Abstract
Iron (Fe) is an essential element for all eukaryotic organisms because it functions as a cofactor in a wide range of biochemical processes. Cells have developed sophisticated mechanisms to tightly control Fe utilization in response to alterations in cellular demands and bioavailability. In response to Fe deficiency, the yeast Saccharomyces cerevisiae activates transcription of the CTH1 and CTH2 genes, which encode proteins that bind to AU-rich elements (AREs) within the 3′ untranslated regions (3′UTRs) of many mRNAs, leading to metabolic reprogramming of Fe-dependent pathways and decreased Fe storage. The precise mechanisms underlying Cth1 and Cth2 function and regulation are incompletely understood. We report here that the Cth1 and Cth2 proteins specifically bind in vivo to AREs located at the 3′UTRs of their own transcripts in an auto- and cross-regulated mechanism that limits their expression. By mutagenesis of the AREs within the CTH2 transcript, we demonstrate that a Cth2 negative-feedback loop is required for the efficient decline in Cth2 protein levels observed upon a rapid rise in Fe availability. Importantly, Cth2 autoregulation is critical for the appropriate recovery of Fe-dependent processes and resumption of growth in response to a change from Fe deficiency to Fe supplementation.
INTRODUCTION
Iron (Fe) is an essential micronutrient for all eukaryotic organisms because it participates as a heme, Fe-S cluster, or oxodiiron cofactor in multiple processes, including oxygen sensing and transport, the tricarboxylic acid (TCA) cycle, mitochondrial respiration, DNA replication and repair, lipid metabolism, and protein translation (1–4). In humans, Fe imbalances underlie many diseases, including hereditary hemochromatosis, Friedreich's ataxia, and aceruloplasminemia (5–8). Despite its abundance, Fe bioavailability is highly restricted due to its low solubility at physiological pH. Indeed, human Fe deficiency is the most widespread nutritional disorder in the world, affecting more than 2 billion people and leading to anemia, mainly in women and children, that is reversed by dietary Fe supplementation (9).
Eukaryotic organisms have developed sophisticated mechanisms to optimize Fe acquisition, distribution, utilization, storage, and mobilization. Multiple regulatory factors are interconnected to coordinate the cellular responses to Fe imbalances due to alterations in Fe bioavailability or changes in metabolic Fe needs. In mammals, the Fe-regulatory proteins IRP1 and IRP2 tightly control cellular Fe metabolism (6, 7, 10). Under Fe-deficient conditions, IRP proteins bind to specific stem-loop RNA structures denoted Fe-responsive elements (IREs) within the 5′ untranslated region (5′UTR) in multiple mRNAs, including the Fe storage protein ferritin, the Fe efflux pump ferroportin, and enzymes, such as mitochondrial aconitase and erythroid aminolevulinic acid synthase, thereby inhibiting their translation. Furthermore, IRP binding to IREs within the 3′UTR of the transferrin receptor transcript promotes its stabilization, leading to an increase in its translation. This coordinated regulation allows Fe-deficient cells to increase uptake of Fe-loaded transferrin and decrease Fe storage in ferritin. When Fe availability resumes, IRP1 acquires an Fe-S cluster that converts it into cytoplasmic aconitase, and FBXL5 protein interacts with IRP2, promoting its ubiquitination and degradation (11–13).
In the yeast Saccharomyces cerevisiae, Fe regulation is controlled by both transcriptional and posttranscriptional mechanisms. When environmental Fe levels are high, the high-affinity Fe acquisition systems are not expressed, and Fe enters the cell through low-affinity transporters. Under these conditions, the Yap5 transcription factor activates the expression of CCC1, encoding a vacuolar Fe transporter that mediates Fe detoxification (7), although it has been demonstrated that Yap5 only partially regulates the expression of CCC1 (14). In response to Fe scarcity, two Fe-responsive transcription factors, Aft1 and Aft2, induce the transcription of a set of genes denoted the Fe regulon, which encodes proteins involved in high-affinity Fe uptake at the plasma membrane, Fe mobilization from vacuolar stores, and metabolic reprogramming of Fe-consuming pathways (15–17). Within this Fe regulon are genes encoding two mRNA-binding proteins, Cth1 and Cth2, characterized by the presence of two tandem zinc fingers (TZFs) of the CX8CX5CX3H type, which are conserved in the mammalian family of tristetraprolin (TTP) mRNA-destabilizing proteins (18–20). While Cth1 is transiently expressed in the early stages of Fe deficiency, Cth2 expression increases during the progress of Fe limitation, reaching its maximum levels when Fe deficiency persists (19, 20). Cth1 and Cth2 bind through their TZFs to specific AU-rich elements (AREs) located in the 3′UTRs of many mRNAs, recruiting the Dhh1 helicase to promote the cytoplasmic 5′-to-3′ turnover of target transcripts (19–21). Cth2 also modulates the 3′-end processing of ARE-containing mRNAs by promoting the degradation of extended transcripts (22, 23). In response to Fe limitation, the ARE-containing 3′UTR of Cth1/Cth2 target mRNAs is sufficient to promote the downregulation of genes that are not regulated by Fe, such as GCN4, in a manner that is fully dependent on AREs and Cth1/Cth2 TZFs (19). Cth1 and Cth2 target transcripts are only partially redundant, since Cth1 promotes the degradation of mRNAs encoding proteins that function in highly Fe-consuming processes, such as mitochondrial respiration, whereas Cth2 exhibits a broader set of targets that includes the TCA cycle and respiration, lipid metabolism, heme synthesis, amino acid biosynthesis, and the Ccc1 vacuolar Fe importer (19, 20). In addition to promoting the downregulation of a wide variety of Fe-dependent processes, Cth1 and Cth2 also function in the activation of essential Fe-dependent enzymes, such as ribonucleotide reductase, which functions in the synthesis of the deoxyribonucleotides necessary for DNA synthesis and repair, by degrading an mRNA encoding the Wtm1 nuclear anchoring protein, facilitating the assembly of an active cytosolic enzyme (24). Cells lacking CTH2 or expressing a nonfunctional CTH2 TZF mutant allele display growth defects under low-Fe conditions that are exacerbated upon deletion of CTH1 (19). Therefore, Cth1 and Cth2 act in concert as molecular rheostats that allow metabolic adaptation to various degrees of Fe limitation. While Cth1 may be involved in adaptation to a transient or modest Fe deficiency, Cth2 likely promotes a generalized metabolic shift to optimize Fe utilization during more severe or prolonged Fe deprivation.
Although Fe-dependent changes in gene expression are crucial for Fe homeostasis and shifts in Fe availability, the mechanisms underlying Cth1/2 function and regulation are incompletely understood. In this study, we demonstrate that CTH1 and CTH2 mRNAs contain functional AREs within their 3′UTRs that function in auto- and cross-regulation for the decay of their respective mRNAs. Our results strongly suggest that this tight control of Cth1 and Cth2 levels is important for the replacement of Cth1 by Cth2 during prolonged Fe limitation and for the rapid decrease in Cth2 levels that is required for the adaptation to Fe supplementation following Fe deficiency.
MATERIALS AND METHODS
Plasmids.
