Abstract
Mitochondrial precursor proteins are synthesized in the cytosol and subsequently imported into mitochondria. The import of mitochondrial intermembrane space proteins is coupled with their oxidative folding and governed by the mitochondrial intermembrane space import and assembly (MIA) pathway. The cytosolic steps that precede mitochondrial import are not well understood. We identified a role for the ubiquitin-proteasome system in the biogenesis of intermembrane space proteins. Interestingly, the function of the ubiquitin-proteasome system is not restricted to conditions of mitochondrial protein import failure. The ubiquitin-proteasome system persistently removes a fraction of intermembrane space proteins under physiological conditions, acting as a negative regulator in the biogenesis of this class of proteins. Thus, the ubiquitin-proteasome system plays an important role in determining the levels of proteins targeted to the intermembrane space of mitochondria.
INTRODUCTION
As many as 800 to 1,000 proteins in a simple eukaryote, Saccharomyces cerevisiae, are required for mitochondria to perform their essential metabolic and regulatory functions in the cell (1–3). The majority of mitochondrial proteins are synthesized on cytosolic ribosomes and targeted to mitochondria. This process involves the main entry gate, formed by the translocase of the outer mitochondrial membrane (TOM) complex. After passing the TOM complex, mitochondrial precursor proteins are directed to their final destinations by specialized translocation machineries and pathways that drive the import of precursor proteins into the mitochondrial matrix, inner mitochondrial membrane, outer mitochondrial membrane, and intermembrane space (IMS) of mitochondria (4–7). The proteins that reside in the IMS are critical for metabolic functions (e.g., energy conversion), mitochondrial biogenesis (e.g., the transport of lipids and proteins and assembly of protein membrane complexes), and regulatory processes at the cellular level (e.g., programmed cell death) (8–12). A large fraction of mitochondrial IMS proteins are imported via the folding-trap mechanism, executed by the mitochondrial intermembrane space import and assembly (MIA) pathway (13–16).
Intermembrane space proteins that follow the MIA pathway are small cysteine-rich proteins that comprise two major families with characteristic twin cysteine motifs: CX3C (small Tim proteins) (17) and CX9C (12, 18, 19). The mechanism utilized by the MIA pathway is unique among mitochondrial transport pathways but also in the context of cellular protein transport machineries (13–16). The most distinctive feature of the MIA pathway is the transfer of disulfide bonds into the incoming precursors, leading to their oxidative folding and to trapping of mature proteins in the IMS (20–24). The specificity of disulfide transfer and oxidative folding is maintained by the recognition by Mia40 of the mitochondrial IMS sorting signal, called MISS/ITS, in the precursor proteins (25, 26). Mia40 serves not only as a receptor on the trans side of the outer mitochondrial membrane but also as an oxidoreductase and folding catalyst (24, 27, 28). Mia40 requires the cooperation of another essential MIA component, the sulfhydryl oxidase Erv1, which is responsible for the oxidation of Mia40 and thereby the availability of Mia40 to transfer disulfide bonds to precursor proteins (20, 29, 30). Erv1 is also needed to maintain the productivity of disulfide transfer and oxidative folding by acting in the ternary complex with Mia40 and a precursor protein (31, 32). Furthermore, the efficiency of this pathway is increased by the recruitment of Mia40 to the sites of the outer membrane translocation of precursor proteins (33) and by the reducing power of the glutathione redox buffer, which eliminates unproductive intermediates in thiol-disulfide exchange reactions (28). Altogether, the events catalyzed by MIA are critical for the mitochondrial localization of precursor proteins into the IMS of mitochondria (18, 20, 29, 34–36). In contrast to mitochondrial biogenesis stages, the fate of IMS proteins following their cytosolic synthesis and prior to mitochondrial uptake has not yet been studied in detail and remains poorly understood. A recent study proposed that IMS precursor proteins are maintained by the cytosolic thioredoxin system in an import-competent state in the cytosol (37).
The present study investigated the cytosolic stage of the biogenesis of IMS proteins. We identified the ubiquitin-proteasome system as the machinery responsible for the clearance of mislocalized IMS proteins. Interestingly, the ubiquitin-proteasome system participates in the biogenesis of IMS proteins under physiological conditions, thereby regulating the influx of IMS precursor proteins into mitochondria.
MATERIALS AND METHODS
Yeast strains, plasmids, and growth conditions.
The Saccharomyces cerevisiae strains used in the present study are listed in Table 1. For the inducible expression of tagged MIA substrates, sequences that encoded Cox12, Cox17, Mic17, and Pet191 were amplified by PCR from yeast genomic DNA. The resulting DNA fragments were cloned into the pESC-URA vector (Agilent), in frame with the FLAG tag. This procedure yielded the pAG1, pAG2, pAG3, and pAG4 plasmids, encoding Pet191, Mic17, Cox12, and Cox17 fusion proteins, respectively, with a FLAG tag at the C terminus and expressed under the control of the GAL10 promoter. Yeast cells were transformed according to the standard procedure. Strains that expressed plasmid-borne Mia40core and cytochrome b2-Mia40core were generated by transforming strain YPH499 with the BG9334 and BG9310 plasmids, respectively (38). In the strain that expressed hMIA40 (yMS555), the MIA40 gene was replaced with the human MIA40 coding sequence by homologous recombination. The strain that expressed the plasmid-borne version of b2-hMIA40 was generated by plasmid shuffling (39). The pYEp96–6HIS-Ub plasmid, encoding 6His-tagged ubiquitin (6His-Ub) under the control of the CUP1 promoter, was also used (40, 41).
Table 1.
Saccharomyces cerevisiae strains used in this study
| Strain (lab ID no.) | Genotype | Reference or source |
|---|---|---|
| YPH499 (524) | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 | 69 |
| mia40-3 (178) | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 mia40::ADE2 [pFL39-mia40-3] | 34 |
| mia40-4 (176) | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 mia40::ADE2 [pFL39-mia40-4] | 34 |
| erv1-2 (235) | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 erv1::ADE2 [pFL39-erv1-2] | 29 |
| b2-hMIA40 (357) | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 mia40::ADE2 [pGB9348 = pFL39-MIA40P-b2-hMIA40-MIA40T] | 39 |
| hMIA40 (555) | MATa ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 mia40::hMIA40 erv1::ADE2 [pGB9011 = pFL39-ERV1P-ERV1-ERV1T] | 39 |
| WCG4a (661) | MATa ura3 his3-11,15 leu2-3,112 | 46 |
| WCG4-11/21a (662) | MATa ura3 his3-11,15 leu2-3,112 pre1-1 pre2-1 | 46 |
| BY4741 (533) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf |
| Δyme1 (671) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YPR024w::kanMX4 | Euroscarf |
| Δrpn13 (647) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YLR421c::kanMX4 | Euroscarf |
| Δirc25 (648) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YLR021w::kanMX4 | Euroscarf |
| Δpoc4 (651) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YPL144w::kanMX4 | Euroscarf |
| Δpre9 (652) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YGR135w::kanMX4 | Euroscarf |
Yeast cells were grown on YPD, YPS, or YPG (1% yeast extract, 2% peptone, and 2% glucose, 2% sucrose, or 3% glycerol, respectively) or on minimal medium that contained 0.17% yeast nitrogen base (YNB) and 0.5% ammonium sulfate, supplemented with appropriate nutrients and a carbon source. For the overproduction of 6His-tagged ubiquitin under the control of the CUP1 promoter, minimal medium was supplemented with 0.1 mM CuSO4. To induce the GAL10 promoter for the expression of pAG1, pAG2, pAG3, and pAG4, 2% galactose was used as the sole carbon source, or 3% glycerol supplemented with 0.2 to 0.5% galactose was used.
Proteasome inhibition.
To prepare yeast cells for MG132 (Enzo Life Sciences) inhibitor treatment, modified YNB minimal medium without ammonium sulfate but with 0.1% proline and 0.003% sodium dodecyl sulfate (SDS) was used as described previously (42). Before the addition of MG132, yeast cells were grown overnight at 19°C to an optical density at 600 nm (OD600) of ∼1.5. The cells were spun down and resuspended in fresh medium. The samples were supplemented with either 75 μM MG132 or a corresponding volume of dimethyl sulfoxide as a solvent.
Cell fractionation.
For fractionation, the equivalent of 30 OD600 units of yeast cells was used. To generate spheroplasts, the cells were incubated in dithiothreitol (DTT) buffer (100 mM Tris-SO4, pH 9.4, and 10 mM DTT) for 15 min at 30°C, followed by 30 min of treatment with 0.05 mg Zymolyase in Zymolyase buffer (1.2 M sorbitol and 20 mM KPi, pH 7.4). The spheroplasts were lysed in homogenization buffer (0.6 M sorbitol, 10 mM Tris-HCl, pH 7.4, and 2 mM phenylmethylsulfonyl fluoride [PMSF]), using a glass Potter homogenizer. The extracts were clarified by centrifugation at 3,000 × g for 5 min at 4°C. Subsequently, a portion was withdrawn as the total fraction (T), and the remaining solution was subjected to further centrifugation at 13,000 × g for 5 min at 4°C. The resulting mitochondrial pellet (P) and postmitochondrial supernatant (S) fractions, together with the total fraction (T), were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by immunolabeling. For each fraction, an equivalent of 0.8 OD600 unit of cell extract was loaded. Fractions T and S were precipitated with 10% trichloroacetic acid and washed with ice-cold acetone prior to solubilization in urea sample buffer (6 M urea, 6% SDS, and 125 mM Tris-HCl, pH 6.8).
Purification of ubiquitinated proteins.
