Abstract
Serial activation of the tyrosine kinases Lck and ZAP-70 initiates signaling downstream of the T cell receptor. We previously reported the structure of an autoinhibited ZAP-70 variant in which two regulatory tyrosine residues (315 and 319) in the SH2-kinase linker were replaced by phenylalanine. We now present a crystal structure of ZAP-70 in which Tyr 315 and Tyr 319 are not mutated, leading to the recognition of a five-residue sequence register error in the SH2-kinase linker of the original crystallographic model. The revised model identifies distinct roles for these two tyrosines. As seen in a recently reported structure of the related tyrosine kinase Syk, Tyr 315 of ZAP-70 is part of a hydrophobic interface between the regulatory apparatus and the kinase domain, and the integrity of this interface would be lost upon engagement of doubly phosphorylated peptides by the SH2 domains. Tyr 319 is not necessarily dislodged by SH2 engagement, which activates ZAP-70 only ∼5-fold in vitro. In contrast, phosphorylation by Lck activates ZAP-70 ∼100-fold. This difference is due to the ability of Tyr 319 to suppress ZAP-70 activity even when the SH2 domains are dislodged from the kinase domain, providing stringent control of ZAP-70 activity downstream of Lck.
INTRODUCTION
Signaling by the T cell receptor (TCR) relies on two nonreceptor tyrosine kinases: the Src family tyrosine kinase Lck and the zeta-chain-associated protein kinase ZAP-70 (Fig. 1) (1). The clustering of coreceptor (CD4 or CD8)-associated Lck with T cell receptors allows Lck to phosphorylate tyrosine residues in immunoreceptor tyrosine-based activation motifs (ITAMs) in the intracellular tails of zeta chains of the TCR complex. Doubly phosphorylated ITAMs in the stimulated TCR complex recruit ZAP-70 to the plasma membrane, where it is phosphorylated by Lck (2, 3). ITAM binding and phosphorylation release ZAP-70 from an autoinhibited state, enabling it to phosphorylate two scaffold proteins, LAT (linker for the activation of T cells) and SLP-76 (Src homology 2 domain-containing leukocyte phosphoprotein of 76 kDa), leading to the recruitment of effector proteins that stimulate T cell activation (4, 5).
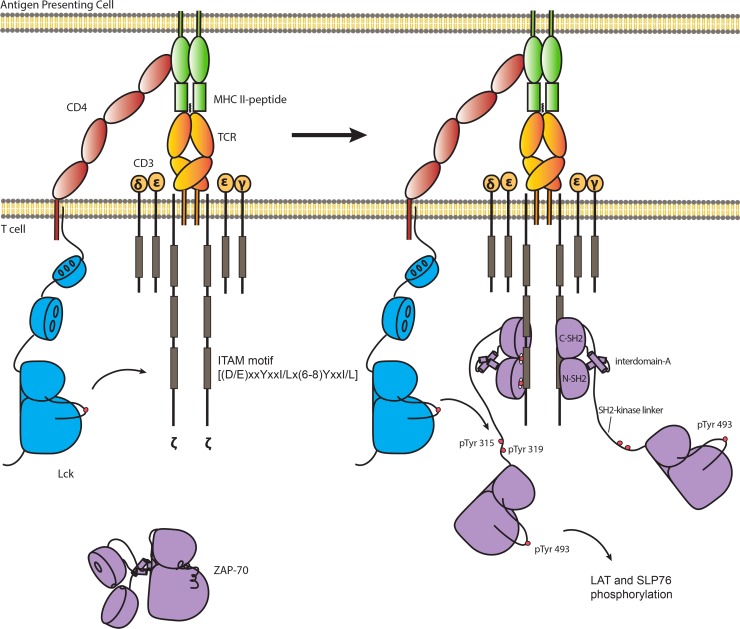
Fig 1.
Phosphorylation of ZAP-70 is required to initiate T cell receptor signaling. TCR signaling is initiated by two tyrosine kinases: the Src family kinase Lck and ZAP-70. Activated Lck phosphorylates ITAMs in the intracellular tails of ζ chains of the TCR complex. Doubly phosphorylated ITAMs recruit ZAP-70 from the cytosol to the plasma membrane. Tyr 315, Tyr 319, and Tyr 493 of ZAP-70 are phosphorylated by Lck to fully activate ZAP-70, enabling ZAP-70 to phosphorylate two scaffold proteins, LAT and SLP-76, leading to the recruitment of effector proteins that stimulate T cell activation.
TCR signaling events in a Jurkat-derived ZAP-70-deficient cell line are completely abolished. Hypomorphic ZAP-70 mutant mice, which have partial defects in TCR signaling, develop autoimmunity or autoimmune disease (6–8). The importance of ZAP-70 for T cell development and activation is further emphasized by the fact that the loss of ZAP-70 function results in severe combined immunodeficiency (SCID) in both humans and mice, which is characterized by a lack of functional peripheral T cells (9–12). The tight association of these diseases with ZAP-70 catalytic activity indicates clearly that ZAP-70 is a potential therapeutic target.
ZAP-70 and its close relative Syk, which is essential for B cell receptor signaling, share a domain architecture that is unique among protein kinases. The N-terminal half of each protein contains two SH2 domains that form a tightly coupled module in which the tandem SH2 domains are connected by a coiled-coil segment known as interdomain A (13, 14). The tandem-SH2 module is connected to the kinase domain by a flexible linker known as interdomain B or the SH2-kinase linker.
The tandem-SH2 modules of ZAP-70 and Syk undergo large conformational changes upon binding to the doubly phosphorylated ITAM peptide (13–16). The relative orientation of the two SH2 domains changes substantially. Key to the generation of this conformational change is the fact that the phosphotyrosine binding site on the N-terminal SH2 domain is located at the interface between the two domains (13). In the absence of the doubly phosphorylated ITAM peptide, the two SH2 domains are splayed apart so that interfacial residues are misaligned at the N-terminal phosphotyrosine binding site (15). The binding of the doubly phosphorylated ITAM peptide reconfigures the site and pulls the two SH2 domains into alignment. The relative rotation of the SH2 domains is coupled to a change in the conformation of the helices in interdomain A (15).
We previously determined a crystallographic model of an inactive variant of full-length ZAP-70 in which two regulatory tyrosine residues in the SH2-kinase linker (residues Tyr 315 and Tyr 319) were both replaced by phenylalanine (we refer to this construct as ZAP-70-FF) (17). The tandem-SH2 module, which is not bound to a phosphorylated ITAM peptide, forms a tightly integrated assembly with the kinase domain, which is in an inactive conformation. One portion of the SH2-kinase linker (spanning residues 313 to 320) is sandwiched between the interdomain A coiled coil and the distal surface of the kinase domain. The formation of this “linker-kinase sandwich” reduces the flexibility of the ZAP-70 kinase domain, impeding the transition between inactive and active conformations. The conformational change in the tandem-SH2 module upon phosphorylated ITAM binding, particularly in interdomain A, is expected to result in the linker-kinase sandwich being dismantled and the tandem-SH2 module being released from the kinase domain. Key aspects of this mechanism have been validated by in vitro and cell-based measurements of the activities of ZAP-70 mutants (18, 19). As we explain below, we have now identified a sequence register error in the original model for the SH2-kinase linker, the implications of which are examined in this paper.
In addition to being activated by the physical release of the tandem-SH2 module from the kinase domain, ZAP-70 is also activated by phosphorylation. In particular, the phosphorylation of Tyr 315 and Tyr 319 in the SH2-kinase linker by Lck is important for ZAP-70 catalytic activity. Mutation of these two tyrosine residues to phenylalanine suppresses ZAP-70 activation as well as ZAP-70-dependent downstream signaling events (20–24). There are other tyrosine residues in ZAP-70 that are phosphorylated upon activation, including Tyr 492 and Tyr 493, in the activation loop of the kinase domain, but our principal focus in this paper is on Tyr 315 and Tyr 319, located in the SH2-kinase linker.
During the course of the investigations reported in this paper, we found that a ZAP-70 construct lacking the tandem-SH2 module but including the SH2-kinase linker can be activated further by phosphorylation. The activity of the isolated kinase domain of ZAP-70 does not increase with phosphorylation, implying that the necessity for phosphorylation is a consequence of an inhibitory action of the SH2-kinase linker. This cannot be understood completely on the basis of the crystallographic model for the ZAP-70-FF construct reported previously by us, because contacts between the SH2-kinase linker and the kinase domain are restricted to the hinge region of the kinase domain in that model, which does not undergo structural changes during the conversion from active to inactive forms.
We now report the determination of the crystal structure of an essentially full-length ZAP-70 construct (missing only 14 residues at the very C terminus) in which both Tyr 315 and Tyr 319 are intact (we refer to this wild-type construct as ZAP-70-YY). The domain assembly of the ZAP-70-YY construct is essentially the same as that reported previously for the ZAP-70-FF construct, with one key difference. Notably, in the ZAP-70-YY structure, the C-terminal part of the SH2-kinase linker adopts a helical conformation and positions the side chain of Tyr 319 so that it interacts with the N lobe of the kinase domain, in a region that undergoes structural changes as the kinase switches between active and inactive conformations. These additional interactions between the SH2-kinase linker and the kinase domain suggest how this segment could suppress the activity of the ZAP-70 kinase domain even when the tandem-SH2 module is displaced. In addition, comparisons of molecular dynamic simulations initiated using the ZAP-70-YY and ZAP-70-FF models indicate that the conformation of the SH2-kinase linker is stable in the ZAP-70-YY model but not in the original ZAP-70-FF model. While this report was being prepared for submission, the structure of the autoinhibited form of Syk was reported (25). Our findings for the conformation of the SH2-kinase linker in ZAP-70, reported below, are consistent with the new Syk structure.