The Escherichia coli DH5α strain was used for the propagation and isolation of plasmids. The CTH1 and CTH2 3′UTRs were cloned into the pRS416-GCN4-No-3′UTR plasmid by using an EcoRI restriction site introduced after the GCN4 termination codon (19) to generate pRS416-GCN4-(CTH2-3′UTR) and pRS416-GCN4-(CTH1-3′UTR) constructs. The GCN4-CTH1-3′UTR fragment was cloned into the pRS415 vector to obtain pRS415-GCN4-(CTH1-3′UTR). Sequences from the CTH1 and CTH2 3′UTRs (see Fig. 3B) were cloned into SmaI-digested pIIIA/MS2-1 vector (25) to obtain pIIIA/MS2-CTH1 and pIIIA/MS2-CTH2. The pRS416-FLAG2-CTH2-C190R, pRS416-FLAG2-CTH2-C223R, pRS416-CTH2p-CTH1-C225R, pRS416-GCN4-(CTH2-3′UTR-AREmt), pRS416-FLAG2-CTH2-AREmt, and pIIIA/MS2-CTH2-AREmt plasmids were generated by using the overlap extension method and the corresponding nonmutagenized plasmids as templates (19, 20). All plasmids containing CTH1 or CTH2 coding sequences, including those with an amino-terminal tag, are expressed under the control of their own promoter unless otherwise indicated. Other plasmids used in this work have been described previously (19, 20, 24, 25). All PCR amplifications were performed with the Phusion polymerase (Finnzymes), and cloned inserts were sequenced.
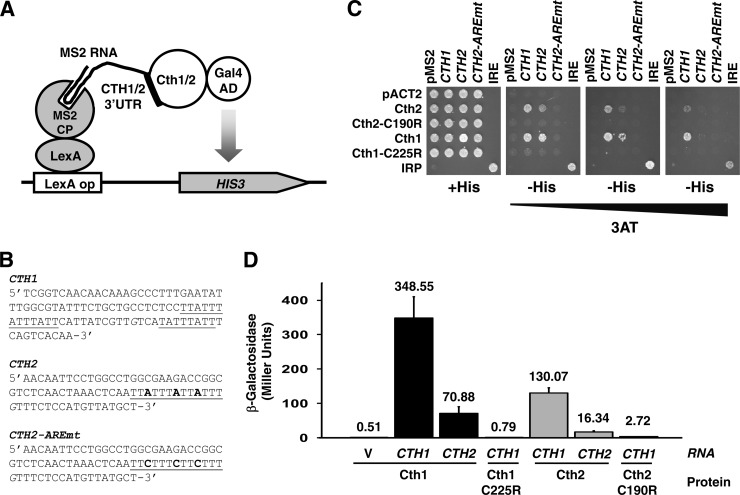
Fig 3.
Cth1 and Cth2 proteins specifically interact with CTH1 and CTH2 mRNAs. (A) Schematic representation of the yeast three-hybrid Cth1/Cth2 and CTH1-CTH2 3′UTR assay. (B) 3′UTR sequences used in the yeast three-hybrid assay. The MS2 RNA was fused to CTH1, CTH2, and CTH2-AREmt. G residues (in italics) were replaced by U residues to minimize premature termination of the RNA polymerase III transcription unit in yeast. Underlined sequences indicate putative AREs. (C) L40coat cells were cotransformed with (9) pIIIA/MS2-1 vector alone or containing the 3′UTR from CTH1, CTH2, or CTH2-AREmt or the IRE as a positive control, and (38) pACT2 vector alone or fused to Cth2, Cth2-C190R, Cth1, Cth1-C225R, and the Fe-responsive element binding protein 1 (IRP) as a positive control. Cells were grown on SC−ura−leu (+His) and SC−ura−leu−his (−His) with increasing concentrations of 3-aminotriazol (3AT). (D) L40coat cells cotransformed with (i) pACT2 vector fused to Cth2, Cth2-C190R, Cth1, or Cth1-C225R and (ii) pIIIA/MS2-1 vector alone or fused to the 3′UTR from CTH1 or CTH2 were grown on SC−leu−ura and assayed for β-galactosidase activity. The error bars indicate standard deviations.
Yeast strains and growth conditions.
The S. cerevisiae strains used in this study were wild-type BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and the isogenic mutant cth1Δ (cth1::KanMX6), cth2Δ (cth2::HisMX6), gcn4Δ (gcn4::KanMX6), cth1Δ cth2Δ (cth1::KanMX4 cth2::HisMX6), and cth1Δ cth2Δ gcn4Δ (cth1::KanMX4 cth2::HisMX6 gcn4::hphB) strains. Yeast cells were grown in synthetic complete medium (SC) or SC lacking specific requirements. The Fe2+-specific chelator bathophenanthroline disulfonic acid disodium (BPS) (Sigma) was used to impose Fe deficiency, and ferrous ammonium sulfate (FAS) (Sigma) was added to create Fe-replete conditions.
To quantify cell growth in liquid media (see Fig. 7), a preculture was grown to exponential phase (A600, ∼0.5 to 0.6) and diluted 200-fold before reinoculation. Cells were grown in SC lacking uracil (SC−ura) or SC−ura with 100 μM BPS (Fe deficient [−Fe]) or reinoculated from SC−ura precultures with 100 μM BPS into SC−ura containing 100 μM FAS (Fe sufficient [+Fe]) or SC−ura without glucose but with 2% glycerol. The optical density was determined over time at 25°C by using a Bioscreen analyzer C (Thermo Labsystems) with a broadband filter (420 to 580 nm) at 30-min intervals over 48 h. Each culture was grown in triplicate, and the average data are shown. The data were corrected for the analyzer by correcting loss of linearity as described previously (26).
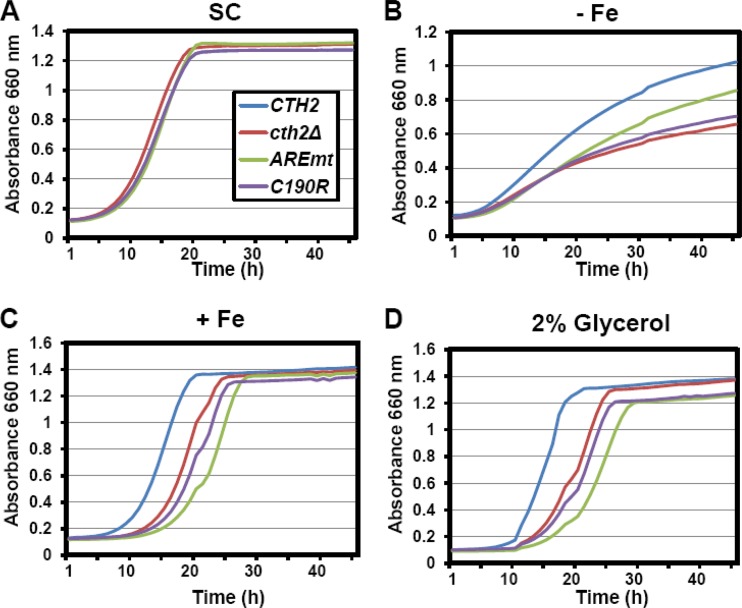
Fig 7.
Cth2 overaccumulation is a growth disadvantage under conditions of fluctuating Fe availability. A cth1Δ cth2Δ double mutant was transformed with pRS416-CTH2 (CTH2), pRS416 (cth2Δ), pRS416-CTH2-AREmt (AREmt), or pRS416-CTH2-C190R (C190R). For SC growth, cells were maintained in SC−ura. For −Fe, +Fe, and 2% glycerol assays, cells were first grown in SC−ura containing 100 μM BPS to induce Fe deficiency and CTH2 expression and then transferred to SC−ura containing either 100 μM BPS (−Fe) or 100 μM FAS (+Fe) or to SC−ura but with 2% glycerol replacing glucose as the sole carbon source. Growth curves were obtained by measuring the absorbance at 660 nm every 30 min for 48 h by using a Bioscreen growth analyzer. A representative experiment of three independent biological replicates with similar results is shown.
Protein analyses.
Total-protein extracts were obtained by glass bead lysis of whole yeast cells in buffer A (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 15 mM EDTA, 0.1% Triton X-100, 2 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) using a Mini Beadbeater (Biospec Products). Protein concentrations were determined by the Coomassie dye-binding assay (Bio-Rad). An equal amount of protein was separated by SDS-PAGE on 12% TGX gels (Bio-Rad) and analyzed by immunoblotting using the indicated antibodies. Flag antibody was purchased from Sigma (F1804-200UG; 080M6035), and Sdh2 antibody was donated by B. Lemire. Primary antibodies were incubated for at least 2 h at room temperature. After washing the membranes, they were incubated with the corresponding secondary antibody (enhanced chemiluminescence [ECL] anti-rabbit or anti-mouse IgG; GE Healthcare), and immunoblots were visualized with SuperSignal chemiluminescent substrate (ThermoScientific).