Cells that produced FLAG-tagged MIA substrates and 6His-Ub were grown overnight in selective minimal medium with 2% galactose and 0.1 mM CuSO4 to mid/late exponential phase at 28°C. The cells were harvested by centrifugation. Alternatively, during the last 3 h of culture, the cells were additionally treated with 75 μM MG132 in fresh medium. The protein purification experiments were performed under native or denaturing conditions. For purification under denaturing conditions, the cells were resuspended in lysis buffer 1 (6 M guanidine hydrochloride, 100 mM KPi, pH 8.0, 10 mM Tris-HCl, pH 8.0, 50 mM iodoacetamide, 5 mM imidazole, 0.1% Triton X-100, 2 mM PMSF, and 75 μM MG132) and disrupted by 10 min of vortexing with glass beads. The solution was clarified by centrifugation, and the supernatant was incubated with Ni-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) for 2 h. The load and unbound samples were withdrawn before and after incubation with the resin. The resin was washed once with lysis buffer 1 and three times with wash buffer 1 (8 M urea, 100 mM KPi, pH 6.4, and 10 mM Tris-HCl, pH 6.4), for 5 min each time. Bound proteins were eluted with Laemmli buffer containing 100 mM DTT and denatured at 65°C for 15 min.
For native purification, the cells were resuspended (15 OD600 units/1 ml) in cold lysis buffer 2 (20 mM Tris-HCl, pH 7.4, 250 mM NaCl, 50 mM iodoacetamide, and 20 mM imidazole for Ni-NTA) and disrupted by use of a cell disrupter apparatus (Constant Systems LDT) at a maximum pressure of 31,000 lb/in2 (2.14 × 108 Pa). Immediately after disruption, the solution was supplemented with 75 μM MG132, 2 mM PMSF, and 0.5% Triton X-100 and incubated on ice for 20 min for further solubilization. Unsolubilized material was removed by centrifugation at 20,000 × g for 15 min, and 1.5 ml of the clarified supernatant was incubated with Ni-NTA or anti-FLAG M2 affinity gel (Sigma-Aldrich) at 4°C for 1 h. The column was washed three times with wash buffer 2 (20 mM Tris-HCl, pH 7.4, 250 mM NaCl, and 20 mM imidazole for Ni-NTA). Proteins bound to the nickel column were eluted by 20 min of incubation with elution buffer 1 (50 mM Tris-HCl, pH 7.4, 250 mM NaCl, and 400 mM imidazole) at room temperature. Proteins bound to the anti-FLAG column were eluted with lysis buffer 1. All of the fractions were denatured in Laemmli buffer that contained 100 mM DTT at 65°C for 15 min prior to SDS-PAGE.
Isolation of mitochondria and import of radiolabeled precursor proteins.
The [35S]methionine-labeled precursor proteins were synthesized using a TNT quick coupled transcription/translation system (Promega). Mitochondria were isolated from a wild-type strain grown at 19°C on YPG medium. A standard protocol was used for the isolation of mitochondria by differential centrifugation (43). The concentration of the resulting crude mitochondrial fraction was adjusted to 10 mg protein/ml in SM buffer (250 mM sucrose and 10 mM morpholinepropanesulfonic acid [MOPS]-KOH, pH 7.2). The import of radiolabeled precursors into isolated yeast mitochondria was performed at 25°C in import buffer (250 mM sucrose, 5 mM MgCl2, 80 mM KCl, 10 mM MOPS-KOH, 5 mM methionine, 10 mM KH2PO4, pH 7.2, and 3% bovine serum albumin [BSA]) supplemented with 2 mM ATP and 2 mM NADH. Import reactions were stopped by placing the mixtures on ice and adding VOA mix (1 μM valinomycin, 20 μM oligomycin, and 8 μM antimycin) to dissipate the inner membrane electrochemical potential. After centrifugation and washing with SM buffer, the samples were denatured in Laemmli buffer containing 50 mM DTT and analyzed by SDS-PAGE. To detect [35S]methionine-labeled proteins, digital autoradiography with Storage Phosphor screens (GE Healthcare) and a PhosphorImager Storm 820 scanner (Amersham Bioscience) was performed.
Protein extracts and Western blotting.
Total protein extracts were prepared by alkaline lysis. The samples were analyzed by reducing SDS-PAGE followed by Western blotting. All of the antibodies used for protein detection were raised in rabbits, with the exception of mouse monoclonal antiubiquitin and anti-FLAG M2 antibodies. Enhanced chemiluminescence signals were detected by use of ImageQuant LAS4010 (GE Healthcare) or X-ray films. For densitometry, ImageQuant TL (GE Healthcare) software was used. The images were processed using Adobe Photoshop CS4. In some figures, nonrelevant gel parts were excised digitally.
RESULTS
Proteins targeted to the IMS are degraded by the proteasome.
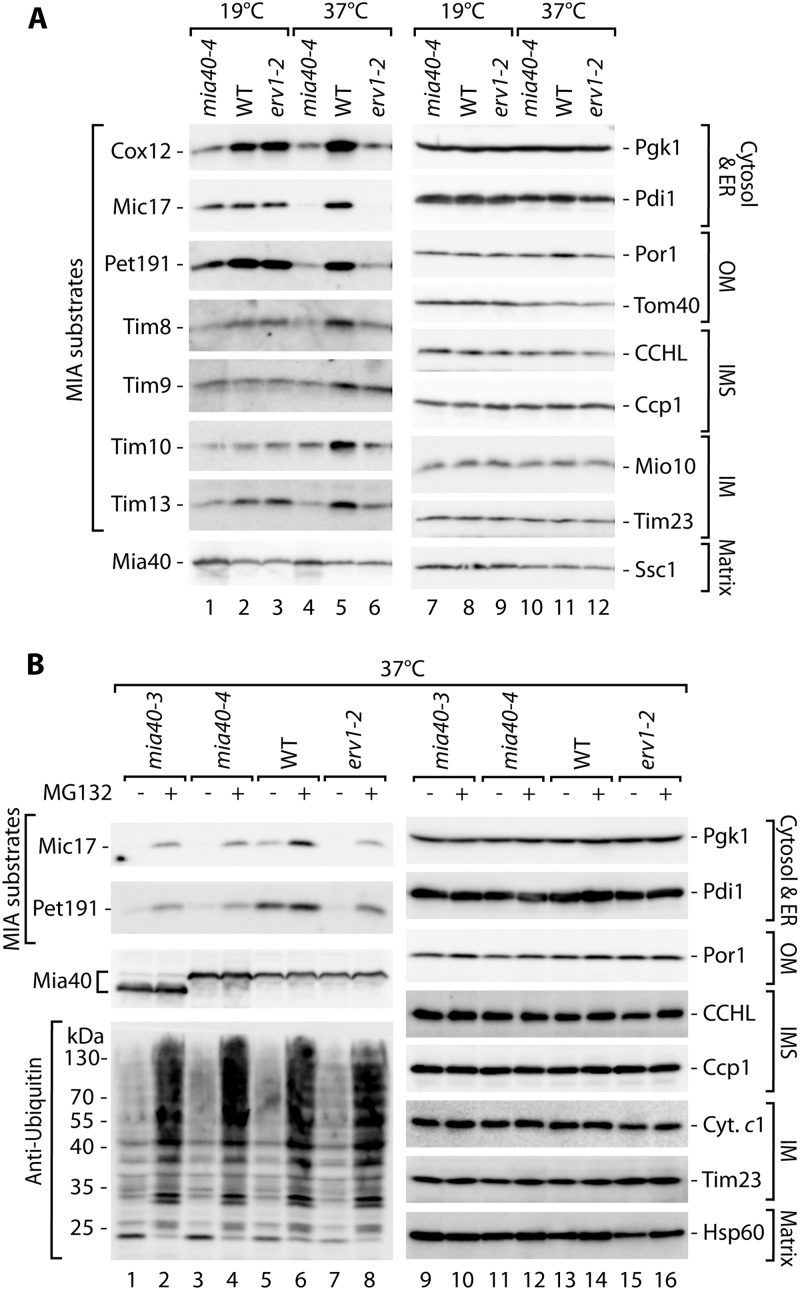
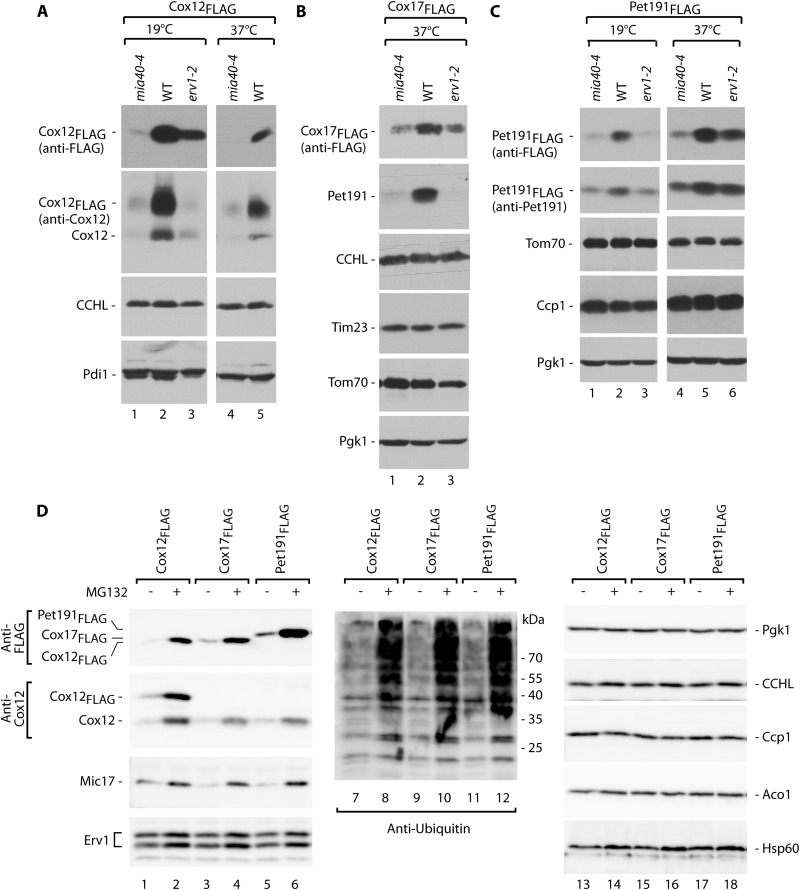
A large body of evidence has demonstrated that a class of IMS proteins does not accumulate in mitochondria in mutants that are defective in the MIA pathway (18, 29, 34). Thus, we investigated whether these proteins are present in other cellular compartments. We utilized temperature-sensitive mutants of the two main components of the MIA pathway, the mia40-4 and erv1-2 strains (32, 34), to determine the steady-state levels of IMS proteins under permissive conditions and after a shift to a higher temperature to induce an import defect in these strains (Fig. 1A). We tested IMS proteins belonging to the two known classes of MIA substrates, i.e., those with the twin CX9C motif (Cox12, Mic17, and Pet191) and small Tim proteins with the twin CX3C motif (Tim8, Tim9, Tim10, and Tim13) (12), in the total protein cellular extracts. Under permissive growth conditions, the proteins were detected in amounts similar to wild-type levels in the case of the erv1-2 strain or at reduced levels in the case of the mia40-4 strain, as expected (Fig. 1A, lanes 1 to 3). Under restrictive conditions, these proteins did not efficiently accumulate in mia40-4 and erv1-2 cells compared with wild-type cells (Fig. 1A, lanes 4 to 6). The control proteins from the cytosol (Pgk1), endoplasmic reticulum (ER) (Pdi1), and mitochondria remained unchanged (Fig. 1A, lanes 7 to 12). These observations raised the possibility that IMS proteins could be subjected to degradation.
