Abstract
FANCJ mutations are linked to Fanconi anemia (FA) and increase breast cancer risk. FANCJ encodes a DNA helicase implicated in homologous recombination (HR) repair of double-strand breaks (DSBs) and interstrand cross-links (ICLs), but its mechanism of action is not well understood. Here we show with live-cell imaging that FANCJ recruitment to laser-induced DSBs but not psoralen-induced ICLs is dependent on nuclease-active MRE11. FANCJ interacts directly with MRE11 and inhibits its exonuclease activity in a specific manner, suggesting that FANCJ regulates the MRE11 nuclease to facilitate DSB processing and appropriate end resection. Cells deficient in FANCJ and MRE11 show increased ionizing radiation (IR) resistance, reduced numbers of γH2AX and RAD51 foci, and elevated numbers of DNA-dependent protein kinase catalytic subunit foci, suggesting that HR is compromised and the nonhomologous end-joining (NHEJ) pathway is elicited to help cells cope with IR-induced strand breaks. Interplay between FANCJ and MRE11 ensures a normal response to IR-induced DSBs, whereas FANCJ involvement in ICL repair is regulated by MLH1 and the FA pathway. Our findings are discussed in light of the current model for HR repair.
INTRODUCTION
Fanconi anemia (FA) is a hereditary disease characterized by growth retardation, reduced fertility, chromosomal instability, and a predisposition to cancer (1). Distinguishing features of FA patients include congenital abnormalities and progressive bone marrow failure, which predispose individuals to acute myeloid leukemia. FA mutant cells of all genetic complementation groups are highly sensitive to agents that induce interstrand cross-links (ICLs) (2). In addition, new evidence demonstrates that the FA pathway has a role in handling DNA damage from endogenous sources (3, 4). Until recently, there were 13 known FA genetic complementation groups (1); however, a new FA gene, designated FANCP/SLX4, and two strong candidate genes, FAN1 and RAD51C, have been discovered (see reference 5 and references therein). Of all the FA genes/candidate genes, FANCM and FANCJ encode DNA motor ATPases believed to play an important role in ICL repair and, more generally, in a robust response to replication stress (6). However, only FANCJ is a bona fide DNA helicase capable of catalytically separating complementary duplex strands (7, 8) and resolving G-quadruplexes (9, 10) in an ATP-dependent manner, the latter of which is important for the role of FANCJ in handling replication stress in G-rich DNA (11). In human cells, FANCJ depletion resulted in elevated mitomycin C (MMC)-induced chromosomal abnormalities, including breakage, chromatid interchanges, triradials, and quadriradials (12, 13). Increased sister chromatid exchange (SCE) was detected in chicken brip1 (fancj) cells (12) as well as FANCJ-deficient human cells (14).
Efficient FANCD2/I monoubiquitination is dependent on FANCM and members of the FA core complex but is not dependent on FANCD1 (BRCA2), FANCJ (BACH1/BRIP1), FANCN (PALB2), FANCO (RAD51C), FANCP (SLX4), or FAN1 (2, 5). FANCD1, FANCJ, and FANCN, but not the FA core and FANCI/D2 complexes, require replication for cross-link association (15). Although FANCJ is not required for FANCD2 monoubiquitination, FANCJ promotes assembly of FANCD2 nuclear foci after exposure to a cross-linking agent (16), suggesting a complex functional relationship between FANCD2 and FANCJ.
FANCJ interacts with BRCA1, a tumor suppressor implicated in double-strand break (DSB) repair (17). FANCJ was identified in a BRCA1-BARD1-TopBP1 complex with MLH1 that was isolated from ionizing radiation (IR)-treated cells and shown to be important for S-phase checkpoint signaling (18). Interaction of FANCJ with MutLα is required for a normal ICL-induced response (19). FANCJ was found to interact directly with Bloom's syndrome helicase (BLM) to help cells cope with replication stress (14). Interestingly, BLM was identified in a BRCA1 genome surveillance complex (BASC) with other DNA repair proteins (MSH2, MSH6, MLH1, RPA, ATM, and RAD50-MRE11-NBS1) (20). BLM was also found to exist in a protein complex (BRAFT) that contained the FA core complex, RPA, and topoisomerase IIIα (TopoIIIα) (21). FANCM acts as a bridge to mediate interaction of the BLM-TopoIIIα complex with the FA core complex (22). The emerging story suggests that interaction of FA proteins with BLM and other DNA repair factors is vital for DNA repair and maintenance of genomic stability.