To validate these conclusions, we used a fluorescence-based approach to measure the recruitment of the adapter protein Grb2 to LAT upon phosphorylation of LAT by ZAP-70. We used this fluorescence assay to monitor the activity of ZAP-70 as a function of phosphorylation by Lck. As mentioned above, a construct of ZAP-70 that includes the SH2-kinase linker and the kinase domain (but not the tandem-SH2 module) has substantially lower activity than the isolated kinase domain and is activated upon phosphorylation by Lck. Taken together, our results reveal additional aspects of the important role played by the SH2-kinase linker in the suppression of the kinase domain activity of ZAP-70 and its release by phosphorylation, resulting in the initiation of T cell signaling.
MATERIALS AND METHODS
Insect cell expression and purification of ZAP-70 constructs.
For the crystallization construct referred to as ZAP-70-YY, DNA encoding human ZAP-70 (residues 1 to 606) followed by a Gly-Ser-Gly linker, a PreScission protease site, and a C-terminal 6-His tag was cloned into pFastBac1 (Invitrogen), using the BamHI and EcoRI restriction sites. The catalytic base Asp 461 in ZAP-70-YY was mutated to asparagine by use of a QuikChange site-directed mutagenesis kit (Stratagene), which was also used for all other point mutations. For the kinase activity assay, a similar cloning strategy was applied to clone the coding sequences for full-length ZAP-70 (residues 1 to 619; ZAP-70-FL), ZAP-70-KD/linker (residues 310 to 619; “linker” refers to the SH2-kinase linker), and ZAP-70-KD (residues 327 to 619), followed by a Gly-Ser-Gly linker, a PreScission protease site, and a C-terminal 6-His tag, into the pFastBac1 (Invitrogen) vector. All ZAP-70 constructs were expressed in baculovirus-infected TriEx Sf9 cells and purified as described previously (17). The final buffer from the gel filtration step was 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM TCEP [tris(2-carboxyethyl)phosphine], and 5% glycerol. Mass spectrometry confirmed the identity of the protein.
Bacterial expression and purification of LAT and Grb2.
Coding sequences for the human LAT cytosolic domain (residues 30 to 233) and full-length Grb2 were amplified by PCR and cloned into a pET-28-derived bacterial expression vector by use of NdeI and XhoI sites. A tobacco etch virus (TEV) protease cleavage site was engineered between the N-terminal 6-His tag and the NdeI site in both constructs. In order to express recombinant LAT and Grb2 in Escherichia coli, the LAT and Grb2 constructs were transformed into the BL21 strain of E. coli. The expression of recombinant LAT was induced by adding 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h at 37°C. The expression of recombinant Grb2 was induced by adding 0.2 mM IPTG for 16 h at 18°C. The bacterial cells were then resuspended in lysis buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 25 mM imidazole, 5 mM β-mercaptoethanol), lysed using a French press, and centrifuged to remove cellular debris at 17,000 rpm for 1 h at 4°C. For LAT purification, the clarified lysate was passed through a Ni-nitrilotriacetic acid (Ni-NTA) column (Qiagen). The recombinant protein was eluted from the column by using a buffer containing 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 250 mM imidazole, and 5 mM β-mercaptoethanol. The eluted LAT protein was incubated overnight with TEV protease to remove the N-terminal His tag. The protein was concentrated by ultrafiltration and further purified by size exclusion chromatography (Superdex 200) to remove aggregated proteins. The final buffer for the gel filtration step was 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 5 mM TCEP. The purification procedure for Grb2 was very similar to that for LAT, except for the final buffer, which included 100 mM arginine to stabilize Grb2 in solution (26).
Preparation of fluorophore-labeled LAT and Grb2.
Alexa Fluor 555 C2 maleimide and Alexa Fluor 647 C2 maleimide were purchased from Invitrogen. LAT and Grb2 were incubated in a 10-fold molar excess of Alexa Fluor 555 C2 maleimide and Alexa Fluor 647 C2 maleimide, respectively, for 2 h at room temperature. Free dyes were separated from protein by size exclusion chromatography (Sephadex G-25).
Fluorescence resonance energy transfer (FRET)-based LAT phosphorylation assay.
The phosphorylation of LAT by ZAP-70 and recruitment of Grb2 in solution were assessed by monitoring the increase of acceptor fluorescence emission (Alexa Fluor 647-labeled Grb2). The reaction buffer contained 20 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 150 mM NaCl, 1 μM Alexa Fluor 555-labeled LAT, 1 μM Alexa Fluor 647-labeled Grb2, and 1 μM kinase. The reaction was triggered by adding ATP to the reaction buffer, and the fluorescence intensity at 670 nm, with excitation at 555 nm, was monitored using a Fluoromax-3 fluorimeter (Horiba Jobin Yvon) at 30°C.
Coupled kinase assay.
The coupled kinase assay was performed using a continuous spectrophotometric method as described previously (18). Briefly, the final reaction buffer contained 20 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 150 mM NaCl, 1 mM phosphoenolpyruvate, 0.3 mg/ml of NADH, 75 U/ml of pyruvate kinase, 105 U/ml of lactate dehydrogenase, 2 mg/ml of poly-Glu/Tyr substrate peptide (Glu/Tyr ratio of 4:1; 5 to 20 kDa), 2 μM tyrosine kinase, and 1 mM ATP. The extent of reaction was monitored as the decrease in absorbance at 340 nm for 30 min at 30°C in a microtiter plate spectrophotometer (SpectraMax).
Crystallization and data collection.
The ZAP-70-YY protein (20 mg/ml, in a buffer containing 100 mM Tris-HCl, pH 9.5, 150 mM NaCl, 5 mM TCEP, and 5% glycerol) was mixed with 5 mM adenylyl imidodiphosphate (AMP-PNP) and 5 mM MgCl2 for 1 h at 4°C. Crystals of ZAP-70-YY were grown using sitting-drop vapor diffusion. A protein stock solution (0.2 μl) was mixed with 0.2 μl reservoir solution (0.1 M Bis-Tris, pH 6.5, 20% polyethylene glycol [PEG] monomethyl ether 5000) by use of a Crystal Phoenix liquid handling system, and the drop was equilibrated at 20°C. The crystals were cryoprotected by the addition of 25% PEG 400 (vol/vol), and X-ray diffraction data were measured at the Advanced Light Source, beamline 8.3.1. Data were integrated and scaled using MOSFLM and Scala via CCP4 (27, 28).
Structure determination and refinement.
The structure of ZAP-70-YY was solved by molecular replacement, using the crystal structure of ZAP-70-FF without the SH2-kinase linker (residues 310 to 327) as the search model. Refinement and model building were performed using REFMAC with TLS refinement and Coot (29–31). In our earlier publication, we noted that electron density maps showed features consistent with an alternate conformation for the SH2-kinase linker (see Materials and Methods in reference 17) that we were unable to model. We now recognize that these features resulted from a five-residue sequence register error in the original model. We manually rebuilt the SH2-kinase linker (residues 303 to 327) of ZAP-70-YY, and the structure was refined to 3.0-Å resolution (the Rfree and R values are 0.288 and 0.214, respectively) (Table 1).
Table 1.
Data collection statistics
| Parameter | Value or description |
|---|---|
| Wavelength (Å) | 1.1159 |
| Space group | P1 |
| Cell dimensions (Å) | a = 48.5, b = 53.2, c = 69.0 |
| Cell angles (°) | α = 106.1, β = 93.3, γ = 104.4 |
| Resolution range (Å) | 30.00–3.0 (3.16–3.00) |
| Rmerge (%)a | 12.2 (44.6) |
| I/σ (I)a | 7.1 (2.4) |
| Completeness (%)a | 93.9 (94.7) |
| Redundancy | 3.0 (3.0) |
| Model refinement statistics | |
| No. of reflections (work/test) | 11,374/579 |
| Rwork/Rfree (%) | 21.4/28.8 |
| RMS deviation bond length (Å) | 0.010 |
| RMS deviation angle (°) | 1.506 |
| Avg temp (B) factor (Å2) | 50.6 |
| Ramachandran plot statistics | |
| Most favored regions (%) | 92.1 |
| Additional allowed regions (%) | 7.7 |
| Disallowed regions (%) | 0.2 |
Statistics for the outer resolution shell are shown in parentheses.
Molecular dynamic simulations.
All molecular dynamic simulations were performed with GROMACS, version 4.5.5 (32), and the Amber ff99SB-ILDN force field (33), the TIP3P water model (34), and periodic boundary conditions. The starting structure for ZAP-70 was taken from Protein Data Bank (PDB) entry 2OZO (17), with all solvent and ATP analogue molecules removed. A ZAP-70-YY model was built by replacing the SH2-kinase linker (residues 310 to 327) of the ZAP-70-FF crystal structure with the SH2-kinase linker (residues 303 to 327) of the ZAP-70-YY structure. Hydrogen atoms were added to the models automatically, using the GROMACS tool pdb2gmx. For the imposition of periodic boundary conditions, protein models were centered in a rhombic dodecahedral box with a minimum distance to the edge of 10 Å. All simulations were performed in 0.15 M NaCl and with a zero net charge of the system. Systems were energy minimized using the steepest descent method until the maximum force was <1,000.0 kJ/mol/nm. Equilibration was then conducted in two steps, with positional restraints applied to all nonhydrogen protein atoms (force constant of 1,000 kJ/mol/nm2). The first step used an NVT ensemble for 100 ps, using a modified Berendsen thermostat (35) with a coupling constant of 0.1 ps to bring the temperature of the system to 300 K. The protein and solvent were coupled separately. All bond lengths were constrained using the P-LINCS algorithm (36), allowing an integration time step of 2 fs. The second phase of equilibration applied an NPT ensemble for 100 ps, using the same temperature coupling conditions and the Parrinello-Rahman barostat (37) applied isotropically to maintain the pressure of the system at 105 Pa. Unrestrained production trajectories were then generated for 50 ns, using the same NPT ensemble conditions. Three replicates of each simulation were generated using different sets of random initial velocities, consistent with a Boltzmann distribution, to start the NVT equilibration step of each replicate. Coordinates and energies were saved every 10 ps for analysis, which was performed using GROMACS tools.
Kinase activity assays in cells.