Fig 1.
MIA precursor proteins are degraded upon mitochondrial import restriction. (A) Cellular protein levels in wild-type (WT) and mia40-4 and erv1-2 conditional mutant strains grown under permissive (19°C) or restrictive (6 h at 37°C) conditions. (B) Cellular protein levels in wild-type (WT), mia40-3, mia40-4, and erv1-2 strains grown for 6 h at a restrictive temperature (37°C), with or without the inhibitor MG132. In both panels, yeast was grown on glycerol as a carbon source. Protein extracts were analyzed by SDS-PAGE and Western blotting. ER, endoplasmic reticulum; OM, outer membrane; IMS, intermembrane space; IM, inner membrane.
We hypothesized that IMS proteins remained in the cytosol because of inefficient import in MIA mutant cells and were degraded by the ubiquitin-proteasome system, a major protein clearance machinery in the eukaryotic cytosol (44). We subjected the well-characterized mia40-3, mia40-4, and erv1-2 MIA mutant strains (32, 34) to treatment with the MG132 proteasome inhibitor (Fig. 1B). Treatment was efficient, as reflected by the accumulation of ubiquitinated proteins (Fig. 1B, lanes 1 to 8, bottom panel). Mic17 and Pet191 were indeed partially rescued in the presence of MG132 in the mia40 and erv1 mutants (Fig. 1B, lanes 1 to 8). The levels of the other proteins from the cytosol, ER, and various mitochondrial compartments remained largely unaffected (Fig. 1B, lanes 9 to 16). Thus, mitochondrial IMS proteins appeared to be cleared from the cytosol under conditions of defective protein import.
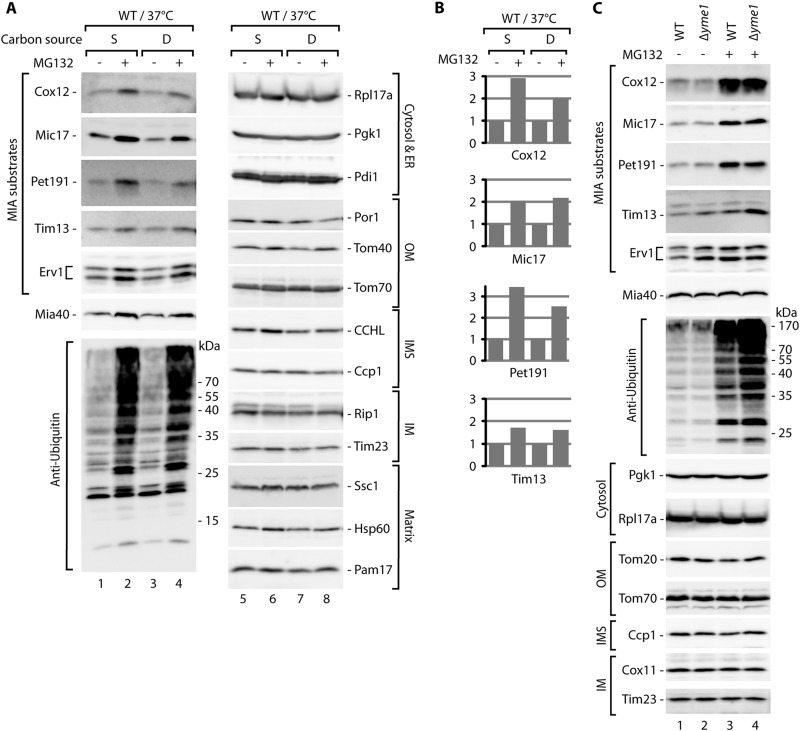
We noticed that the levels of MIA substrate proteins were also increased by MG132 treatment of the wild-type strain (Fig. 1B, lanes 5 and 6). Additionally, the Mia40 protein appeared to be slightly sensitive to proteasomal inhibition. Mia40 is a CX9C protein that is similar to one class of its substrates. However, Mia40 is attached to the presequence and transported to the mitochondria via the presequence translocase TIM23, independent of the MIA pathway (38). These two facts raised the possibility that the clearance process does not occur only in situations of defective protein import via the MIA pathway. We assessed this possibility by using a wild-type strain grown on fermentative carbon sources that result in slower mitochondrial biogenesis (Fig. 2A). We observed a substantial increase in the steady-state levels of MIA substrate proteins (Fig. 2A, lanes 1 to 4, and B) after MG132 treatment. Mia40 and Erv1, another component of the MIA pathway and a substrate of the MIA pathway (16), were moderately increased by MG132 treatment (Fig. 2A, lanes 1 to 4). The efficiency of MG132 treatment was confirmed by the increased levels of ubiquitinated protein species (Fig. 2A, lanes 1 to 4, bottom panel). Other control proteins from various cellular compartments were unaffected (Fig. 2A, lanes 5 to 8).
Fig 2.
MIA precursor proteins are stabilized by proteasome inhibition. (A) The wild-type (WT) strain was grown in minimal medium with either 2% sucrose (S) or 2% glucose (D) at 19°C, followed by treatment with MG132 for 5 h at 37°C. (B) Relative quantification results for the MIA substrate proteins tested in panel A. The signal of the wild-type strain was set to 1 for each carbon source. (C) The Δyme1 mutant and the corresponding wild-type (WT) strain were grown in minimal medium with 2% sucrose at 19°C. MG132 treatment occurred for 6 h at 37°C. (A and C) Total protein extracts were analyzed by SDS-PAGE followed by Western blotting. ER, endoplasmic reticulum; OM, outer membrane; IMS, intermembrane space; IM, inner membrane.
The possibility exists that MG132 exerts a nonspecific effect on protein quality control and the clearance system present inside mitochondria, specifically in the IMS. Thus, we decided to test a strain with the yme1 deletion (Fig. 2C). The Yme1 protein forms a core of the i-AAA protease, which is located in the inner mitochondrial membrane, with its active site facing the IMS. It is involved in the processing and turnover of proteins localized in the inner membrane and the IMS (45). We compared the effects of MG132 in the wild-type and Δyme1 strains and concluded that the effect of MG132 was not mediated by Yme1, because the increases in the accumulation of IMS proteins with MG132 treatment were similar in Δyme1 and wild-type cells (Fig. 2C, lanes 1 to 4). Additionally, we also observed moderate increases in the levels of Tim13 and Erv1 in Δyme1 cells compared with the wild-type strain in the absence of MG132 (Fig. 2C, lane 1 versus lane 2), indicating an additional level of quality control inside mitochondria that involves Yme1. Notably, Mia40 was less affected in this experiment than in the experiment documented in Fig. 2A. The variability in the response of Mia40 to proteasomal inhibition is likely attributable to its intrinsically high import efficiency via the TIM23 pathway, resulting in shorter exposure to the proteasome in the cytosol. Importantly, the results of this experiment and subsequent experiments with the proteasomal mutant strains (Fig. 3) demonstrated that an increase in MIA pathway components was not responsible for the elevation in MIA substrate levels upon proteasomal inhibition. Altogether, the data indicated that MG132 inhibited the clearance of IMS precursor proteins prior to their import into mitochondria.
