Abstract
Arkadia is a RING domain E3 ubiquitin ligase that activates the transforming growth factor β (TGF-β) pathway by inducing degradation of the inhibitor SnoN/Ski. Here we show that Arkadia contains three successive SUMO-interacting motifs (SIMs) that mediate noncovalent interaction with poly-SUMO2. We identify the third SIM (VVDL) of Arkadia to be the most relevant one in this interaction. Furthermore, we provide evidence that Arkadia can function as a SUMO-targeted ubiquitin ligase (STUBL) by ubiquitinating SUMO chains. While the SIMs of Arkadia are not essential for SnoN/Ski degradation in response to TGF-β, we show that they are necessary for the interaction of Arkadia with polysumoylated PML in response to arsenic and its concomitant accumulation into PML nuclear bodies. Moreover, Arkadia depletion leads to accumulation of polysumoylated PML in response to arsenic, highlighting a requirement of Arkadia for arsenic-induced degradation of polysumoylated PML. Interestingly, Arkadia homodimerizes but does not heterodimerize with RNF4, the other STUBL involved in PML degradation, suggesting that these two E3 ligases do not act synergistically but most probably act independently during this process. Altogether, these results identify Arkadia to be a novel STUBL that can trigger degradation of signal-induced polysumoylated proteins.
INTRODUCTION
Ubiquitin ligases are important regulators that modulate signaling pathways. Arkadia is a member of the RING finger ubiquitin ligase superfamily that promotes activation of the transforming growth factor β (TGF-β) signaling pathway. Upon activation of TGF-β signaling, Arkadia binds to phosphorylated Smad2/3 (1–3) and induces degradation of the Smad7 inhibitor (4) and the SnoN/Ski repressors (1, 2, 5), enabling transcription of TGF-β target genes.
E3 ubiquitin ligases catalyze covalent fixation of an ubiquitin protein on the lysine residue of their specific substrate through a sequential enzymatic reaction that involves an E1 activating enzyme and an E2 conjugating enzyme. Multiple rounds of this reaction can also lead to polyubiquitination of the substrate, since ubiquitin contains many lysine residues (Lys residues 11, 29, 33, 48, and 63) that can be targeted for subsequent attachment of ubiquitin. The most well characterized functional consequence of polyubiquitination is the proteasome-dependent degradation of substrates modified by ubiquitin chains linked through lysine 48. Other ubiquitin-like protein modifiers (UBL), such as the small ubiquitin-like modifier (SUMO), can also be conjugated to proteins through a related enzymatic reaction. In most cases, sumoylation occurs on a lysine residue in the consensus modification site Ψ-Lys-X-Glu (where Ψ is hydrophobic residues). In vertebrates, proteins can be sumoylated by SUMO1, SUMO2, or SUMO3, which are three paralogues of SUMO sharing similarity. SUMO2 and SUMO3 are highly related and can also be sumoylated because they contain an internal SUMO consensus modification site that is missing on SUMO1. This results in the generation of poly-SUMO2/3 chains on SUMO-modified substrates (6). Poly-SUMO2/3 chains can nonetheless be terminated by incorporation of SUMO1, and mixed poly-SUMO chains have also been described (7). Sumoylation and polysumoylation are posttranslational modifications induced by a stress signal. While mono- or multisumoylation of many substrates has been widely characterized, it is still unclear which substrates are polysumoylated and what the physiological consequences of this modification are. Polysumoylation is induced in response to heat shock (8) or trivalent arsenic (henceforth referred to as arsenic) treatment (9). In response to heat shock, a large panel of polysumoylated substrates has been identified in mammals (10, 11), with the most well characterized so far being poly(ADP-ribose) polymerase 1 (PARP-1) (12). In contrast, the response to arsenic induces polysumoylation of one specific substrate, the promyelocytic leukemia protein (PML) (9). Indeed, it is well established that arsenic induces accumulation of PML in nuclear bodies, where it undergoes SUMO modification and subsequent ubiquitination and degradation by the proteasome. In acute promyelocytic leukemia (APL), PML is fused to retinoic acid receptor α (RARα), creating an oncogenic fusion protein, PML-RARα, that initiates acute leukemia by impeding differentiation along the myeloid lineage. Remarkably, arsenic also induces sumoylation and subsequent degradation of PML-RARα (13–17), and this leads to clinical remissions (18). Yet, the mechanism by which sumoylated PML is degraded in response to arsenic treatment has remained puzzling for a long time. The identification of RING E3 ubiquitin ligase complexes containing a SUMO-interacting motif (SIM) in the yeast Saccharomyces cerevisiae has led to the hypothesis that SUMO-targeted ubiquitin ligase (STUBL) could specifically induce degradation of SUMO-modified proteins (reviewed in references 19 to 23). In mammals, RNF4 is the only STUBL described so far (24), and it has been shown to play a critical role in the degradation of PML and PML-RARα by specifically recognizing and ubiquitinating arsenic-induced polysumoylated PML (9, 25–28).
In the present work, we report that RNF111/Arkadia also functions as a STUBL involved in the arsenic-induced PML degradation. Notably, we identified three successive SIMs localized in the N-terminal region of Arkadia that display a strong affinity for poly-SUMO2 and show that the third SIM, VVDL, is the most relevant for this binding. We further demonstrate that, in response to arsenic, Arkadia interacts with polysumoylated PML and relocalizes to PML nuclear bodies in a SIM-dependent manner. Finally, depletion of Arkadia impairs the subsequent degradation of polysumoylated PML in nuclear bodies, indicating that it might act in concert with RNF4 in this process. However, since we could not detect any heterodimerization or synergistic effect between Arkadia and RNF4, our results suggest that they may act in parallel and/or sequentially during this process. Altogether, our results not only identify a new inducible mechanism of action for Arkadia but also suggest that it constitutes a new STUBL that degrades polysumoylated substrates.
MATERIALS AND METHODS
Plasmids and constructions.
The following plasmids were previously described. The hemagglutinin (HA)-tagged Smad2 (HA-Smad2) and HA-Smad3 plasmids were gifts from C. Hill, the HA-Smad7 plasmid was a gift from J. Wrana, the HA-SnoN plasmid (29) was a gift from K. Luo, and the HA-Ski, 6Myc-SnoN, and 6Myc-Ski plasmids (30) were gifts from K. Miyazono. The CAGA12-Luc plasmid was described in reference 31, and the Flag-Ark wild type (Flag-Ark-wt; mouse, short isoform) and Flag-Ark-RING* (C931A mutant, mouse, short isoform) plasmids were gifts from S. C. Lin (32). The Flag-RNF4 (rat cDNA) plasmid was a gift from J. J. Palvimo (33). pcDNA3-HA-SUMO2-wt and pcDNA3-HA-SUMO2-K11R were gifts from A. Sharrocks (6) and have been described in reference 34. GFP-PML-IV-wt in an enhanced green fluorescent protein (EGFP) vector (Clontech) and GFP-PML-IV-3KR in pcDNA3 were gifts from David Bazett-Jones (35).
All the Flag-Ark constructs described were cloned in the pBICEP-CMV2 (Sigma) vector, where the Flag tag is a 3× Flag. Flag-Ark-wt and Flag-Ark-RING* were subcloned at NotI/BamHI in pBICEP-CMV2 (Sigma). SIM mutants Flag-Ark-SIM1* (V298A, V300A, and I301A), Flag-Ark-SIM2* (V324A, I326A, and V327A), Flag-Ark-SIM3* (V380A, V381A, and L383A), Flag-Ark-SIM13*, Flag-Ark-SIM23*, and Flag-Ark-SIM123* were generated by site-directed mutagenesis on pBICEP-CMV2 Flag-Ark-wt using a QuikChange kit (Stratagene). Flag-Ark-SIM123*-RING* was generated by subcloning of the EcoRI-EcoRV insert from Flag-Ark-SIM123* into Flag-Ark-RING*. Flag-Ark-1-400 and Flag-Ark-1-400-SIM123* were generated by PCR on Flag-Ark-wt and Flag-Ark-SIM123* and subcloned at NotI-SalI in pBICEP-CMV2. Flag-Ark-280-400 and GST-Ark-280-400 wt or SIM mutants were generated by PCR on Flag-Ark-wt or Flag-Ark-SIM mutants and subcloned at EcoRI/SalI in pBICEP-CMV2 or pGEX4T3 (GE Healthcare). Flag-Ark-400-Cter-RING* was generated by PCR on Flag-Ark-RING* and subcloned at EcoRI/BamHI in pBICEP-CMV2. GST-Ark-665-Cter was cloned at EcoRI/SalI in pGEX4T3 from PCR on Flag-Ark-wt. Amino-terminal SUMO1–glutathione S-transferase (GST) and SUMO3-GST fusions where cloned as NcoI-HindIII fragments into pET16 (Novagen). GST-RNF4 corresponds to the human cDNA cloned in pGEX-2T (GE Healthcare). HA-PML-III was cloned in pSG5 (Stratagene) into the EcoRI site. GFP-PML-IV-wt cloned into the pBabe-puro lentiviral vector (Cell Biolabs) was used for infection.
Cell culture, treatment, and transfection and luciferase assay.
HeLa, HEK293, and HT-1080 cells were cultured in Dulbecco modified Eagle medium with 10% fetal calf serum. Cells were induced at the indicated times with 2 ng/ml TGF-β1 (PreproTech). The HT-1080-GFP-PML stable cell line was generated by infection of the ecotrope HT1080-E14 cell line (a gift from Eugene Kandel, Roswell Park Cancer Institute, NY) (36, 37) with pBabe-puro green fluorescent protein-labeled PML (GFP-PML) retrovirus. Arsenic (As2O3; A1010; Sigma) was resuspended in NaOH (0.1 M), and cell treatment was performed at a final concentration of 1 μM. Transient transfections of plasmids were performed using Lipofectamine 2000 according to the manufacturer's instruction. Small interfering RNA (siRNA) transfection in the HT1080 cell line was performed with the Dharmafect-1 reagent using 50 nM (final concentration) Thermo Scientific Dharmacon siRNA according to the manufacturer's instruction. The following siRNAs from Thermo Scientific were used: siRNA for Ark (siArk; SMARTpool; MQ-007002-01), individual siArk(1) (D-007002-03) and siArk(2) (D-007002-04), individual nontargeting siRNA (siNT; D-001810-02), and siRNA for RNF4 (siRNF4; SMARTpool; M-006557-03).