The cell-based kinase activity assays were carried out as described before (17, 19). The expression constructs for FLAG-tagged LAT and full-length Lck have been described previously (19). Kidney epithelial 293T cells were transiently transfected in 24-well plates by using Lipofectamine and Plus reagents (Invitrogen) in accordance with the manufacturer's instructions. Cells were lysed by resuspension in 2× concentrated SDS sample buffer (100 μl per well of 293T cells). Cellular debris was removed by ultracentrifugation (30 min, 386,000 × g). Supernatants were analyzed by immunoblotting with an antibody against phosphotyrosine (monoclonal antibody 4G10; Upstate Biotechnology). The expression levels of LAT-FLAG, ZAP-70, and Lck were assessed by probing with respective antibodies, which have been described previously (19).
Protein structure accession number.
Structural coordinates and structure factors of ZAP-70-YY have been deposited in the RCSB Protein Data Bank (PDB) with the PDB code of 4K2R.
RESULTS AND DISCUSSION
Crystal structure of full-length ZAP-70 with the wild-type SH2-kinase linker sequence.
An essentially full-length ZAP-70 construct (ZAP-70-YY) (residues 1 to 606; lacking only 14 residues at the very C terminus) with Tyr 315 and Tyr 319 both intact was crystallized in complex with the poorly hydrolyzable ATP analog AMP-PNP, and X-ray diffraction data were measured to a 3.0-Å resolution. In order to facilitate expression and purification of the unphosphorylated form of the protein, we introduced a mutation (Asp 461 to asparagine) that inactivates the kinase domain, as was done to obtain the ZAP-70-FF protein. The structure was determined by molecular replacement, using the ZAP-70-FF model without the SH2-kinase linker (residues 310 to 327) as the search model. The electron density for residues 303 to 327 in the SH2-kinase linker was clearly resolved in electron density maps calculated using crystallographic data for the ZAP-70-YY construct (Fig. 2A). This allowed modeling of the backbone and side chains for this segment of the structure. As in the ZAP-70-FF model, two regions of the molecule appeared to be disordered and were not modeled. These were the N-terminal portion of the SH2-kinase linker (residues 258 to 302) and a portion of the activation loop of the kinase domain (residues 484 to 500).
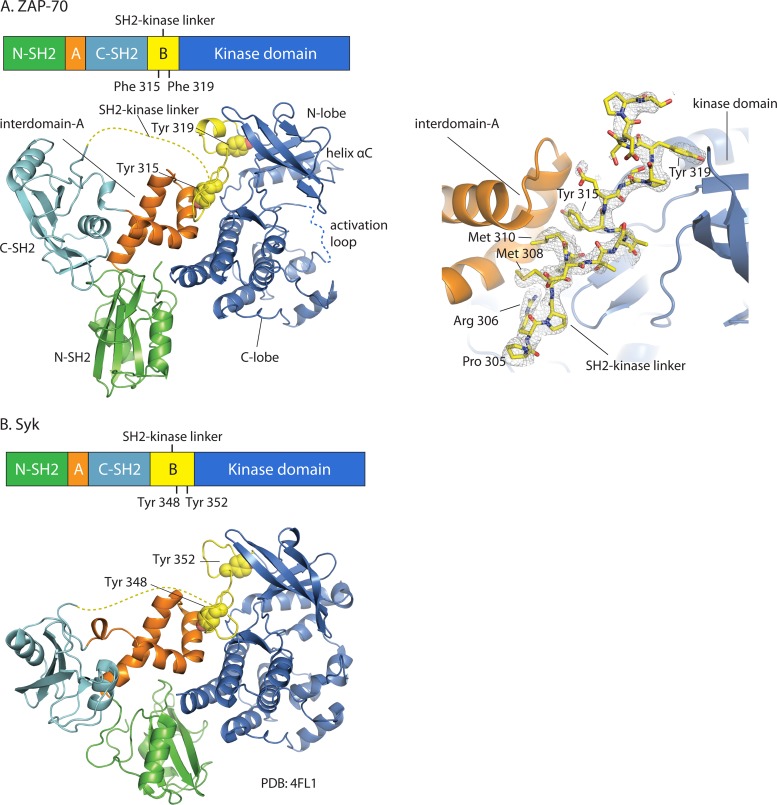
Fig 2.
Overall structural organization of autoinhibited ZAP-70 and Syk. Domain organizations and crystal structures are shown for ZAP-70 (this work) (A) and Syk (PDB code 4FL2) (B). The two crystal structures are superimposed around the C lobes of the kinase domains. Disordered regions are depicted as dotted lines. The side chains of Tyr 315 and Tyr 319 in ZAP-70 and Tyr 348 and Tyr 352 in Syk are shown as spheres. A simulated-annealing composite omit map (A, right panel) (residues 305 to 323, contoured at 1.2 σ) is shown for the SH2-kinase linker of ZAP-70 (gray mesh representation).
Overall comparison of crystallographic models for the ZAP-70-YY and ZAP-70-FF constructs.
We superimposed our ZAP-70-YY model and the ZAP-70-FF model, using the C lobe of the kinase domain as a reference. The ZAP-70-YY model is essentially the same as that reported previously for the ZAP-70-FF construct, with a root mean square (RMS) deviation of 0.4 Å in the positions of C-α atoms over 516 residues (excluding 25 and 18 residues in the SH2-kinase linker of the ZAP-70-YY and ZAP-70-FF models, respectively). The tandem-SH2 module is docked onto the distal surface of the catalytic kinase domain to form a butterfly-shaped structure, with the phosphotyrosine binding sites facing away from the catalytic site (Fig. 2A). The SH2-kinase linker is packed between the kinase domain and the interdomain A coiled coil to form an interface which is quite similar to the linker-kinase sandwich interface described earlier for the ZAP-70-FF construct. The ZAP-70-YY model corresponds to an inactive conformation of ZAP-70. As in the ZAP-70-FF model reported earlier, the conformation of the kinase domain is very similar to that of autoinhibited forms of the Src family kinases Hck and c-Src (38, 39).
The most striking difference between the crystallographic models for the ZAP-70-YY and ZAP-70-FF constructs concerns the conformation of the ordered portion of the SH2-kinase linker. Tyr/Phe 315 and Tyr/Phe 319 in the SH2-kinase linker are located at different positions in the two models. In the original ZAP-70-FF model, Phe 315 and Phe 319 pack closely with each other and with other hydrophobic residues in the interdomain A coiled-coil region and the hinge region of the kinase domain to form a compact hydrophobic core, the linker-kinase sandwich. In contrast, in the ZAP-70-YY model, only Tyr 315 is located in the linker-kinase sandwich. Tyr 315 is in the position occupied by Phe 319 in the previous model. Another hydrophobic residue in the SH2-kinase linker, Met 310, occupies the position where Phe 315 was modeled previously. The net result of these changes is that the structure of the linker-kinase sandwich is virtually unchanged, except that a tyrosine has been replaced by a methionine, with small adjustments in the protein backbone to accommodate this structural difference.
As noted earlier, the new crystallographic model for the ZAP-70-YY construct is similar to that reported very recently for Syk (Fig. 2B) (25). We superimposed our ZAP-70-YY model and the full-length structure of Syk, using the C lobe of the kinase domain as a reference. Notably, most of the interdomain A segment (residues 119 to 155 in ZAP-70 and residues 124 to 160 in Syk) and the aligned region in the SH2-kinase linker (residues 308 to 321 in ZAP-70 and residues 341 to 354 in Syk), including the two conserved tyrosine residues (Tyr 315 and Tyr 319 in ZAP-70 and Tyr 348 and Tyr 352 in Syk), are closely aligned in the superimposed structures, with RMS deviations of 0.6 Å and 0.5 Å, respectively, in the positions of C-α atoms. In contrast, the tandem-SH2 domains of ZAP-70 and Syk are less closely aligned.
Tyr 319 is in an entirely new position in the new ZAP-70 model compared to our original one. The C-terminal part of the SH2-kinase linker in the ZAP-70-YY model forms a short helix and positions the side chain of Tyr 319 into the N lobe of the kinase domain, where it packs against the side chains of residues in helix αC, a key regulatory element in the kinase domain (Fig. 3). The contacts between the C-terminal end of the SH2-kinase linker (residues 318 to 327) and the kinase segment, including those made by Tyr 319, bury a surface area of ∼823 Å2. The formation of the new interface between the SH2-kinase linker and the N lobe of the kinase domain suggests that the SH2-kinase linker might be capable of stabilizing the inactive conformation of the kinase domain on its own, without necessarily requiring the tandem-SH2 module.
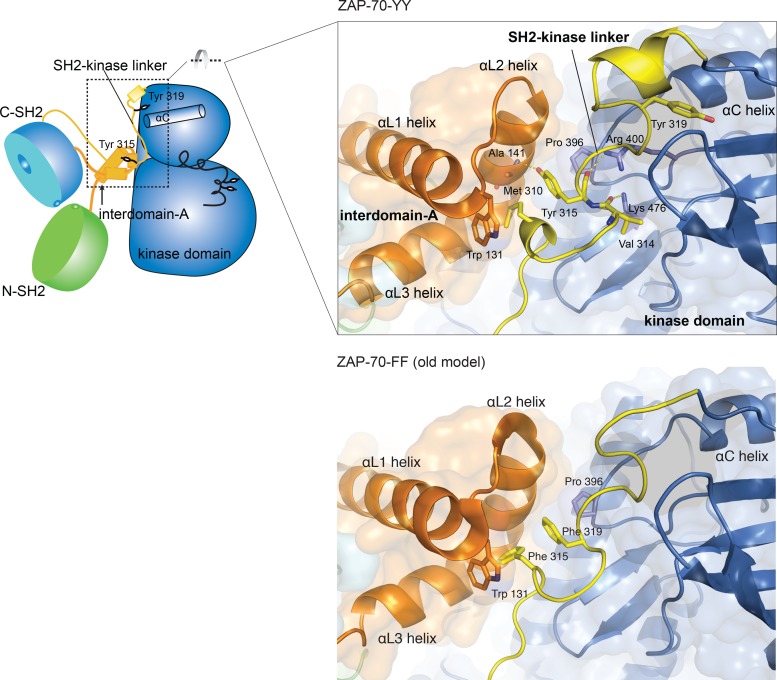
Fig 3.