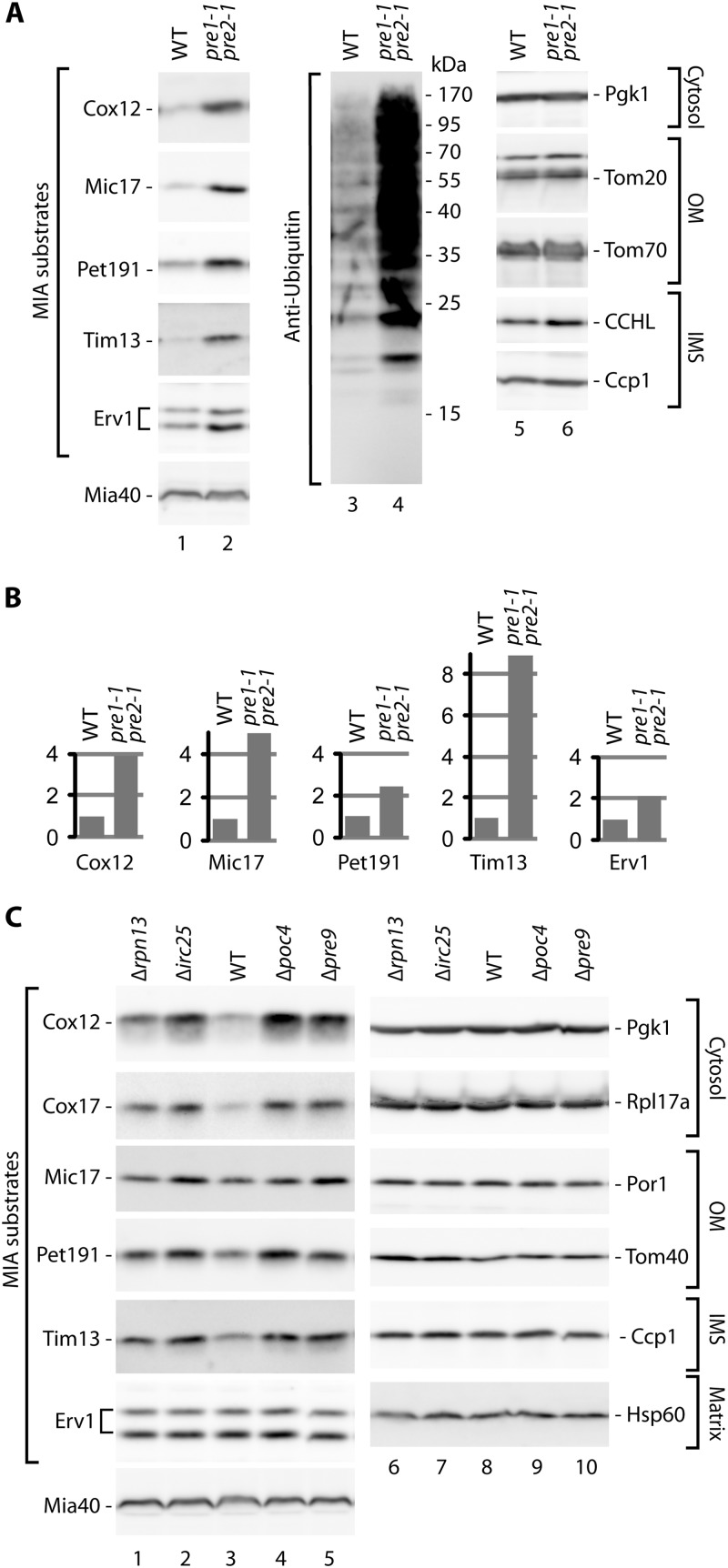
Fig 3.
The accumulation of MIA precursor proteins is increased in mutants with reduced proteasome activity. (A) The pre1-1 pre2-1 double proteasome mutant and corresponding wild-type (WT) strain were grown at 19°C. (B) Relative quantification results for selected proteins tested in panel A. The signal from the WT strain was set to 1 for each protein tested. (C) The proteasome-deficient Δrpn13, Δirc25, Δpoc4, and Δpre9 deletion strains and corresponding wild-type (WT) strain were grown at 19°C. (A and C) Yeast was grown on glycerol as a carbon source. Total protein extracts were analyzed by SDS-PAGE followed by Western blotting with specific antibodies, as indicated. OM, outer membrane; IMS, intermembrane space; IM, inner membrane.
To further confirm that MG132-sensitive degradation was indeed attributable to the proteasome, we examined a strain with genetically impaired proteasomal function, the pre1-1 pre2-1 strain, in which the two essential subunits are defective (46). We assessed the levels of the IMS proteins Cox12, Mic17, Pet191, Tim13, and Erv1 (Fig. 3A). These proteins accumulated more efficiently in pre1-1 pre2-1 cells than in wild-type cells (Fig. 3A, lanes 1 and 2, and B). As expected, the pre1-1 pre2-1 mutant cells expressed higher levels of ubiquitinated protein species (Fig. 3A, lanes 3 and 4). The levels of proteins from the cytosol and mitochondria, including Mia40, were unchanged (Fig. 3A, lanes 1, 2, 5, and 6).
We additionally tested deletion mutants of nonessential proteasome subunits (Rpn13 and Pre9) and proteasome assembly factors (Irc25 and Poc4) that exhibit proteasomal dysfunction (47–49) (Fig. 3C). The proteasomal mutant cells expressed increased levels of MIA substrates compared with the wild-type strain (Fig. 3C, lanes 1 to 5). Control proteins from the cytosol and mitochondria remained unaffected (Fig. 3C, lanes 6 to 10). Thus, our experiments demonstrated that the proteins targeted to the IMS were rescued by defects in the proteasome caused by genetic changes. These findings excluded the possibility of off-target inhibition by MG132. In summary, the proteasome degrades a fraction of MIA-dependent mitochondrial proteins under physiological conditions, where the import of proteins is not defective.
Intermembrane space proteins are ubiquitinated prior to degradation.
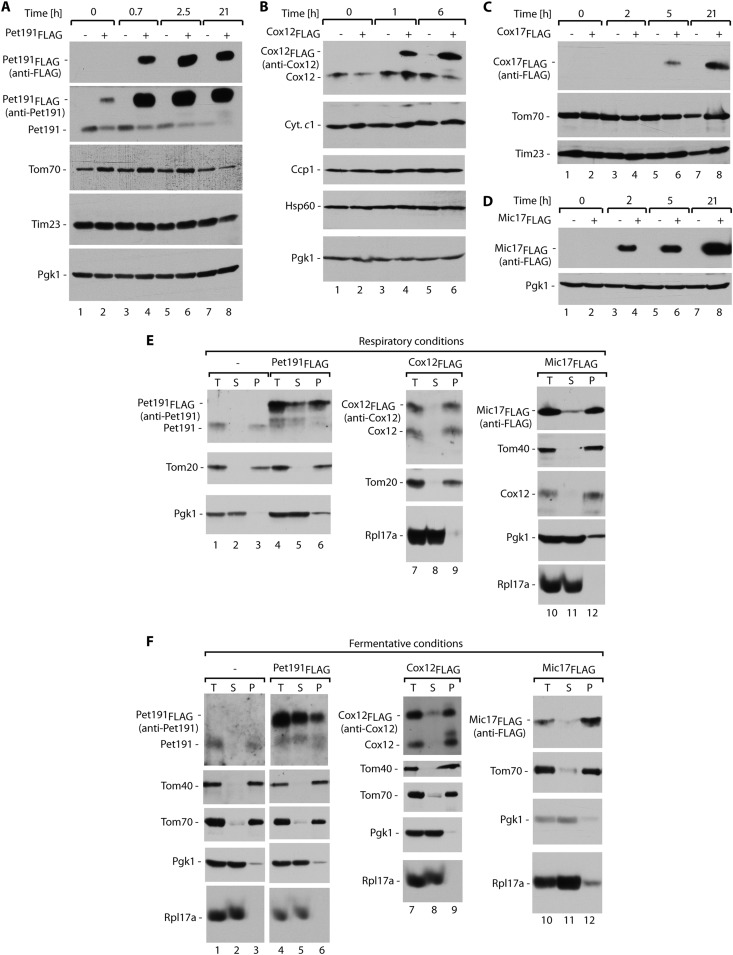
One step that precedes the degradation of proteins by the proteasome is ubiquitination (44, 50, 51). We reasoned that a fraction of IMS proteins should be found in ubiquitinated forms. The sensitivity of specific antibodies against IMS proteins did not permit the investigation of a pool of native proteins dispersed in species of various sizes. Therefore, we constructed fusion proteins of Pet191, Cox12, Cox17, and Mic17 with a FLAG tag at the C terminus and placed them under the control of an inducible GAL10 promoter. All of the fusion proteins were expressed efficiently within a few hours of galactose induction, with Cox17FLAG being the slowest one (Fig. 4A to D). In the case of Pet191, an increase in the production of Pet191FLAG was reproducibly accompanied by a decrease in the steady-state levels of native Pet191 (Fig. 4A), which was expected if these two proteins utilize common mitochondrial biogenesis machineries. The expression of the fusion proteins did not significantly affect the levels of other mitochondrial proteins and cytosolic Pgk1.
Fig 4.
Model FLAG fusion proteins are targeted to mitochondria. Total protein extracts of yeast cells producing Pet191FLAG (A), Cox12FLAG (B), Cox17FLAG (C), or Mic17FLAG (D) were analyzed after the indicated induction times. The expression of Pet191FLAG, Cox12FLAG, Cox17FLAG, or Mic17FLAG was induced in selective minimal medium that contained 3% glycerol and 0.5% galactose at 19°C (A, C, and D) or 24°C (B). (E and F) Subcellular localization of Pet191FLAG, Cox12FLAG, and Mic17FLAG. (E) Cells were grown in minimal medium that contained 3% glycerol with 0.2% galactose at 19°C. (F) Cells were grown in minimal medium that contained 2% galactose at 19°C. For all panels, the samples were analyzed by SDS-PAGE and Western blotting. T, total; S, postmitochondrial supernatant; P, mitochondrial pellet.