For luciferase assay, HEK293 cells were transfected with 0.2 μg CAGA12-Luc, 0.05 μg TK-Renilla, and increasing amounts of Arkadia constructs up to 0.25 μg. TGF-β was added 24 h after transfection for 8 h before lysis in passive lysis buffer (Promega). Sequential measurements of luciferase and Renilla activity were performed using a dual-luciferase reporter system (Promega). Luciferase activities were normalized to Renilla activities.
Western blotting and immunoprecipitations.
Whole-cell extracts were prepared from 6-well plates by direct lysis in Laemmli's sample buffer (60 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.01% bromophenol blue), followed by sonication and boiling before loading. Western blotting was performed using standard procedures. The following antibodies were used for either Western blotting, immunoprecipitation, or immunofluorescence, as indicated: anti-Flag (Sigma), antihemagglutinin (anti-HA; Roche), anti-myc (Roche), anti-GFP (Abcam), anti-SUMO2/3 (ab3742; Abcam), anti-PML (H-238; Santa Cruz), anti-Ark (anti-RNF111 [M05]; Abnova), and rabbit anti-RNF4 (a kind gift from J. J. Palvimo).
Immunoprecipitations were performed using either whole-cell extract in lysis buffer (20 mM Tris, pH 7.5, 300 mM NaCl, 5 mM EDTA, 1% NP-40, 10% glycerol, protease inhibitors [Roche], 20 mM N-ethylmaleimide [NEM]) or nuclear extracts. Nuclear extracts were produced by successive extraction of the cytoplasmic fraction with hypotonic buffer (20 mM HEPES, pH 7.5, 10 mM NaCl, 0.2 mM EDTA, 20% glycerol, 1.5 mM MgCl2, 0.1% Triton X-100, 25 mM NaF, 25 mM glycerol phosphate, 1 mM dithiothreitol [DTT], 20 mM NEM), followed by extraction of the nuclear fraction with hypertonic buffer (500 mM NaCl). The nuclear extract was then diluted to 300 mM NaCl with hypotonic buffer prior to immunoprecipitation. Binding was performed overnight with 5 μg of the corresponding antibody coupled to 20 μl of a protein G-Sepharose bead slurry. After 3 washings with 300 mM NaCl buffer, proteins were eluted with Laemmli's sample buffer. For transfected cells, immunoprecipitations were performed in lysis buffer containing 300 mM NaCl.
Immunofluorescence.
For immunofluorescence, cells were fixed in paraformaldehyde (4%) for 10 min. Permeabilization and blocking were performed at the same time for 30 min in phosphate-buffered saline (PBS) buffer containing 0.3% bovine serum albumin and 0.3% Triton X-100. Primary and secondary antibodies (Alexa) were incubated successively for 1 h in blocking buffer. Washings were performed in PBS–0.1% Triton X-100, with the last wash containing DAPI (4′,6-diamidino-2-phenylindole) at 0.1 μg/ml. Coverslips were then mounted onto glass slides in Prolong Gold antifade reagent (Life Technologies) and analyzed on an Olympus microscope using cellF software or on a Nipkow confocal microscope using Metamorph software.
Generation of recombinant proteins and GST pulldown.
GST-labeled proteins (GST proteins) were transformed into Escherichia coli strain BL21, and proteins were produced overnight at 37°C after induction with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) during exponential growth. Proteins were extracted in 50 mM Tris-HCl (pH 8), 150 mM NaCl, 25% glycerol, and protease inhibitors (Roche) supplemented with 1 mg/ml lysozyme and 0.5% NP-40. For GST pulldown, 5 μg of each GST protein was conjugated to 20 μl of a glutathione-Sepharose bead slurry (GE Healthcare), and the conjugated proteins were incubated at 4°C overnight on a wheel with HEK293 cell lysate or with 1 μg of recombinant poly-SUMO2 chains (Boston Biochem) in lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% NP-40, 10% glycerol, 20 mM NEM, protease inhibitors [Roche]). The beads were washed 3 times with lysis buffer, and proteins were eluted with Laemmli's sample buffer and analyzed by Western blotting.
In vitro ubiquitination assay.
Flag-Arkadia or Flag-RNF4 constructs were transfected in HEK293 cells in 6-well plates, and Flag-tagged proteins were immunoprecipitated as described above in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 8], 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, protease inhibitors [Roche]) to avoid binding of endogenous poly-SUMO and washed 3 times with RIPA buffer, 2 times with 500 mM NaCl buffer to remove any specific binding, and 2 times with 1× ubiquitination buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM DTT) to equilibrate. Ten microliters of agarose-bound Flag-proteins was then added to 200 ng of recombinant poly-SUMO2 (Boston Biochem) on ice for 30 min. Ubiquitination assays were then performed in the presence of 100 ng UbcH5b, 100 ng UbcH5c, 5 μg HA-Ub, 50 ng UBE1, and 1× energy-regenerating solution containing ATP and MgCl2 (all reagents were from Boston Biochem) in ubiquitination buffer for 45 min at 37°C. Reactions were stopped by the addition of Laemmli's sample buffer and boiling, and the reaction mixtures were analyzed by Western blotting.
RESULTS
Arkadia interacts with SUMO through its three SIMs.
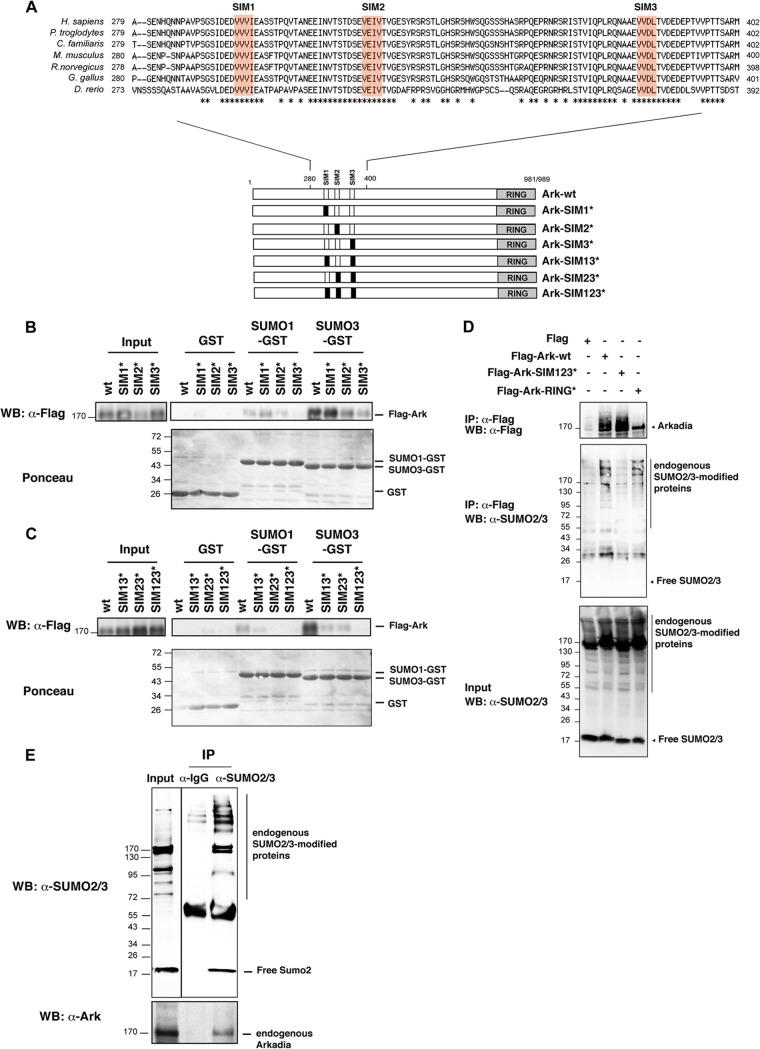
Inspection of the human Arkadia amino acid sequence revealed the presence of three putative SIMs characterized by the sequence of four amino acids V/L/I, V/L/I, X, and V/L/I or V/L/I, X, V/L/I, and V/L/I, suggesting that Arkadia could interact noncovalently with SUMO. The three SIMs lie within a region encompassing amino acids 280 to 400 and are highly conserved among vertebrate species (Fig. 1A). Using GST pulldown with SUMO1-GST and SUMO3-GST, we confirmed that Arkadia binds to both SUMO1 and SUMO3, although we noticed that Arkadia has more affinity for SUMO3 than SUMO1 (Fig. 1B and C). To assess the requirement of the three putative SIMs in this interaction, we generated individual (SIM1*, SIM2*, SIM3*), double (SIM13*, SIM23*), and triple (SIM123*) mutants by replacement of the three hydrophobic residues of the SIMs with alanine, which is known to disrupt SIM binding to SUMO (38) (Fig. 1A). GST pulldown experiments indicated that SIM3 is the most critical for SUMO binding, while SIM2 and SIM1 mutations have a milder effect and no effect, respectively (Fig. 1B). However, mutation of the three SIMs was necessary for complete abrogation of Arkadia binding to SUMO3 (Fig. 1C), indicating that the three SIMs are functional for Arkadia interaction with SUMO. We then sought to determine if Arkadia interacts with SUMO-modified proteins in vivo by attempting to coimmunoprecipitate endogenous SUMO2/3 with either Flag-Ark-wt, the SIM123 mutant, or the RING mutant in HEK293 cells (Fig. 1D). Note that Arkadia is unstable as a result of autoubiquitination, and therefore, ubiquitinated forms of Arkadia can be observed when the RING domain is intact. Endogenous SUMO2/3-modified proteins spread from 55 kDa to 200 kDa, while free SUMO2/3 migrated at 17 kDa (Fig. 1D, input). We found that SUMO2/3-modified proteins coimmunoprecipitated with Flag-Ark-wt, whereas mutation of the three SIMs of Arkadia (Flag-Ark-SIM123*) abolished this interaction. Mutation of the RING domain (Flag-Ark-RING*) did not affect Arkadia binding to SUMO2/3-modified proteins. Interestingly, free SUMO2/3 did not immunoprecipitate with Flag-Ark, suggesting that Arkadia interacts with only SUMO-modified proteins. Finally, we also confirmed that endogenous Arkadia coimmunoprecipitated with endogenous SUMO-modified proteins (Fig. 1E). Altogether, these results demonstrate that Arkadia interacts with SUMO2-modified proteins through its three SIMs.