Comparison of SH2-kinase linkers in the new, ZAP-70-YY model (top) and the original, ZAP-70-FF model (bottom).
Hydrophobic interactions and hydrogen bonds stabilize the linker-kinase sandwich.
The linker-kinase sandwich interface in the ZAP-70-YY model buries ∼1,111 Å2, ∼1,044 Å2, and ∼806 Å2 of surface area on the kinase domain, interdomain A, and the SH2-kinase linker, respectively. The hydrophobic side chains of Met 310 and Tyr 315 from the SH2-kinase linker and of Trp 131 from interdomain A are packed closely at the center of the linker-kinase sandwich. Pro 396 from the αC-β4 loop of the kinase domain packs against the side chain of Tyr 315 (Fig. 3).
The linker-kinase sandwich is further stabilized by a number of hydrogen bonds. The side chains of Arg 400 and Lys 476 in the kinase domain form hydrogen bonds with the backbone carbonyl oxygens of Tyr 315 and Val 314, respectively. The hydroxyl oxygen of the Tyr 315 side chain forms a hydrogen bond with the carbonyl oxygen of Ala 141 in interdomain A (Fig. 3). Despite the presence of these hydrogen bonds, the interior of the linker-kinase sandwich is generally hydrophobic, so phosphorylation of Tyr 315 would disrupt this assembly.
The linker-kinase sandwich described here for the ZAP-70-YY model is similar in its general organization to that described originally for the ZAP-70-FF model, but the changes in position of Tyr 315 and Tyr 319 lead to differences in the detailed nature of the hydrophobic contacts (Fig. 3). In the original ZAP-70-FF model, a set of perpendicular aromatic-aromatic interactions between Phe/Tyr 315 and Phe/Tyr 319 in the SH2-kinase linker and Trp 131 in the interdomain A coiled-coil region form the heart of this assembly (Fig. 3). In the ZAP-70-YY model, Met 310 replaces Phe/Tyr 315 and Phe/Tyr 315 moves to the position where Phe/Tyr 319 is in the ZAP-70-FF model. As a consequence of this replacement, Tyr 319 in the ZAP-70-YY model is positioned ∼12 Å away, and its side chain is inserted into the N lobe of the kinase domain.
Intramolecular interactions between the SH2-kinase linker and the kinase domain stabilize the inactive conformation of the kinase domain.
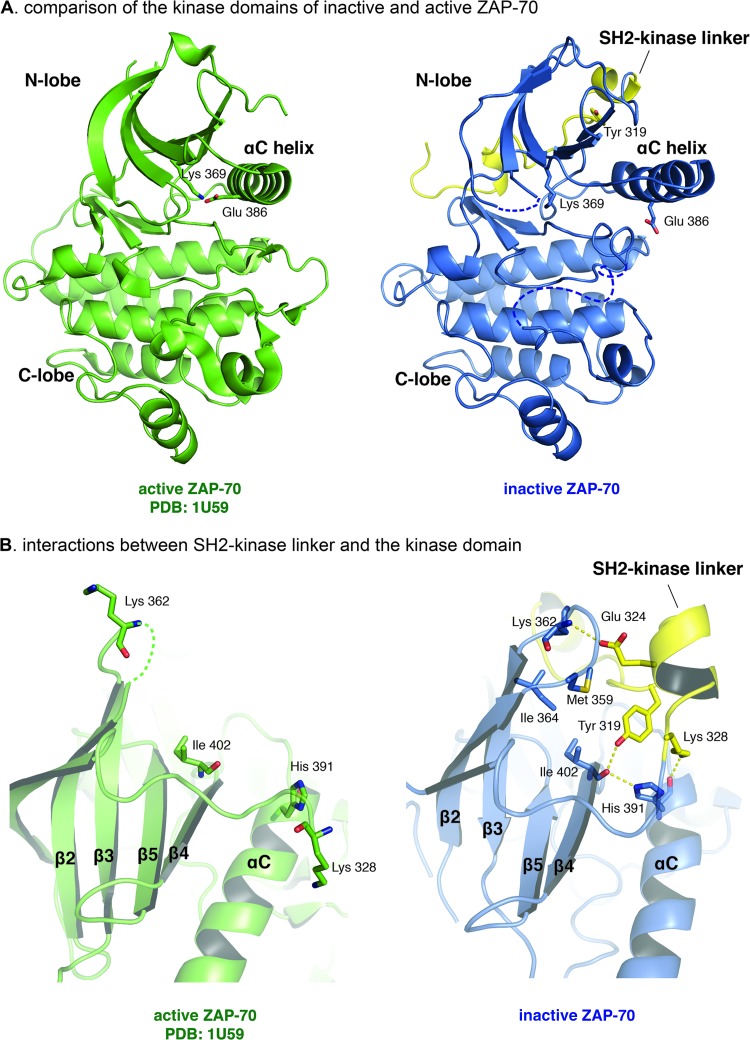
The conformation of the kinase domain in the ZAP-70-YY model is essentially the same as that described earlier for the ZAP-70-FF model. This inactive conformation of the kinase domain in the ZAP-70-YY model appears to be stabilized by intramolecular interactions between the SH2-kinase linker and the N lobe of the kinase domain (Fig. 4). Residues Pro 322 to Lys 326 in the SH2-kinase linker adopt a helical conformation and position the side chain of Tyr 319 into the space between helix αC and the main body of the N lobe of the kinase domain. The carbonyl oxygen of Ile 402, which is located at the beginning of the β4 strand, forms hydrogen bonds with the side chains of Tyr 319, located in the SH2-kinase linker, and His 391, which is located at the end of the αC helix. The side chains of Glu 324 and Lys 328, which are in the SH2-kinase linker, form additional hydrogen bonds with the carbonyl oxygen of Lys 362 and the carbonyl oxygen of His 391, in the N lobe of the kinase domain, respectively. As a consequence of this hydrogen bond network, the position of the αC helix appears to be stabilized in an inactive conformation.
Fig 4.
Interactions between the SH2-kinase linker and the kinase domain stabilize the inactive conformation of the kinase domain. (A) Comparison of the kinase domains of inactive and active ZAP-70. (B) Interactions between the SH2-kinase linker and the kinase domain. (Left) Active ZAP-70 kinase domain (PDB code 1U59). (Right) Inactive ZAP-70 kinase domain with the SH2-kinase linker.
The hydrogen bond network that holds helix αC in place is further stabilized by a hydrophobic cluster formed by Tyr 319 and Pro 318 in the SH2-kinase linker and Met 359, Ile 402, and Ile 364 in the kinase domain. If the SH2-kinase linker is released from the body of the kinase domain, as we expect would happen upon phosphorylation of Tyr 315 and Tyr 319, both the hydrogen bond network and the hydrophobic cluster will be dismantled. The consequences of this dismantling can be understood by examining the structure of the active conformation of the ZAP-70 kinase domain, in which the SH2-kinase linker is absent (40). Loss of the interactions with the SH2-kinase linker is correlated with a rotation of the side chain of His 391 in the αC helix (Fig. 4). This rotation releases restraints on the movement of the αC helix, enabling the kinase to switch to the active conformation.
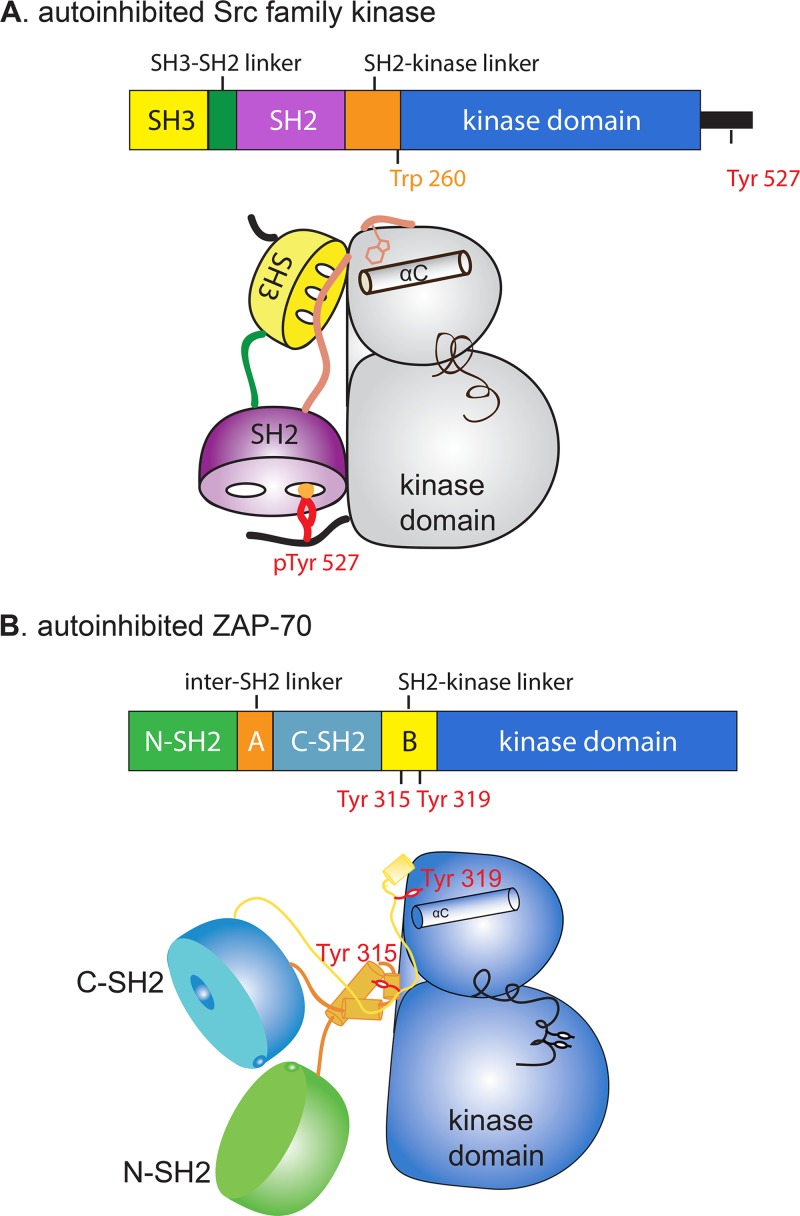
The mechanism by which the SH2-kinase linker inhibits ZAP-70, as described above, bears some resemblance to certain aspects of the autoinhibition of Src family kinases (Fig. 5). In the Src family kinases, the side chain of Trp 260 (chicken c-Src numbering) is inserted between the αC helix and the main body of the kinase domain, thereby stabilizing the inactive conformation. In ZAP-70, the residue corresponding to Trp 260 in c-Src is Leu 330, but this residue does not appear to be critical for the suppression of kinase activity (18). Indeed, as we now show, it is the side chain of Tyr 319 in the SH2-kinase linker of ZAP-70 that plays a role analogous to that of Trp 260 in c-Src.