RNA analyses.
Total yeast RNA was isolated with a modified hot-phenol method (27). For RNA blots, approximately 30 μg of RNA per lane was separated in formaldehyde-agarose gels and blotted onto nylon membranes (Hybond N; Amersham Biosciences). PCR-amplified fragments were gel purified and radiolabeled with [α-32P]dCTP to be used as probes in hybridizations in PSE buffer (300 mM NaH2PO4-Na2HPO4, pH 7.2, 7% SDS, 1 mM EDTA) at 65°C. Actin (ACT1) was used as a loading control. Signal intensities were quantified using a Fujifilm BAS-1500 phosphorimager.
For quantitative real-time PCR (qRT-PCR), genomic DNA was eliminated using Turbo DNA-free (Ambion), and cDNA synthesis was performed according to the recommendations for the SuperScript III first-strand synthesis system (Invitrogen). qRT-PCR was performed using iQ SYBR green Supermix on a Bio-Rad iQ5 real-time PCR detection system. The amplification conditions consisted of a starting step of denaturation at 95°C for 3 min, followed by 39 cycles of 95°C for 10 min, 55°C for 10 min, and 72°C for 30 min. After 10 min at 95°C to terminate the reaction, the PCR plate was maintained at 4°C until its analysis. The results were normalized to ACT1 and analyzed by the 2−ΔCT method as previously described (28). Statistical significance was determined using Student's t test.
Succinate dehydrogenase assays.
Mitochondrion-enriched fractions were prepared from exponential yeast cultures at an optical density at 600 nm (OD600) of 0.8 as described previously (29). Succinate dehydrogenase (SDH) assays were performed with p-iodonitrotetrazolium violet as an artificial electron acceptor for the SDH complex. Protein extracts were incubated in 300 μl of succinate buffer (10 mM succinic acid in 50 mM phosphate buffer, pH 7.4) with 100 μl of p-iodonitrotetrazolium violet solution (2.5 mg of p-iodonitrotetrazolium violet in 50 mM phosphate buffer, pH 7.4). Reactions were terminated with 1 ml of stop solution (10 g of trichloroacetic acid in 100 ml of ethyl acetate-ethanol [1:1 {vol/vol}]), and the absorbance of the supernatant was measured at 490 nm. Error bars represent the standard deviations of duplicate determinations of four independent assays.
Miscellaneous methods.
Yeast three-hybrid and β-galactosidase assays were performed as previously described (25, 27).
RESULTS
Autoregulation of Cth2 expression through a negative-feedback loop.
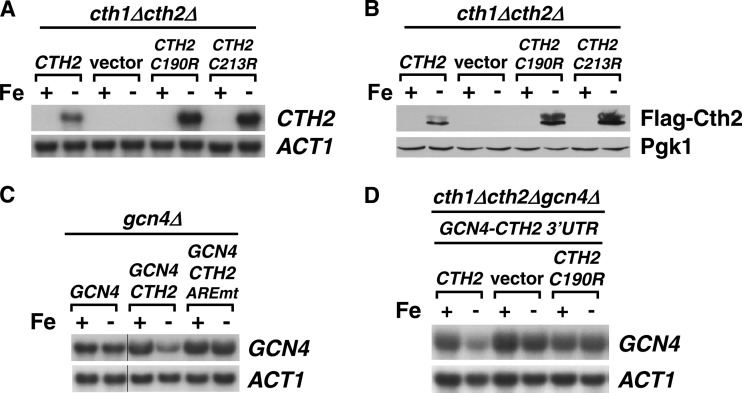
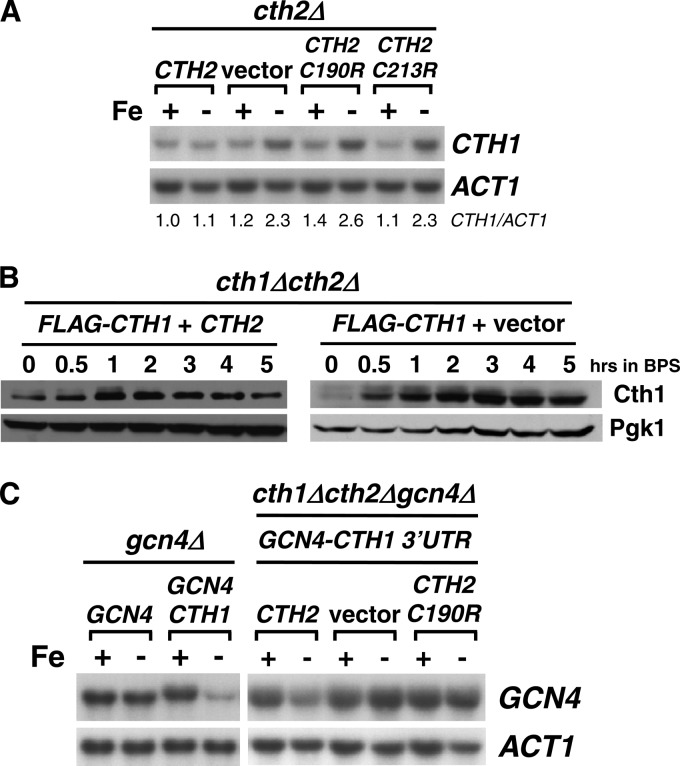
In response to Fe deficiency, yeast cells increase the expression of Cth2 mRNA and protein, which facilitates metabolic adaptation by targeted mRNA turnover (19, 21, 24, 30). Mutagenesis of specific cysteine residues (C190R and C213R) within Cth2 TZF domains abrogates the Cth2 capacity for binding and degradation of ARE-containing mRNAs (19, 21). We observed that CTH2 alleles mutagenized within the TZF coding region (C190R and C213R alleles) display increased steady-state transcript and protein levels under low-Fe conditions that are achieved by addition of the Fe2+-specific chelator BPS compared to wild-type CTH2 (Fig. 1A and B). Interestingly, the CTH2 mRNA 3′UTR sequence contains a putative ARE beginning 46 nucleotides (nt) after the CTH2 translation termination codon. Previous studies have shown that mutagenesis of this ARE in a reporter transcript containing the CTH2 3′UTR leads to a significant increase in transcript stability and steady-state levels (22). Therefore, we decided to explore the possibility that Cth2 regulates its mRNA abundance by binding to and degrading its own mRNA. To test this hypothesis, we first ascertained whether the CTH2 3′UTR was able to promote the low-Fe-dependent degradation of the transcript encoded by a reporter gene that is not regulated in response to Fe deficiency (19). For this purpose, the CTH2 3′UTR was fused to the GCN4 coding sequence, and the steady-state levels of GCN4-(CTH2-3′UTR) chimeric mRNA were measured under +Fe and −Fe conditions (Fig. 1C). Whereas wild-type GCN4 mRNA levels are not significantly altered by Fe availability, GCN4-(CTH2-3′UTR) mRNA is downregulated in response to Fe limitation (Fig. 1C). Importantly, mutagenesis of the 5′-UUAUUUAUUAUUU-3′ ARE in CTH2-3′UTR to 5′-UUCUUUCUUCUUU-3′ completely abrogates the Fe-dependent downregulation of GCN4-(CTH2-3′UTR) mRNA (Fig. 1C). Moreover, the decrease in GCN4-(CTH2-3′UTR) transcript in response to Fe deficiency is dependent on a functional Cth2 protein. As shown in Fig. 1D, the Fe deprivation-dependent decrease in GCN4-(CTH2-3′UTR) steady-state mRNA levels is abolished in cells that lack CTH2 or express the nonfunctional CTH2-C190R allele. Taken together, these results demonstrate that the ARE sequence found in the CTH2 3′UTR is necessary and sufficient to induce the Cth2 and Fe starvation-dependent downregulation of GCN4 mRNA and strongly suggest that Cth2 utilizes a negative-feedback loop that limits its own expression.