Fig 1.
Arkadia interacts noncovalently with SUMO through three SIMs. (A) Arkadia contains three SIMs in the region from amino acids 280 to 400. SIMs are localized at position 300 (SIM1, VVVI), position 326 (SIM2, VEIV), and position 382 (SIM3, VVDL) of the human protein. The RING domain is localized in the C-terminal region. Sequence alignment of the region from amino acids 280 to 400 of Arkadia orthologues from vertebrate species indicates that the three SIMs are highly conserved throughout evolution. Asterisks indicate conserved amino acids. The different Arkadia mutants generated are represented in the lower panel, where the mutated motifs are indicated in black. (B and C) Arkadia interacts with SUMO through its three SIMs. HEK293 cells were transfected with the indicated Flag-Ark constructs, and equal amounts of whole-cell lysates were subjected to GST pulldown experiments with immobilized GST, SUMO1-GST, or SUMO3-GST. The Arkadia protein was detected by Western blotting (WB) with anti-Flag antibody before (5%) and after pulldown of the lysate. Blots were stained with Ponceau S prior to immunostaining in order to control the amount of GST proteins in the experiment. (D) The three SIMs of Arkadia are required for interaction with SUMO2 modified proteins in vivo. HEK293 cells were transfected with empty vector, Flag-Ark-wt, Flag-Ark-SIM123* mutated in the SIM, or Flag-Ark-RING* mutated in the RING. Flag immunoprecipates from whole-cell extracts were analyzed by Western blotting using anti-Flag and anti-SUMO2/3 antibodies. The corresponding lysates (input) were analyzed using anti-SUMO2/3 antibody. IP, immunoprecipitation. (E) Arkadia and SUMO2 interact at an endogenous level. Whole-cell extracts of HeLa cells were immunoprecipitated with anti-rabbit IgG (control) or anti-SUMO2/3 and analyzed by Western blotting along with input (10% whole-cell lysate) using anti-SUMO2/3 and anti-Ark antibodies. Numbers to the left of the gels are molecular masses (in kDa).
SIMs of Arkadia are not involved in TGF-β-induced SnoN/Ski degradation.
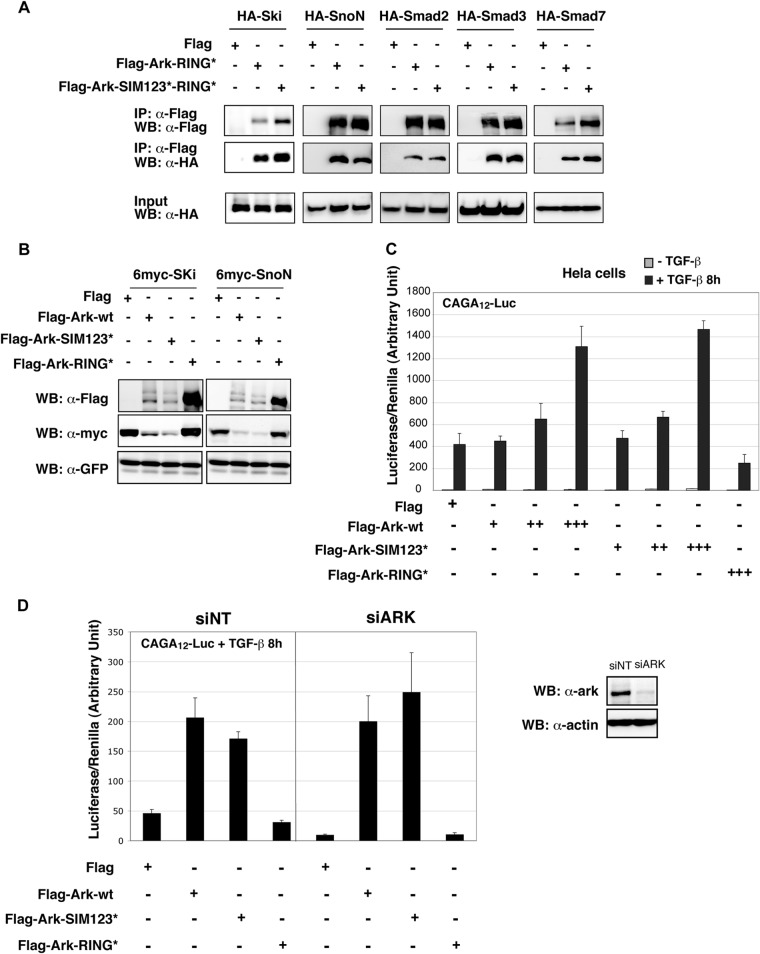
Arkadia is an essential component required for the activation of the TGF-β pathway (39, 40). We and others have previously shown that upon TGF-β stimulation, Arkadia binds to phosphorylated Smad2/3 and induces degradation of SnoN/Ski, thereby enabling induction of the Smad-dependent transcription (1, 2, 5). This function requires the C-terminal RING domain of Arkadia, which mediates ubiquitination of SnoN/Ski. To determine whether the highly conserved SIMs are also involved in this function, we first investigated the effect of their mutation on the ability of Arkadia to interact with its regulators and substrates in the TGF-β pathway. To this aim, we compared binding of HA-tagged Smad2, Smad3, SnoN, Ski, and Smad7 to Flag-Ark-wt or Flag-Ark mutated in the SIMs by coimmunoprecipitation. Experiments were carried out in HEK293 cells that have an active TGF-β pathway generated by an autocrine signal. Of note, we used the RING-mutated form of Arkadia (RING*) in order to avoid degradation of the substrates. Our results indicated that mutation of the SIMs, which abolished binding to SUMO2, did not affect binding of Arkadia to any of its known partners in the TGF-β pathway (Fig. 2A). Moreover, coexpression experiments indicated that mutation of the SIMs did not impair Arkadia's ability to induce degradation of SnoN or Ski, while mutation of the RING domain did (Fig. 2B). Interestingly, as described in Fig. 1D, Arkadia autoubiquitination was still observed when the SIMs were mutated but was abolished when the RING domain was mutated, indicating that the SIMs are not required for efficient autoubiquitination and degradation of Arkadia. Finally, we were unable to observe a significant difference in Arkadia's ability to induce a Smad-dependent luciferase reporter derived from the PAI-1 promoter (CAGA12-Luc) when the SIMs were mutated, while the RING mutation disabled this Arkadia function (Fig. 2C). In order to rule out the possibility that the lack of an effect of the SIM mutation was not due to interference with endogenous wild-type Arkadia, we performed the same experiment in cells in which Arkadia had previously been silenced with an siRNA. As shown in Fig. 2D, we were unable to detect any significant difference between wild-type Arkadia and the SIM mutant. In view of these observations, we concluded that the SIMs are dispensable for Arkadia-induced degradation of SnoN/Ski and the attendant activation of TGF-β signaling.
Fig 2.
Arkadia SIMs are not essential for TGF-β signaling activation. (A) SIMs are not necessary for the interaction of Arkadia with its different partners in the TGF-β pathway. HEK293 cells were cotransfected with the indicated combination of Flag-Ark-RING* or Flag-Ark-SIM123*-RING* and plasmids expressing HA-tagged Ski, SnoN, Smad2, Smad3, and Smad7. Flag immunoprecipates were analyzed by Western blotting using anti-Flag and anti-HA antibodies. The corresponding whole-cell lysates (input) were analyzed using anti-HA antibody. (B) The SIMs of Arkadia are not necessary for SnoN and Ski degradation. HEK293 cells were transfected with 6-myc–SnoN or 6-myc–Ski together with Flag-Ark-wt, Flag-Ark-SIM123*, or Flag-Ark-RING* and along with a GFP expression vector as an internal control for transfection. Whole-cell lysates were analyzed with anti-Flag, anti-myc, and anti-GFP antibodies. (C and D) Arkadia SIMs are not necessary for Smad-dependent transcription on a synthetic reporter. HeLa cells were transfected with CAGA12-Luc together with TK-Renilla and Flag-Ark-wt, Flag-Ark-SIM123*, or Flag-Ark-RING*, as indicated. TGF-β was added or not 8 h before lysis and measurement of luciferase and Renilla activities. Experiments were done in triplicate and repeated three times. (D) Cells were transfected with a nontarget siRNA (siNT) or siRNA targeting Arkadia (siArk) 48 h prior to plasmid transfection. Arkadia knockdown efficiency was evaluated by Western blotting on whole-cell extracts using anti-Ark antibody, as shown on the right. Actin was used as a loading control.
Arkadia specifically binds and ubiquitinates poly-SUMO2 in a SIM-dependent manner.