Fig 5.
Cartoon representations of an autoinhibited Src family kinase and autoinhibited ZAP-70.
To summarize, our crystallographic model of the ZAP-70-YY construct reveals a direct role in autoinhibition for the SH2-kinase linker, distinct from the clamping action of the linker-kinase sandwich segment on the hinge region of the kinase domain. On the basis of the ZAP-70-FF model, we had proposed that the formation of the linker-kinase sandwich results in interactions that reduce the flexibility of the kinase domain, thereby reducing activity. The overall structure of the linker-kinase sandwich and the nature of the interface formed with the hinge region of the kinase domain are essentially unchanged in our new structure, although the positions of Tyr 315 and Tyr 319 have been altered.
The revised model for the SH2-kinase linker is appropriate for the ZAP-70-FF variant as well.
We replaced the SH2-kinase linker (residues 310 to 327) in the original ZAP-70-FF model with the structure of the corresponding segment from the new ZAP-70-YY model, replacing Tyr 315 and Tyr 319 with phenylalanine. Ten rounds of restrained refinement were done using REFMAC (30) against crystallographic data for the ZAP-70-FF construct. After the refinement, the Rfree values were 28.0% and 26.7% for the original and new ZAP-70-FF models, respectively (the corresponding R values are 20.4% and 19.5%, respectively). The resulting electron density maps suggest that the model for the SH2-kinase linker derived from the new analysis of the ZAP-70-YY data is consistent with the crystallographic data for the ZAP-70-FF construct, suggesting that the detailed conformation of the SH2-kinase linker was not modeled correctly in the original analysis. This is also consistent with a new report on Syk which shows that a Syk construct corresponding to ZAP-70-FF adopts a structure similar to that of ZAP-70-YY in the SH2-kinase linker region (25).
The original crystallographic analysis of the ZAP-70-FF construct and the analysis of the new structure of the ZAP-70-YY construct were both done at moderate resolutions (2.6 Å and 3.0 Å, respectively). Although our new model for the SH2-kinase linker appears to be a better fit to the crystallographic data in both cases, the resolution of the X-ray data, particularly for our new structure, does not permit completely reliable positioning of side chains for the SH2-kinase linker. We therefore turned to molecular dynamic simulations to assess which of the two models, the original or the new one, is more stable in terms of the interactions made by the SH2-kinase linker.
We assessed the stability of the SH2-kinase linker in the three models by generating three sets of molecular dynamic trajectories. All sets of trajectories were initiated using models for ZAP-70 derived from the original model of ZAP-70-FF that were identical except for the conformation of the SH2-kinase linker. In the first set of trajectories, the conformation of the linker in the initial model was taken from the original model (the ZAP-70-FF model), whereas in the second set the structure of the SH2-kinase linker in the initial model was taken from the model for ZAP-70-YY (the ZAP-70-YY model). In the third set, the initial model was identical to the second initial model, except that Tyr 315 and Tyr 319 were replaced by phenylalanine residues (the ZAP-70-FF-new model). A 20-residue flexible linker composed of repeated Gly-Ser-Gly motifs was used to connect the tandem-SH2 module and the N-terminal part of the crystallographic models, instead of the SH2-kinase linker. The actual linker connecting the tandem-SH2 unit to the crystallographically resolved part of the SH2-kinase linker is longer (48 residues) and is expected to be flexible. The shorter Gly-Ser linker was used in the model for computational expediency and was not expected to affect the conformation of the ordered portion of the SH2-kinase linker. The activation loop of ZAP-70, which is partially disordered in the crystal structures, was modeled based on the crystal structure of c-Src (PDB accession no. 2SRC).
For each of the three starting models, we generated three independent molecular dynamic trajectories, each lasting 50 ns (see Materials and Methods). The C lobe of the kinase domain is the least flexible part of the ZAP-70 structure. During the course of our simulations, the RMS deviation of C-α atom positions in the C lobe, with respect to the initial model, remained less than 1.0 Å for all trajectories (Fig. 6B). These values are within the range seen for stable domains in molecular dynamic trajectories over comparable time scales.
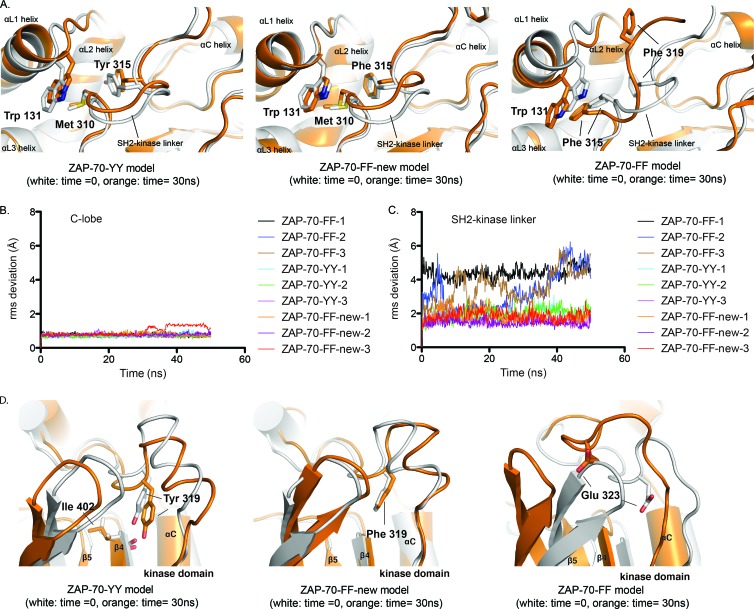
Fig 6.
The conformation of the SH2-kinase linker in the ZAP-70-YY model is energetically favorable. (A) Overlays of instantaneous structures of the SH2-kinase linker at ∼30 ns from molecular dynamic trajectories obtained using the C lobe of the kinase domain as a reference for the alignment. (Left) ZAP-70-YY model. (Middle) ZAP-70-FF-new model. (Right) ZAP-70-FF model. (B) RMS deviations for C-α atoms in the C lobe of the kinase domain with respect to the initial positions. (C) RMS deviations for C-α atoms in the SH2-kinase linker with respect to the initial positions, using the C lobe of the kinase domain as a reference for the alignment. (D) Overlays of instantaneous structures of the N lobes of the kinase domains at ∼30 ns from molecular dynamic trajectories obtained using the C lobe of the kinase domain as a reference for the alignment. (Left) ZAP-70-YY model. (Middle) ZAP-70-FF-new model. (Right) ZAP-70-FF model.
The two sets of trajectories with the new SH2-kinase linker conformation (ZAP-70-YY and ZAP-70-FF-new) showed that the conformation of the SH2-kinase linker was stable over the length of the simulation (Fig. 6A, left and middle panels). Using the C lobe of the kinase domain as a frame of reference, the RMS deviation of the C-α atom positions in the SH2-kinase linker (residues 306 to 316, a span of residues that is common to both models) oscillated around 1.9 Å, 2.0 Å, and 1.6 Å for the three trajectories with the initial model of ZAP-70-YY and around 1.8 Å, 1.5 Å, and 1.9 Å for the three trajectories with the initial model of ZAP-70-FF-new (Fig. 6C).
In contrast, the conformation of the SH2-kinase linker was unstable in trajectories initiated from the original model of ZAP-70-FF. In all three trajectories, the linker-kinase sandwich was partially or completely dismantled, with either Tyr 315 or Tyr 319 (both modeled as phenylalanine) popping out of the hydrophobic cluster at the heart of the interface (Fig. 6A, right panel). Using the C lobe of the kinase domain as a frame of reference, the RMS deviation of C-α atom positions in the SH2-kinase linker (residues 311 to 320) over the length of the simulation increased either abruptly (ZAP-70-FF-1) or gradually (ZAP-70-FF-2 and ZAP-70-FF-3) to values larger than 5 Å (Fig. 6C). Notably, the original model positioned the side chain of Glu 323 near helix αC, a position occupied instead by Tyr/Phe 319 in the ZAP-70-YY and ZAP-70-FF-new models. While the position of Tyr/Phe 319 near helix αC was stable in the set of trajectories initiated from models of ZAP-70-YY and ZAP-70-FF-new (Fig. 6D, left and middle panels), in the set of trajectories with the initial model of ZAP-70-FF, Glu 323 underwent a large displacement away from its initial position (Fig. 6D, right panels).
In conclusion, the molecular dynamic trajectories were consistent with the new models (ZAP-70-YY and ZAP-70-FF-new) for the SH2-kinase linker being energetically favorable in the autoinhibited conformation of ZAP-70.
Autoinhibition of ZAP-70 is fully released upon phosphorylation by Src family kinases.
The new model for the SH2-kinase linker suggests that release of autoinhibition requires both the dismantling of the linker-kinase sandwich and the release of the interactions between the SH2-kinase linker (including Tyr 319) and the helix αC region of the kinase domain. These two sets of interactions appear to be relatively uncoupled in structural terms, indicating that disruption of the linker-kinase sandwich might be insufficient to completely activate the kinase.
