Abstract
The glucocorticoid receptor (GR) regulates adaptive transcriptional programs that alter metabolism in response to stress. Network properties that allow GR to tune gene expression to match specific physiologic demands are poorly understood. We analyzed the transcriptional consequences of GR activation in murine lungs deficient for KLF15, a transcriptional regulator of amino acid metabolism that is induced by glucocorticoids and fasting. Approximately 7% of glucocorticoid-regulated genes had altered expression in Klf15-knockdown (Klf15−/−) mice. KLF15 formed coherent and incoherent feed-forward circuits with GR that correlated with the expression dynamics of the glucocorticoid response. Coherent feed-forward gene regulation by GR and KLF15 was characterized by combinatorial activation of linked GR-KLF15 regulatory elements by both factors and increased GR occupancy, while expression of KLF15 reduced GR occupancy at the incoherent target, MT2A. Serum deprivation, which increased KLF15 expression in a GR-independent manner in vitro, enhanced glucocorticoid-mediated induction of feed-forward targets of GR and KLF15, such as the loci for the amino acid-metabolizing enzymes proline dehydrogenase and alpha-aminoadipic semialdehyde synthase. Our results establish feed-forward architecture as an organizational principle for the GR network and provide a novel mechanism through which GR integrates signals and regulates expression dynamics.
INTRODUCTION
Endogenous glucocorticoids (GCs) regulate crucial adaptive responses to diverse stressors in vertebrates (1), while synthetic GC-like compounds are widely used clinically in the treatment of immune-mediated disease (2). The genomic effects of GCs are mediated through their binding to the glucocorticoid receptor (GR), which, in response to ligand, induces temporally dynamic transcriptional programs that modulate myriad physiologic processes, including inflammation and metabolism (1). GR regulates gene expression through associating directly and indirectly with specific regions of DNA, leading to altered recruitment and activity of RNA polymerase II-associated transcriptional complexes (3). Chromatin structure, allostery, receptor phosphorylation, and restricted coregulator activity are all implicated in contributing to the induction of tissue-specific gene expression patterns by GR (4–9). However, the mechanisms through which temporally dynamic regulation of gene expression is achieved by GR (10) and the integration of nonligand signals by GR to tune transcriptional programs within a single cell type are not well understood.
One ancient regulatory mechanism that is employed by prokaryotes and eukaryotes alike to integrate signals and confer temporal control to gene expression is the feed-forward transcription circuit, which is defined by a primary factor regulating the expression of a second factor, with the two factors together regulating the expression of additional genes (11). Two basic subtypes of feed-forward circuits have been described. In the coherent feed-forward motif, factor X induces (or represses) factor Y, and both factors induce (or repress) a third target, while incoherent circuits are characterized by factors X and Y having opposing effects on the expression of a third gene (12). Coherent feed-forward circuits can confer slow-on, fast-off kinetics to the expression of downstream targets, while incoherent feed-forward loops are capable of fold change signal discrimination and can bestow distinct timing to peak transcriptional responses (11, 13). Feed-forward circuits thus represent a potential mechanism through which to organize the GR-regulated transcriptome into subunits that integrate different signals and exhibit distinct expression dynamics. Although numerous transcription factors are directly induced by GR (14), the role of feed-forward circuits in mediating transcriptional programs in response to GR activation is not well established.
KLF15 is one of several Kruppel-like, zinc finger transcription factors (KLFs) whose expression is directly induced by GR (14, 15). KLF15 has been shown to exert transcriptional control over amino acid, lipid, and glucose metabolism (16–18), and is also implicated in the induction of proatrophic targets in skeletal muscle in response to GCs (19). However, the genome-wide role of KLF15 as a downstream regulator of GR signaling and the molecular characteristics of GR-KLF15 gene programming are unknown. In this study, we used expression profiling of wild-type and Klf15-knockout (Klf15−/−) mice to define a pulmonary gene set that requires KLF15 for normal transcriptional responses to GCs. We used promoter analysis and chromatin immunoprecipitation to determine the regulatory mechanisms that underpin GR-KLF15 cross talk. Our data indicate that GR and KLF15 form coherent and incoherent feed-forward circuits, providing a novel mechanism for temporal control and signal integration by GR.
MATERIALS AND METHODS
Reagents and antibodies.
Dexamethasone (dex) sodium phosphate (American Regent, Inc., Shirley, NY) diluted in sterile phosphate-buffered saline (PBS) was used for mouse microarray experiments. The dexamethasone 21-phosphate disodium salt (D1159) used for mouse reverse transcription-quantitative PCR (RT-qPCR) experiments, the dexamethasone (D1756) used for cell culture studies, and mifepristone (RU-486; M8046) were purchased from Sigma-Aldrich. The following antibodies were used for Western blot analyses: anti-KLF15 (ab81604) and anti-beta actin (ab75186) from Abcam and enhanced chemiluminescence (ECL) sheep anti-mouse IgG–horseradish peroxidase (HRP; 95017-332) and ECL donkey anti-rabbit IgG–HRP (95017-330) from VWR. Antibodies used for chromatin immunoprecipitation (ChIP) included anti-GR (N-499; a generous gift from Keith Yamamoto), anti-KLF15 (ab81064), and anti-FLAG (F1804) from Sigma-Aldrich. The KLF15 full-length expression plasmid (pcDNA-KLF15), the KLF15-expressing adenovirus (Ad-KLF15), and the green fluorescent protein (GFP)-expressing adenovirus (Ad-GFP) control have been described previously (20). Small interfering RNA (siRNA) studies were performed using the ON-TARGETplus SMARTpool siRNA reagent against human KLF15 (siKLF15; L-006975-00-0005) and the ON-TARGETplus nontargeting siRNA control reagent (siCtrl; D-001810-10-05), obtained from Dharmacon.
Treatment of animals with dexamethasone and processing of lung tissue.
All protocols concerning animal use were approved by the Institutional Animal Care and Use Committee for National Jewish Health or Case Western Reserve University and conducted in strict accordance with the National Institutes of Health guidelines for the care and use of laboratory animals (21). Mice were housed in a temperature- and humidity-controlled facility with a 12-h light/12-h dark cycle and ad libitum access to water and standard laboratory rodent chow. Klf15−/− mice have been previously described (22). Studies were performed with two independent cohorts of sex-matched (6- to 8-week-old) wild-type (WT) and Klf15−/− mice on a pure C57BL/6 background. For microarray experiments, dex was dissolved in sterile PBS and administered via intraperitoneal injection at a final concentration of 2.5 mg/kg of body weight in the early phase of the light cycle. For RT-qPCR validation, dex was dissolved in sterile saline and administered as indicated above. Equivalent doses of PBS or saline were administered to vehicle controls. At 4 or 8 h following injections, mice were euthanized and lung tissue was immediately excised, rinsed in PBS, and homogenized in TRIzol (Life Technologies) or placed in RNAlater (Life Technologies) and subsequently homogenized in TRIzol. Total RNA was extracted and purified from the resulting lysates using a PureLink RNA minikit (Life Technologies) according to the manufacturer's instructions.
Microarray.
Biologic quadruplicate RNA samples were obtained from wild-type and Klf15−/− mice treated with dex or vehicle for 4 and 8 h. An Agilent 2100 bioanalyzer was used to assess RNA quality, and fluorescent linear amplification kits were used to label and amplify RNA according to the manufacturer's protocols (Agilent). Equal amounts of Cy3-labeled target were hybridized to whole mouse genome 4×44K ink-jet arrays (Agilent) for 14 h. Raw signal intensities were extracted, and the data set was normalized using the quantile normalization method (23). Three samples were excluded as technical outliers. A one-way analysis of variance linear model was used to estimate the mean log2 fold changes and false discovery rate (FDR) values as described previously (24). Computations were performed using the R package limma functions in the Bioconductor software package (25, 26). For determining biologically relevant effects on gene expression, genes were identified as differentially regulated on the basis of having an FDR of <0.05 and a fold change of at least 50% between treatment conditions or genotype (i.e., a <0.66 or >1.5 difference between conditions). For construction of GR-KLF15-dependent Venn diagrams, if multiple probes for a gene were included on the array, each probe meeting the FDR and fold change criteria was generally counted. Gene ontology (GO) analysis and functional clustering were performed using the DAVID bioinformatics resource (27, 28). A given ontology term within each gene set was considered to be overrepresented on the basis of the P value and if ∼10% or more of the genes within the set were associated with that term.
Quantitative PCR.
An OpenArray real-time PCR platform (Life Technologies) was used for high-throughput qPCR. Purified total RNA (1 μg) was reverse transcribed using the high-capacity cDNA reverse transcription kit and protocol from Life Technologies. The resulting cDNA was diluted 1:10 with water and subjected to 14 cycles of preamplification using a custom pooled assay mix and preamplification master mix from Life Technologies, according to the manufacturer's protocol. The preamplification products were diluted 1:20 and analyzed using a customized OpenArray template and the OpenArray platform according to the manufacturer's protocol (Life Technologies). This generates standard RT-qPCR curves and threshold cycle (CT) values; results were normalized to those for RPL19 following the ΔΔCT method as described previously (29).