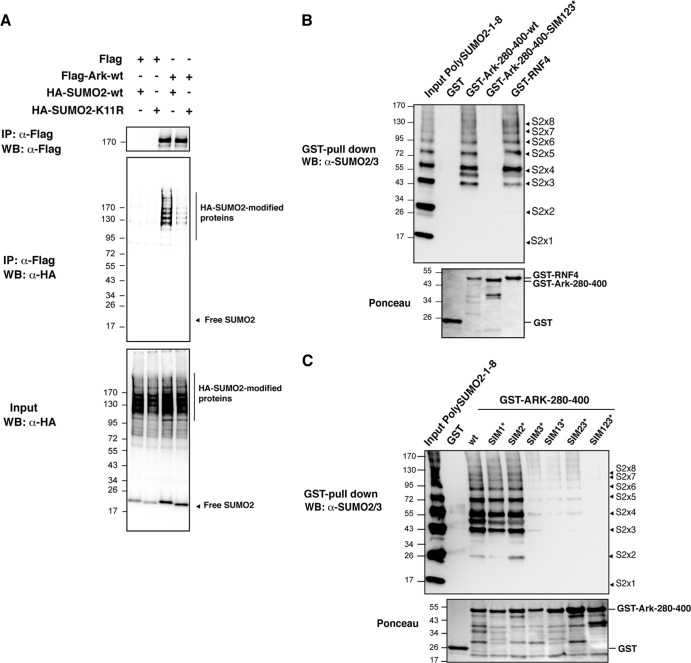
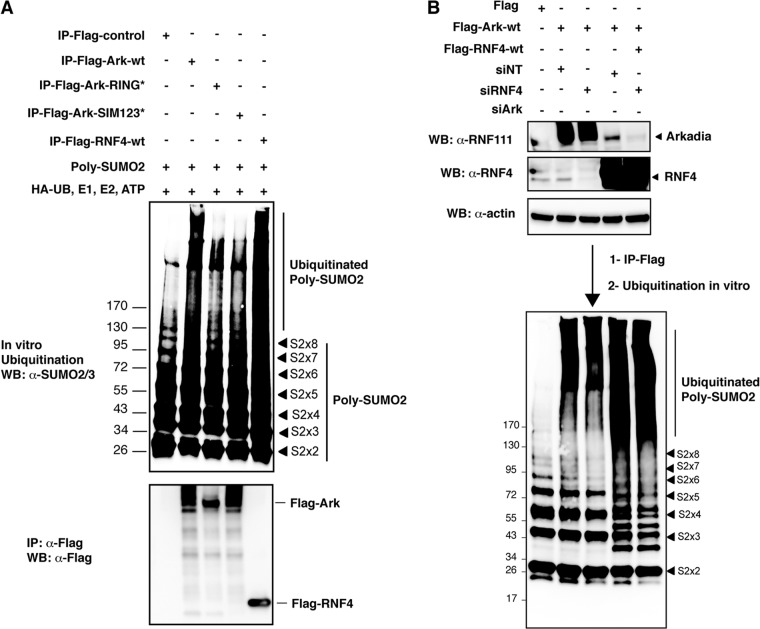
It has been proposed that successive SIMs generate a preferential interface for interaction with SUMO chains, with each SIM binding one SUMO (41). Recently, the E3 ubiquitin ligase RNF4 was shown to interact specifically with poly-SUMO through four consecutive SIMs (9). To establish whether the SIMs of Arkadia could also generate a preferential binding for SUMO chains, we compared the ability of Flag-Ark to coimmunoprecipitate with wild-type HA-SUMO2 or HA-SUMO2-K11R mutated in the SUMO consensus that cannot form SUMO2 polymers (Fig. 3A). The HA-SUMO2-K11R mutant displays a similar global sumoylation pattern as HA-SUMO2-wt, likely because most detected SUMO conjugates are mono- or multisumoylated (Fig. 3A, input). However, we found that Flag-Ark coimmunoprecipitated significantly more sumoylated proteins in the presence of HA-SUMO2-wt than in the presence of HA-SUMO2-K11R, suggesting that Arkadia preferentially binds polysumoylated proteins in vitro. We then performed in vitro GST pulldown of recombinant SUMO2 chains of various sizes (from monomers to octamers) with the region from amino acids 280 to 400 of Arkadia containing either wild-type (GST-Ark-280-400-wt) or mutated (GST-Ark-280-400-SIM123*) SIMs. As expected, we found that Arkadia interacted with poly-SUMO2 chains through its SIMs, since the interaction could be observed with Ark-280-400-wt but not with Ark-280-400-SIM123* (Fig. 3B). More importantly, the three SIMs of Arkadia preferentially interact with trimers to octamers of SUMO2, whereas interaction with SUMO2 monomers or dimers was not detectable, similar to what was observed for RNF4. In order to define which SIM contributes to poly-SUMO2 binding, we then performed pulldown of poly-SUMO2 with GST-Ark-280-400 mutated in each SIM individually or in combination (Fig. 3C). We found that SIM3 is the most critical SIM involved in the poly-SUMO2 interaction. Additional mutation of SIM1 or SIM2 had little effect on binding efficiency, although mutation of the three SIMs is nonetheless required for complete abolition of this interaction. These results clearly demonstrate the preferential binding of Arkadia to SUMO2 chains and further suggest that poly-SUMO2 could be a target for the E3 ubiquitin ligase activity of Arkadia. To address this hypothesis, we performed an in vitro ubiquitination assay using recombinant poly-SUMO2 in the presence or absence of Flag-purified wild-type or mutant Arkadia. Flag-RNF4 was used as a positive control (Fig. 4A). The SUMO2/3 immunoblot revealed that a smear of higher-molecular-mass species was specifically detected in the presence of wild-type Arkadia or RNF4 but not in the presence of Arkadia mutated in its RING domain or in its 3 SIMs, indicating that Arkadia induces ubiquitination of poly-SUMO2 in a RING- and SIM-dependent manner and might therefore constitute a new STUBL. Notably, our results also reveal that Arkadia displays weaker STUBL activity than RNF4, presumably because it is a more complex protein. Finally, to rule out the possibility that Arkadia STUBL activity is not due to copurification of RNF4 and vice versa, we performed an in vitro ubiquitination assay with Arkadia purified from cells that had been depleted or not of RNF4 and, conversely, with RNF4 purified from cells that had been depleted or not of Arkadia (Fig. 4B). Under these experimental conditions, we could not observe any significant and reproducible decrease in the STUBL activity of either E3, indicating that Arkadia and RNF4 have distinct STUBL functions. Altogether, these results show that Arkadia binding to poly-SUMO2 leads to ubiquitination of the SUMO2 chain itself, suggesting that Arkadia might constitute a novel STUBL.
Fig 3.
Arkadia interacts specifically with polysumoylated proteins and poly-SUMO2 chains through its SIMs. (A) Arkadia interacts with poly-SUMO2 chains in vitro. HEK293 cells were transfected with empty vector or Flag-Ark-wt, along with HA-SUMO2-wt or the HA-SUMO2-K11R mutant, which cannot form poly-SUMO2 chains. Flag immunoprecipates from nuclear extracts were analyzed by Western blotting using anti-Flag and anti-HA antibodies. The corresponding lysates before immunoprecipitation were analyzed using anti-HA antibody. (B) Arkadia interacts with poly-SUMO2 chains in vitro. Recombinant poly-SUMO2 chains (1 to 8 SUMO2 molecules, as shown by the arrowheads) were subjected to GST pulldown experiments with immobilized GST, GST-Ark-280-400-wt, GST-Ark-280-400-SIM123*, or GST-RNF4. Poly-SUMO2 proteins in the input (25%) and after pulldown were detected with anti-SUMO2/3 antibody by Western blotting. The blot was stained with Ponceau S prior to immunostaining in order to control the amount of GST proteins in the experiment. (C) SIM3 is the most critical SIM for poly-SUMO interaction. The experiment was performed as described for panel B with the indicated GST-Ark-280-400 SIM mutants. Numbers to the left of the gels are molecular masses (in kDa).
Fig 4.
(A) Arkadia ubiquitinates poly-SUMO2 chains in a SIM- and RING-dependent manner. HEK293 cells were transfected with either Flag empty vector, Flag-Ark-wt, Flag-Ark-RING*, Flag-Ark-SIM123*, or Flag-RNF4 as a positive control. The different Flag constructs were purified on anti-Flag agarose beads, and 10 μl of Flag-purified proteins was incubated in the presence of recombinant poly-SUMO2 chains for 30 min on ice. In vitro ubiquitination was then performed for 45 min at 37°C in the presence of HA-Ub, E1 (UBE1), E2 (UbcH5b and UbcH5c), and ATP. Ubiquitinated poly-SUMO2 was visualized by Western blotting as a shift of higher-molecular-mass species using anti-SUMO2/3 antibody. The relative level of Flag-purified proteins used in the assay was evaluated in parallel by Western blotting using an anti-Flag antibody, as shown in the lower blot. (B) Arkadia and RNF4 independently ubiquitinate poly-SUMO2 chains. HEK293 cells were transfected with siArk or siRNF4. At 48 h after siRNA transfection, cells were transfected with Flag-Ark-wt or Flag-RNF4-wt, as indicated. At 24 h after plasmid transfection, cell lysates were extracted and Arkadia and RNF4 expression were evaluated by Western blotting using anti-Ark and anti-RNF4 as well as antiactin as a loading control. The rest of the lysate was purified with anti-Flag agarose beads. Ten microliters of Flag-purified proteins was used for in vitro ubiquitination assay of poly-SUMO2 chains, as described for panel A. Ubiquitinated poly-SUMO2 was visualized by Western blotting as a shift of higher-molecular-mass species using anti-SUMO2/3 antibody. Numbers to the left of the gels are molecular masses (in kDa).
Arkadia binds to polysumoylated PML and relocalizes to PML nuclear bodies in response to arsenic.