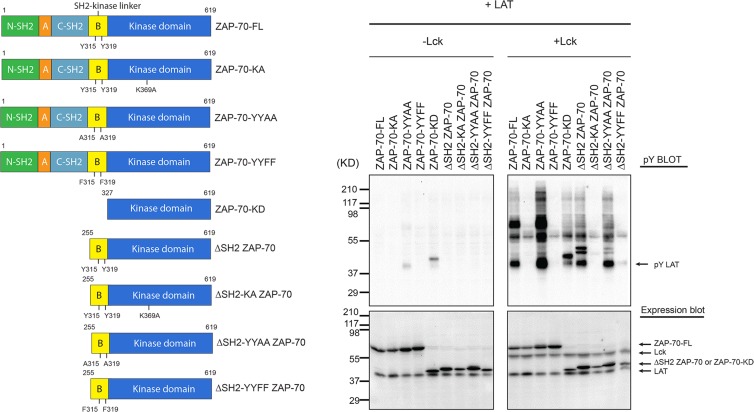
In order to test this hypothesis, we first measured the effect of deleting the tandem-SH2 module upon the ability of ZAP-70 to phosphorylate LAT in cells (Fig. 7). To do this, we used a system in which HEK 293T cells were transiently transfected with different constructs of ZAP-70 and LAT-FLAG, with or without cotransfection of Lck, as described previously (19). The data showed that, as observed previously, robust phosphorylation of full-length ZAP-70 required cotransfection with Lck. Deletion of the tandem-SH2 module of ZAP-70, leaving the full-length SH2-kinase linker and the kinase domain, resulted in a construct (ΔSH2 ZAP-70; residues 255 to 619) that also required the presence of Lck for LAT phosphorylation. A similar construct in which Tyr 315 and Tyr 319 were mutated to phenylalanine (ΔSH2-YYFF ZAP-70) showed no activity toward LAT, even in the presence of Lck (Fig. 7). Two catalytically inactive constructs (ZAP-70-KA and ΔSH2-KA ZAP-70; “KA” indicates the K369A mutation) were used as negative controls in the cell-based experiments. These data suggest that phosphorylation by Lck of Tyr 315 and Tyr 319 in the SH2-kinase linker is necessary for the activation of ZAP-70. The data also indicate that the SH2-kinase linker is able to suppress the kinase activity of ZAP-70 even in the absence of the tandem-SH2 module.
Fig 7.
In vivo kinase activity of ZAP-70 in HEK 293T cells. (Left) Domain architectures of various ZAP-70 truncation constructs. (Right) HEK 293 cells were transiently transfected with various constructs of ZAP-70 and LAT-FLAG together, with or without Lck, as indicated at the top. Cells were lysed in 2× SDS-PAGE sample buffer and analyzed by immunoblotting with an antibody against phosphotyrosine (monoclonal antibody 4G10). The expression levels of LAT-FLAG, ZAP-70, and Lck were assessed by probing with specific antibodies.
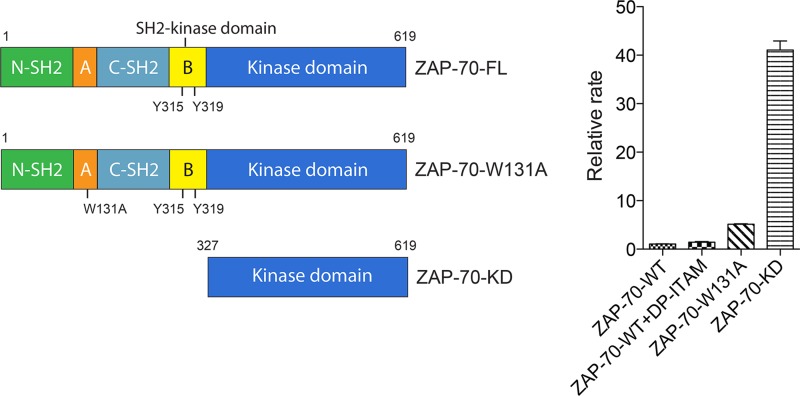
We also tested the catalytic activity of ZAP-70 in vitro, using two different readouts of activity. In the first set of experiments, we used a heterologous peptide substrate (poly-Glu/Tyr) and a coupled-enzyme assay that measures ATP consumption (18). The purpose of this set of experiments was to compare the activity of full-length ZAP-70 to that of the isolated kinase domain, because the activity of the latter construct was not measured in our previous studies. The constructs tested included full-length wild-type ZAP-70 (ZAP-70-FL), the construct with the W131A mutation in the context of ZAP-70-FL (ZAP-70-W131A), and the kinase domain alone (ZAP-70-KD; residues 327 to 619) (Fig. 8, left panel). The W131A mutant was chosen for study because its mutation disrupts the linker-kinase sandwich (18). Each of these constructs was expressed using a recombinant baculovirus system and purified for measurement of kinase activity.
Fig 8.
In vitro catalytic activity of ZAP-70 determined by coupled-kinase assay. (Left) Domain architectures of various ZAP-70 truncation constructs. (Right) Relative rates of phosphorylation of LAT by various ZAP-70 constructs.
In order to estimate the relative values of the catalytic efficiencies of the different constructs, we measured the rates of reaction for fixed tyrosine kinase and substrate concentrations (2 μM ZAP-70 and 2 mg/ml of the poly-Glu/Tyr substrate peptide). In agreement with our previous results, when autoinhibited full-length ZAP-70 was mixed with excess doubly phosphorylated ITAM peptide (5 μM), the in vitro catalytic activity was increased by a factor of 1.4 ± 0.1 compared to that of full-length ZAP-70 without the doubly phosphorylated ITAM peptide (Fig. 8, right panel). Introduction of the W131A mutation increased the catalytic activity by a factor of 5.2 ± 0.1 (Fig. 8, right panel). Notably, the isolated kinase domain (ZAP-70-KD), in which inhibition by the SH2-kinase linker was completely absent, exhibited the highest level of kinase activity, with the reaction velocity increased by a factor of 41.1 ± 1.6 compared to that of full-length ZAP-70 (Fig. 8, right panel), suggesting that the isolated kinase domain was not inhibited in its activity. Trp 131 is located at the heart of the linker-kinase sandwich (Fig. 3), and the moderate level of activation seen for the W131A mutant demonstrates that disruption of the linker-kinase sandwich is insufficient for complete release of autoinhibition.
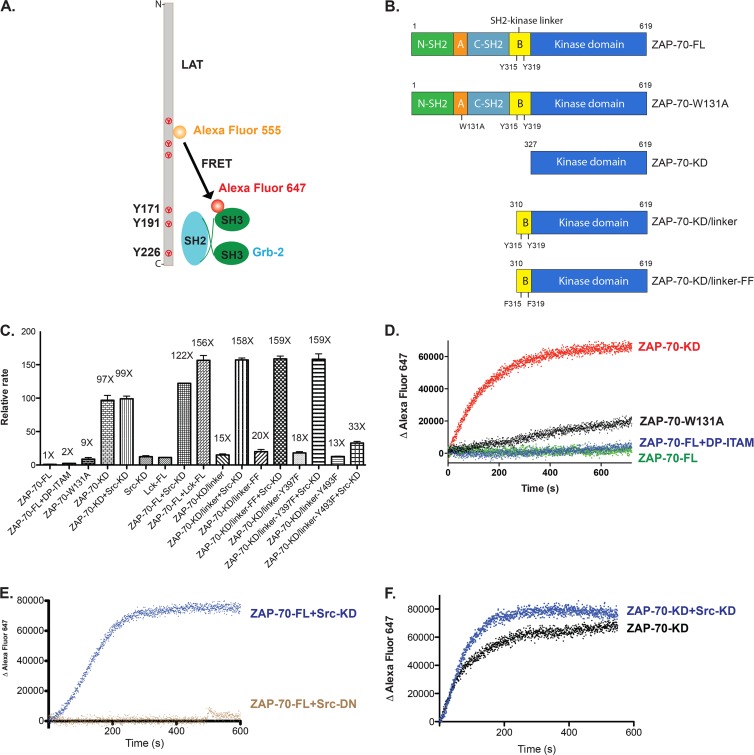
In order to determine the catalytic activity of ZAP-70 toward its physiological substrate, LAT, we set up an in vitro FRET-based assay. Briefly, two fluorophores, Alexa Fluor 555 and Alexa Fluor 647, were used to label LAT and Grb2 (growth factor receptor-bound protein 2), respectively, through thiol-maleimide cross-linking (Fig. 9A). The intracellular domain (residues 30 to 233) of LAT is intrinsically disordered and contains six tyrosine residues that are phosphorylated by ZAP-70 (41). Grb2 is composed of two SH3 domains flanking one SH2 domain, and it binds to phosphorylated tyrosines at positions 171, 191, and 226 in LAT, with a nanomolar to submicromolar binding affinity for each site (42). Recruitment of Grb2 to phosphorylated LAT brings the donor (Alexa Fluor 555) and acceptor (Alexa Fluor 647) fluorophores into close proximity, and the increase in acceptor fluorescence intensity due to FRET is assumed to reflect the amount of phosphorylated LAT (Fig. 9A). Due to the presence of multiple phosphorylation sites in LAT, with various distances to the fluorescent dye, the relationship between the observed FRET value and the extent of phosphorylation may not be strictly proportional. We have not investigated this relationship further and have simply used the rate of increased acceptor fluorescence intensity as an indicator of the extent of reaction. The final concentration of all proteins in the FRET assay (tyrosine kinases, LAT, and Grb2) was 1 μM, unless otherwise noted.
Fig 9.