For standard RT-qPCR, 1 μg of purified total RNA was reverse transcribed using Moloney murine leukemia virus reverse transcriptase (Promega) primed with random primers (Life Technologies) and then diluted 1:5 prior to qPCR analysis using Fast SYBR green qPCR master mix (Life Technologies). Expression levels for different conditions were obtained as described by comparing the mean CT value for each gene relative to the mean RPL19CT value (the ΔΔCT method). For repressed genes (i.e., a ΔΔCT value of <0.0), the relative fold change is depicted graphically as −(2x), where x is the absolute value of ΔΔCT, as indicated in the text and legends. Samples were generally analyzed in biologic triplicate and technical duplicate, with obvious outliers excluded. The primer pairs used in the reported experiments generated single products on the basis of melting curve analysis and generally had amplification efficiencies of greater than 90%. Primer sequences are shown in Table S5 in the supplemental material.
Plasmid construction and bioinformatic analysis.
ENCODE data indicating GR ChIP-sequencing (ChIP-seq) peaks previously defined in A549 cells (14, 30) were visualized in the UCSC genome browser at the KLF15, PRODH, AASS, and MT2A loci. Genomic areas ranging from ∼0.5 to 2.5 kb and containing GR ChIP-seq peak regions were amplified by PCR, shuttled into pCR2.1-TOPO (Life Technologies), and subsequently introduced into the PGL3 promoter vector (Life Technologies). Each region was scanned with the MatInspector software tool to identify close matches to sequence consensuses for GR and Kruppel-like factor (KLF) binding sites. Cloning primers and the sequences of putative GR and KLF15 binding sites are delineated in Table S5 and Fig. S1 in the supplemental material. All plasmids will be made available through Addgene.
Cell culture and transfections.
A549 and Beas-2B cells were grown in high-glucose Dulbecco modified Eagle medium (DMEM) containing l-glutamine and supplemented with penicillin-streptomycin (pen-strep) and 5% or 10% fetal bovine serum (FBS; HyClone), respectively. Cells were maintained in 5% CO2 at 37°C. For reporter assays, ∼4 × 104 cells were plated per well in 48-well dishes. The next day, cells were transfected as indicated in the Results with firefly luciferase plasmids and the Renilla luciferase (RL) expression vector pSV40-RL (Promega) at a ratio of 10:1 or with a combination of firefly luciferase plasmids, pcDNA-KLF15 (or pcDNA control), and pSV40-RL at a ratio of 5:5:1 by adding 50 μl of Lipofectamine 2000 (Life Technologies) per plasmid DNA (400 ng total) complex to cells cultured in 250 μl of DMEM containing pen-strep and FBS. For siRNA studies, cells were cotransfected with firefly and Renilla luciferase plasmids in combination with a 25 nM concentration of siKLF15 (or siCtrl) using Lipofectamine RNAiMAX transfection reagent according to the manufacturer's protocol (Life Technologies). Approximately 18 h later, cells were treated with 100 nM dex or vehicle in fresh complete medium or serum-free medium, as indicated for starvation experiments, for 8 h. Cells were then lysed, and firefly and Renilla luciferase activities were measured using the dual-luciferase reporter system (Promega) according to the manufacturer's protocol. Luminescence was detected from 10 μl lysate using an Infinite M1000 plate reader (Tecan). Relative luciferase activity was obtained after normalization of firefly to Renilla luciferase activity. Each experiment was performed in biologic quadruplicate and repeated at least once with qualitatively similar results. P values indicated in the figure legends were calculated using t tests and nonparametric tests.
ChIP-qPCR.
A549 and Beas-2B cells were grown to confluence in 100-mm dishes and treated with 1 μM dex or vehicle (ethanol) for 1 h. Cross-linking was achieved by adding 37% formaldehyde directly to the culture medium to a final concentration of 1% and incubating for 3 (A549 cells) or 7 (Beas-2B cells) min at room temperature. Formaldehyde was quenched with 125 mM glycine for 10 min at 4°C, after which cells were washed in ice-cold Dulbecco's phosphate-buffered saline (DPBS) for 5 min at 4°C. Cells were scraped off dishes in ice-cold immunoprecipitation (IP) lysis buffer (50 mM HEPES-KOH at pH 7.4, 1 mM EDTA, 150 mM NaCl, 10% glycerol, 0.5% Triton X-100) supplemented with protease inhibitor cocktail (PIC; Roche) and nutated for 30 min at 4°C to lyse. Nuclei were collected by centrifugation (600 × g for 5 min at 4°C) and resuspended in ice-cold radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl at pH 8.0, 1 mM EDTA, 150 mM NaCl, 5% glycerol, 0.1% sodium deoxycholate, 0.1% SDS, 1% Triton X-100) supplemented with PIC. Samples were sonicated with a Diagenode Bioruptor at high power in 30-s bursts separated by 30-s incubations in ice water for a total of 24 (A549 cells) or 20 (Beas-2B cells) min. Lysates were cleared by centrifugation (maximum speed for 15 min at 4°C), and supernatants were used for immunoprecipitation. Protein G magnetic Dynabeads (Invitrogen) were preincubated with 12 μg/sample GR (N499) or FLAG antibodies plus bovine serum albumin (BSA) in RIPA buffer for 1 h at 4°C and then washed twice with ice-cold RIPA buffer, prior to nutating in the presence of cleared lysates containing sheared chromatin for 2 h at 4°C in RIPA buffer supplemented with PIC and BSA. Beads were collected and subjected to 4 washes with ice-cold RIPA buffer containing 500 mM NaCl, followed by 4 washes with ice-cold LiCl buffer (20 mM Tris at pH 8.0, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate). Beads were then incubated in TE (Tris-EDTA) containing 0.7% SDS and 200 μg/ml proteinase K (Life Technologies) for 3 h at 55°C, and cross-links were reversed for 16 h at 65°C. ChIP DNA was purified with a ChIP DNA clean and concentrator kit (Zymo) using 7 volumes of DNA binding buffer, followed by elution in 50 μl elution buffer. Analysis of DNA obtained by ChIP was performed using qPCR amplification and SYBR green detection. Occupancy for a given factor/region under a specific experimental condition was measured as the difference between the CT value for the specific region relative to the geometric mean of the CT values for three control regions not predicted to occupy either factor and is thus on a log2 scale. qPCR analysis of input DNA established that control and experimental primer efficiencies were well matched (generally <0.5 cycle difference) using input dilutions designed to match ChIP samples. Assays were generally performed in biologic quadruplicate and repeated at least once with qualitatively similar results. P values were calculated using nonparametric tests to compare data, as indicated in the figure legends. The sequences of primers used to amplify specific test and control regions of ChIP DNA are provided in Table S5 in the supplemental material.
Endogenous KLF15 ChIP was performed as described above with the following modifications: Beas-2B cells grown to confluence in complete medium with serum were transferred to fresh serum-free medium for 4 h, followed by treatment for an additional 4 h with 1 μM dex in serum-free medium; ∼12 μg of anti-KLF15 antibody was used for each immunoprecipitation reaction.
Western blotting.
For protein expression studies, A549 and Beas-2B cells were plated on 100-mm dishes in phenol red-free DMEM containing 10% charcoal-dextran-stripped FBS (Life Technologies) and pen-strep. Approximately 18 h later, cells were treated with dex (100 nM for A549 cells; 1 μM for Beas-2B cells) or vehicle (ethanol), as indicated in the figure legends, for 4 h. Western blotting and protein detection were performed following standard protocols.
Microarray data accession number.
Raw data and additional experimental details are available at the Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE44695.
RESULTS
GR-KLF15-dependent transcriptional programming.