Fig 1.
Cth2 autoregulates expression through a negative-feedback loop involving binding to its own ARE and promoting mRNA decay. (A) A cth1Δ cth2Δ double mutant was transformed with pRS416-CTH2 (CTH2), pRS416 (vector), pRS416-CTH2-C190R (CTH2-C190R), or pRS416-CTH2-C213R (CTH2-C213R) plasmid. Cells were grown for 8 to 9 h in SC−ura containing 100 μM FAS (+Fe) or 100 μM BPS (−Fe), and RNA was extracted and analyzed for CTH2 expression by RNA blotting. Relative quantitation of CTH2 and ACT1, CTH2-C190R and ACT1, and CTH2-C213R/ACT1 mRNAs under −Fe conditions was 1.0, 1.9, and 1.7, respectively. (B) A cth1Δ cth2Δ double mutant was transformed with pRS416-FLAG2-CTH2 (CTH2), pRS416 (vector), pRS416-FLAG2-CTH2-C190R (CTH2-C190R), or pRS416-FLAG2-CTH2-C213R (CTH2-C213R) plasmid. The cells were grown as described for panel A. Protein was extracted and analyzed by immunoblotting using anti-FLAG antibody. (C) A gcn4Δ strain was transformed with pRS416-GCN4 (GCN4), pRS416-GCN4-(CTH2-3′UTR) (GCN4-CTH2), or pRS416-GCN4-(CTH2-3′UTR-AREmt) (GCN4-CTH2-AREmt). Cells were grown and analyzed for GCN4 mRNA expression levels as described for panel A. (D) A cth1Δ cth2Δ gcn4Δ triple mutant transformed with pRS415-GCN4-(CTH2-3′UTR) was cotransformed with pRS416-CTH2 (Cth2), pRS416 (vector), or pRS416-CTH2-C190R (C190R). Cells were grown in SC−ura−leu and analyzed as for panel C. Actin (ACT1) was used as a loading control in all RNA-blotting assays, and Pgk1 was used as a loading control in all immunoblot assays.
Cth2 protein regulates the expression levels of CTH1 mRNA.
Our previous studies demonstrated that the TZF-containing protein Cth1 cooperates with Cth2 in the metabolic adaptation to Fe deficiency via targeted mRNA degradation (19, 20). In fact, the CTH1 promoter region contains two Fe-responsive elements that are essential for the Aft1/Aft2-dependent activation of CTH1 transcription upon Fe limitation (20). Despite this, only modest elevation in the steady-state CTH1 mRNA levels is observed when cells are grown under low-Fe conditions, and only a transient increase in Cth1 protein levels is detected (19, 20). Intriguingly, we have observed that CTH1 steady-state mRNA levels increase in cells that do not express CTH2 or that express a nonfunctional Cth2 protein mutated at Cys190 or Cys213 (Fig. 2A). Furthermore, while transient expression of a FLAG epitope-tagged Cth1 protein was previously observed in wild-type cells during Fe deficiency (20), FLAG-Cth1 protein levels are induced by Fe deficiency and sustained in cells that do not express Cth2 (Fig. 2B). Given that the CTH1 3′ region contains three putative AREs located 53, 80, and 159 nt after the CTH1 termination codon, we hypothesized that CTH1 mRNA could be a target for Cth2-mediated degradation under Fe deficiency conditions. To test whether the CTH1-3′UTR confers Cth2-dependent Fe-regulated mRNA decay, the CTH1 3′ region was fused to the GCN4 coding sequence and the steady-state levels of GCN4-(CTH1-3′UTR) mRNA were assayed under +Fe and −Fe conditions. As observed for the CTH2 3′UTR, the CTH1 3′UTR promotes the downregulation of GCN4 mRNA upon Fe deficiency in a manner that is fully dependent on the presence of a functional Cth2 protein (Fig. 2C). Taken together, these results demonstrate that the CTH1 3′UTR is necessary and sufficient to induce the Cth2 and Fe starvation-dependent downregulation of GCN4 mRNA and strongly suggest that Cth2 promotes the downregulation of CTH1 expression under Fe-deficient conditions.
Fig 2.
Cth2 downregulates CTH1 mRNA levels in response to Fe deficiency. (A) A cth2Δ mutant was transformed with pRS416-CTH2 (CTH2), pRS416 (vector), pRS416-CTH2-C190R (C190R), or pRS416-CTH2-C213R (C213R). Cells were grown and analyzed for CTH1 mRNA expression levels as described for Fig. 1A. Relative quantitation of CTH1 and ACT1 mRNA levels is shown in reference to CTH2-expressing cells under +Fe conditions. (B) A cth1Δ cth2Δ double mutant was cotransformed with pRS415-FLAG2-CTH1 plus pRS416-CTH2 (FLAG-CTH1 + CTH2) or pRS415-FLAG2-CTH1 plus pRS416 (FLAG-CTH1 + vector). BPS (100 μM) was added to exponential-phase cells growing in SC−ura−leu, and aliquots were analyzed for Flag2-Cth1 protein expression as described for Fig. 1B. (C) A gcn4Δ mutant strain was transformed with pRS416-GCN4 (GCN4) or pRS416-GCN4-(CTH1-3′UTR) (GCN4-CTH1) plasmid, and a cth1Δ cth2Δ gcn4Δ triple mutant transformed with pRS415-GCN4-(CTH1-3′UTR) (GCN4-CTH1 3′UTR) and cotransformed with pRS416-CTH2 (CTH2), pRS416 (vector), or pRS416-CTH2-C190R (CTH2-C190R). gcn4Δ and cth1Δ cth2Δ gcn4Δ transformants were grown in SC−ura and SC−ura−leu, respectively, containing 100 μM FAS (+Fe) or 100 μM BPS (−Fe) and analyzed for GCN4 mRNA expression levels as described for Fig. 1A. Actin (ACT1) was used as a loading control in all RNA-blotting assays, and Pgk1 was used as a loading control in all immunoblot assays.
Cth1 and Cth2 bind both CTH1 and CTH2 mRNAs.
The results shown here indicate that both mRNA binding by Cth2 protein and cis-acting AREs are essential for the downregulation of both CTH1 and CTH2 mRNAs. Additional experimental evidence supports the notion that Cth1 also downregulates the CTH1 and CTH2 mRNAs. First, it has been previously reported that the disruption of the CTH1 gene results in the accumulation of CTH2 mRNA (31). By using a CTH2 promoter-CTH1 fusion gene construct and a CTH2-C190R allele that eliminates Cth2-dependent regulation, we observed that CTH2 mRNA levels are lower in cells that express a functional CTH1 allele than in cth1Δ mutants or cells expressing the nonfunctional CTH1-C225R allele, strongly suggesting that the Cth1 protein downregulates CTH2 mRNA (data not shown). Moreover, using a cth1Δ cth2Δ gcn4Δ strain that expresses either a wild-type Cth1 protein or one in which Cys225 was mutated to arginine from the CTH2 promoter, and a GCN4-(CTH1-3′UTR) fusion gene, we observed that a functional Cth1 protein specifically promotes the downregulation of a GCN4 mRNA containing the CTH1 3′UTR, suggesting that Cth1 protein also autoregulates its own mRNA levels via its 3′UTR (data not shown).