To be considered suitable models for our studies, FLAG fusions should have characteristics that are similar to those of native proteins. First, we examined the subcellular localization of Pet191FLAG, Cox12FLAG, and Mic17FLAG. They were found in the mitochondrial fraction (Fig. 4E and F), similar to other mitochondrial proteins (e.g., Tom20, Tom40, Tom70, and Cox12), and in contrast to cytosolic Pgk1 and Rpl17a. Only in the case of Pet191FLAG was a considerable pool also present in the cytosolic fraction, and this was more abundant under fermentative growth conditions (compare Fig. 4E and F, lanes 4 to 6). Native Pet191 was efficiently localized in the mitochondria (Fig. 4E and F, lanes 1 to 3). Thus, only Pet191FLAG was partially mislocalized to the cytosol, but Cox12FLAG and Mic17FLAG efficiently accumulated in the mitochondria.
Second, the fusion proteins should depend on the MIA pathway. We produced the fusion proteins Cox12FLAG, Cox17FLAG, and Pet191FLAG in the mia40-4 and erv1-2 mutants and grew the cells under permissive and restrictive conditions (Fig. 5A to C). The levels of fusion proteins were decreased in the mia40-4 and erv1-2 mutants, similar to the case for the native Cox12 and Pet191 proteins (Fig. 5A and B). Other cellular and mitochondrial marker proteins remained unaffected (Fig. 5A to C).
Fig 5.
FLAG-tagged MIA substrate proteins respond to oxidative folding restrictions and are stabilized by proteasomal inhibition, mimicking their endogenous counterparts. (A) Total protein extracts of wild-type (WT), mia40-4, and erv1-2 yeast cells producing Cox12FLAG. Expression was induced in selective minimal medium with 3% glycerol and 0.5% galactose at 19°C or 37°C for 5 h. (B) Total protein extracts of wild-type (WT), mia40-4, and erv1-2 yeast cells producing Cox17FLAG. Expression was induced in selective minimal medium with 3% glycerol and 0.5% galactose at 37°C for 5 h. (C) Total protein extracts of wild-type (WT), mia40-4, and erv1-2 cells producing Pet191FLAG. Expression of Pet191FLAG was induced in selective minimal medium with 3% glycerol and 0.5% galactose at 19°C or 37°C for 40 min. (D) Cox12FLAG, Cox17FLAG, and Pet191FLAG were induced in minimal medium that contained 2% galactose and subsequently repressed in minimal medium that contained 2% glucose, with or without MG132 inhibitor, for 4.5 h at 28°C. For all panels, total protein extracts were analyzed by SDS-PAGE followed by Western blotting with specific antibodies, as indicated.
Third, FLAG fusion proteins should respond to inhibition of the proteasome. Cox12FLAG, Cox17FLAG, and Pet191FLAG accumulated more efficiently in cells under conditions of proteasome inhibition induced by MG132, similar to native Cox12, Mic17, and ubiquitin-bound protein species, in contrast to cytosolic and mitochondrial marker proteins (Fig. 5D). We concluded that our FLAG fusions that were derived from MIA substrate proteins behaved similarly to native IMS proteins and thus were suitable models for studying ubiquitination.
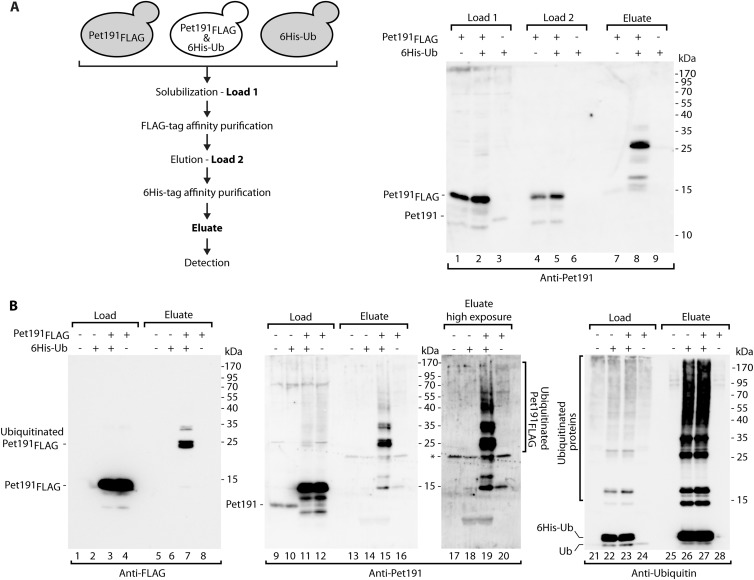
To study protein ubiquitination, our model substrates were coexpressed with 6His-Ub for affinity purification of protein species covalently bound to ubiquitin (41). First, we performed purification using an anti-FLAG column followed by Ni-NTA purification, thus enriching FLAG-containing proteins followed by enriching proteins modified with 6His-Ub (Fig. 6A, left panel). The eluate contained higher-molecular-weight species that were specifically enriched via double-affinity purification only for the extracts that contained Pet191FLAG and 6His-Ub, as expected for Pet191FLAG modified with ubiquitin (Fig. 6A, lanes 7 to 9). We then addressed the ubiquitination of Pet191FLAG under conditions of proteasomal inhibition. Total protein extracts from cells that produced Pet191FLAG or 6His-Ub were subjected to affinity purification with Ni-NTA to enrich ubiquitinated species (Fig. 6B). In the eluates, anti-FLAG and anti-Pet191 antibodies detected species that represented the model Pet191FLAG protein modified by one or more ubiquitin molecules (Fig. 6B, lanes 7, 15, and 19). The species were not found in the eluates derived from the cells that did not express Pet191FLAG or 6His-Ub (Fig. 6B). Pulldown via 6His-Ub was efficient, as reflected by immunodecoration against ubiquitin (Fig. 6B, lanes 21 to 28). Thus, under both physiological conditions and proteasomal inhibition conditions, we detected Pet191FLAG modified by ubiquitin.
Fig 6.
Pet191 is ubiquitinated in vivo. Strains that produced Pet191FLAG and/or 6His-tagged ubiquitin (6His-Ub) or were transfected with empty vectors were subjected to affinity purification from total cell extracts. (A) Affinity purification of Pet191FLAG under native conditions, followed by another affinity purification of 6His-tagged ubiquitinated proteins under denaturing conditions. The diagram illustrates the experimental design. Loads 1 and 2, 1%; eluate, 100%. (B) Affinity purification of 6His-tagged ubiquitinated proteins. During the last 3 h of growth, the cells were treated with MG132. Purification was performed under denaturing conditions. Load for anti-FLAG and anti-Pet191 blots, 1%; load for antiubiquitin blot, 3%; eluate, 100%. For both panels, the samples were analyzed by reducing SDS-PAGE and immunolabeling. *, nonspecific band.
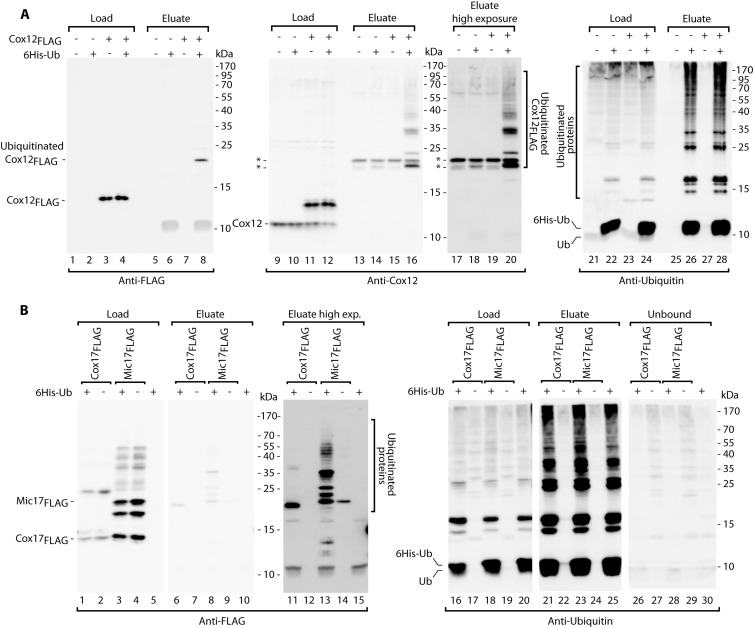
A substantial amount of Pet191FLAG was found in the cytosolic pool (Fig. 4E and F, lanes 4 to 6), and the mislocalization could render Pet191FLAG more prone to ubiquitination. In contrast, other fusion proteins, i.e., Cox12FLAG and Mic17FLAG, were efficiently localized in the mitochondria (Fig. 4E and F, lanes 7 to 12) and thus only transiently present in the cytosol, a feature typical of native IMS proteins. Thus, the ubiquitination of Cox12FLAG was tested under conditions of proteasome inhibition via the affinity purification of 6His-Ub (Fig. 7A). The FLAG-containing monoubiquitin species was detected in the eluate fraction (Fig. 7A, lane 8). Additionally, the Cox12-specific antibody allowed the detection of several other species with multiple ubiquitin moieties attached to Cox12FLAG (Fig. 7A, lanes 16 and 20). The presence of these species was fully dependent on the presence of Cox12FLAG and 6His-Ub, confirming that they represented Cox12FLAG bound to ubiquitin. Antiubiquitin immunodecoration indicated that the proteins that were bound to 6His-Ub were highly enriched in the eluate (Fig. 7A, lanes 25 to 28). We also tested Cox17FLAG and Mic17FLAG for ubiquitination (Fig. 7B). Even in the absence of the proteasomal inhibitor, we detected higher-molecular-weight species with at least one ubiquitin molecule attached to the model proteins in the eluate fractions (Fig. 7B, lanes 6 to 15). Altogether, our model proteins behaved like the native MIA substrates and underwent covalent modification with ubiquitin prior to their degradation.