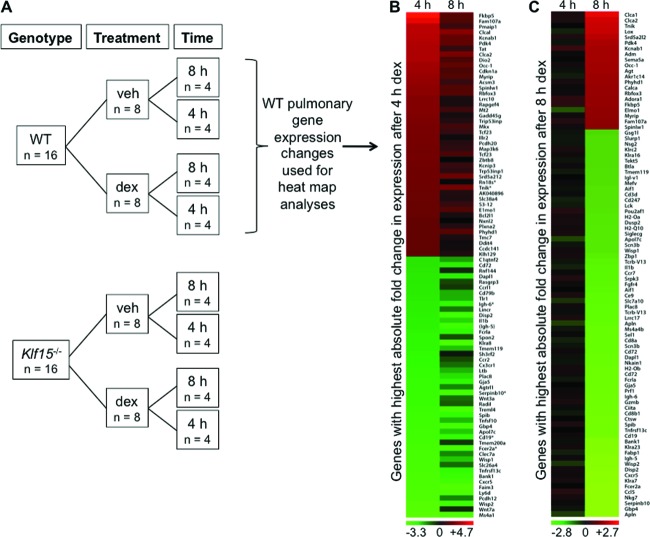
To examine the role of the GC-inducible transcription factor KLF15 in mediating the transcriptional response to GCs, we performed array-based expression profiling in whole lungs harvested from WT and Klf15−/− mice 4 or 8 h after intraperitoneal injection with the synthetic GC, dex, or saline vehicle. A schematic of the microarray study is shown in Fig. 1A. We focused on the lung since pulmonary disease is a major clinical target for GCs (31). Over 5,000 genes exhibited significant expression changes in response to any duration (4 or 8 h) of systemic dex treatment, based on an FDR of <0.05 and a fold change in expression of 50% between treatment conditions. Heat map analysis revealed that the changes in gene expression induced by dex in WT mice were temporally dynamic. Specifically, of the 90 transcripts with the greatest absolute fold changes in expression level after 4 h of dex treatment (in comparison to the expression level with vehicle alone), 78 had expression levels that were closer to the baseline after 8 h of dex treatment (visualized as a heat map in Fig. 1B). In contrast, each of the 90 genes with the greatest absolute fold changes in expression after 8 h of dex treatment (Fig. 1C) exhibited smaller changes in expression at the earlier (4 h) dex treatment time point. One possible explanation for these temporally dynamic changes in gene expression, which are similar to results observed in other timed analyses of responses to GCs (10, 32), is that transcription factors directly regulated by GR at early time points, such as KLF15, may mediate downstream transcriptional responses to GCs.
Fig 1.
Activation of GR signaling causes temporally dynamic changes in pulmonary gene expression. (A) Schematic illustrating the experimental design used to generate whole murine lung samples for microarray expression profiling. RNA was purified from lungs harvested from WT and Klf15−/− mice 4 or 8 h after intraperitoneal injection with 2.5 mg/kg dex or PBS vehicle (veh). (B and C) Heat maps illustrating gene expression changes identified by microarray after both 4 and 8 h of dex treatment in WT mouse lungs. (B) The heat map was generated using the 90 transcripts with the greatest absolute fold change in expression after 4 h of dex treatment; expression changes for this gene set after both 4 and 8 h are shown. (C) The heat map was generated with genes exhibiting the greatest absolute fold change in expression after 8 h of dex treatment, and expression changes after both 4 and 8 h are again shown. For both heat maps, genes with expression levels in the lowest quartile across the microarrays as a whole were excluded from analysis.
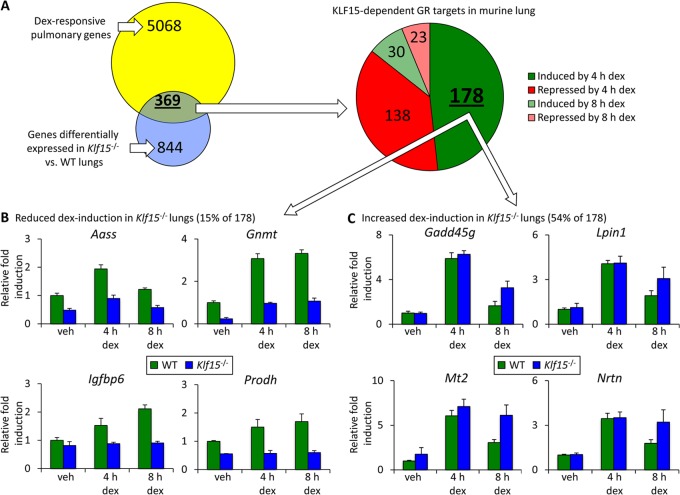
Our analysis of the pulmonary transcriptome in Klf15−/− mice with and without dex treatment supported this notion. Using the same FDR and fold change criteria outlined above, we found that 844 transcripts were differentially expressed in Klf15−/− mouse lungs compared to wild-type mouse lungs across drug treatment conditions. Of the KLF15-regulated gene set, 369 genes, representing ∼7% of all GR-regulated targets that we identified in the lung, were also regulated by dex (Fig. 2A). High-throughput qPCR validation for ∼95 of the genes regulated by GC signaling, KLF15, or both showed expression patterns that were qualitatively similar to the corresponding microarray data for almost all of the tested genes (see Table S1 in the supplemental material). Together, these findings delineate an extensive pulmonary gene network whose expression is regulated by both GR and KLF15.
Fig 2.
KLF15-dependent transcriptional responses to glucocorticoid. (A) WT and Klf15−/− mice were injected with 2.5 mg/kg of dex, and lungs were harvested after 4 and 8 h for microarray expression analysis. The Venn diagram illustrates genes that were differentially expressed in response to dex, genes that were differentially expressed in WT versus Klf15−/− mice (either under control conditions or with dex injection), and the intersection of these gene sets. The pie chart depicting gene number is based on the primary transcriptional response to dex in the GR-KLF15-dependent gene subset defined in the Venn diagram. Genes were defined as exhibiting differential expression by genotype or with dex treatment on the basis of an FDR of <0.05 and a fold change in expression of at least 50% between treatment conditions or genotype. (B and C) Representative examples of microarray data for specific genes that exhibited reduced (B) or potentiated (C) expression after dex treatment in Klf15−/− mouse lungs compared to WT mouse lungs. Fold change, based on the microarray data, indicates the expression level compared to that for the WT vehicle-treated control, which was normalized to 1 for each gene and condition. Error bars indicate the normalized standard deviation for the biologic replicates from each condition calculated from array intensity values.
Next, we investigated the temporal characteristics of the response to dex within the GR-KLF15 gene subset. The majority of the genes in this subset showed a significant change in expression after 4 h of dex treatment, which was relatively evenly split between transcripts that were induced and transcripts that were repressed by dex (Fig. 2A). Since genes induced by GCs at early time points are frequently regulated directly by GR binding (14), we focused our subsequent analysis on dex-induced, KLF15-dependent genes (see Table S2 in the supplemental material). Within this subset, we observed two dominant patterns of GR-KLF15 dependency. The first major pattern was defined by those genes exhibiting reduced dex induction in Klf15−/− mouse lungs compared to WT mouse lungs and accounted for ∼15% of the targets within the dex-induced, KLF15-dependent gene set. Representative examples of genes that follow this pattern, which is consistent with an inductive role for KLF15, are illustrated in Fig. 2B. Ontologic analysis of this gene set supported a biologic function for this pattern of GR-KLF15 interaction, as ∼60% of the genes in this subset are implicated in amino acid metabolism or mitochondrial function, indicative of highly significant enrichment for specific cellular processes (see Table S3 in the supplemental material).
The second mode of cooperative interaction occurred much more frequently, contributing to the regulation of greater than half of the GR-inducible, KLF15-dependent subset. This mode assumed the form of genes exhibiting greater dex induction in Klf15−/− mouse lungs than WT mouse lungs (Fig. 2C), consistent with a repressive role for KLF15, which has previously been reported to act as either an activator or a repressor (19, 33), depending on the promoter context. The GR-induced, KLF15-repressed gene set was strongly enriched for genes implicated in metal binding and targets of N-glycosylation (see Table S4 in the supplemental material), among other overrepresented processes, again suggesting that functional significance is associated with specific patterns of GR-KLF15 dependency.
GR and KLF15 cross talk is consistent with both coherent and incoherent feed-forward circuitry.
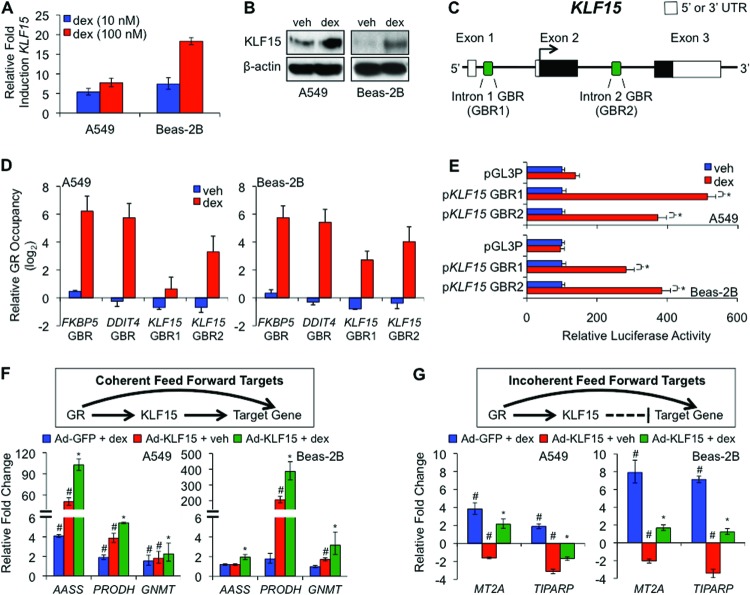
The lung contains many cell types, rendering it difficult to characterize biochemically GR-KLF15 cross talk in the lung in vivo. Therefore, to explore further the mechanistic basis for GR-KLF15 dependency, we used two well-established culture models of human airway epithelium, A549 and Beas-2B cells. In both cell types, treatment with dex induced KLF15 mRNA (Fig. 3A) and protein (Fig. 3B). Next, based on ENCODE GR ChIP-seq data from A549 cells that are accessible through the UCSC genome browser (14, 30), we performed ChIP assays, which showed that GR binds two intronic glucocorticoid binding regions (GBRs) in the KLF15 locus (Fig. 3C and D) in both A549 and Beas-2B cells. Reporters containing these regions were induced in both A549 and Beas-2B cells by dex in transfection assays (Fig. 3E); the GBR in the first intron had previously been shown to respond to GR in other cell types (34). Together these data indicate that KLF15 is directly induced by GR signaling in A549 and Beas-2B cells through two glucocorticoid response elements (GREs).