Taken together, these results suggest that both the Cth1 and Cth2 proteins auto- and cross-regulate the expression of their mRNAs and predict that both proteins should specifically bind to the AREs contained within the 3′UTRs of both the CTH1 and CTH2 mRNAs. To test this possibility, the in vivo interaction between the Cth1 or Cth2 protein and the CTH1 or CTH2 mRNA-derived sequences was evaluated by using the yeast three-hybrid assay, a genetic surrogate for detecting protein-RNA interactions (25) (Fig. 3A). For this purpose, sequences from the 3′UTRs of CTH1 and CTH2 mRNAs containing potential AREs, and the same CTH2 3′ region with a mutagenized ARE (Fig. 3B), were fused to DNA encoding bacteriophage MS2 RNA. Moreover, the coding sequences of CTH1, CTH1-C225R, CTH2, or CTH2-C190R were fused in frame to the Gal4 transactivation domain. Plasmids were cotransformed into the L40coat yeast strain, which expresses a protein fusion between the LexA DNA-binding domain and the MS2 coat protein, and growth was assayed in medium lacking histidine. The IRP/IRE system of mRNA-binding protein and mRNA target sequence was used as a positive control for growth in the absence of histidine, which is indicative of protein-RNA interactions. As shown by growth in medium without histidine, Cth1 and Cth2, but not Cth1-C225R and Cth2-C190R proteins, interact with sequences derived from CTH1 and CTH2 mRNAs but not with CTH2 mRNA with a mutagenized ARE (Fig. 3C). Given that the L40coat strain expresses the LacZ gene under the control of the LexA DNA-binding domain, we quantitatively confirmed the interaction between the Cth1/Cth2 proteins and the CTH1 and CTH2 mRNAs by β-galactosidase assays (Fig. 3D). Taken together, these results indicate that the Cth1 and Cth2 proteins specifically bind in vivo to both CTH1 and CTH2 mRNAs, and this binding promotes their downregulation in a tight auto- and cross-regulated mechanism that interconnects the expression of both genes.
CTH2 mRNA downregulation is important for rapid extinction of Cth2 expression promoted by a switch from Fe deficiency to Fe supplementation.
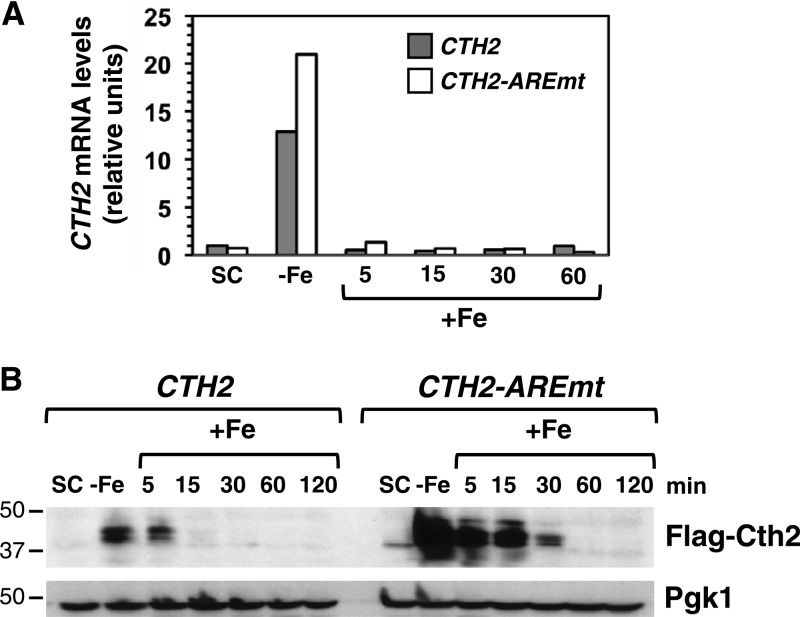
We have shown here that under Fe-deficient conditions CTH2-C190R and CTH2-C213R mRNA and protein levels are higher than those displayed by wild-type CTH2 (Fig. 1), likely due to the lack of Cth2-mediated autodegradation. Based on this observation, we hypothesized that this autoregulation strategy would be important for the downregulation of CTH2 expression that should occur upon Fe addition to an Fe-deficient medium. To test this hypothesis, yeast cells expressing either FLAG-CTH2 or the FLAG-CTH2-AREmt allele were grown under Fe sufficiency conditions (SC-ura), and Fe bioavailability was decreased by addition of 100 μM BPS to chelate extracellular Fe and activate CTH2 expression. After 6 h of growth under Fe limitation, 100 μM FAS was added to restore Fe-replete conditions, and the cells were grown for a further 2 h. As suggested by our previous results with CTH2-TZF mutants (Fig. 1), the analysis of CTH2 and CTH2-AREmt mRNA levels by qRT-PCR indicated that the ARE functions in the control of CTH2 mRNA levels in response to Fe deficiency (Fig. 4A). After Fe readdition, both CTH2 and CTH2-AREmt mRNA levels rapidly decreased. Importantly, CTH2-AREmt mRNA abundance was higher than that observed for CTH2 transcript during early times after Fe addition (Fig. 4A), strongly suggesting that the integrity of the CTH2 ARE is essential for the rapid downregulation of its expression that occurs upon Fe repletion.
Fig 4.
The CTH2 mRNA ARE regulates CTH2 expression levels. (A) A cth1Δ cth2Δ double-mutant strain was transformed with pRS416-FLAG2-CTH2 or pRS416-FLAG2-CTH2-AREmt. Cells were grown in SC−ura (SC), and then 100 μM BPS was added for 6 h (−Fe). After Fe deficiency, 100 μM FAS was added for 2 h (+Fe). mRNAs were extracted from the SC and −Fe and at different time points after Fe addition, reverse transcribed, and quantified by qRT-PCR using specific primers for CTH2 mRNA detection. A representative experiment is shown. Actin expression was used to normalize mRNA levels. (B) A cth1Δ cth2Δ double-mutant strain was transformed with pRS416-FLAG2-CTH2 or pRS416-FLAG2-CTH2-AREmt. Cells were grown as described for panel A, and aliquots were collected at the indicated times and analyzed by immunoblotting using anti-FLAG antibody. Pgk1 was used as a loading control. Molecular masses in kDa are shown on the left. Cells from the same aliquots were used for both mRNA and protein determinations.
To test whether the CTH2 ARE is important for the downregulation of steady-state Cth2 protein levels, we determined Flag-Cth2 protein levels in these cells. Consistent with the increase in Cth2-TZF mutant protein levels that we observed during Fe limitation (Fig. 1), mutagenesis of the CTH2 ARE leads to a significant increase in Cth2 protein abundance compared to cells expressing wild-type CTH2 (Fig. 4B). Furthermore, Cth2 protein expressed from the wild-type gene was not detectable after 15 min of Fe readdition, whereas in cells that express the CTH2-AREmt allele, Cth2 protein was still detected 30 min after Fe addition (Fig. 4B). Taken together, these results demonstrate that the CTH2 ARE is required to limit Cth2 expression levels under low-Fe conditions and to allow more efficient downregulation of both CTH2 mRNA and protein when cells are switched from Fe-deficient to Fe-supplemented growth conditions.
Cth2 downregulation upon Fe supplementation is important for the reemergence of Fe-dependent processes.