Fig 7.
MIA precursor proteins are ubiquitinated in vivo. (A) Strains that produced Cox12FLAG and/or 6His-tagged ubiquitin (6His-Ub) or were transfected with empty vectors were subjected to affinity purification of 6His-tagged ubiquitinated proteins. During the final 3 h of growth, the cells were treated with MG132. Purification was performed under denaturing conditions. Load for anti-FLAG and anti-Cox12 blots, 1%; load for antiubiquitin blot, 9%; eluate, 100%. (B) Strains that produced Cox17FLAG or Mic17FLAG together with 6His-tagged ubiquitin or empty control vectors were subjected to affinity purification of 6His-tagged ubiquitinated proteins under native conditions. Load and unbound lanes, 7%; eluate, 100%. For both panels, the samples were analyzed by reducing SDS-PAGE and Western blotting with the indicated antibodies. *, nonspecific bands.
The proteasome exerts an inhibitory effect on IMS protein biogenesis.
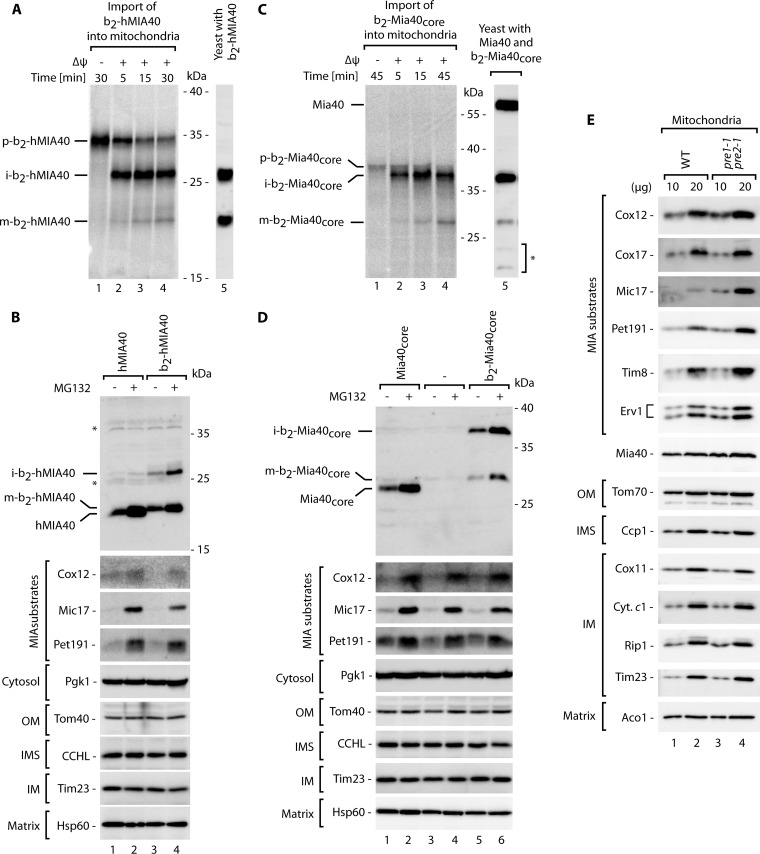
Are the proteins that are rescued from the ubiquitin-proteasome system imported into mitochondria to expand the pool of properly localized and functional mitochondrial IMS proteins? To answer this question, we used a fusion protein that expresses a characteristic of MIA substrates but whose localization in mitochondria is easy to detect in total protein cellular extracts. Mia40 is a protein that belongs to the same family of CX9C proteins as its substrate proteins, but in yeast it contains a cleavable mitochondrial targeting signal for the TIM23 import pathway. Our data suggested that Mia40, similar to its substrates, could also be subjected to ubiquitin-proteasome control (Fig. 1B and 2A), but its intrinsically high mitochondrial import efficiency results in a small time frame for the ubiquitin-proteasome system to act in the cytosol. Interestingly, in higher eukaryotes, Mia40 is a substrate of the MIA pathway, in contrast to the case in yeast (38, 39, 52). We used a fusion of human Mia40 (hMIA40) attached to the bipartite presequence of cytochrome b2. The presequence of cytochrome b2 serves to localize the mature protein in the IMS and is cleaved by two mitochondrial peptidases: the mitochondrial matrix peptidase MPP, which generates an intermediate form, and the inner membrane peptidase IMP, which generates a soluble mature form in the IMS of mitochondria (53). When we imported b2-hMIA40 into isolated mitochondria, we noticed the formation of the intermediate form followed by a slower generation of the mature form (Fig. 8A, lanes 1 to 4). These forms migrated exactly as the forms observed when b2-hMIA40 was expressed in yeast (Fig. 8A, lane 5). Thus, we established a model protein that natively utilizes the MIA pathway but whose presence inside mitochondria can easily be monitored by the appearance of the processed forms in total cellular extracts.
Fig 8.
IMS proteins accumulate in mitochondria upon inhibition of proteasome activity. The intermediate and mature forms of radiolabeled b2-hMIA40 (A) and b2-Mia40core (C) proteins generated during import into the isolated mitochondria were compared with total protein extracts derived from the cells that expressed b2-hMIA40 and b2-Mia40core. Import was performed in the presence or absence of an inner mitochondrial membrane potential (Δψ), as indicated. Import reactions and total protein extracts were analyzed by autoradiography (lanes 1 to 4) and immunolabeling with Mia40-specific antibodies (lanes 5). Yeast strains that produced hMIA40 or b2-hMIA40 (B) or Mia40core or b2-Mia40core (D) were grown for 5 h at 37°C, with or without MG132. (E) Steady-state levels of proteins in mitochondria isolated from the pre1-1 pre2-1 mutant and corresponding wild-type strain grown in sucrose-containing medium at 19°C. (B, D, and E) protein extracts were analyzed by SDS-PAGE followed by Western blotting. p, precursor; i, intermediate; m, mature; OM, outer membrane; IMS, intermembrane space; IM, inner membrane. *, nonspecific bands.
We subjected the strains that expressed hMIA40 and b2-hMIA40 to treatment with MG132 (Fig. 8B). With hMIA40, we confirmed that the human protein was indeed subjected to clearance by the proteasome (Fig. 8B, lanes 1 and 2). In the case of b2-hMIA40, we observed a significant increase in the mitochondrion-localized intermediate and mature forms in the presence of MG132 (Fig. 8B, lanes 3 and 4). These results demonstrated that inhibition of the proteasome was accompanied by the efficient import of rescued b2-hMIA40 into mitochondria. We used a similar approach to study Mia40core and b2-Mia40core. Mia40core is a conserved functional domain of yeast Mia40 that lacks a native presequence of yeast Mia40 and is imported via the MIA pathway, thus resembling hMIA40 (38). b2-Mia40core was imported into the mitochondria, and proteolytic cleavage generated species that migrated exactly like the intermediate and mature forms observed when b2-Mia40core was expressed in yeast (Fig. 8C). The levels of the intermediate and mature forms of Mia40core and b2-Mia40core increased upon treatment of the cells with MG132, similar to the typical MIA substrates (Fig. 8D). Notably, the intermediate forms of both b2-hMIA40 and b2-Mia40core cannot be released from mitochondria because they are anchored in the inner mitochondrial membrane (38). Thus, the increases in the levels of intermediate membrane-bound forms of both b2-hMIA40 and b2-Mia40core upon proteasomal inhibition led to the conclusion that the proteasome was inhibited during the stage that preceded mitochondrial protein import. Thus, inhibition of the proteasome expands the pool of MIA substrates that can accumulate productively in mitochondria.
To further validate our findings, we analyzed the levels of native IMS proteins in mitochondria isolated from the pre1-1 pre2-1 proteasomal mutant strain and its corresponding wild-type strain. The analysis of the steady-state levels of the proteins confirmed an increase in the mitochondrial accumulation of the MIA substrates in the proteasome-deficient mutant, whereas levels of the other mitochondrial proteins remained unchanged (Fig. 8E). These findings demonstrated that a defect in proteasomal activity resulted in an increase in the mitochondrial abundance of MIA substrates. In summary, the newly synthesized MIA substrate proteins were subjected to two processes, namely, mitochondrial protein import and proteasomal degradation, that act in parallel to regulate the levels of mature IMS proteins.
DISCUSSION
Cellular proteins that are misfolded, aggregated, or damaged undergo degradation, and this process is a ubiquitous and essential element of cellular quality control. Furthermore, protein degradation is used to regulate the half-life and turnover of proteins that play major roles in regulatory processes in various aspects of cell function. The degradation of cytosolic proteins and ER proteins following their retrograde transport to the cytosol is governed by the ubiquitin-proteasome system (44, 50, 51, 54–59).
The present study identified an unexpected role of cytosolic degradation in the biogenesis of mitochondrial proteins. We demonstrated that the proteins directed to the mitochondrial IMS are substrates of ubiquitin-proteasome control in a systemic manner. This conclusion is based on the following lines of evidence. The proteins targeted to the mitochondrial IMS were rescued by the use of the proteasomal inhibitor MG132 and by a defect in proteasomal function that was caused by genetic changes. The latter observation excludes the possibility that the effect exerted by MG132 was attributable to off-target inhibition of other proteases present inside the mitochondria or elsewhere. Proteins that are selected for proteasomal degradation are marked by the covalent attachment of ubiquitin, a small and conserved protein modifier. The attachment of ubiquitin results from a reaction cascade completed by ubiquitin ligase enzymes that transfer ubiquitin on substrate proteins to build a chain of polyubiquitin (44, 50, 51, 57). We identified ubiquitinated species of precursor proteins targeted to the IMS that were present in cells even under conditions where the proteasome was fully active. Thus, we established a role for the ubiquitin-proteasome system in the turnover of this class of mitochondrial proteins.