Fig 3.
GR and KLF15 cross talk in culture models of airway cells. (A) qPCR analysis of KLF15 mRNA expression in A549 and Beas-2B cells treated as indicated. Relative fold induction reflects normalized expression levels with dex treatment compared to that for vehicle-treated samples. Bars indicate means + SDs. (B) Western blot analysis for KLF15 and β-actin (loading control) protein in dex and vehicle-treated A549 and Beas-2B cells. (C) Schematic diagram of relative genomic locations of two putative GBRs identified within the human KLF15 locus. (D) ChIP-qPCR analysis of GR occupancy at KLF15 GBR1 and GBR2 in cells treated as indicated. FKBP5 and DDIT4 contain well-established functional GBRs that were included as positive controls for IP efficiency. Relative GR occupancy was calculated on the basis of the difference between the CT value for the indicated target in comparison to the geometric mean of CT values for 3 controls from regions that are not associated with GR occupancy. Bars indicate means + SDs expressed on a log2 scale. (E) Luciferase activity of KLF15 GBR1 and GBR2 reporter constructs transiently transfected into A549 (top) and Beas-2B (bottom) cells prior to treatment with dex or vehicle, as indicated. dex-induced activation of each reporter is expressed relative to its activity in vehicle-treated cells and was normalized to that for a control simian virus 40-Renilla reporter. Bars represent means + SDs. *, P < 0.05. The pGL3-promoter (pGL3P) is the parent vector from which the KLF15-GBR reporters were constructed. (F and G) (Top) Schematics depict the putative structural organization of generic GR-KLF15 coherent (F) and incoherent (G) feed-forward circuits; (bottom) bar graphs represent changes of expression of the indicated genes determined by qPCR in A549 and Beas-2B cells infected with a KLF15-overexpressing (Ad-KLF15) or control (Ad-GFP) adenovirus overnight (17 h) prior to treatment with dex (1 μM) or vehicle for 4 h. Transcript levels are expressed relative to those for Ad-GFP plus vehicle-treated controls, with repression visualized as a negative relative fold change. Bars indicate means + SDs. *, expression with Ad-KLF15 and dex treatment together is different (P ≤ 0.05) from that with either dex or Ad-KLF15 treatment alone, on the basis of nonparametric analysis; #, expression with Ad-KLF15 or dex treatment is different (P ≤ 0.05) from that under control conditions.
We next asked whether specific targets of the GR-KLF15 axis that we had identified in murine lung were regulated by GR and KLF15 in the cell line models. To accomplish this, we used adenoviral transduction to overexpress KLF15 (Ad-KLF15) in A549 and Beas-2B cells prior to treatment with dex or vehicle for 4 h. This approach allowed us to distinguish the transcriptional responses to GR and KLF15 individually and in combination and also to determine whether the effects of KLF15 overexpression in the cell lines were concordant with the effects of KLF15 deficiency in mice. Gene expression was assayed by RT-qPCR, and relative fold change was determined by comparing transcript levels of each group to those of vehicle-treated cells infected with a control virus (Ad-GFP). Although there were some differences between the cell lines, dex-activated GR expression and adenovirus-mediated KLF15 overexpresssion were both (in the absence of steroid) sufficient to significantly stimulate the transcription of AASS, GNMT, and PRODH in at least one of the two cell lines (Fig. 3F, bottom), while the combination of both inputs led to induction of even greater expression of these genes (P ≤ 0.05). These data show that GR and KLF15 increase the expression of AASS, GNMT, and PRODH both individually and in concert, as occurs in type I coherent feed-forward gene regulation (Fig. 3F, schematic diagram), one of eight potential feed-forward architectures that has been described (12). We also identified two genes, MT2A and TIPARP, which were induced by dex and repressed by Ad-KLF15 (P ≤ 0.05) (Fig. 3G, bottom), with combinatorial treatment resulting in an intermediate level of expression for both genes (P ≤ 0.05). These responses are consistent with the regulatory logic of a type 1 incoherent feed-forward circuit in which GR and KLF15 have opposing effects on the expression of downstream targets (Fig. 3G, schematic diagram). When considered together with the in vivo expression patterns for these genes (Fig. 2; see Table S2 in the supplemental material), these data suggest that A549 and Beas-2B cells can be used to model GR-KLF15 cross talk at selected genes. These data also support the hypothesis that KLF15 regulates transcriptional responses to GCs through formation of both coherent and incoherent feed-forward circuits with GR.
Composite GR-KLF15 response elements mediate feed-forward gene regulation.
To determine whether GR and KLF15 directly regulate presumptive coherent and incoherent feed-forward targets, we again used published GR ChIP-seq data (14) to identify likely GBRs within the PRODH, AASS, and MT2A loci. Analysis of these 0.6- to 2.5-kb genomic regions with MatInspector identified both consensus GR and consensus KLF binding sequences that we reasoned might provide combinatorial regulation by GR and KLF15 (Fig. 4A; see Fig. S1 in the supplemental material). These regions were thus used to generate luciferase reporters that were transfected into both airway cell lines in combination with a KLF15 expression plasmid (pcDNA-KLF15) or control vector. Reporter activity was then assayed after 8 h of dex or vehicle treatment, allowing us to determine if the putative regulatory region(s) was capable of responding to GR and/or KLF15. As shown in Fig. 4B, we found that discrete regulatory regions from each of the tested loci responded to both dex and KLF15. For example, the region spanning GBR2 of PRODH exhibited a small but significant activation in response to dex treatment, while forced expression of KLF15 in the absence of steroid elicited a robust 5- to 10-fold increase in reporter activity. Exposure to dex in the presence of constitutive KLF15 expression resulted in even greater activity of the PRODH GBR2 reporter than either treatment alone, indicating that this region is sufficient to confer responsiveness to simultaneous input from both transcription factors. Similarly, AASS GBR1 showed a modest induction with dex treatment: ∼5-fold induction with transfection of pcDNA-KLF15 and 10- to 20-fold induction with both dex and KLF15. In contrast, transfection of pcDNA-KLF15 resulted in a decrease in dex-mediated induction of the MT2A reporter in Beas-2B cells, where endogenous MT2A expression was robustly repressed by KLF15 (Fig. 3). Thus, although some differences in response magnitude were observed (e.g., in A549 cells, the AASS GBR1 reporter [Fig. 4B] was more responsive to dex and KLF15 than the endogenous gene [Fig. 3], while the MT2A reporter was not repressed by KLF15 in A549 cells), the responses of the GBR-containing reporters for PRODH, AASS, and MT2A were generally consistent with the expression patterns of the corresponding endogenous genes.
Fig 4.
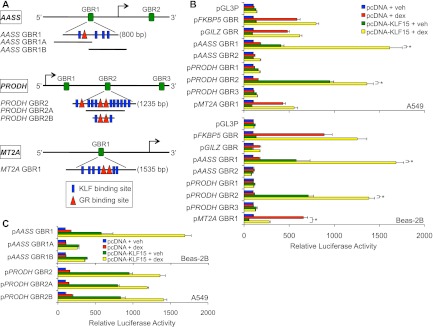
Composite response elements mediate feed-forward gene regulation by GR and KLF15. (A) Schematic diagram illustrating relative genomic locations of putative GR and KLF15 binding sites within the indicated genes that were used to generate reporter constructs and design primers for ChIP-qPCR. (B and C) Luciferase activity of the indicated reporter constructs transiently transfected into A549 and Beas-2B cells in combination with a KLF15 expression plasmid (pcDNA-KLF15) or control vector (pcDNA) prior to treatment with 100 nM dex or vehicle. Bars indicate mean reporter activation (+SD) relative to the activity of each construct in pcDNA- plus vehicle-treated controls. P values of <0.05, based on nonparametric analysis, were calculated between the conditions indicated by asterisks and the corresponding brackets.
We performed several control experiments to test the specificity of GR-KLF15 coregulation. Reporters containing GREs from the canonical GR targets FKBP5 and GILZ showed stronger baseline responses to dex (29, 35) and only modest or absent coregulation by KLF15 (Fig. 4B). Moreover, regions from the PRODH and AASS loci that span other putative GBRs and/or KLF15 binding regions were not functional under our treatment conditions (Fig. 4), indicating that the mere possession of conserved binding sequences does not necessitate a functional response element. Taken together, these data provide additional evidence that KLF15 forms both coherent and incoherent feed-forward circuits with GR.