Cth2 protein is responsible for the downregulation of approximately 90 ARE-containing mRNAs encoding proteins that participate in Fe-dependent processes, including respiration and heme synthesis, and proteins that function in Fe compartmentalization, such as the Ccc1 vacuolar Fe importer (19, 21, 32). Consequently, we ascertained whether the sustained Cth2 protein levels observed after the readdition of Fe in cells expressing the CTH2-AREmt allele impacted the expression of Cth2 target genes at both the mRNA and protein levels. The mRNA levels of Cth2 targets, including succinate dehydrogenase subunit 2 (SDH2) and ferrochelatase (HEM15), were determined by qRT-PCR under the same conditions used to determine CTH2 mRNA levels in Fig. 4A, which include Fe-sufficient conditions, Fe-deficient conditions, and Fe-deficient conditions followed by Fe supplementation. We observed that in the CTH2-AREmt yeast strain, both mRNAs were efficiently downregulated in response to Fe deficiency (Fig. 5A and B). Furthermore, transcript levels for both Cth2 targets progressively increased after Fe addition to Fe-deficient cells (Fig. 5A and B). Importantly, the elevation of Cth2 target mRNA levels upon Fe supply is defective in cells that express CTH2-AREmt (Fig. 5A and B), suggesting that the increased levels of Cth2 protein in these cells (Fig. 4B) delays the recovery of Cth2 target mRNA levels due to sustained degradation. To further assess whether this decrease in Cth2 target transcript abundance correlates with protein levels, Sdh2 protein abundance was measured by immunoblotting. Whereas no significant differences were observed for Fe-sufficient conditions (Fig. 5C), CTH2-AREmt cells exhibited a delay in the recovery of Sdh2 protein levels upon Fe supplementation compared to cells expressing the wild-type CTH2 allele (Fig. 5D), closely inversely correlating with the abundance of FLAG-Cth2 protein levels (Fig. 4B). We ascertained whether these defects in mRNA and protein recovery observed in cells expressing the CTH2-AREmt allele upon Fe readdition had an effect on the enzymatic activity of Cth2-regulated targets. Succinate dehydrogenase activity from yeast cells expressing either the CTH2 or CTH2-AREmt allele was measured under conditions of Fe sufficiency or Fe deficiency and 4 h after supplementation with exogenous Fe. Upon Fe recovery, cells expressing the CTH2-AREmt allele exhibited a nearly 50% decrease in succinate dehydrogenase activity compared to cells expressing wild-type CTH2 (Fig. 6). Taken together, these results strongly suggest that the ARE-dependent downregulation of CTH2 mRNA and protein levels that occurs upon an increase in Fe availability is important for the rapid recovery of Fe-dependent processes that are targets of Cth2 protein under Fe-deficient conditions.
Fig 5.
Dysregulation of Cth2 targets in cells with a CTH2 allele defective in autoregulation. (A and B) A cth1Δ cth2Δ double-mutant strain transformed with pRS416-FLAG2-CTH2 or pRS416-FLAG2-CTH2-AREmt plasmid was grown for Fe sufficiency (SC), deficiency (−Fe), or deficiency followed by Fe supplementation (+Fe) as described for Fig. 4A. mRNAs were extracted and analyzed by qRT-PCR using specific primers to amplify SDH2 (A) and HEM15 (B). A representative experiment is shown. Actin expression was used to normalize mRNA levels. (C and D) A cth1Δ cth2Δ double-mutant strain transformed with pRS416-FLAG2-CTH2 or pRS416-FLAG2-CTH2-AREmt plasmid was grown as described for Fig. 4A. Samples were analyzed by immunoblotting using polyclonal antibodies against Sdh2 and Pgk1. Molecular masses in kDa are shown on the left. Different sections from the same immunoblot are shown in panel D.
Fig 6.
Recovery of succinate dehydrogenase activity is compromised after Fe supplementation in CTH2-AREmt cells. Mitochondrial SDH activity was measured in a cth1Δ cth2Δ double-mutant strain transformed with pRS416-CTH2 or pRS416-CTH2-AREmt plasmid. Cells were grown as described for Fig. 4. Mitochondrion-enriched extracts for SDH analyses were obtained by differential centrifugation. The analyses were performed with four biological replicates, and each sample was analyzed in duplicate. The error bars represent the standard deviations. The asterisk indicates statistically significant differences by analysis of variance (ANOVA) between yeast cells expressing CTH2 or CTH2-AREmt after Fe readdition (P < 0.0005).
Cth2 negative-feedback regulation is important for growth during alterations in Fe bioavailability.
To ascertain whether the defects in Cth2 target regulation observed in cells lacking Cth2 autodegradation impact cell physiology, the growth of cells expressing the CTH2 or CTH2-AREmt allele under distinct conditions of Fe availability was evaluated, using cth2Δ mutants and cells expressing the CTH2-C190R nonfunctional protein as controls for these assays. No growth difference was observed among the four strains grown in SC (Fig. 7A). Cells expressing wild-type CTH2 exhibit lower growth rates under Fe deficiency than under Fe-sufficient conditions (Fig. 7A and B). As previously reported, yeast cells lacking CTH2 or expressing the CTH2-C190R nonfunctional allele display a significant growth defect under low-Fe conditions compared to cells expressing wild-type CTH2 (Fig. 7B) (19). Interestingly, yeast cells expressing the CTH2-AREmt allele also show a growth defect under low-Fe conditions, although it is not as pronounced as the defect observed for cth2Δ and CTH2-C190R cells (Fig. 7B). Similar results are obtained when growth is assayed in solid medium (data not shown). The growth defect of CTH2-AREmt cells under low Fe suggests that an excessive accumulation of Cth2 protein is deleterious for cell growth. Consistent with this observation, previous studies suggested that Cth2 overexpression can be toxic to S. cerevisiae (21, 31), which may actually reflect an inability to cope with Fe limitation.
To determine whether CTH2 autoregulation is important for cell growth when cellular Fe availability rapidly changes, cells were grown under Fe-deficient conditions (100 μM BPS) and then Fe supplemented by adding 100 μM FAS. Cells lacking a functional CTH2 (cth2Δ or CTH2-C190R) exhibited a significant delay in resuming growth compared to wild-type cells (Fig. 7C), suggesting that the optimization of Fe utilization by Cth2 during Fe scarcity is also important for growth recovery when Fe availability increases. Importantly, CTH2-AREmt-expressing cells display a more severe delay in growth recovery when transferred from Fe-deficient to Fe-sufficient conditions (Fig. 7C), consistent with the delay in upregulation of Fe-dependent processes shown in Fig. 5 and 6. As respiration is an Fe-dependent process and some Cth2 targets encode proteins involved in respiration, these strains were evaluated with respect to cell growth when transferred from Fe deficiency to medium containing a nonfermentable carbon source (glycerol), for which respiration is indispensable. Under these conditions, cells lacking a functional Cth2 protein (cth2Δ or CTH2-C190R) show a significant delay in growth compared to cells expressing wild-type CTH2 (Fig. 7D). However, cells expressing CTH2-AREmt show a more severe delay when transferred to respiratory conditions (Fig. 7D). Taken together, these results support the hypothesis that maintaining appropriate concentrations of Cth2 is important during Fe scarcity and in response to Fe supplementation. Moreover, the rapid shutdown of Cth2 expression, mediated at least in part by CTH2 autoregulation, is required for resuming growth in response to changes in cellular Fe demands and Fe bioavailability.
DISCUSSION
Eukaryotic cells regulate gene expression at transcriptional and posttranscriptional levels to rapidly respond to environmental changes. In S. cerevisiae, the Aft1 and Aft2 transcription factors promote the transcriptional activation of genes encoding proteins that participate in extracellular Fe acquisition, Fe mobilization and recycling, and metabolic remodeling when Fe bioavailability is low (15–17). Furthermore, during Fe deficiency, a decrease in the levels of some Fe-dependent metabolites, including heme and α-isopropylmalate, leads to downregulation in the transcription of genes that function in respiration and leucine synthesis, respectively (33). In addition to this transcriptional regulation mediated by Fe-responsive metabolites, the metabolic reprogramming observed in response to low Fe is also posttranscriptionally controlled by the mRNA-binding proteins Cth1 and Cth2 (19, 20). Both the Cth1 and Cth2 proteins bind ARE-containing mRNAs encoding proteins that contain Fe as a cofactor, participate in Fe-dependent processes, or function in Fe storage. Through the generation of chimeric mRNAs, ARE mutagenesis, and in vivo protein-RNA interaction assays, we demonstrate here that Cth1 and Cth2 proteins specifically bind functional AREs within the 3′UTRs of CTH1 and CTH2 transcripts, promoting autologous and heterologous mRNA degradation.