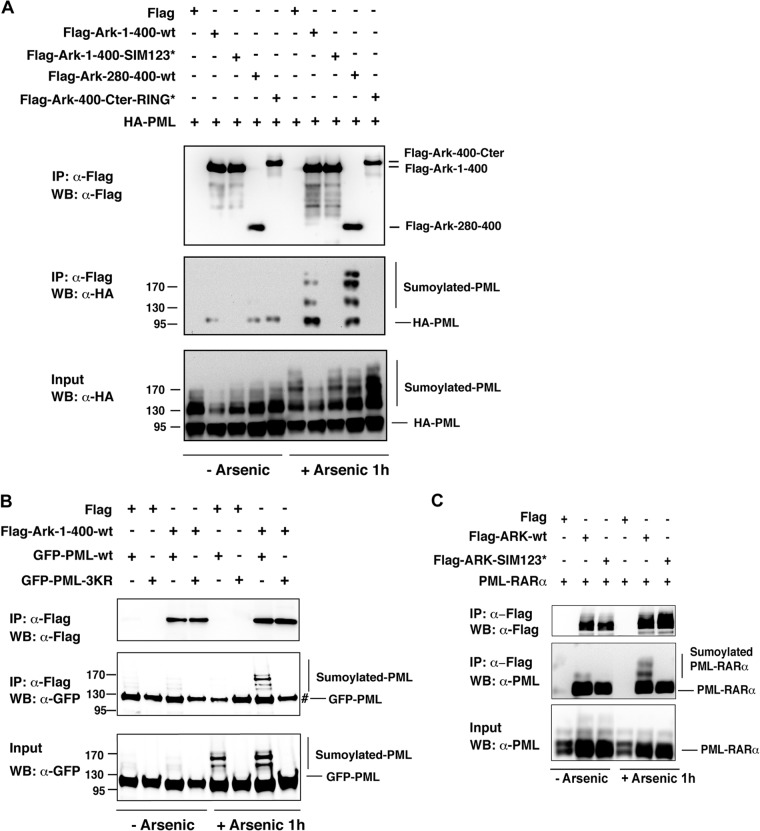
Modification of substrates with SUMO chains occurs in response to a cellular stress, and to date the best-characterized substrate for such modification is PML, which undergoes polysumoylation in response to arsenic treatment, which is commonly used in APL therapy (9, 27). In cultured cells, PML or the PML moiety of the oncogenic fusion protein PML-RARα found in APL is rapidly polysumoylated and accumulates in PML nuclear bodies upon arsenic treatment (at 1 h), and this modification results in subsequent degradation of PML or PML-RARα (9, 14, 25, 27, 28). This observation prompted us to investigate whether Arkadia could interact with arsenic-induced polysumoylated PML by coimmunoprecipitation of HA-tagged PML with different Flag-Ark mutants in HEK293 cells. We confirmed that high-molecular-mass polysumoylated PML species accumulate after 1 h of arsenic treatment (Fig. 5A, input). More importantly, we were able to detect a specific interaction of sumoylated PML with Arkadia (Fig. 5A, IP: α-Flag). As predicted, this association was observed with the N-terminal portion of Arkadia corresponding to the region from amino acids 1 to 400 or amino acids 280 to 400 that contains intact SIMs but not with the corresponding region mutated in the SIMs or with the remaining fragment of Arkadia containing amino acids 400 to the C terminus (Fig. 5A). Moreover, no binding with polysumoylated PML was observed with the PML-3KR mutated in the three SUMO acceptor sites (27) (Fig. 5B). Interestingly, we also found that wild-type Arkadia interacted with arsenic-induced polysumoylated PML-RARα, whereas Arkadia mutated in the SIMs did not (Fig. 5C). Note that some nonspecific binding could be observed with unmodified PML or PML-RARα in these different experiments (Fig. 5A to C), raising the possibility that Arkadia also binds unmodified PML. Nevertheless, our results demonstrate that robust binding to arsenic-induced SUMO-modified PML or PML-RARα is observed only when the SIMs of Arkadia are intact, indicating that Arkadia specifically binds these sumoylated substrates via its SIMs.
Fig 5.
Arkadia interacts with sumoylated PML through its SIMs. (A and B) Arkadia specifically interacts with SUMO-modified PML through its SIMs. HEK293 cells were transfected with the indicated Flag-Ark constructs together with HA-PML (A) or with GFP-PML-wt or GFP-PML-3KR mutated in the 3 SUMO consensus sites (B). Cells were treated or not with arsenic for 1 h. Flag immunoprecipates were analyzed by Western blotting using anti-Flag and anti-HA (A) or anti-GFP (B) antibody. The corresponding whole-cell lysates (input) were analyzed using anti-HA (A) or anti-GFP (B) antibody. #, nonspecific binding of unmodified GFP-PML under all conditions. Numbers to the left of the gels are molecular masses (in kDa). (C) Arkadia specifically interacts with SUMO-modified PML-RARα through its SIMs. HEK293 cells were transfected with Flag-Ark-wt or Flag-Ark-SIM123* together with PML-RARα. Cells were treated or not with arsenic for 1 h. Flag immunoprecipates were analyzed by Western blotting using anti-Flag and anti-PML antibodies. The corresponding whole-cell lysates (input) were analyzed using anti-PML antibodies.
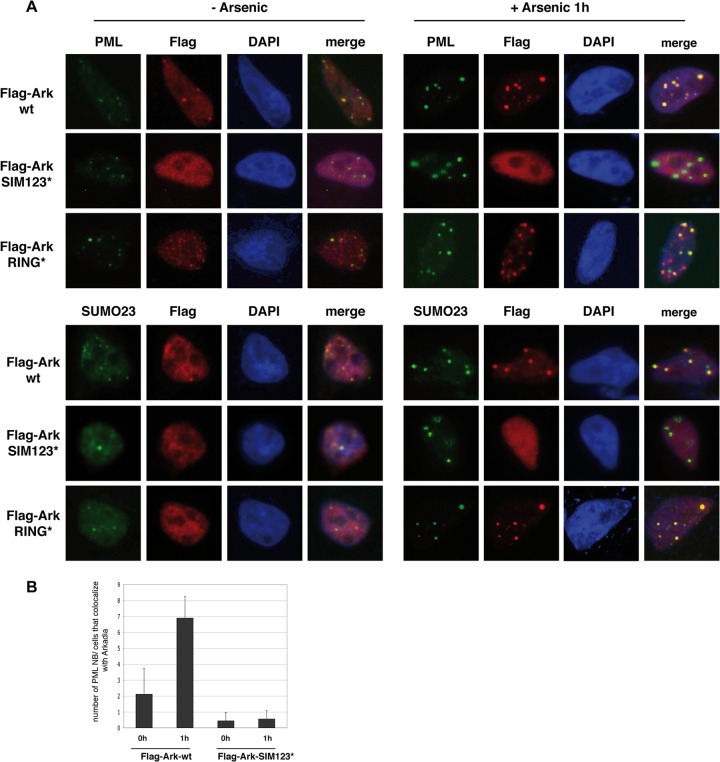
We then investigated whether the specific binding of Arkadia to polysumoylated PML could lead to accumulation of Arkadia in PML nuclear bodies. Consistent with previous reports (9, 14, 25, 27), accumulation of sumoylated PML after 1 h of arsenic treatment induced an increase in PML nuclear body number and intensity in HeLa cells, as gauged by immunofluorescence staining of endogenous PML and SUMO2/3 (Fig. 6A). We found that transfected Flag-Ark-wt colocalized with endogenous PML and SUMO2/3 in PML nuclear bodies and that this colocalization was dramatically increased after 1 h of arsenic treatment, when PML nuclear bodies increased in number and intensity (Fig. 6A, Flag-Ark-wt, and B). Mutation of the SIMs of Arkadia completely abolished this colocalization, whereas mutation of the RING domain had no effect (Fig. 6A, Flag-Ark-SIM123* and Flag-Ark-RING*, and B). These results indicate that arsenic-induced binding of Arkadia to SUMO-modified PML redirects Arkadia to PML nuclear bodies in a SIM-dependent manner. Conversely, we could not observe any significant effect of overexpressed SIM-mutated Arkadia on PML nuclear body number or size upon 1 h of arsenic treatment, suggesting that Arkadia is not required for nuclear body formation and that it does not act as a dominant negative ligase to prevent accumulation of SUMO-modified PML in PML nuclear bodies (Fig. 6A, Flag-Ark-SIM123*). A similar observation was made for RING-mutated Arkadia, confirming that Arkadia E3 ubiquitin ligase activity is not involved in the recruitment of SUMO-modified PML to PML nuclear bodies (Fig. 6A, Flag-Ark-RING*). Altogether, these results demonstrate that, upon arsenic treatment, the SIMs of Arkadia act as a localization signal that targets Arkadia to PML nuclear bodies through its interaction with polysumoylated PML.
Fig 6.
SIMs relocalize Arkadia to PML nuclear bodies in response to arsenic. (A) HeLa cells transfected with Flag-Ark-wt, Flag-Ark-SIM123*, or Flag-Ark-RING* were treated or not with arsenic for 1 h before fixation. Cells were analyzed by immunofluorescence microscopy using anti-Flag antibody (red) to visualize Arkadia and anti-PML antibody or anti-SUMO2/3 antibody (green) to visualize endogenous PML or SUMO2/3. DNA was stained with DAPI (blue). (B) PML nuclear bodies (NB) that colocalize with Flag-Ark-wt or Flag-Ark-SIM123* were visualized with anti-PML staining, and the numbers of PML nuclear bodies in 30 transfected cells were counted.
Arkadia is involved in arsenic-induced degradation of polysumoylated PML.