To characterize further the regulation of AASS and PRODH by GR and KLF15, we generated additional reporter constructs spanning portions of the regions that we had defined above to be responsive to both GR and KLF15 (Fig. 4A). We cotransfected Beas-2B cells with these reporters and the constitutive KLF15 expression vector (or an empty vector control). We subsequently assayed relative luciferase activity following treatment with dex or vehicle for 8 h. For PRODH, we defined an ∼300-bp region containing 2 putative GR binding sites and 2 putative KLF15 binding sites that exhibited activity similar to that of the parent reporter construct (Fig. 4C). For AASS, we created two reporters that split the GR-KLF15 response region and separated several putative KLF binding sequences from a GR binding sequence. Intriguingly, each of the two subregions was activated by KLF15 but did not respond to dex (Fig. 4C). As the subregions contained 20 bp of overlap, which is larger than canonical GR binding sequences (36), these data indicate that regulation of AASS likely occurs through a complex composite GR-KLF15 response element.
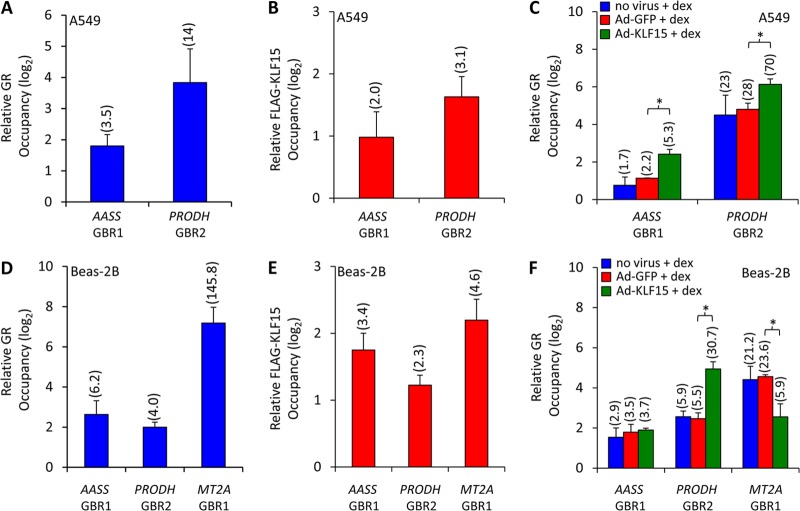
To establish that regulation of the AASS and PRODH loci by GR and KLF15 was associated with both factors binding to chromatin at the corresponding endogenous sites, we performed ChIP assays. Antibodies against GR and FLAG were used to immunoprecipitate factor-bound chromatin in A549 and Beas-2B cells treated with dex for 1 h (Fig. 5A) or infected with adenovirus overexpressing FLAG-tagged KLF15 (Fig. 5B), respectively. Both manipulations resulted in enrichment of PRODH GBR2 and AASS GBR1 among purified chromatin products subjected to qPCR, indicating the capacity for in vivo occupancy by both factors. We similarly tested whether the GR-KLF15-responsive region of the MT2A locus was bound by GR and KLF15 in Beas-2B cells and found enrichment for both factors (Fig. 5D and E). This establishes that AASS, PRODH, and MT2A are regulated directly by KLF15 and GR, a fundamental requirement of feed-forward architecture. Moreover, when Ad-KLF15-infected cells were given a 1-h pulse of dex, a significant increase in AASS GBR1 and PRODH GBR2 enrichment was observed among GR-immunoprecipitated chromatin relative to that in cells treated with dex alone (or in combination with control Ad-GFP), indicating that GR binding is potentiated in the presence of KLF15 (Fig. 5C). In contrast, binding of MT2A by GR was reduced by Ad-KLF15 infection (Fig. 5F). These data thus provide a mechanistic basis for feed-forward regulation by GR and KLF15 and indicate that the genomic context determines whether KLF15 potentiates or reduces GR binding within chromatin.
Fig 5.
KLF15 and GR bind composite GR-KLF15 response elements. (A and B) ChIP analysis of GR (A) and FLAG-KLF15 (B) occupancy of AASS GBR1 and PRODH GBR2 sites in dex-treated and Ad-KLF15-infected A549 cells, respectively. Factor occupancy is expressed relative to that of three negative-control sites, as described for Fig. 3. Data represent means + SDs expressed on a log2 scale; values in parentheses indicate fold enrichment, on a nonlogarithmic basis, of occupied versus nonoccupied control sites. (C) GR occupancy of the indicated sites determined by ChIP in dex-treated A549 cells following incubation with Ad-KLF15, control (Ad-GFP), or no virus, as indicated. P values of <0.05, based on nonparametric analysis, were calculated between the conditions indicated by asterisks and the corresponding brackets. (D to F) ChIP analysis of AASS GBR1, PRODH GBR2, and MT2A GBR1 occupancy in Beas-2B cells, performed and analyzed as described above for panels A to C.
Signal integration by GR and KLF15.
The output of feed-forward transcriptional circuits can be modulated by two inputs that independently regulate the two constituent transcription factors (12), providing a potential mechanism for signal integration. To determine whether feed-forward regulation by GR and KLF15 integrates signals to alter transcription, we analyzed the effects of serum starvation, which has been shown to induce KLF15 expression in other cell types (37), on the expression of genes within the GR-KLF15 network. First, we analyzed the effect of 4 h of serum deprivation on KLF15 expression in A549 and Beas-2B cells, which resulted in a robust induction of KLF15 mRNA (Fig. 6A). This response did not depend on GR, as KLF15 expression was similarly activated by serum deprivation in the presence of the GR antagonist RU-486 (Fig. 6A). Serum deprivation also modestly increased AASS and PRODH expression in both cell types, and the effect of serum starvation on all three genes—KLF15, AASS, and PRODH—was significantly augmented by dex (Fig. 6B). These results are consistent with the combinatorial effects of serum starvation and glucocorticoid signaling on regulation of genes within the GR-KLF15 network. Moreover, ChIP of GR and endogenous KLF15 under conditions of serum deprivation and dex treatment revealed simultaneous occupancy of the PRODH and AASS loci by both factors (Fig. 6C and D); the GR-KLF15-responsive region of the MT2A locus was similarly co-occupied. These data corroborate the ChIP data that we obtained with adenoviral-mediated KLF15 overexpression and serve to formally establish feed-forward regulation of these targets by GR and KLF15.
Fig 6.
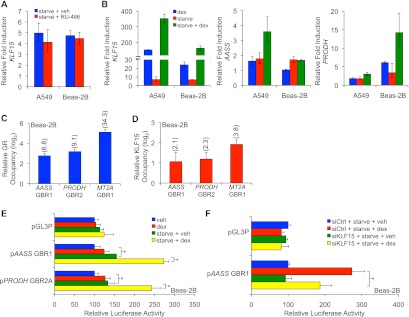
Signal integration by GR-KLF15 coherent feed-forward circuitry. (A) KLF15 mRNA expression following serum starvation in the presence or absence of the GR antagonist RU-486 (100 nM) in A549 and Beas-2B cells, as measured by qPCR. Bars represent fold induction (mean + SD) compared to that for cells treated with vehicle in complete medium. (B) qPCR analysis of KLF15, AASS, and PRODH mRNA expression in airway cells transferred to fresh complete or serum-free medium for 4 h prior to treatment with dex or vehicle for an additional 4 h. Transcript levels are expressed relative to that for cells receiving vehicle in complete medium. Bars indicate means + SDs. (C and D) ChIP-qPCR analysis of GR (C) and endogenous KLF15 (D) occupancy of AASS GBR1, PRODH GBR2, and MT2A GBR1 sites in Beas-2B cells that were serum starved for 4 h prior to treatment with dex for an additional 4 h. Factor occupancy is expressed relative to that of three negative-control sites, as described for Fig. 3. Data represent means + SDs expressed on a log2 scale; values in parentheses indicate fold enrichment, on a nonlogarithmic basis, of occupied versus nonoccupied control sites, as described for Fig. 5. (E) Luciferase activity of the indicated reporters transiently transfected into Beas-2B cells prior to treatment with dex or vehicle in serum-free or complete medium. Bars indicate mean reporter activation (+ SD) relative to the activity of each construct in controls treated with vehicle in complete medium. *, P < 0.05. (F) Luciferase activity of the AASS GBR1 reporter cotransfected into Beas-2B cells with KLF15 or control siRNA prior to serum starvation in the presence or absence of dex. Reporter activity (mean + SD) is expressed relative to that in serum-starved cells treated with vehicle and siCtrl. *, P < 0.05.