We previously demonstrated by chromatin immunoprecipitation experiments and CTH1 promoter fusion analysis that Aft1 and Aft2 specifically activate the transcription of CTH1 mRNA during Fe deficiency (20). However, steady-state CTH1 transcript levels are not detectably increased in Fe-deficient cells incubated in 100 μM BPS for 8 to 9 h (19) (Fig. 2A, CTH2). The increase in CTH1 or GCN4-(CTH1-3′UTR) mRNA levels in cells lacking a functional Cth2 or Cth1 protein, respectively, suggests that the lack or very weak elevation of CTH1 transcript in response to low Fe is due to a combination of Aft1/2-mediated transcriptional activation coupled to active CTH1 mRNA degradation by both Cth1 and Cth2 proteins (Fig. 2 and data not shown). At the protein level, wild-type cells display a modest and transient increase in Cth1 protein 2 h after imposition of Fe deficiency (20). Importantly, when CTH2 is absent, Cth1 protein levels robustly accumulate over a time course of several hours of Fe limitation. These results suggest that during the progression of Fe deficiency, Cth2 protein levels increase and accelerate CTH1 mRNA decay, thus restricting Cth1 expression to the initial stages of Fe limitation and leaving Cth2 as the dominant regulator during severe or prolonged Fe deficiency. Given that the Cth1 and Cth2 proteins have only slightly overlapping target mRNAs (20), we propose that this cross-regulation could establish the priority by which Fe-dependent processes are downregulated during the progression in severity of Fe deficiency. At the beginning of Fe deficiency, respiration and other processes might be primarily targeted for downregulation by Cth1 (20). When Fe deficiency persists, Cth2 would lead the downregulation of other Fe-dependent processes and vacuolar Fe storage, in addition to respiration. Previous studies (31) and our results also indicate that Cth1 protein downregulates CTH2 mRNA, although this is a relatively modest effect, perhaps due to the low CTH1 expression levels compared to CTH2 under low-Fe conditions.
Although both CTH1 and CTH2 contribute to yeast adaptation to Fe deficiency, cells lacking a functional Cth2 protein exhibit a pronounced growth defect under Fe-limited conditions, whereas no growth defect has been observed for cth1Δ cells (19). A role for CTH1 in cell growth under low Fe has been observed only in the cth1Δ cth2Δ double mutant (19). While the primary reason for Cth2 predominance in the response to Fe deficiency may be high expression levels, it could also be due to the larger number of transcripts whose degradation under Fe deficiency is Cth2 dependent (20). Previous results demonstrated that Cth2 overexpression inhibits growth, suggesting that this may be due to excessive degradation of transcripts encoding proteins important for cell growth (21, 31). Cth2 toxicity can be rescued by removing its ability to bind and degrade mRNAs, since Cth2 TZF mutagenesis or deletion of genes implicated in the mechanism of Cth2-mediated mRNA turnover, such as the RNA helicase gene DHH1, rescues growth (21, 31). Here, we show how CTH2 transcripts lacking a functional ARE program a robust increase in Cth2 protein levels under low-Fe conditions compared to wild-type cells. This elevated expression of Cth2 is harmful for the cell, as shown by the growth defect displayed by the CTH2-AREmt mutants under low-Fe conditions and the delay in growth recovery during the adaptation to Fe supplementation. Several reasons, including the excessive degradation of specific mRNAs and the titration of proteins required for global mRNA turnover, could be responsible for these defects. Further studies are necessary to decipher the mechanism underlying the defects associated with cells that overexpress a functional Cth2 protein.
We have shown that Cth2 autoregulation limits CTH2 expression levels, which prevents excessive accumulation that could be deleterious to cell growth, but why do cells simultaneously transcribe and degrade CTH2 mRNA instead of simply decreasing its transcription rate? Previous studies have shown that simultaneous activation of both transcription and degradation permits more rapid and sensitive changes in gene expression that are advantageous in response to environmental stresses (34, 35). In mammals, an analogous mRNA-binding protein, TTP, specifically binds in vivo to AREs within the 3′UTRs of many transcripts, stimulating their delivery and degradation in processing bodies (18, 36). Recent studies indicate that TTP-mediated regulation of gene expression plays an important role in tumorigenesis (37). It has been proposed that TTP binds its own transcript and promotes mRNA degradation in an autoregulatory pathway that limits TTP synthesis (38, 39). This feedback-regulatory loop could facilitate a rapid return to a resting, nonactivated state upon removal of the activation signal (38). To explore the physiological relevance of this regulatory loop in yeast, we investigated the contribution of Cth2 autoregulation to the adaptation of Fe-deficient cells to a rapid increase in Fe availability. We first observed that when cells are transferred from Fe-deficient to Fe-sufficient conditions, Cth2 protein rapidly disappears, perhaps to facilitate the reemergence of respiration and other Fe-dependent processes repressed by Cth2 in order to enhance energy production and cell growth and to allow storage of Fe in excess of cellular needs. Importantly, in the absence of ARE-mediated downregulation of CTH2 mRNA, Cth2 protein levels are further sustained after Fe addition. As a consequence, we observed that CTH2-AREmt cells exhibit prolonged repression of Fe-dependent processes during the shift to Fe sufficiency, as shown by the decreased mRNA, protein, and enzymatic activities of Cth2 targets, which leads to a significant delay in cell growth recovery compared to wild-type cells.
Taken together, the results described here demonstrate that the expression of the yeast RNA-binding proteins Cth1 and Cth2 during Fe deficiency is tightly controlled in a coordinated manner through simultaneous transcriptional and posttranscriptional mechanisms, as we depict in a summarized model (Fig. 8). Degradation of CTH1 mRNA triggered by Cth2 allows the replacement of Cth1 by Cth2 protein during the progress of Fe deficiency, whereas Cth2 autoregulation enhances the response of Fe-deficient cells to a rapid increase in Fe availability by decreasing Cth2 levels and permitting the activation of Fe-dependent processes crucial for optimal cell growth. As Fe deficiency is a common human nutritional deficiency that is treated with Fe supplements, these studies underscore the potential complex metabolic rearrangements that must occur in response to fluctuations in the availability of this essential metal.
Fig 8.
CTH1 and CTH2 auto- and cross-regulation model. CTH1 and CTH2 mRNAs are themselves posttranscriptionally regulated by Cth1 and Cth2 proteins, creating an auto- and trans-regulatory loop that facilitates adaptation to rapid changes in Fe bioavailability. This auto- and trans-regulation occurs through ARE sequences within the 3′ UTRs of CTH1 and CTH2 mRNAs, which promote their destabilization. In the absence of AREs, both auto- and trans-regulation are impaired, and Cth1/Cth2 mRNAs and proteins accumulate abnormally, causing delays in cell adaptation to changing environments.
ACKNOWLEDGMENTS
This work was supported by a postdoctoral fellowship from the Spanish Ministry of Economy and Competitiveness to M.M.-P., grant AGL2011-29099 from the Spanish Ministry of Economy and Competitiveness to S.P., National Institutes of Health Predoctoral Fellowship FDK081304A to S.V.V., and National Institutes of Health grant GM48140 to D.J.T.
We thank members of the Thiele and Puig laboratories for helpful suggestions and critically reading the manuscript. We are also grateful to Marvin Wickens, Andy Dancis, Emmanuel Lesuisse, and Bernard Lemire for plasmids and antibodies.