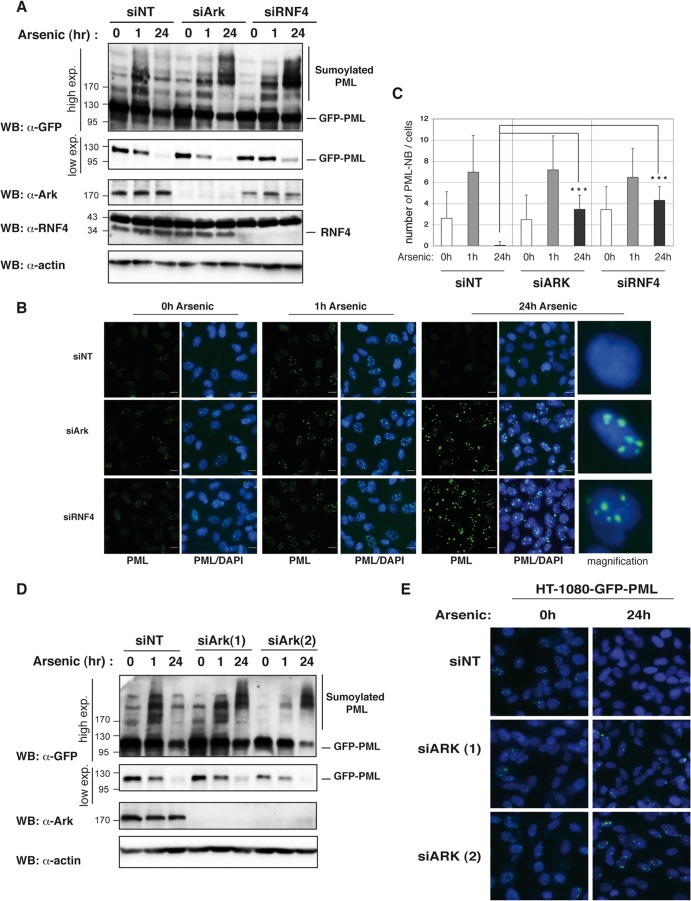
The rapid polysumoylation of PML or PML-RARα in response to arsenic treatment is followed by their proteasomal degradation, a process that is known to require the STUBL RNF4 (9, 27). The observation that Arkadia is a poly-SUMO-specific E3 ubiquitin ligase that binds polysumoylated PML in response to arsenic thus raised the possibility that it is a new STUBL involved in proteasomal degradation of PML or PML-RARα. To probe this possibility, we generated an HT1080-GFP-PML stable cell line that expresses low levels of GFP-PML in order to follow the kinetics of PML sumoylation and degradation in response to arsenic after depletion of endogenous Arkadia. Cells transfected with either control (siNT), Arkadia (siArk), or RNF4 (siRNF4) siRNA were exposed to arsenic for different times, and the fate of PML was evaluated by either Western blotting using anti-GFP antibody or GFP-based fluorescence microscopy (Fig. 7). As observed in HeLa cells, PML sumoylation was clearly detected 1 h after arsenic treatment in HT1080-GFP-PML cells transfected with control siRNA (siNT), as evidenced by the shift of higher-molecular-mass species in Western blots (Fig. 7A). Of note, we confirmed that this shift was associated with the concomitant accumulation of GFP-PML in PML nuclear bodies by immunofluorescence (Fig. 7B and C). Interestingly, both PML sumoylation and PML accumulation in nuclear bodies were still observed 1 h after arsenic treatment when Arkadia was depleted, confirming our earlier observation (Fig. 6A) that Arkadia does not significantly affect this process. In agreement with previous studies, Western blot analysis of the HT1080-GFP-PML cells treated with the control siRNA indicated that sumoylated PML is completely degraded 24 h after arsenic treatment and this is associated with a dramatic decrease of the unmodified PML protein level (Fig. 7A). The degradation of PML was also detectable by immunofluorescence assay, since a complete decline in the fluorescence intensity of GFP-PML was observed after 24 h of arsenic exposure (Fig. 7B). Strikingly, we observed that depletion of Arkadia had an effect similar to that of RNF4 depletion after 24 h of arsenic exposure, in that it led to an accumulation of polysumoylated PML detectable as supershifted higher-molecular-mass species of PML by Western blotting (Fig. 7A). This effect was associated with a strong accumulation of GFP-PML in large nuclear bodies 24 h after exposure to arsenic, as opposed to its degradation in the corresponding control siRNA (Fig. 7B and C). Neither effect occurred as a result of off-target effects, as similar results were obtained with two different individual siRNAs targeting Arkadia by Western blotting (Fig. 7D) or by immunofluorescence assay (Fig. 7E). Arkadia depletion had no effect on the stability of unmodified PML in the absence of arsenic treatment, which confirmed that this E3 ubiquitin ligase is specifically involved in the degradation of the polysumoylated form of PML that accumulates after arsenic treatment.
Fig 7.
Arkadia is involved in arsenic-induced degradation of sumoylated PML. (A) Depletion of Arkadia inhibits arsenic-induced degradation of sumoylated PML. HT1080-GFP-PML cells were transfected with a nontargeting siRNA (siNT) and siRNA SMARTpool targeting Arkadia (siArk) or RNF4 (siRNF4). Cells were exposed to arsenic at the indicated times. Whole-cell lysates were analyzed by Western blotting with anti-Ark, anti-RNF4, anti-GFP, and antiactin antibodies. A high exposure (exp.) and a low exposure of the same anti-GFP Western blot are shown. (B) Depletion of Arkadia inhibits arsenic-induced degradation of PML nuclear bodies. HT1080-GFP-PML cells were transfected with siNT, siArk, or siRNF4. Cells were exposed to arsenic at the indicated times before fixation, permeabilization, and DAPI staining. GFP-PML (green) expression and cellular localization were then analyzed by GFP-based fluorescence with the same laser intensity for each condition and are represented alone or merged with DAPI (blue). Bars, 20 μm. (C) Quantification of the number of PML nuclear bodies visualized with anti-PML staining in 30 cells under the indicated conditions, as described for panel B. (D and E) The same experiments described for panels A and B, respectively, performed with two individual siRNAs targeting Arkadia, siArk(1), and siArk(2). Numbers to the left of the gels are molecular masses (in kDa).
Arkadia and RNF4 act independently in arsenic-induced degradation of polysumoylated PML.
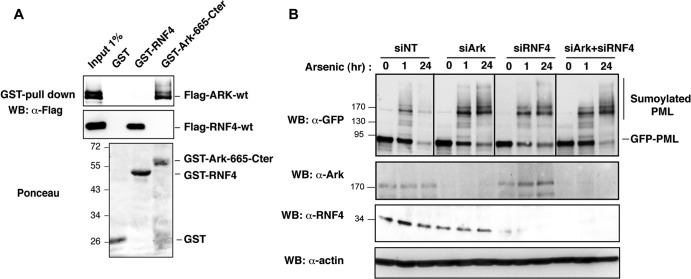
RING finger proteins usually function as dimers, and RNF4 has been shown to homodimerize (42, 43). We then sought to determine whether Arkadia and RNF4 could interact through their RING domain and function as a heterodimer. We performed GST pulldown of Flag-Ark-wt or Flag-RNF4 with recombinant full-length GST-RNF4 or GST-Ark-665-Cter, which contains the RING C-terminal (Cter) region (Fig. 8A). We confirmed that GST-RNF4 pulled down Flag-RNF4. Likewise, we found that GST-Ark-665-Cter pulled down Flag-Ark-wt, demonstrating that Arkadia can also homodimerize, most probably through its RING domain. However, no binding of Flag-Ark-wt or Flag-RNF4 was observed with GST-RNF4 or GST-Ark-665-Cter, respectively, indicating that Arkadia and RNF4 cannot form heterodimers. Consistent with this finding, we were unable to detect an in vitro interaction between Arkadia and RNF4, even after arsenic treatment (data not shown), arguing against the possibility that these E3 ligases could form physical complexes. In support of this finding, we also observed that combined depletion of Arkadia and RNF4 under limited siRNA conditions (partial depletion) does not have a significant synergistic effect on degradation of polysumoylated PML compared to depletion of each one singly, indicating that these two E3 ligases might work independently rather than synergistically during this process (Fig. 8B).
Fig 8.
Arkadia and RNF4 have independent STUBL functions. (A) Arkadia and RNF4 homodimerize but do not form heterodimers. HEK293 cells were transfected with Flag-Ark-wt or Flag-RNF4-wt constructs, and equal amounts of whole-cell lysates were subjected to GST pulldown experiments with immobilized GST, GST-RNF4, or GST-Ark-665-Cter. Arkadia and RNF4 proteins were detected by Western blotting with anti-FLAG antibody before (5%) and after pulldown of the lysate. Blots were stained with Ponceau S prior to immunostaining in order to control the amount of GST proteins in the experiment. (B) Arkadia and RNF4 are independently involved in arsenic-induced degradation of PML. HT1080-GFP-PML cells were transfected with a limiting amount of siRNA, siNT (60 nM), siArk (50 nM), or siRNF4 (10 nM) or with a combination of siArk (50 nM) and siRNF4 (10 nM). At 48 h after transfection, cells were exposed or not to arsenic for 1 h or 24 h. Whole-cell lysates were analyzed by Western blotting with anti-GFP, anti-Ark, anti-RNF4, and antiactin antibodies. Numbers to the left of the gels are molecular masses (in kDa).
DISCUSSION
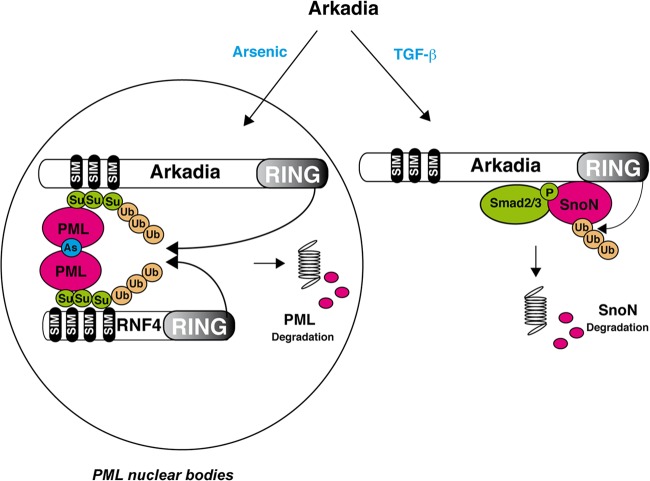
Arkadia is an E3 ubiquitin ligase whose role has so far been restricted to its requirement in the TGF-β signaling response. In this pathway, Arkadia acts as a signal-dependent E3 ubiquitin ligase for SnoN and Ski (2). Here, we demonstrate that Arkadia is also involved in another signal-dependent degradation process to coordinate degradation of PML in response to arsenic treatment. Interestingly, in the TGF-β pathway, the degradation signal for SnoN/Ski is triggered by binding of Arkadia to phospho-Smad2, which accumulates upon TGF-β induction, whereas in this new model (Fig. 9), the degradation signal is induced by binding of Arkadia to polysumoylated PML, which accumulates after arsenic treatment. Indeed, in the present work, we have shown that Arkadia interacts with SUMO-modified PML after arsenic treatment and is redirected to PML nuclear bodies in a SIM-dependent manner, where it is required for the subsequent degradation of polysumoylated PML.