Next, we asked whether serum deprivation modulated the expression of the GR-KLF15-dependent AAAS and PRODH reporters. Similar to the induction of the corresponding endogenous genes, we found that serum deprivation modestly induced the AASS and PRODH reporters and that a combination of dex and serum deprivation led to still greater induction of reporter activity (Fig. 6E). To confirm that KLF15 is responsible for the inducing effects of serum deprivation on the reporters, we used siRNA to knock down KLF15 expression and achieved reasonable knockdown efficiency in Beas-2B cells (see Fig. S2 in the supplemental material). Although knockdown of KLF15 did not reduce the induction of the corresponding endogenous genes (see Fig. S2 in the supplemental material), likely secondary to compensatory induction of other metabolic regulators, in conjunction with the broad general effects of serum deprivation on GR activity (38), KLF15 knockdown reduced AASS luciferase reporter induction in Beas-2B cells subjected to serum deprivation with and without dex cotreatment (Fig. 6F). This indicates that activation of KLF15 through GR-independent signaling can act in combination with GCs to augment transcription, thus establishing a signal integration function for GR-KLF15 feed-forward circuitry.
Presumptive GR-KLF15 regulatory logic correlates with expression dynamics.
Our data from the two cell culture models definitively establish that GR and KLF15 regulate PRODH, AASS, and MT2A through feed-forward circuitry. These genes are representative of the two major GR-inducible, KLF15-dependent gene subsets that we observed in the microarray analysis (Fig. 2; see Table S2 in the supplemental material). Since specific feed-forward architectures can confer different temporal characteristics to client gene regulation (11), we next asked whether the temporal properties of the dex response in WT animals depended on the underlying effect of KLF15 deficiency on gene expression after 8 h of dex treatment (i.e., enhanced or reduced expression in Klf15−/− animals in comparison to that in WT animals). To accomplish this, we formulated data from Table S2 in the supplemental material as a 2-by-2 contingency table (Table 1) and performed a Fisher exact test. This test indicated that the categorization of dex-induced genes by whether expression was higher after 4 or 8 h of dex treatment in WT mice was strongly influenced (P < 0.00001) by the primary effect of KLF15 deficiency on gene expression after 8 h of GC treatment. Thus, glucocorticoid-induced, KLF15-dependent genes, grouped by whether the primary effect of KLF15 is inductive (coherent) or repressive (incoherent), display responses to dex that are statistically distinguishable on a temporal basis, supporting a role for circuit architecture in modulating the dynamics of the gene expression response to glucocorticoids.
Table 1.
2 × 2 contingency table based on categorizing GR-induced, KLF15-dependent transcripts
| Categorya | No. of transcripts with relative levels of expression in dex-treated WT mice for whichb: |
||
|---|---|---|---|
| 4 h < 8 h | 4 h > 8 h | Row total | |
| Increased induction by dex in Klf15−/− vs WT mice | 4 | 96 | 100 |
| Reduced induction by dex in Klf15−/− vs WT mice | 13 | 13 | 26 |
| Column total | 17 | 109 | 126 |
Transcripts in these categories had either increased expression (expression in Klf15−/− mice was >1.5 times that in WT mice) or decreased expression (expression in WT mice was >1.5 times that in Klf15−/− mice) in Klf15−/− mice after 8 h of dex compared to that in WT mice, as measured by microarray.
Transcripts in these categories had significant changes induced by dex after 4 h and are grouped according to whether the absolute expression measured by microarray was higher after 4 or 8 h of dex treatment.
DISCUSSION
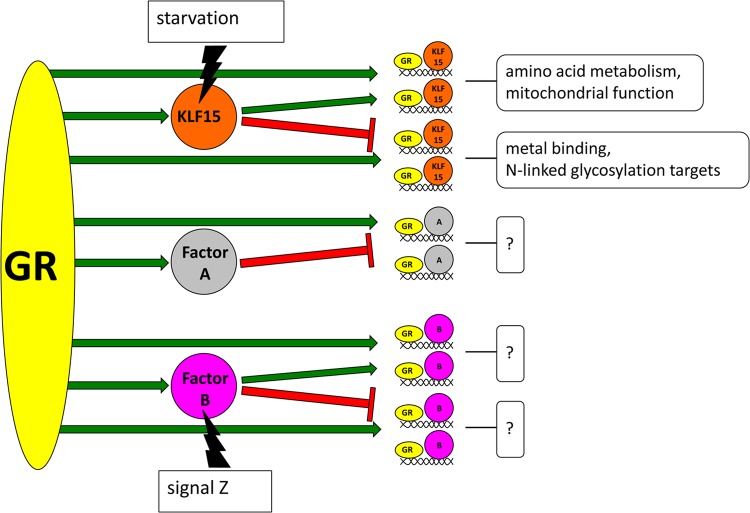
GR signaling is required for crucial adaptive responses to a wide range of physiologic stimuli, necessitating precise control and organization of subordinate transcription programs. Our data establish that a surprisingly large fraction (∼7%) of GC-regulated genes in the lung requires the presence of a single, direct GR target, KLF15, for correct expression responses to GC treatment. Within this GR-KLF15-dependent subset, the in vivo expression of GC-inducible genes followed patterns consistent with both coherent and incoherent feed-forward circuitries, the two primary subtypes of this archetypal regulatory system. We confirmed the basic characteristics of feed-forward gene regulation by GR and KLF15 using cell culture models, in which GR and KLF15 had combinatorial inductive effects on coherent targets and opposing effects on the expression of incoherent targets. These effects were mediated through enhanced recruitment of GR to chromatin by KLF15 at the PRODH and AASS loci, while KLF15 reduced GR binding at the incoherent target, MT2A. Taken together, our data establish that GR and KLF15 form feed-forward circuits that are correlated with the timing and magnitude of expression changes induced by GC signaling. Thus, temporal control of GC signaling not only is achieved through pulsatile characteristics of endogenous hormone secretion (39) but also is associated with the underlying architecture of GR-regulated transcriptional circuits. Our results also support a model in which feed-forward circuitry organizes GR-regulated transcriptional responses into subprograms (Fig. 7).
Fig 7.
Model of feed-forward gene regulation and signal integration by GR. In this model, GR directly regulates the expression of transcription factors such as KLF15, factor A, and factor B. These target transcription factors subsequently regulate discrete groups of GR target genes in combination with GR, thus forming feed-forward circuits that control expression dynamics for subsets of the greater GR-regulated transcriptome. As shown here, factor A forms incoherent feed-forward circuits with GR, while factor B and KLF15 form both coherent and incoherent feed-forward circuits. Target gene groups regulated by specific feed-forward circuit types are predicted to have linked physiologic functions, illustrated by boxed question marks and exemplified by the unique gene ontology terms (e.g., amino acid metabolism, metal binding) that were overrepresented within the coherent and incoherent GR-KLF15-regulated gene groups. Feed-forward circuits also allow GR to integrate nonligand signals, illustrated here by starvation and signal Z, to alter transcriptional outputs.
Feed-forward circuits have been intensely studied through both mathematical modeling and direct experimentation, generally in single-cell organisms, which are readily amenable to both detailed analyses of transcriptional kinetics and genetic manipulation of circuit components. These approaches have established that feed-forward architectures can confer specific properties to client gene expression dynamics (12). In our work, we assayed changes in steady-state gene expression in response to dex treatment at just two time points, an approach that offered only limited information with respect to the spectrum of transcriptional dynamics within the GR-KLF15 network. Despite this limitation, our data showed a very strong correlation between presumptive incoherent feed-forward architecture (i.e., genes that follow the pattern shown in Fig. 2C) and the likelihood that higher expression in wild-type mice occurred after 4 h, rather than 8 h, of dex treatment. More detailed temporal analysis of gene expression, including assaying changes in transcriptional rates, will provide additional insight into the dynamics of GR-mediated gene regulation within the GR-KLF15 feed-forward network.
In addition to regulating expression dynamics, feed-forward circuits can act as signal integrators that alter client gene expression in response to combinatorial inputs. Our data support such a function for GR-KLF15 circuitry. GR-independent induction of KLF15 through serum deprivation was associated with enhanced expression of feed-forward targets such as AASS and PRODH in response to dex, while KLF15 knockdown reduced the inductive effect of serum starvation on the activity of pAASS-GBR1, one of the GR-KLF15-responsive luciferase reporters that we generated. A requirement for two signals to fully activate PRODH, AASS, and other genes that regulate amino acid metabolism, which were highly overrepresented among coherent feed-forward GR-KLF15 expression profiles, provides a potential mechanism to constrain the induction of metabolically costly gene programming by GCs to appropriate physiologic circumstances. In that regard, mTOR signaling has been reported to modulate the catabolic effects of GCs on skeletal muscle (19, 40). It remains to be determined whether mTOR and other nutrition-sensitive signaling pathways function through GR-KLF15 feed-forward logic to regulate amino acid catabolism and energetics in the lung and other GR target organs.