Footnotes
Published ahead of print 25 March 2013
REFERENCES
- 1. Lill R. 2009. Function and biogenesis of iron-sulphur proteins. Nature 460:831–838 [DOI] [PubMed] [Google Scholar]
- 2. Lill R, Hoffmann B, Molik S, Pierik AJ, Rietzschel N, Stehling O, Uzarska MA, Webert H, Wilbrecht C, Muhlenhoff U. 2012. The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochim. Biophys. Acta 1823:1491–1508 [DOI] [PubMed] [Google Scholar]
- 3. Lill R, Muhlenhoff U. 2008. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 77:669–700 [DOI] [PubMed] [Google Scholar]
- 4. Ozer A, Bruick RK. 2007. Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one? Nat. Chem. Biol. 3:144–153 [DOI] [PubMed] [Google Scholar]
- 5. Ganz T, Nemeth E. 2011. The hepcidin-ferroportin system as a therapeutic target in anemias and iron overload disorders. Hematology Am. Soc. Hematol. Educ. Program 2011:538–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hentze MW, Muckenthaler MU, Galy B, Camaschella C. 2010. Two to tango: regulation of mammalian iron metabolism. Cell 142:24–38 [DOI] [PubMed] [Google Scholar]
- 7. Kaplan JC. 2011. The 2012 version of the gene table of monogenic neuromuscular disorders. Neuromuscul. Disord. 21:833–861 [DOI] [PubMed] [Google Scholar]
- 8. Sheftel A, Stehling O, Lill R. 2010. Iron-sulfur proteins in health and disease. Trends Endocrinol. Metab. 21:302–314 [DOI] [PubMed] [Google Scholar]
- 9. Baynes RD, Bothwell TH. 1990. Iron deficiency. Annu. Rev. Nutr. 10:133–148 [DOI] [PubMed] [Google Scholar]
- 10. Rouault TA. 2006. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2:406–414 [DOI] [PubMed] [Google Scholar]
- 11. Pantopoulos K. 2004. Iron metabolism and the IRE/IRP regulatory system: an update. Ann. N. Y. Acad. Sci. 1012:1–13 [DOI] [PubMed] [Google Scholar]
- 12. Salahudeen AA, Thompson JW, Ruiz JC, Ma HW, Kinch LN, Li Q, Grishin NV, Bruick RK. 2009. An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 326:722–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vashisht AA, Zumbrennen KB, Huang X, Powers DN, Durazo A, Sun D, Bhaskaran N, Persson A, Uhlen M, Sangfelt O, Spruck C, Leibold EA, Wohlschlegel JA. 2009. Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326:718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pimentel C, Vicente C, Menezes RA, Caetano S, Carreto L, Rodrigues-Pousada C. 2012. The role of the Yap5 transcription factor in remodeling gene expression in response to Fe bioavailability. PLoS One 7:e37434 doi:10.1371/journal.pone.0037434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kaplan CD, Kaplan J. 2009. Iron acquisition and transcriptional regulation. Chem. Rev. 109:4536–4552 [DOI] [PubMed] [Google Scholar]
- 16. Philpott CC, Protchenko O. 2008. Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot. Cell 7:20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sanvisens N, Puig S. 2011. Causes and consequences of nutritional iron deficiency in living organisms, p 245–276 In Merkin TC. (ed), Biology of starvation in humans and other organisms. Nova Science Publishers, Inc., New York, NY [Google Scholar]
- 18. Carballo E, Lai WS, Blackshear PJ. 1998. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281:1001–1005 [DOI] [PubMed] [Google Scholar]
- 19. Puig S, Askeland E, Thiele DJ. 2005. Coordinated remodeling of cellular metabolism during iron deficiency through targeted mRNA degradation. Cell 120:99–110 [DOI] [PubMed] [Google Scholar]
- 20. Puig S, Vergara SV, Thiele DJ. 2008. Cooperation of two mRNA-binding proteins drives metabolic adaptation to iron deficiency. Cell Metab. 7:555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pedro-Segura E, Vergara SV, Rodriguez-Navarro S, Parker R, Thiele DJ, Puig S. 2008. The Cth2 ARE-binding protein recruits the Dhh1 helicase to promote the decay of succinate dehydrogenase SDH4 mRNA in response to iron deficiency. J. Biol. Chem. 283:28527–28535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ciais D, Bohnsack MT, Tollervey D. 2008. The mRNA encoding the yeast ARE-binding protein Cth2 is generated by a novel 3′ processing pathway. Nucleic Acids Res. 36:3075–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Prouteau M, Daugeron MC, Seraphin B. 2008. Regulation of ARE transcript 3′ end processing by the yeast Cth2 mRNA decay factor. EMBO J. 27:2966–2976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sanvisens N, Bano MC, Huang M, Puig S. 2011. Regulation of ribonucleotide reductase in response to iron deficiency. Mol. Cell 44:759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. SenGupta DJ, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. 1996. A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl. Acad. Sci. U. S. A. 93:8496–8501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Warringer J, Blomberg A. 2003. Automated screening in environmental arrays allows analysis of quantitative phenotypic profiles in Saccharomyces cerevisiae. Yeast 20:53–67 [DOI] [PubMed] [Google Scholar]
- 27. Puig S, Lee J, Lau M, Thiele DJ. 2002. Biochemical and genetic analyses of yeast and human high affinity copper transporters suggest a conserved mechanism for copper uptake. J. Biol. Chem. 277:26021–26030 [DOI] [PubMed] [Google Scholar]
- 28. Rossignol T, Ding C, Guida A, d'Enfert C, Higgins DG, Butler G. 2009. Correlation between biofilm formation and the hypoxic response in Candida parapsilosis. Eukaryot. Cell 8:550–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pastor MM, Proft M, Pascual-Ahuir A. 2009. Mitochondrial function is an inducible determinant of osmotic stress adaptation in yeast. J. Biol. Chem. 284:30307–30317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vergara SV, Puig S, Thiele DJ. 2011. Early recruitment of AU-rich element-containing mRNAs determines their cytosolic fate during iron deficiency. Mol. Cell. Biol. 31:417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thompson MJ, Lai WS, Taylor GA, Blackshear PJ. 1996. Cloning and characterization of two yeast genes encoding members of the CCCH class of zinc finger proteins: zinc finger-mediated impairment of cell growth. Gene 174:225–233 [DOI] [PubMed] [Google Scholar]
- 32. Li L, Bagley D, Ward DM, Kaplan J. 2008. Yap5 is an iron-responsive transcriptional activator that regulates vacuolar iron storage in yeast. Mol. Cell. Biol. 28:1326–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ihrig J, Hausmann A, Hain A, Richter N, Hamza I, Lill R, Muhlenhoff U. 2010. Iron regulation through the back door: iron-dependent metabolite levels contribute to transcriptional adaptation to iron deprivation in Saccharomyces cerevisiae. Eukaryot. Cell 9:460–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Perez-Ortin JE, Alepuz PM, Moreno J. 2007. Genomics and gene transcription kinetics in yeast. Trends Genet. 23:250–257 [DOI] [PubMed] [Google Scholar]
- 35. Shalem O, Dahan O, Levo M, Martinez MR, Furman I, Segal E, Pilpel Y. 2008. Transient transcriptional responses to stress are generated by opposing effects of mRNA production and degradation. Mol. Syst. Biol. 4:223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carballo E, Lai WS, Blackshear PJ. 2000. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95:1891–1899 [PubMed] [Google Scholar]
- 37. Rounbehler RJ, Fallahi M, Yang C, Steeves MA, Li W, Doherty JR, Schaub FX, Sanduja S, Dixon DA, Blackshear PJ, Cleveland JL. 2012. Tristetraprolin impairs myc-induced lymphoma and abolishes the malignant state. Cell 150:563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brooks SA, Connolly JE, Rigby WF. 2004. The role of mRNA turnover in the regulation of tristetraprolin expression: evidence for an extracellular signal-regulated kinase-specific, AU-rich element-dependent, autoregulatory pathway. J. Immunol. 172:7263–7271 [DOI] [PubMed] [Google Scholar]
- 39. Tchen CR, Brook M, Saklatvala J, Clark AR. 2004. The stability of tristetraprolin mRNA is regulated by mitogen-activated protein kinase p38 and by tristetraprolin itself. J. Biol. Chem. 279:32393–32400 [DOI] [PubMed] [Google Scholar]