Phosphorylation of ZAP-70 by Src family kinases fully activates ZAP-70 in vitro. (A) In vitro FRET-based approach to measure the catalytic activity of ZAP-70 for its physiological substrate, LAT. (B) Domain architectures of various ZAP-70 truncation constructs. (C) Relative rates of phosphorylation of LAT by ZAP-70 or Src family kinases. Src-KD, isolated c-Src kinase domain; Lck-FL, full-length Lck; DP-ITAM, doubly phosphorylated ITAM peptide. (D) Time course of phosphorylation of LAT as measured by the recruitment of Grb2. Red, isolated ZAP-70 kinase domain; black, ZAP-70-W131A; blue, full-length ZAP-70 bound to an excess amount of doubly phosphorylated ITAM peptide; green, full-length ZAP-70. (E) Time course of phosphorylation of LAT as measured by the recruitment of Grb2. Blue, full-length ZAP-70 plus Src kinase domain; brown, full-length ZAP-70 plus Src kinase domain with D386N mutation. (F) Time course of phosphorylation of LAT as measured by the recruitment of Grb2. Black, isolated ZAP-70 kinase domain; blue, isolated ZAP-70 kinase domain plus Src kinase domain.
We found, using this FRET-based LAT phosphorylation assay, that full-length ZAP-70 (ZAP-70-FL) exhibits very low kinase activity toward LAT. The kinase activity increased approximately 2-fold when excess doubly phosphorylated ITAM peptide was added (Fig. 9B to D). The W131A mutation in the context of full-length ZAP-70 increased the kinase activity toward LAT by a factor of ∼9 (Fig. 9B to D). ZAP-70-KD showed a robust kinase activity in this assay, with a reaction rate ∼100-fold higher than that observed for full-length ZAP-70 (Fig. 9B to D).
Phosphorylation of Tyr 315 and Tyr 319 of ZAP-70 by Src family kinases is expected to release autoinhibition by the SH2-kinase linker and would therefore be expected to fully activate ZAP-70. We found that when full-length autoinhibited ZAP-70 was phosphorylated by c-Src or Lck, the activity of full-length ZAP-70 toward LAT was increased to a level comparable to that for the isolated kinase domain. Src family kinases by themselves are inefficient at phosphorylating LAT when present at the same concentration (1 μM) (Fig. 9C). Catalytically inactive c-Src (D386N) did not increase the activity of full-length ZAP-70 (Fig. 9E), indicating that phosphorylation of ZAP-70 by Src family kinases is responsible for the activation of ZAP-70.
In our in vitro biochemical assays, both Src and Lck fully activated ZAP-70. Because the Src and Lck kinase domains are similar (68% identity, based on 254 residues) and the expression level of the recombinant c-Src kinase domain in E. coli is much higher than that of the Lck kinase domain (15 to 20 mg/liter versus 0.1 mg/liter of purified protein) (43), we used the c-Src kinase domain for most of our in vitro experiments.
We expected that the SH2-kinase linker would inhibit the kinase domain even if the SH2 domains were removed. To test that hypothesis, a ZAP-70 construct lacking the tandem-SH2 module and the disordered portion of the SH2-kinase linker (ZAP-70-KD/linker; residues 310 to 619) was expressed and purified (Fig. 9B). The activity of this construct was similar to that of the W131A mutant (that is, it was ∼10-fold more active than unphosphorylated full-length ZAP-70), but its activity was ∼10-fold lower than that of the isolated kinase domain (Fig. 9C).
Several tyrosine residues in ZAP-70 have been reported to be phosphorylated by Src family kinases, including Tyr 315, Tyr 319, and Tyr 493. In order to determine which tyrosine residues were phosphorylated in our in vitro phosphorylation assay, we digested the phosphorylated ZAP-70 protein samples with trypsin and analyzed the resulting peptides by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Four sites of robust phosphorylation were identified in the SH2-kinase linker and the kinase domain: Tyr 315, Tyr 319, Tyr 397, and Tyr 493. Tyr 397 is located in the loop connecting the αC helix and the β4 strand, and Tyr 493 is located in the activation loop.
A variant of ZAP-70-KD/linker in which Tyr 397 was mutated to phenylalanine was also fully activated by Src family kinase phosphorylation (Fig. 9C). Notably, a variant of ZAP-70-KD/linker in which Tyr 493, in the activation loop, was mutated to phenylalanine could not be activated further by Src family kinase phosphorylation (Fig. 9C). These results suggest that phosphorylation of ZAP-70 on Tyr 493 is necessary for activation when the SH2-kinase linker is present. In contrast, the activity of the isolated kinase domain of ZAP-70 was not increased further upon phosphorylation by c-Src (Fig. 9F). We interpret these results to mean that the isolated kinase domain of ZAP-70 is free to adopt the active conformation, as indicated by the crystal structure of the unphosphorylated kinase domain (40).
We also mutated both Tyr 315 and Tyr 319 to phenylalanine in the ZAP-70-KD/linker construct (ZAP-70-KD/linker-FF) (Fig. 9B), and the resulting construct had a similar level of kinase activity as ZAP-70-KD/linker (Fig. 9C). Notably, both ZAP-70-KD/linker and ZAP-70-KD/linker-FF were fully activated by Src family kinase phosphorylation (Fig. 9C). We presume that once Tyr 493, located in the activation loop of the ZAP-70 kinase domain, is phosphorylated, the inhibition by the SH2-kinase linker is overcome. Note that this construct (ZAP-70-KD/linker) has only the C-terminal portion of the SH2-kinase linker, in contrast to the ΔSH2 ZAP-70 construct used in the cell-based assay shown in Fig. 7. In contrast to the ΔSH2-YYFF ZAP-70 construct, which was inactive in cells even in the presence of Lck, the phosphorylation of the ZAP-70-KD/linker protein by c-Src resulted in recovery of full activity toward LAT (Fig. 9C). This discrepancy may be due to the lack of phosphatases in the in vitro assay. Competition with the dephosphorylation reaction might amplify small differences in the rate of activation loop phosphorylation.
Conclusions.
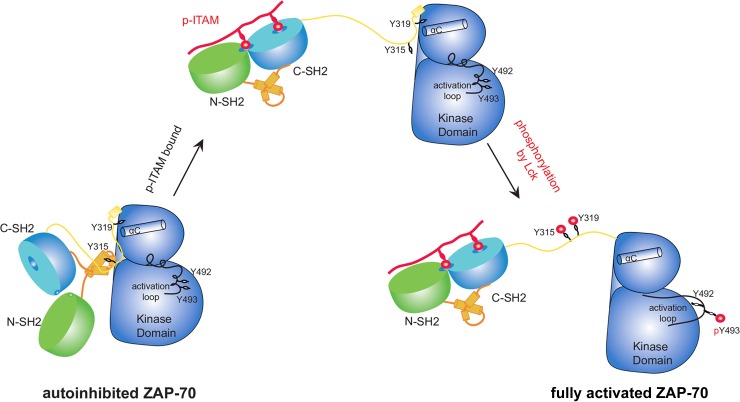
The structure of ZAP-70-YY shows that Tyr 315 and Tyr 319 participate in the formation of two distinct inhibitory clamps on the kinase domain. The engagement of a doubly phosphorylated ITAM peptide by the tandem-SH2 domains of ZAP-70 would not only recruit ZAP-70 to the plasma membrane, where Lck is located, but also induce a substantial conformational change that releases the tandem-SH2 module. This conformational change would facilitate the phosphorylation of Tyr 315 and Tyr 319 by Lck, thereby preventing redocking of the tandem-SH2 module on the kinase domain. We presume that the release of the tandem-SH2 module and the phosphorylation of both Tyr 315 and Tyr 319 within the SH2-kinase linker make the kinase domain more flexible and that this would facilitate phosphorylation of the activation loop. Given the high degree of sequence conservation between ZAP-70 and Syk, a similar autoinhibition mechanism involving the SH2-kinase linker is expected to underlie the regulation of Syk. This has now been confirmed by the structure of the autoinhibited form of full-length Syk, which was published just prior to submission of this report (25).
In summary, our biochemical and structural studies suggest that when ZAP-70 is recruited to doubly phosphorylated ITAMs in the TCR ζ chain, it is not completely activated until it is phosphorylated by Src family kinases such as Lck (Fig. 10). This model is consistent with previously reported observations that binding of ZAP-70 to constitutively phosphorylated TCR-ζ is insufficient for complete ZAP-70 activation in thymocytes (44). The presence of such TCR-ζ-associated inactive forms of ZAP-70 may be important for the priming of rapid TCR signaling after TCR activation.
Fig 10.
Model of ZAP-70 activation by doubly phosphorylated ITAM peptide and Lck phosphorylation.
ACKNOWLEDGMENTS
We thank the staff at beamline 8.3.1 (ALS, Berkeley, CA) for their excellent support and Tony Iavarone at the QB3 mass spectrometry facility for mass spectrometry support. We also thank Qi Wang, Jonathan Winger, and members of the Kuriyan laboratory for many helpful and inspiring discussions.
This work was supported in part by NIH grant PO1 AI091580 to A.W. and J.K. Q.Y. is supported by the Cancer Research Institute-Irvington Institute Fellowship Program.