Two factors forming both coherent and incoherent feed-forward circuits, as we have shown here for GR and KLF15 cross talk, were observed only rarely in detailed analyses of circuit architecture in lower organisms (11, 41). Thus, although ChIP data are required to definitively classify additional members of the GR-induced, KLF15-dependent gene sets as feed-forward targets, the unusual coupling of both circuit structures to GR signaling is likely to reflect distinct physiologic requirements for the expression of genes subordinate to either syntax. This notion is supported by the differing ontologies and expression dynamics of the presumptive coherent and incoherent GR-KLF15-dependent gene sets. Whether the incoherent GR circuit provides for fold change discrimination in response to increasing levels of hormone, a distinguishing feature of incoherent feed-forward gene regulation (42), remains to be determined. Fold change discrimination and other properties of feed-forward circuitry may also contribute to differences in the dose-response to GCs that have been observed for the expression of individual GR-regulated genes (43, 44).
Kruppel-like factors are increasingly recognized as important mediators of vital physiologic processes and are frequently regulated by nuclear receptors (45–48). Relatively little is known about how KLF family members achieve transcriptional specificity, as the information content provided by their preferred DNA binding sequences, the CACCC box and variants (49, 50), is relatively limited. Our work indicates that specificity for KLF15 is conferred in part through composite elements containing both CACCC boxes and GBRs (51), which adds to a growing number of examples of combinatorial gene regulation by nuclear receptors and the KLF family (52–55). Nuclear receptor signaling may thus impart specificity to the KLF family through a bipartite paradigm in which individual nuclear receptors both induce specific KLF family members and coregulate their transcriptional activity. This process is likely to depend on cofactors both for nuclear receptors and for KLF family members, such as p300 (56, 57), as well as the underlying chromatin structure of potential target genes. Coregulator activity, chromatin structure, and other cell type-specific programming are also likely to selectively tune the output of GR-KLF15 feed-forward circuits, providing a possible explanation for the differences in the magnitude of AASS and PRODH induction that we observed in A549 versus Beas-2B cells following dex treatment and KLF15 transduction (Fig. 3F and G).
In addition to its profound role in normal physiology, GR is widely targeted in the clinic by synthetic ligands to treat immune-mediated disease (2). Our results suggest that dysregulated expression of selected GR targets, such as KLF15, could mediate altered transcriptional responses to GC treatment for substantial subsets of the GR-regulated transcriptome. In this model, individual GR targets serve as feed-forward nodes that influence both therapeutic efficacy and the development of catabolic side effects that frequently complicate GC-based therapies. For example, reduced food intake increases KLF15 expression (58), potentially predisposing malnourished patients to deleterious GC-induced catabolic effects in the lung and other tissues. An expanded understanding of the circuit architecture and systems biology of the GR signaling network thus holds promise for improving the development and application of GC-based therapies in the clinic.
Supplementary Material
ACKNOWLEDGMENTS
We thank Stephen Tapscott and Keith Yamamoto for their comments and insights. Samantha Cooper provided invaluable advice on the ChIP methodology. Rebecca Barbeau and David Erle provided expert assistance in planning and conducting the microarray experiments, which were performed at the Sandler Asthma Basic Research (SABRE) Center Functional Genomics Core Facility.
This work was supported by NIH grant P30HL101294 through American Recovery and Reinvestment Act funds and by grant R01HL109557 (to A.N.G.).
Footnotes
Published ahead of print 18 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01474-12.
REFERENCES
- 1. Sapolsky RM, Romero LM, Munck AU. 2000. How do glucocorticoids influence stress responses? Integrating permissive, suppressive, stimulatory, and preparative actions. Endocrinol. Rev. 21:55–89 [DOI] [PubMed] [Google Scholar]
- 2. Cole TJ. 2006. Glucocorticoid action and the development of selective glucocorticoid receptor ligands. Biotechnol. Annu. Rev. 12:269–300 [DOI] [PubMed] [Google Scholar]
- 3. Nicolaides NC, Galata Z, Kino T, Chrousos GP, Charmandari E. 2010. The human glucocorticoid receptor: molecular basis of biologic function. Steroids 75:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen W, Dang T, Blind RD, Wang Z, Cavasotto CN, Hittelman AB, Rogatsky I, Logan SK, Garabedian MJ. 2008. Glucocorticoid receptor phosphorylation differentially affects target gene expression. Mol. Endocrinol. 22:1754–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. 2011. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat. Genet. 43:264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. 2009. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 324:407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gross KL, Cidlowski JA. 2008. Tissue-specific glucocorticoid action: a family affair. Trends Endocrinol. Metab. 19:331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee DY, Northrop JP, Kuo MH, Stallcup MR. 2006. Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. J. Biol. Chem. 281:8476–8485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luo M, Simons SS., Jr 2009. Modulation of glucocorticoid receptor induction properties by cofactors in peripheral blood mononuclear cells. Hum. Immunol. 70:785–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. John S, Johnson TA, Sung MH, Biddie SC, Trump S, Koch-Paiz CA, Davis SR, Walker R, Meltzer PS, Hager GL. 2009. Kinetic complexity of the global response to glucocorticoid receptor action. Endocrinology 150:1766–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alon U. 2007. Network motifs: theory and experimental approaches. Nat. Rev. Genet. 8:450–461 [DOI] [PubMed] [Google Scholar]
- 12. Mangan S, Alon U. 2003. Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. U. S. A. 100:11980–11985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kaplan S, Bren A, Dekel E, Alon U. 2008. The incoherent feed-forward loop can generate non-monotonic input functions for genes. Mol. Syst. Biol. 4:203 doi:10.1038/msb.2008.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, Myers RM. 2009. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 19:2163–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bagamasbad P, Ziera T, Borden SA, Bonett RM, Rozeboom AM, Seasholtz A, Denver RJ. 2012. Molecular basis for glucocorticoid induction of the Kruppel-like factor 9 gene in hippocampal neurons. Endocrinology 153:5334–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haldar SM, Jeyaraj D, Anand P, Zhu H, Lu Y, Prosdocimo DA, Eapen B, Kawanami D, Okutsu M, Brotto L, Fujioka H, Kerner J, Rosca MG, McGuinness OP, Snow RJ, Russell AP, Gerber AN, Bai X, Yan Z, Nosek TM, Brotto M, Hoppel CL, Jain MK. 2012. Kruppel-like factor 15 regulates skeletal muscle lipid flux and exercise adaptation. Proc. Natl. Acad. Sci. U. S. A. 109:6739–6744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gray S, Feinberg MW, Hull S, Kuo CT, Watanabe M, Sen-Banerjee S, DePina A, Haspel R, Jain MK. 2002. The Kruppel-like factor KLF15 regulates the insulin-sensitive glucose transporter GLUT4. J. Biol. Chem. 277:34322–34328 [DOI] [PubMed] [Google Scholar]
- 18. Gray S, Wang B, Orihuela Y, Hong EG, Fisch S, Haldar S, Cline GW, Kim JK, Peroni OD, Kahn BB, Jain MK. 2007. Regulation of gluconeogenesis by Kruppel-like factor 15. Cell Metab. 5:305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shimizu N, Yoshikawa N, Ito N, Maruyama T, Suzuki Y, Takeda S, Nakae J, Tagata Y, Nishitani S, Takehana K, Sano M, Fukuda K, Suematsu M, Morimoto C, Tanaka H. 2011. Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell Metab. 13:170–182 [DOI] [PubMed] [Google Scholar]
- 20. Haldar SM, Lu Y, Jeyaraj D, Kawanami D, Cui Y, Eapen SJ, Hao C, Li Y, Doughman YQ, Watanabe M, Shimizu K, Kuivaniemi H, Sadoshima J, Margulies KB, Cappola TP, Jain MK. 2010. Klf15 deficiency is a molecular link between heart failure and aortic aneurysm formation. Sci. Transl. Med. 2:26ra26 doi:10.1126/scitranslmed.3000502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. National Research Council 1996. Guide for the care and use of laboratory animals. National Academies Press, Washington, DC [Google Scholar]
- 22. Fisch S, Gray S, Heymans S, Haldar SM, Wang B, Pfister O, Cui L, Kumar A, Lin Z, Sen-Banerjee S, Das H, Petersen CA, Mende U, Burleigh BA, Zhu Y, Pinto YM, Liao R, Jain MK. 2007. Kruppel-like factor 15 is a regulator of cardiomyocyte hypertrophy. Proc. Natl. Acad. Sci. U. S. A. 104:7074–7079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bolstad BM, Irizarry RA, Astrand M, Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193 [DOI] [PubMed] [Google Scholar]