Fig 9.
Model for the E3 ubiquitin ligase activity of Arkadia induced by arsenic versus TGF-β. Upon TGF-β induction, interaction of Arkadia with phosphorylated Smad2 triggers degradation of SnoN, whereas upon arsenic treatment, interaction of Arkadia with poly-SUMO in PML nuclear bodies induces degradation of polysumoylated PML together with RNF4.
Arkadia is a novel poly-SUMO-targeted ubiquitin ligase.
Our finding that Arkadia can bind and ubiquitinate SUMO2 chains in a SIM- and RING-dependent manner suggests that Arkadia is a new member of the STUBL family that targets sumoylated proteins to degradation. STUBL is a new family of RING finger ubiquitin ligases, initially identified in the yeast Saccharomyces cerevisiae, that includes Slx5 and Slx8 (reviewed in references 19 to 23). Slx5 and Slx8 function as heterodimers that bind SUMO through one SIM and ubiquitinate the substrate through the RING domain of Slx8. Although the identity of the SUMO-modified substrates has not been established, depletion of either Slx5 or Slx8 leads to accumulation of sumoylated proteins (44), and these STUBLs are required for genome stability in yeast (21). In mammals, over 200 RING-containing proteins have been reported, but the only STUBL characterized so far is RNF4, which contains four SIMs that generate preferential binding to poly-SUMO chains, suggesting that in mammals, STUBLs specifically target poly-SUMO-modified substrates for degradation (9). Our finding that the RING finger E3 ubiquitin ligase Arkadia also contains three successive SIMs in its N-terminal region raised the possibility that it constitutes a new poly-SUMO-specific ubiquitin ligase. In support of this hypothesis, we found that Arkadia is able to bind specifically poly-SUMO2 via its three SIMs and displays the strongest affinity to poly-SUMO2 trimers or longer chains compared to its affinity to mono-SUMO or dimers of SUMO, as described for RNF4 (9). Interestingly, one of these SIMs appears to be more critical for this interaction, suggesting that poly-SUMO binding is mainly driven by the VVDL SIM3 motif. Furthermore, our finding that Arkadia can also ubiquitinate poly-SUMO2 chains in a SIM- and RING-dependent manner demonstrates that Arkadia contains STUBL activity. We noticed, however, that Arkadia STUBL activity in vitro seems to be less efficient than RNF4 activity, presumably because it is a more complex protein than RNF4 and might be subjected to regulation. Importantly, we identified polysumoylated PML to be a new substrate that binds Arkadia in response to arsenic and found that this interaction occurs only when the SIMs are intact. Finally, we showed that Arkadia is necessary for the degradation of polysumoylated PMLs that have accumulated in response to arsenic treatment. Degradation of polysumoylated PMLs is the most well characterized poly-SUMO-dependent degradation process in mammalian cells. Since RNF4 has been shown in previous work to be also involved in this degradation process (9, 25, 27, 28), this raised the question of the entangled mechanism of action of these two STUBLs. Our results indicate that both E3 ligases are required for this degradation, suggesting that they are not redundant in this process. However, we could not detect any interaction of Arkadia with RNF4 in mammalian cells like that shown for the STUBL Slx5 and Slx8 in yeast. Moreover, Arkadia and RNF4 do not seem to act synergistically in the degradation of sumoylated PML, suggesting that they might, rather, work independently in this process. Arsenic-induced sumoylation of PML is a rapid process that is achieved within 1 h, whereas degradation of SUMO-modified PML is a longer process that takes up to 24 h to be completed (9). It is therefore possible that this mechanism of degradation is a multistep process that would involve two E3 ligases in parallel and/or sequentially. One explanation could be that Arkadia could act as an E4 to potentiate the degradation process driven by RNF4. Another possibility is that Arkadia and RNF4 ubiquitinate polysumoylated PML through different ubiquitin chain linkages that would both be necessary for degradation of PML. Indeed, it has recently been shown that Arkadia can ubiquitinate the μ2 subunit of clathrin adaptor 2 through the unconventional lysine 27 linkage (45). Hence, our identification of the requirement for Arkadia in the arsenic-induced SUMO-dependent degradation of PML adds up to the possibility that it is a crucial STUBL in the mechanism by which arsenic exerts its chemotherapeutic effect on APL.
Relevance of Arkadia binding to poly-SUMO in TGF-β pathway regulation.
In our study, we sought to determine whether the STUBL function of Arkadia could also be involved in TGF-β signal transduction. Our results indicate that the SIMs of Arkadia are dispensable for SnoN/Ski interaction and degradation and their mutation does not significantly impair the ability of Arkadia to induce a TGF-β-dependent synthetic reporter derived from the PAI-1 promoter. Our results further indicate that, unlike mutation of the RING domain, mutation of the SIMs does not have a dominant negative effect on Arkadia function in the TGF-β pathway. While SnoN has been shown to be sumoylated on lysines 50 and 383 (46, 47), our results suggest that sumoylation of SnoN is not required for effective degradation by Arkadia upon TGF-β signaling. This is in agreement with the finding that sumoylation of SnoN does not affect its metabolic stability or its ability to repress TGF-β signaling, as shown by the analysis of a loss-of-function lysine mutant or a gain-of-function SnoN-SUMO fusion (46, 47). However, it is not yet clear if some particular stress conditions can induce polysumoylation of SnoN, and it is possible that Arkadia binding to poly-SUMO might nonetheless be involved in the fine-tuning of some TGF-β responses under such conditions. Conversely, it is also possible that stimuli increasing general polysumoylation delocalize Arkadia from its substrate in the TGF-β pathway and therefore exert an indirect negative effect on the pathway. Consistent with this, we found that arsenic treatment, by generating a new polysumoylated target for Arkadia, delocalizes Arkadia to PML nuclear bodies.
Arkadia binding to poly-SUMO reveals new potential functions for Arkadia.
Polysumoylation is a newly identified posttranslational modification, and our identification of Arkadia as an E3 that can target poly-SUMO-modified proteins might also contribute to a better understanding of the functional consequences of such modification in mammals. Polysumoylation appears to be specifically induced by different cellular stresses which are known to generate multiple alterations that affect genome integrity, thereby perturbing cell homeostasis. Interestingly, this could provide an additional antitumor function for Arkadia, besides its tumor suppressor role described in TGF-β signaling (48). In mammalian cells, heat shock induces changes in the polysumoylation state of more than 300 proteins (10, 11), which may thus constitute as many new potential targets for Arkadia. Furthermore, RNF4 STUBL activity has recently been involved in the cellular response to DNA damage through its binding with the sumoylated mediator of DNA damage checkpoint (MDC1) (49–51). In light of these observations, one would surmise that Arkadia could also function to guard the genome by eliminating such sumoylated proteins accumulated under stress conditions. Whether Arkadia or RNF4 can target indifferently all these polysumoylated proteins or shows some preferential binding to such substrates remains to be defined.
ACKNOWLEDGMENTS
We thank E. Kandel for the HT1080-E14 cell line, D. Bazett-Jones for GFP-PML-IV-wt and GFP-PML-IV-3KR, and J. J. Palvimo for Flag-RNF4 and anti-RNF4 antibody. We thank A. Sharrocks for HA-SUMO2-wt and HA-SUMO2-K11R. We thank members of the laboratory for useful discussions and comments on the manuscript. We thank Romain Morichon at the IF65 platform for confocal microscopy analysis and the rotation student Numa Lauront.
This work was funded by grants to L.L. from Marie Curie Actions (ERG FP7), la Ligue Nationale Contre le Cancer (LNCC), and l'Association pour la Recherche sur le Cancer (ARC). This work was also supported by grants to A.A. from ARC programs and grants to A.D. from the LNCC, Odyssey-RE, and l'ANR. Y.E. and H.N.-K. were supported by the MRT.