Footnotes
Published ahead of print 25 March 2013
REFERENCES
- 1. Au-Yeung BB, Deindl S, Hsu LY, Palacios EH, Levin SE, Kuriyan J, Weiss A. 2009. The structure, regulation, and function of ZAP-70. Immunol. Rev. 228:41–57 [DOI] [PubMed] [Google Scholar]
- 2. Sloan-Lancaster J, Presley J, Ellenberg J, Yamazaki T, Lippincott-Schwartz J, Samelson LE. 1998. ZAP-70 association with T cell receptor zeta (TCRzeta): fluorescence imaging of dynamic changes upon cellular stimulation. J. Cell Biol. 143:613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sloan-Lancaster J, Zhang W, Presley J, Williams BL, Abraham RT, Lippincott-Schwartz J, Samelson LE. 1997. Regulation of ZAP-70 intracellular localization: visualization with the green fluorescent protein. J. Exp. Med. 186:1713–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang W, Sloan-Lancaster J, Kitchen J, Trible RP, Samelson LE. 1998. LAT: the ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell 92:83–92 [DOI] [PubMed] [Google Scholar]
- 5. Bubeck Wardenburg J, Fu C, Jackman JK, Flotow H, Wilkinson SE, Williams DH, Johnson R, Kong G, Chan AC, Findell PR. 1996. Phosphorylation of SLP-76 by the ZAP-70 protein-tyrosine kinase is required for T-cell receptor function. J. Biol. Chem. 271:19641–19644 [DOI] [PubMed] [Google Scholar]
- 6. Sakaguchi N, Takahashi T, Hata H, Nomura T, Tagami T, Yamazaki S, Sakihama T, Matsutani T, Negishi I, Nakatsuru S, Sakaguchi S. 2003. Altered thymic T-cell selection due to a mutation of the ZAP-70 gene causes autoimmune arthritis in mice. Nature 426:454–460 [DOI] [PubMed] [Google Scholar]
- 7. Siggs OM, Miosge LA, Yates AL, Kucharska EM, Sheahan D, Brdicka T, Weiss A, Liston A, Goodnow CC. 2007. Opposing functions of the T cell receptor kinase ZAP-70 in immunity and tolerance differentially titrate in response to nucleotide substitutions. Immunity 27:912–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hsu LY, Tan YX, Xiao Z, Malissen M, Weiss A. 2009. A hypomorphic allele of ZAP-70 reveals a distinct thymic threshold for autoimmune disease versus autoimmune reactivity. J. Exp. Med. 206:2527–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van Oers NS, Weiss A. 1995. The Syk/ZAP-70 protein tyrosine kinase connection to antigen receptor signalling processes. Semin. Immunol. 7:227–236 [DOI] [PubMed] [Google Scholar]
- 10. Elder ME, Lin D, Clever J, Chan AC, Hope TJ, Weiss A, Parslow TG. 1994. Human severe combined immunodeficiency due to a defect in ZAP-70, a T cell tyrosine kinase. Science 264:1596–1599 [DOI] [PubMed] [Google Scholar]
- 11. Elder ME, Skoda-Smith S, Kadlecek TA, Wang F, Wu J, Weiss A. 2001. Distinct T cell developmental consequences in humans and mice expressing identical mutations in the DLAARN motif of ZAP-70. J. Immunol. 166:656–661 [DOI] [PubMed] [Google Scholar]
- 12. Matsuda S, Suzuki-Fujimoto T, Minowa A, Ueno H, Katamura K, Koyasu S. 1999. Temperature-sensitive ZAP70 mutants degrading through a proteasome-independent pathway. Restoration of a kinase domain mutant by Cdc37. J. Biol. Chem. 274:34515–34518 [DOI] [PubMed] [Google Scholar]
- 13. Hatada MH, Lu X, Laird ER, Green J, Morgenstern JP, Lou M, Marr CS, Phillips TB, Ram MK, Theriault K, Zoller MJ, Karas JL. 1995. Molecular basis for interaction of the protein tyrosine kinase ZAP-70 with the T-cell receptor. Nature 377:32–38 [DOI] [PubMed] [Google Scholar]
- 14. Futterer K, Wong J, Grucza RA, Chan AC, Waksman G. 1998. Structural basis for Syk tyrosine kinase ubiquity in signal transduction pathways revealed by the crystal structure of its regulatory SH2 domains bound to a dually phosphorylated ITAM peptide. J. Mol. Biol. 281:523–537 [DOI] [PubMed] [Google Scholar]
- 15. Folmer RH, Geschwindner S, Xue Y. 2002. Crystal structure and NMR studies of the apo SH2 domains of ZAP-70: two bikes rather than a tandem. Biochemistry 41:14176–14184 [DOI] [PubMed] [Google Scholar]
- 16. Grucza RA, Futterer K, Chan AC, Waksman G. 1999. Thermodynamic study of the binding of the tandem-SH2 domain of the Syk kinase to a dually phosphorylated ITAM peptide: evidence for two conformers. Biochemistry 38:5024–5033 [DOI] [PubMed] [Google Scholar]
- 17. Deindl S, Kadlecek TA, Brdicka T, Cao X, Weiss A, Kuriyan J. 2007. Structural basis for the inhibition of tyrosine kinase activity of ZAP-70. Cell 129:735–746 [DOI] [PubMed] [Google Scholar]
- 18. Deindl S, Kadlecek TA, Cao X, Kuriyan J, Weiss A. 2009. Stability of an autoinhibitory interface in the structure of the tyrosine kinase ZAP-70 impacts T cell receptor response. Proc. Natl. Acad. Sci. U. S. A. 106:20699–20704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brdicka T, Kadlecek TA, Roose JP, Pastuszak AW, Weiss A. 2005. Intramolecular regulatory switch in ZAP-70: analogy with receptor tyrosine kinases. Mol. Cell. Biol. 25:4924–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gong Q, Jin X, Akk AM, Foger N, White M, Gong G, Bubeck Wardenburg J, Chan AC. 2001. Requirement for tyrosine residues 315 and 319 within zeta chain-associated protein 70 for T cell development. J. Exp. Med. 194:507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Di Bartolo V, Mege D, Germain V, Pelosi M, Dufour E, Michel F, Magistrelli G, Isacchi A, Acuto O. 1999. Tyrosine 319, a newly identified phosphorylation site of ZAP-70, plays a critical role in T cell antigen receptor signaling. J. Biol. Chem. 274:6285–6294 [DOI] [PubMed] [Google Scholar]
- 22. Magnan A, Di Bartolo V, Mura AM, Boyer C, Richelme M, Lin YL, Roure A, Gillet A, Arrieumerlou C, Acuto O, Malissen B, Malissen M. 2001. T cell development and T cell responses in mice with mutations affecting tyrosines 292 or 315 of the ZAP-70 protein tyrosine kinase. J. Exp. Med. 194:491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu J, Zhao Q, Kurosaki T, Weiss A. 1997. The Vav binding site (Y315) in ZAP-70 is critical for antigen receptor-mediated signal transduction. J. Exp. Med. 185:1877–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Williams BL, Irvin BJ, Sutor SL, Chini CC, Yacyshyn E, Bubeck Wardenburg J, Dalton M, Chan AC, Abraham RT. 1999. Phosphorylation of Tyr319 in ZAP-70 is required for T-cell antigen receptor-dependent phospholipase C-gamma1 and Ras activation. EMBO J. 18:1832–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grädler U, Schwarz D, Dresing V, Musil D, Bomke J, Frech M, Greiner H, Jakel S, Rysiok T, Muller-Pompalla D, Wegener A. 2013. Structural and biophysical characterization of the Syk activation switch. J. Mol. Biol. 425:309–333 [DOI] [PubMed] [Google Scholar]
- 26. Guilloteau JP, Fromage N, Ries-Kautt M, Reboul S, Bocquet D, Dubois H, Faucher D, Colonna C, Ducruix A, Becquart J. 1996. Purification, stabilization, and crystallization of a modular protein: Grb2. Proteins 25:112–119 [DOI] [PubMed] [Google Scholar]
- 27. Leslie AGW, Powell HR. 2007. Processing diffraction data with Mosflm. Evolving Methods Macromol. Crystallogr. 245:11 [Google Scholar]
- 28. Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 67:235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Winn MD, Murshudov GN, Papiz MZ. 2003. Macromolecular TLS refinement in REFMAC at moderate resolutions. Methods Enzymol. 374:300–321 [DOI] [PubMed] [Google Scholar]
- 30. Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 53:240–255 [DOI] [PubMed] [Google Scholar]
- 31. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 32. Hess B, Kutzner C, van der Spoel D, Lindahl E. 2008. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4:435–447 [DOI] [PubMed] [Google Scholar]
- 33. Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. 2010. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78:1950–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. 1983. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79:926–935 [Google Scholar]
- 35. Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. 1984. Molecular-dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690 [Google Scholar]
- 36. Hess B. 2008. P-LINCS: a parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 4:116–122 [DOI] [PubMed] [Google Scholar]
- 37. Parrinello M, Rahman A. 1981. Polymorphic transitions in single-crystals—a new molecular-dynamics method. J. Appl. Phys. 52:7182–7190 [Google Scholar]
- 38. Sicheri F, Moarefi I, Kuriyan J. 1997. Crystal structure of the Src family tyrosine kinase Hck. Nature 385:602–609 [DOI] [PubMed] [Google Scholar]
- 39. Xu W, Harrison SC, Eck MJ. 1997. Three-dimensional structure of the tyrosine kinase c-Src. Nature 385:595–602 [DOI] [PubMed] [Google Scholar]
- 40. Jin L, Pluskey S, Petrella EC, Cantin SM, Gorga JC, Rynkiewicz MJ, Pandey P, Strickler JE, Babine RE, Weaver DT, Seidl KJ. 2004. The three-dimensional structure of the ZAP-70 kinase domain in complex with staurosporine: implications for the design of selective inhibitors. J. Biol. Chem. 279:42818–42825 [DOI] [PubMed] [Google Scholar]
- 41. Sangani D, Venien-Bryan C, Harder T. 2009. Phosphotyrosine-dependent in vitro reconstitution of recombinant LAT-nucleated multiprotein signalling complexes on liposomes. Mol. Membr. Biol. 26:159–170 [DOI] [PubMed] [Google Scholar]
- 42. Houtman JC, Higashimoto Y, Dimasi N, Cho S, Yamaguchi H, Bowden B, Regan C, Malchiodi EL, Mariuzza R, Schuck P, Appella E, Samelson LE. 2004. Binding specificity of multiprotein signaling complexes is determined by both cooperative interactions and affinity preferences. Biochemistry 43:4170–4178 [DOI] [PubMed] [Google Scholar]
- 43. Seeliger MA, Nagar B, Frank F, Cao X, Henderson MN, Kuriyan J. 2007. c-Src binds to the cancer drug imatinib with an inactive Abl/c-Kit conformation and a distributed thermodynamic penalty. Structure 15:299–311 [DOI] [PubMed] [Google Scholar]
- 44. van Oers NS, Killeen N, Weiss A. 1996. Lck regulates the tyrosine phosphorylation of the T cell receptor subunits and ZAP-70 in murine thymocytes. J. Exp. Med. 183:1053–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]