- 24. Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B 57:289–300 [Google Scholar]
- 25. Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3. doi:10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- 26. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80 doi:10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang da W, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, Lempicki RA. 2009. Extracting biological meaning from large gene lists with DAVID. Curr. Protoc. Bioinformatics Chapter 13:Unit 13.11. doi:10.1002/0471250953.bi1311s27 [DOI] [PubMed] [Google Scholar]
- 28. Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57 [DOI] [PubMed] [Google Scholar]
- 29. Gerber AN, Masuno K, Diamond MI. 2009. Discovery of selective glucocorticoid receptor modulators by multiplexed reporter screening. Proc. Natl. Acad. Sci. U. S. A. 106:4929–4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. ENCODE Project Consortium 2011. A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 9:e1001046 doi:10.1371/journal.pbio.1001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barnes PJ. 2006. Corticosteroids: the drugs to beat. Eur. J. Pharmacol. 533:2–14 [DOI] [PubMed] [Google Scholar]
- 32. Masuno K, Haldar SM, Jeyaraj D, Mailloux CM, Huang X, Panettieri RA, Jr, Jain MK, Gerber AN. 2011. Expression profiling identifies Klf15 as a glucocorticoid target that regulates airway hyperresponsiveness. Am. J. Respir. Cell Mol. Biol. 45:642–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Otteson DC, Liu Y, Lai H, Wang C, Gray S, Jain MK, Zack DJ. 2004. Kruppel-like factor 15, a zinc-finger transcriptional regulator, represses the rhodopsin and interphotoreceptor retinoid-binding protein promoters. Invest. Ophthalmol. Vis. Sci. 45:2522–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Asada M, Rauch A, Shimizu H, Maruyama H, Miyaki S, Shibamori M, Kawasome H, Ishiyama H, Tuckermann J, Asahara H. 2011. DNA binding-dependent glucocorticoid receptor activity promotes adipogenesis via Kruppel-like factor 15 gene expression. Lab. Invest. 91:203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hubler TR, Scammell JG. 2004. Intronic hormone response elements mediate regulation of FKBP5 by progestins and glucocorticoids. Cell Stress Chaperones 9:243–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. So AY, Cooper SB, Feldman BJ, Manuchehri M, Yamamoto KR. 2008. Conservation analysis predicts in vivo occupancy of glucocorticoid receptor-binding sequences at glucocorticoid-induced genes. Proc. Natl. Acad. Sci. U. S. A. 105:5745–5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dmitriev P, Petrov A, Ansseau E, Stankevicins L, Charron S, Kim E, Bos TJ, Robert T, Turki A, Coppee F, Belayew A, Lazar V, Carnac G, Laoudj D, Lipinski M, Vassetzky YS. 2011. The Kruppel-like factor 15 as a molecular link between myogenic factors and a chromosome 4q transcriptional enhancer implicated in facioscapulohumeral dystrophy. J. Biol. Chem. 286:44620–44631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ren R, Oakley RH, Cruz-Topete D, Cidlowski JA. 2012. Dual role for glucocorticoids in cardiomyocyte hypertrophy and apoptosis. Endocrinology 153:5346–5360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Conway-Campbell BL, Pooley JR, Hager GL, Lightman SL. 2012. Molecular dynamics of ultradian glucocorticoid receptor action. Mol. Cell. Endocrinol. 348:383–393 [DOI] [PubMed] [Google Scholar]
- 40. Latres E, Amini AR, Amini AA, Griffiths J, Martin FJ, Wei Y, Lin HC, Yancopoulos GD, Glass DJ. 2005. Insulin-like growth factor-1 (IGF-1) inversely regulates atrophy-induced genes via the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway. J. Biol. Chem. 280:2737–2744 [DOI] [PubMed] [Google Scholar]
- 41. Eichenberger P, Fujita M, Jensen ST, Conlon EM, Rudner DZ, Wang ST, Ferguson C, Haga K, Sato T, Liu JS, Losick R. 2004. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2:e328 doi:10.1371/journal.pbio.0020328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goentoro L, Shoval O, Kirschner MW, Alon U. 2009. The incoherent feedforward loop can provide fold-change detection in gene regulation. Mol. Cell 36:894–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. He Y, Szapary D, Simons SS., Jr 2002. Modulation of induction properties of glucocorticoid receptor-agonist and -antagonist complexes by coactivators involves binding to receptors but is independent of ability of coactivators to augment transactivation. J. Biol. Chem. 277:49256–49266 [DOI] [PubMed] [Google Scholar]
- 44. Reddy TE, Gertz J, Crawford GE, Garabedian MJ, Myers RM. 2012. The hypersensitive glucocorticoid response specifically regulates period 1 and expression of circadian genes. Mol. Cell. Biol. 32:3756–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Frigo DE, Sherk AB, Wittmann BM, Norris JD, Wang Q, Joseph JD, Toner AP, Brown M, McDonnell DP. 2009. Induction of Kruppel-like factor 5 expression by androgens results in increased CXCR4-dependent migration of prostate cancer cells in vitro. Mol. Endocrinol. 23:1385–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nakajima Y, Akaogi K, Suzuki T, Osakabe A, Yamaguchi C, Sunahara N, Ishida J, Kako K, Ogawa S, Fujimura T, Homma Y, Fukamizu A, Murayama A, Kimura K, Inoue S, Yanagisawa J. 2011. Estrogen regulates tumor growth through a nonclassical pathway that includes the transcription factors ERbeta and KLF5. Sci. Signal. 4:ra22 doi:10.1126/scisignal.2001551 [DOI] [PubMed] [Google Scholar]
- 47. McConnell BB, Yang VW. 2010. Mammalian Kruppel-like factors in health and diseases. Physiol. Rev. 90:1337–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Avci HX, Lebrun C, Wehrle R, Doulazmi M, Chatonnet F, Morel MP, Ema M, Vodjdani G, Sotelo C, Flamant F, Dusart I. 2012. Thyroid hormone triggers the developmental loss of axonal regenerative capacity via thyroid hormone receptor alpha1 and Kruppel-like factor 9 in Purkinje cells. Proc. Natl. Acad. Sci. U. S. A. 109:14206–14211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Miller IJ, Bieker JJ. 1993. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol. Cell. Biol. 13:2776–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S. 2008. Kruppel-like transcription factors: a functional family. Int. J. Biochem. Cell Biol. 40:1996–2001 [DOI] [PubMed] [Google Scholar]
- 51. Schule R, Muller M, Otsuka-Murakami H, Renkawitz R. 1988. Cooperativity of the glucocorticoid receptor and the CACCC-box binding factor. Nature 332:87–90 [DOI] [PubMed] [Google Scholar]
- 52. Wade HE, Kobayashi S, Eaton ML, Jansen MS, Lobenhofer EK, Lupien M, Geistlinger TR, Zhu W, Nevins JR, Brown M, Otteson DC, McDonnell DP. 2010. Multimodal regulation of E2F1 gene expression by progestins. Mol. Cell. Biol. 30:1866–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Velarde MC, Iruthayanathan M, Eason RR, Zhang D, Simmen FA, Simmen RC. 2006. Progesterone receptor transactivation of the secretory leukocyte protease inhibitor gene in Ishikawa endometrial epithelial cells involves recruitment of Kruppel-like factor 9/basic transcription element binding protein-1. Endocrinology 147:1969–1978 [DOI] [PubMed] [Google Scholar]
- 54. Zhang D, Zhang XL, Michel FJ, Blum JL, Simmen FA, Simmen RC. 2002. Direct interaction of the Kruppel-like family (KLF) member, BTEB1, and PR mediates progesterone-responsive gene expression in endometrial epithelial cells. Endocrinology 143:62–73 [DOI] [PubMed] [Google Scholar]
- 55. Grunewald M, Johnson S, Lu D, Wang Z, Lomberk G, Albert PR, Stockmeier CA, Meyer JH, Urrutia R, Miczek KA, Austin MC, Wang J, Paul IA, Woolverton WL, Seo S, Sittman DB, Ou XM. 2012. Mechanistic role for a novel glucocorticoid-KLF11 (TIEG2) protein pathway in stress-induced monoamine oxidase A expression. J. Biol. Chem. 287:24195–24206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bonnefond A, Lomberk G, Buttar N, Busiah K, Vaillant E, Lobbens S, Yengo L, Dechaume A, Mignot B, Simon A, Scharfmann R, Neve B, Tanyolac S, Hodoglugil U, Pattou F, Cave H, Iovanna J, Stein R, Polak M, Vaxillaire M, Froguel P, Urrutia R. 2011. Disruption of a novel Kruppel-like transcription factor p300-regulated pathway for insulin biosynthesis revealed by studies of the c.-331 INS mutation found in neonatal diabetes mellitus. J. Biol. Chem. 286:28414–28424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang XL, Herzog B, Waltner-Law M, Hall RK, Shiota M, Granner DK. 2004. The synergistic effect of dexamethasone and all-trans-retinoic acid on hepatic phosphoenolpyruvate carboxykinase gene expression involves the coactivator p300. J. Biol. Chem. 279:34191–34200 [DOI] [PubMed] [Google Scholar]
- 58. Jeyaraj D, Scheer FA, Ripperger JA, Haldar SM, Lu Y, Prosdocimo DA, Eapen SJ, Eapen BL, Cui Y, Mahabeleshwar GH, Lee HG, Smith MA, Casadesus G, Mintz EM, Sun H, Wang Y, Ramsey KM, Bass J, Shea SA, Albrecht U, Jain MK. 2012. Klf15 orchestrates circadian nitrogen homeostasis. Cell Metab. 15:311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.