The ubiquitin-proteasome system has been implicated in the degradation of mature mitochondrial proteins, mainly located in the outer membrane (45, 60–63). The ubiquitin-proteasome system was previously demonstrated to degrade immature cytochrome c and EndoG, two proteins targeted to the IMS (64, 65). Our study identified the entire class of internal mitochondrial proteins, the MIA substrates, as being controlled by the ubiquitin-proteasome system. Interestingly, model MIA substrates that failed to properly fold and assemble inside mitochondria, such as Tim9 and Tim10, were degraded by mitochondrial Yme1 (66). Additionally, the Ups1 and Ups2 proteins were subjected to mitochondrial quality control when they were not assembled with the partner protein Mdm35 (67). Thus, both cellular compartments, the cytosol and IMS, are equipped with quality control systems to clear immature IMS proteins.
Importantly, our results demonstrate that IMS proteins are under persistent control by the ubiquitin-proteasome system under conditions in which the protein import machinery and biogenesis pathways are fully active. This could indicate that the mitochondrial import of MIA substrate proteins is rather slow, allowing the ubiquitin-proteasome system to compete for these targets. This interpretation is based on our observation that the MIA substrate proteins are more accessible to proteasomal activity when the cells are grown on a fermentative carbon source (i.e., under conditions where there is less need for mitochondrial biogenesis). Further support comes from the analysis of Mia40. Despite being a good substrate for the ubiquitin-proteasome system, Mia40 is degraded less apparently than MIA substrates because in yeast the mitochondrial targeting signal of Mia40 is likely very efficient at directing the protein to mitochondria. Indeed, changing the native signal of Mia40 slows its import and results in improved proteasomal accessibility. Interestingly, inhibition of the proteasome renders proteins that are efficiently imported into the IMS. Further research is needed to mechanistically dissect this stage. The proteasome-rescued proteins could be imported into mitochondria in ubiquitinated forms. However, based on size restrictions imposed by the outer mitochondrial membrane translocase TOM, we favor a scenario that implicates a role for deubiquitinating enzymes in the cytosol (68) in preparing IMS protein precursors to enter mitochondria.
The ubiquitin-proteasome system acts as negative regulator of the early cytosolic stages of IMS protein biogenesis. However, a function of the ubiquitin-proteasome system in maintaining protein homeostasis under circumstances in which mitochondria and, specifically, the IMS lose their integrity can be postulated. In general, the fact that the presence of IMS proteins is tightly controlled by the ubiquitin-proteasome system raises the issues of whether and how mitochondrial precursor proteins contribute to cellular proteostasis and its failure under conditions of defective clearance mechanisms and/or a decrease in mitochondrial protein import. However, even under physiological conditions, the need for mitochondrial IMS proteins may vary depending on metabolic requirements. The mechanism that involves the ubiquitin-proteasome system provides the means to rapidly adjust mitochondrial protein biogenesis at the posttranslational stage in response to various physiological needs.
ACKNOWLEDGMENTS
Research in the Chacinska laboratory was supported by the Foundation for Polish Science-Welcome Programme, an EMBO installation grant, and grants from the Ministry of Science and Higher Education (N N301 298337) and the National Science Centre in Poland (2011/02/B/NZ2/01402). P.B., A.G., and M.E.S. were supported by stipends from the Foundation for Polish Science-Welcome Programme, cofinanced by the European Union within the European Regional Development Fund.
We thank Bernard Guiard and Teresa Zoladek for discussions and materials.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 18 March 2013
REFERENCES
- 1. Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schönfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C. 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. U. S. A. 100:13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Prokisch H, Scharfe C, Camp DG, Xiao W, David L, Andreoli C, Monroe ME, Moore RJ, Gritsenko MA, Kozany C, Hixson KK, Mottaz HM, Zischka H, Ueffing M, Herman ZS, Davis RW, Meitinger T, Oefner PJ, Smith RD, Steinmetz LM. 2004. Integrative analysis of the mitochondrial proteome in yeast. PLoS Biol. 2:e160 doi:10.1371/journal.pbio.0020160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Perocchi F, Jensen LJ, Gagneur J, Ahting U, von Mering C, Bork P, Prokisch H, Steinmetz LM. 2006. Assessing systems properties of yeast mitochondria through an interaction map of the organelle. PLoS Genet. 2:e170 doi:10.1371/journal.pgen.0020170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dolezal P, Likic V, Tachezy J, Lithgow T. 2006. Evolution of the molecular machines for protein import into mitochondria. Science 313:314–318 [DOI] [PubMed] [Google Scholar]
- 5. Neupert W, Herrmann JM. 2007. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76:723–749 [DOI] [PubMed] [Google Scholar]
- 6. Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. 2009. Importing mitochondrial proteins: machineries and mechanisms. Cell 138:628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schmidt O, Pfanner N, Meisinger C. 2010. Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 11:655–667 [DOI] [PubMed] [Google Scholar]
- 8. Wasilewski M, Scorrano L. 2009. The changing shape of mitochondrial apoptosis. Trends Endocrinol. Metab. 20:287–294 [DOI] [PubMed] [Google Scholar]
- 9. Herrmann JM, Riemer J. 2010. The intermembrane space of mitochondria. Antioxid. Redox Signal. 13:1341–1358 [DOI] [PubMed] [Google Scholar]
- 10. Martinou JC, Youle RJ. 2011. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev. Cell 21:92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nunnari J, Suomalainen A. 2012. Mitochondria: in sickness and in health. Cell 148:1145–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vögtle FN, Burkhart JM, Rao S, Gerbeth C, Hinrichs J, Martinou JC, Chacinska A, Sickmann A, Zahedi RP, Meisinger C. 2012. Intermembrane space proteome of yeast mitochondria. Mol. Cell. Proteomics 11:1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Endo T, Yamano K, Kawano S. 2010. Structural basis for the disulfide relay system in the mitochondrial intermembrane space. Antioxid. Redox Signal. 13:1359–1373 [DOI] [PubMed] [Google Scholar]
- 14. Sideris DP, Tokatlidis K. 2010. Oxidative protein folding in the mitochondrial intermembrane space. Antioxid. Redox Signal. 13:1189–1204 [DOI] [PubMed] [Google Scholar]
- 15. Herrmann JM, Riemer J. 2012. Mitochondrial disulfide relay: redox-regulated protein import into the intermembrane space. J. Biol. Chem. 287:4426–4433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stojanovski D, Bragoszewski P, Chacinska A. 2012. The MIA pathway: a tight bond between protein transport and oxidative folding in mitochondria. Biochim. Biophys. Acta 1823:1142–1150 [DOI] [PubMed] [Google Scholar]
- 17. Koehler CM. 2004. The small Tim proteins and the twin Cx3C motif. Trends Biochem. Sci. 29:1–4 [DOI] [PubMed] [Google Scholar]
- 18. Gabriel K, Milenkovic D, Chacinska A, Müller J, Guiard B, Pfanner N, Meisinger C. 2007. Novel mitochondrial intermembrane space proteins as substrates of the MIA import pathway. J. Mol. Biol. 365:612–620 [DOI] [PubMed] [Google Scholar]
- 19. Longen S, Bien M, Bihlmaier K, Kloeppel C, Kauff F, Hammermeister M, Westermann B, Herrmann JM, Riemer J. 2009. Systematic analysis of the twin cx(9)c protein family. J. Mol. Biol. 393:356–368 [DOI] [PubMed] [Google Scholar]
- 20. Mesecke N, Terziyska N, Kozany C, Baumann F, Neupert W, Hell K, Herrmann JM. 2005. A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 121:1059–1069 [DOI] [PubMed] [Google Scholar]
- 21. Grumbt B, Stroobant V, Terziyska N, Israel L, Hell K. 2007. Functional characterization of Mia40p, the central component of the disulfide relay system of the mitochondrial intermembrane space. J. Biol. Chem. 282:37461–37470 [DOI] [PubMed] [Google Scholar]
- 22. Müller JM, Milenkovic D, Guiard B, Pfanner N, Chacinska A. 2008. Precursor oxidation by Mia40 and Erv1 promotes vectorial transport of proteins into the mitochondrial intermembrane space. Mol. Biol. Cell 19:226–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Banci L, Bertini I, Cefaro C, Ciofi-Baffoni S, Gallo A, Martinelli M, Sideris DP, Katrakili N, Tokatlidis K. 2009. MIA40 is an oxidoreductase that catalyzes oxidative protein folding in mitochondria. Nat. Struct. Mol. Biol. 16:198–206 [DOI] [PubMed] [Google Scholar]
- 24. Tienson HL, Dabir DV, Neal SE, Loo R, Hasson SA, Boontheung P, Kim SK, Loo JA, Koehler CM. 2009. Reconstitution of the Mia40-Erv1 oxidative folding pathway for the small Tim proteins. Mol. Biol. Cell 20:3481–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Milenkovic D, Ramming T, Müller JM, Wenz LS, Gebert N, Schulze-Specking A, Stojanovski D, Rospert S, Chacinska A. 2009. Identification of the signal directing Tim9 and Tim10 into the intermembrane space of mitochondria. Mol. Biol. Cell 20:2530–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sideris DP, Petrakis N, Katrakili N, Mikropoulou D, Gallo A, Ciofi-Baffoni S, Banci L, Bertini I, Tokatlidis K. 2009. A novel intermembrane space-targeting signal docks cysteines onto Mia40 during mitochondrial oxidative folding. J. Cell Biol. 187:1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Banci L, Bertini I, Cefaro C, Cenacchi L, Ciofi-Baffoni S, Felli IC, Gallo A, Gonnelli L, Luchinat E, Sideris D, Tokatlidis K. 2010. Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import. Proc. Natl. Acad. Sci. U. S. A. 107:20190–20195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bien M, Longen S, Wagener N, Chwalla I, Herrmann JM, Riemer J. 2010. Mitochondrial disulfide bond formation is driven by intersubunit electron transfer in Erv1 and proofread by glutathione. Mol. Cell 37:516–528 [DOI] [PubMed] [Google Scholar]