Footnotes
Published ahead of print 25 March 2013
REFERENCES
- 1. Le Scolan E, Zhu Q, Wang L, Bandyopadhyay A, Javelaud D, Mauviel A, Sun L, Luo K. 2008. Transforming growth factor-beta suppresses the ability of Ski to inhibit tumor metastasis by inducing its degradation. Cancer Res. 68:3277–3285 [DOI] [PubMed] [Google Scholar]
- 2. Levy L, Howell M, Das D, Harkin S, Episkopou V, Hill CS. 2007. Arkadia activates Smad3/Smad4-dependent transcription by triggering signal-induced SnoN degradation. Mol. Cell. Biol. 27:6068–6083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mavrakis KJ, Andrew RL, Lee KL, Petropoulou C, Dixon JE, Navaratnam N, Norris DP, Episkopou V. 2007. Arkadia enhances Nodal/TGF-beta signaling by coupling phospho-Smad2/3 activity and turnover. PLoS Biol. 5:e67 doi:10.1371/journal.pbio.0050067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Koinuma D, Shinozaki M, Komuro A, Goto K, Saitoh M, Hanyu A, Ebina M, Nukiwa T, Miyazawa K, Imamura T, Miyazono K. 2003. Arkadia amplifies TGF-beta superfamily signalling through degradation of Smad7. EMBO J. 22:6458–6470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nagano Y, Mavrakis KJ, Lee KL, Fujii T, Koinuma D, Sase H, Yuki K, Isogaya K, Saitoh M, Imamura T, Episkopou V, Miyazono K, Miyazawa K. 2007. Arkadia induces degradation of SnoN and c-Ski to enhance transforming growth factor-beta signaling. J. Biol. Chem. 282:20492–20501 [DOI] [PubMed] [Google Scholar]
- 6. Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276:35368–35374 [DOI] [PubMed] [Google Scholar]
- 7. Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, Hay RT, Lamond AI, Mann M, Vertegaal AC. 2008. In vitro identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vitro strategy. Mol. Cell. Proteomics 7:132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Saitoh H, Hinchey J. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275:6252–6258 [DOI] [PubMed] [Google Scholar]
- 9. Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. 2008. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10:538–546 [DOI] [PubMed] [Google Scholar]
- 10. Bruderer R, Tatham MH, Plechanovova A, Matic I, Garg AK, Hay RT. 2011. Purification and identification of endogenous polySUMO conjugates. EMBO Rep. 12:142–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. 2009. System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2:ra24 doi:10.1126/scisignal.2000282 [DOI] [PubMed] [Google Scholar]
- 12. Martin N, Schwamborn K, Schreiber V, Werner A, Guillier C, Zhang XD, Bischof O, Seeler JS, Dejean A. 2009. PARP-1 transcriptional activity is regulated by sumoylation upon heat shock. EMBO J. 28:3534–3548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen GQ, Zhu J, Shi XG, Ni JH, Zhong HJ, Si GY, Jin XL, Tang W, Li XS, Xong SM, Shen ZX, Sun GL, Ma J, Zhang P, Zhang TD, Gazin C, Naoe T, Chen SJ, Wang ZY, Chen Z. 1996. In vitro studies on cellular and molecular mechanisms of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia: As2O3 induces NB4 cell apoptosis with downregulation of Bcl-2 expression and modulation of PML-RAR alpha/PML proteins. Blood 88:1052–1061 [PubMed] [Google Scholar]
- 14. Lallemand-Breitenbach V, Zhu J, Puvion F, Koken M, Honore N, Doubeikovsky A, Duprez E, Pandolfi PP, Puvion E, Freemont P, de The H. 2001. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 193:1361–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Muller S, Miller WH, Jr, Dejean A. 1998. Trivalent antimonials induce degradation of the PML-RAR oncoprotein and reorganization of the promyelocytic leukemia nuclear bodies in acute promyelocytic leukemia NB4 cells. Blood 92:4308–4316 [PubMed] [Google Scholar]
- 16. Shao W, Fanelli M, Ferrara FF, Riccioni R, Rosenauer A, Davison K, Lamph WW, Waxman S, Pelicci PG, Lo Coco F, Avvisati G, Testa U, Peschle C, Gambacorti-Passerini C, Nervi C, Miller WH., Jr 1998. Arsenic trioxide as an inducer of apoptosis and loss of PML/RAR alpha protein in acute promyelocytic leukemia cells. J. Natl. Cancer Inst. 90:124–133 [DOI] [PubMed] [Google Scholar]
- 17. Zhu J, Koken MH, Quignon F, Chelbi-Alix MK, Degos L, Wang ZY, Chen Z, de The H. 1997. Arsenic-induced PML targeting onto nuclear bodies: implications for the treatment of acute promyelocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 94:3978–3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang ZY, Chen Z. 2008. Acute promyelocytic leukemia: from highly fatal to highly curable. Blood 111:2505–2515 [DOI] [PubMed] [Google Scholar]
- 19. Geoffroy MC, Hay RT. 2009. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat. Rev. Mol. Cell Biol. 10:564–568 [DOI] [PubMed] [Google Scholar]
- 20. Heideker J, Perry JJ, Boddy MN. 2009. Genome stability roles of SUMO-targeted ubiquitin ligases. DNA Repair (Amst.) 8:517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nagai S, Davoodi N, Gasser SM. 2011. Nuclear organization in genome stability: SUMO connections. Cell Res. 21:474–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Perry JJ, Tainer JA, Boddy MN. 2008. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem. Sci. 33:201–208 [DOI] [PubMed] [Google Scholar]
- 23. Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN. 2007. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 26:4089–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sun H, Leverson JD, Hunter T. 2007. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 26:4102–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Geoffroy MC, Jaffray EG, Walker KJ, Hay RT. 2010. Arsenic-induced SUMO-dependent recruitment of RNF4 into PML nuclear bodies. Mol. Biol. Cell 21:4227–4239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hakli M, Karvonen U, Janne OA, Palvimo JJ. 2005. SUMO-1 promotes association of SNURF (RNF4) with PML nuclear bodies. Exp. Cell Res. 304:224–233 [DOI] [PubMed] [Google Scholar]
- 27. Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de The H. 2008. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 10:547–555 [DOI] [PubMed] [Google Scholar]
- 28. Percherancier Y, Germain-Desprez D, Galisson F, Mascle XH, Dianoux L, Estephan P, Chelbi-Alix MK, Aubry M. 2009. Role of SUMO in RNF4-mediated promyelocytic leukemia protein (PML) degradation: sumoylation of PML and phospho-switch control of its SUMO binding domain dissected in living cells. J. Biol. Chem. 284:16595–16608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. He J, Tegen SB, Krawitz AR, Martin GS, Luo K. 2003. The transforming activity of Ski and SnoN is dependent on their ability to repress the activity of Smad proteins. J. Biol. Chem. 278:30540–30547 [DOI] [PubMed] [Google Scholar]
- 30. Akiyoshi S, Inoue H, Hanai J, Kusanagi K, Nemoto N, Miyazono K, Kawabata M. 1999. c-Ski acts as a transcriptional co-repressor in transforming growth factor-beta signaling through interaction with Smads. J. Biol. Chem. 274:35269–35277 [DOI] [PubMed] [Google Scholar]
- 31. Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. 1998. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17:3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu W, Rui H, Wang J, Lin S, He Y, Chen M, Li Q, Ye Z, Zhang S, Chan SC, Chen YG, Han J, Lin SC. 2006. Axin is a scaffold protein in TGF-beta signaling that promotes degradation of Smad7 by Arkadia. EMBO J. 25:1646–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moilanen AM, Poukka H, Karvonen U, Hakli M, Janne OA, Palvimo JJ. 1998. Identification of a novel RING finger protein as a coregulator in steroid receptor-mediated gene transcription. Mol. Cell. Biol. 18:5128–5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang SH, Sharrocks AD. 2010. The SUMO E3 ligase activity of Pc2 is coordinated through a SUMO interaction motif. Mol. Cell. Biol. 30:2193–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dellaire G, Ching RW, Dehghani H, Ren Y, Bazett-Jones DP. 2006. The number of PML nuclear bodies increases in early S phase by a fission mechanism. J. Cell Sci. 119:1026–1033 [DOI] [PubMed] [Google Scholar]
- 36. Kandel ES, Nudler E. 2002. Template switching by RNA polymerase II in vitro. Evidence and implications from a retroviral system. Mol. Cell 10:1495–1502 [DOI] [PubMed] [Google Scholar]
- 37. Levenson VV, Lausch E, Kirschling DJ, Broude EV, Davidovich IA, Libants S, Fedosova V, Roninson IB. 1999. A combination of genetic suppressor elements produces resistance to drugs inhibiting DNA replication. Somat. Cell Mol. Genet. 25:9–26 [DOI] [PubMed] [Google Scholar]
- 38. Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. 2004. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U. S. A. 101:14373–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Episkopou V, Arkell R, Timmons PM, Walsh JJ, Andrew RL, Swan D. 2001. Induction of the mammalian node requires Arkadia function in the extraembryonic lineages. Nature 410:825–830 [DOI] [PubMed] [Google Scholar]
- 40. Niederlander C, Walsh JJ, Episkopou V, Jones CM. 2001. Arkadia enhances nodal-related signalling to induce mesendoderm. Nature 410:830–834 [DOI] [PubMed] [Google Scholar]
- 41. Mukhopadhyay D, Ayaydin F, Kolli N, Tan SH, Anan T, Kametaka A, Azuma Y, Wilkinson KD, Dasso M. 2006. SUSP1 antagonizes formation of highly SUMO2/3-conjugated species. J. Cell Biol. 174:939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liew CW, Sun H, Hunter T, Day CL. 2010. RING domain dimerization is essential for RNF4 function. Biochem. J. 431:23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Plechanovova A, Jaffray EG, McMahon SA, Johnson KA, Navratilova I, Naismith JH, Hay RT. 2011. Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat. Struct. Mol. Biol. 18:1052–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Z, Jones GM, Prelich G. 2006. Genetic analysis connects SLX5 and SLX8 to the SUMO pathway in Saccharomyces cerevisiae. Genetics 172:1499–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mizutani A, Saitoh M, Imamura T, Miyazawa K, Miyazono K. 2010. Arkadia complexes with clathrin adaptor AP2 and regulates EGF signalling. J. Biochem. 148:733–741 [DOI] [PubMed] [Google Scholar]
- 46. Hsu YH, Sarker KP, Pot I, Chan A, Netherton SJ, Bonni S. 2006. Sumoylated SnoN represses transcription in a promoter-specific manner. J. Biol. Chem. 281:33008–33018 [DOI] [PubMed] [Google Scholar]
- 47. Wrighton KH, Liang M, Bryan B, Luo K, Liu M, Feng XH, Lin X. 2007. Transforming growth factor-beta-independent regulation of myogenesis by SnoN sumoylation. J. Biol. Chem. 282:6517–6524 [DOI] [PubMed] [Google Scholar]
- 48. Sharma V, Antonacopoulou AG, Tanaka S, Panoutsopoulos AA, Bravou V, Kalofonos HP, Episkopou V. 2011. Enhancement of TGF-{beta} signaling responses by the E3 ubiquitin ligase Arkadia provides tumor suppression in colorectal cancer. Cancer Res. 71:6438–6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Galanty Y, Belotserkovskaya R, Coates J, Jackson SP. 2012. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 26:1179–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Luo K, Zhang H, Wang L, Yuan J, Lou Z. 2012. Sumoylation of MDC1 is important for proper DNA damage response. EMBO J. 31:3008–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yin Y, Seifert A, Chua JS, Maure JF, Golebiowski F, Hay RT. 2012. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 26:1196–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]