20–92. Appl. Environ. Microbiol. 78: 4902–4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tsuji H, Moriyama K, Nomoto K, Akaza H. 2012. Identification of an enzyme system for daidzein-to-equol conversion in Slackia sp. strain NATTS. Appl. Environ. Microbiol. 78: 1228–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- 19. Park HY, Kim M, Han J. 2011. Stereospecific microbial production of isoflavanones from isoflavones and isoflavone glucosides. Appl. Microbiol. Biotechnol. 91: 1173–1181 [DOI] [PubMed] [Google Scholar]
- 20. Setchell KD, Clerici C, Lephart ED, Cole SJ, Heenan C, Castellani D, Wolfe BE, Nechemias-Zimmer L, Brown NM, Lund TD, Handa RJ, Heubi JE. 2005. S-Equol, a potent ligand for estrogen receptor β, is the exclusive enantiomeric form of the soy isoflavone metabolite produced by human intestinal bacterial flora. Am. J. Clin. Nutr. 81: 1072–1079 [DOI] [PubMed] [Google Scholar]
- 21. Vogel-Scheel J, Alpert C, Engst W, Loh G, Blaut M. 2010. Requirement of purine and pyrimidine synthesis for colonization of the mouse intestine by Escherichia coli. Appl. Environ. Microbiol. 76: 5181–5187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ochman H, Gerber AS, Hartl DL. 1988. Genetic applications of an inverse polymerase chain reaction. Genetics 120: 621–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Case CL, Rodriguez JR, Mukhopadhyay B. 2009. Characterization of an NADH oxidase of the flavin-dependent disulfide reductase family from Methanocaldococcus jannaschii. Microbiology 155: 69–79 [DOI] [PubMed] [Google Scholar]
- 24. Yokoyama S, Oshima K, Nomura I, Hattori M, Suzuki T. 2011. Complete genomic sequence of the equol-producing bacterium Eggerthella sp. strain YY7918, isolated from adult human intestine. J. Bacteriol. 193: 5570–5571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kavanagh KL, Jornvall H, Persson B, Oppermann U. 2008. The SDR superfamily: functional and structural diversity within a family of metabolic and regulatory enzymes. Cell. Mol. Life Sci. 65: 3895–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Voskuil MI, Chambliss GH. 1998. The −16 region of Bacillus subtilis and other gram-positive bacterial promoters. Nucleic Acids Res. 26: 3584–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hubbard PA, Liang X, Schulz H, Kim JJ. 2003. The crystal structure and reaction mechanism of Escherichia coli 2,4-dienoyl-CoA reductase. J. Biol. Chem. 278: 37553–37560 [DOI] [PubMed] [Google Scholar]
- 28. Dym O, Eisenberg D. 2001. Sequence-structure analysis of FAD-containing proteins. Protein Sci. 10: 1712–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Buckel W, Golding BT. 2006. Radical enzymes in anaerobes. Annu. Rev. Microbiol. 60: 27–49 [DOI] [PubMed] [Google Scholar]
- 30. Collins MD, Farrow JA, Phillips BA, Kandler O. 1983. Streptococcus garvieae sp. nov. and Streptococcus plantarum sp. nov. J. Gen. Microbiol. 129: 3427–3431 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.