- 29. Rissler M, Wiedemann N, Pfannschmidt S, Gabriel K, Guiard B, Pfanner N, Chacinska A. 2005. The essential mitochondrial protein Erv1 cooperates with Mia40 in biogenesis of intermembrane space proteins. J. Mol. Biol. 353:485–492 [DOI] [PubMed] [Google Scholar]
- 30. Banci L, Bertini I, Calderone V, Cefaro C, Ciofi-Baffoni S, Gallo A, Kallergi E, Lionaki E, Pozidis C, Tokatlidis K. 2011. Molecular recognition and substrate mimicry drive the electron-transfer process between MIA40 and ALR. Proc. Natl. Acad. Sci. U. S. A. 108:4811–4816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stojanovski D, Milenkovic D, Müller JM, Gabriel K, Schulze-Specking A, Baker MJ, Ryan MT, Guiard B, Pfanner N, Chacinska A. 2008. Mitochondrial protein import: precursor oxidation in a ternary complex with disulfide carrier and sulfhydryl oxidase. J. Cell Biol. 183:195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Böttinger L, Gornicka A, Czerwik T, Bragoszewski P, Loniewska-Lwowska A, Schulze-Specking A, Truscott KN, Guiard B, Milenkovic D, Chacinska A. 2012. In vivo evidence for cooperation of Mia40 and Erv1 in the oxidation of mitochondrial proteins. Mol. Biol. Cell 23:3957–3969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. von der Malsburg K, Müller JM, Bohnert M, Oeljeklaus S, Kwiatkowska P, Becker T, Loniewska-Lwowska A, Wiese S, Rao S, Milenkovic D, Hutu DP, Zerbes RM, Schulze-Specking A, Meyer HE, Martinou JC, Rospert S, Rehling P, Meisinger C, Veenhuis M, Warscheid B, van der Klei IJ, Pfanner N, Chacinska A, van der Laan M. 2011. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev. Cell 21:694–707 [DOI] [PubMed] [Google Scholar]
- 34. Chacinska A, Pfannschmidt S, Wiedemann N, Kozjak V, Sanjuán Szklarz LK, Schulze-Specking A, Truscott KN, Guiard B, Meisinger C, Pfanner N. 2004. Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J. 23:3735–3746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Allen S, Balabanidou V, Sideris DP, Lisowsky T, Tokatlidis K. 2005. Erv1 mediates the Mia40-dependent protein import pathway and provides a functional link to the respiratory chain by shuttling electrons to cytochrome c. J. Mol. Biol. 353:937–944 [DOI] [PubMed] [Google Scholar]
- 36. Terziyska N, Lutz T, Kozany C, Mokranjac D, Mesecke N, Neupert W, Herrmann JM, Hell K. 2005. Mia40, a novel factor for protein import into the intermembrane space of mitochondria is able to bind metal ions. FEBS Lett. 579:179–184 [DOI] [PubMed] [Google Scholar]
- 37. Durigon R, Wang Q, Ceh Pavia E, Grant CM, Lu H. 2012. Cytosolic thioredoxin system facilitates the import of mitochondrial small Tim proteins. EMBO Rep. 13:916–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chacinska A, Guiard B, Müller JM, Schulze-Specking A, Gabriel K, Kutik S, Pfanner N. 2008. Mitochondrial biogenesis, switching the sorting pathway of the intermembrane space receptor Mia40. J. Biol. Chem. 283:29723–29729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sztolsztener ME, Brewinska A, Guiard B, Chacinska A. 2013. Disulfide bond formation: sulfhydryl oxidase ALR controls mitochondrial biogenesis of human MIA40. Traffic 14:309–320 [DOI] [PubMed] [Google Scholar]
- 40. Morvan J, Froissard M, Haguenauer-Tsapis R, Urban-Grimal D. 2004. The ubiquitin ligase Rsp5p is required for modification and sorting of membrane proteins into multivesicular bodies. Traffic 5:383–392 [DOI] [PubMed] [Google Scholar]
- 41. Stawiecka-Mirota M, Pokrzywa W, Morvan J, Zoladek T, Haguenauer-Tsapis R, Urban-Grimal D, Morsomme P. 2007. Targeting of Sna3p to the endosomal pathway depends on its interaction with Rsp5p and multivesicular body sorting on its ubiquitylation. Traffic 8:1280–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu C, Apodaca J, Davis LE, Rao H. 2007. Proteasome inhibition in wild-type yeast Saccharomyces cerevisiae cells. Biotechniques 42:158, 160,, 162 [DOI] [PubMed] [Google Scholar]
- 43. Meisinger C, Pfanner N, Truscott KN. 2006. Isolation of yeast mitochondria. Methods Mol. Biol. 313:33–39 [DOI] [PubMed] [Google Scholar]
- 44. Ciechanover A. 2012. Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Biochim. Biophys. Acta 1824:3–13 [DOI] [PubMed] [Google Scholar]
- 45. Anand R, Langer T, Baker MJ. 2013. Proteolytic control of mitochondrial function and morphogenesis. Biochim. Biophys. Acta 1833:195–204 [DOI] [PubMed] [Google Scholar]
- 46. Heinemeyer W, Gruhler A, Möhrle V, Mahé Y, Wolf DH. 1993. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chymotryptic activity and degradation of ubiquitinated proteins. J. Biol. Chem. 268:5115–5120 [PubMed] [Google Scholar]
- 47. Velichutina I, Connerly PL, Arendt CS, Li X, Hochstrasser M. 2004. Plasticity in eucaryotic 20S proteasome ring assembly revealed by a subunit deletion in yeast. EMBO J. 23:500–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. 2008. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453:481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kusmierczyk AR, Kunjappu MJ, Funakoshi M, Hochstrasser M. 2008. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat. Struct. Mol. Biol. 15:237–244 [DOI] [PubMed] [Google Scholar]
- 50. Finley D. 2009. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78:477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Husnjak K, Dikic I. 2012. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu. Rev. Biochem. 81:291–322 [DOI] [PubMed] [Google Scholar]
- 52. Hofmann S, Rothbauer U, Mühlenbein N, Baiker K, Hell K, Bauer MF. 2005. Functional and mutational characterization of human MIA40 acting during import into the mitochondrial intermembrane space. J. Mol. Biol. 353:517–528 [DOI] [PubMed] [Google Scholar]
- 53. Chacinska A, van der Laan M, Mehnert CS, Guiard B, Mick DU, Hutu DP, Truscott KN, Wiedemann N, Meisinger C, Pfanner N, Rehling P. 2010. Distinct forms of mitochondrial TOM-TIM supercomplexes define signal-dependent states of preprotein sorting. Mol. Cell. Biol. 30:307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Goldberg AL. 2003. Protein degradation and protection against misfolded or damaged proteins. Nature 426:895–899 [DOI] [PubMed] [Google Scholar]
- 55. Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. 2004. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature 429:841–847 [DOI] [PubMed] [Google Scholar]
- 56. Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. 2008. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell 132:101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Deshaies RJ, Joazeiro CA. 2009. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78:399–434 [DOI] [PubMed] [Google Scholar]
- 58. Varshavsky A. 2011. The N-end rule pathway and regulation by proteolysis. Protein Sci. 20:1298–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Walter P, Ron D. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334:1081–1086 [DOI] [PubMed] [Google Scholar]
- 60. Escobar-Henriques M, Westermann B, Langer T. 2006. Regulation of mitochondrial fusion by the F-box protein Mdm30 involves proteasome-independent turnover of Fzo1. J. Cell Biol. 173:645–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Margineantu DH, Emerson CB, Diaz D, Hockenbery DM. 2007. Hsp90 inhibition decreases mitochondrial protein turnover. PLoS One 2:e1066 doi:10.1371/journal.pone.0001066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cohen MM, Leboucher GP, Livnat-Levanon N, Glickman MH, Weissman AM. 2008. Ubiquitin-proteasome-dependent degradation of a mitofusin, a critical regulator of mitochondrial fusion. Mol. Biol. Cell 19:2457–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Karbowski M, Youle RJ. 2011. Regulating mitochondrial outer membrane proteins by ubiquitination and proteasomal degradation. Curr. Opin. Cell Biol. 23:476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pearce DA, Sherman F. 1997. Differential ubiquitin-dependent degradation of the yeast apo-cytochrome c isozymes. J. Biol. Chem. 272:31829–31836 [DOI] [PubMed] [Google Scholar]
- 65. Radke S, Chander H, Schäfer P, Meiss G, Krüger R, Schulz JB, Germain D. 2008. Mitochondrial protein quality control by the proteasome involves ubiquitination and the protease Omi. J. Biol. Chem. 283:12681–12685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Baker MJ, Mooga VP, Guiard B, Langer T, Ryan MT, Stojanovski D. 2012. Impaired folding of the mitochondrial small TIM chaperones induces clearance by the i-AAA protease. J. Mol. Biol. 424:227–239 [DOI] [PubMed] [Google Scholar]
- 67. Potting C, Wilmes C, Engmann T, Osman C, Langer T. 2010. Regulation of mitochondrial phospholipids by Ups1/PRELI-like proteins depends on proteolysis and Mdm35. EMBO J. 29:2888–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Komander D, Clague MJ, Urbé S. 2009. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10:550–563 [DOI] [PubMed] [Google Scholar]
- 69. Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]