Although FA mutant cells are characterized by hypersensitivity to ICL agents, the role of FANCJ and downstream members of the pathway may be to facilitate homologous recombination (HR) repair of DSBs that arise during the processing of ICLs or at broken replication forks or that are induced directly by IR or certain chemical agents. Early events in DSB repair are break recognition and strand resection to promote HR repair in replicating cells (23, 24). An early responder is the MRE11-RAD50-NBS1 (MRN) complex, which senses DSBs and binds to DNA ends. MRN along with CtIP initiates end resection to remove 50 to 100 nucleotides from the 5′ terminus of the DNA, which provides a favorable DNA substrate for nucleases and helicases to execute long-strand resection. Two protein complexes, BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN, can perform long-strand resection during DSB repair (25). The recent demonstration that FANCJ and BLM interact (14) led us to explore if FANCJ might interact with other DSB repair proteins involved at an early stage of damage detection and processing. In this work, we describe a FANCJ protein interaction with a key player of DSB repair, namely, the MRE11 complex. MRE11 recruits FANCJ to DSBs, and in turn, FANCJ modulates MRE11 nuclease activity at DNA ends. Our results are discussed in light of the dynamic interplay among FANCJ and proteins engaged in early steps of DNA damage detection and processing.
MATERIALS AND METHODS
Proteins.
Recombinant FLAG-tagged FANCJ (7), MRE11 (26), MRN complex (27), and DDX11 (ChlR1) (28) were purified as previously described. Exonuclease III (ExoIII) was purchased from Promega.
Site-directed mutagenesis.
The MRE11-H129N mutation was introduced into the mCherry-MRE11 DNA construct by use of a Stratagene QuikChange site-directed mutagenesis kit according to the manufacturer's instructions, using the following primers: forward, GTTTAGTATTCATGGCAATAATGACGATCCCACAGGGGCAG; and reverse, CTGCCCCTGTGGGATCGTCATTATTGCCATGAATACTAAAC. Desired mutations in constructs were confirmed by direct DNA sequencing using the purified plasmids.
Cell culture and transfection.
HeLa cells and FA-J (EUFA30-F) mutant and corresponding wild-type cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, and 1% l-glutamine at 37°C in 5% CO2. Immortalized fibroblast lines FA-A (PD220) and FA-D2 (PD20) and their respective corrected cells, provided by the Fanconi Anemia Research Fund, were grown in the same medium as HeLa cells, but supplemented with 0.2 mg/ml puromycin. The HCC1937 line, which carries a hypomorphic mutation in BRCA1, and the corresponding BRCA1-reconstituted cells were grown in RPMI 1640 supplemented with 10% FBS and 1% penicillin-streptomycin.
For FANCJ or MRE11 knockdown, a FANCJ-specific small interfering RNA (siRNA) or MRE11 siRNA (100 nM; Dharmacon) was transfected into HeLa cells by use of Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. After two transfections on consecutive days, cells were harvested 48 h after the second transfection.
Green fluorescent protein (GFP)-vector, GFP-FANCJ, mCherry-vector, or mCherry-MRE11 plasmid DNA (2 μg) was used to transfect different cell lines, using Lipofectamine 2000 (Invitrogen). FA-J null cells were transfected with plasmid DNA encoding GFP-FANCJ or with vector by use of a Nucleofector kit (Lonza) according to the manufacturer's protocol.
Coimmunoprecipitation experiments.
Coimmunoprecipitation experiments were performed as described previously (14). Antibodies for Western blot detection were a rabbit polyclonal FANCJ antibody (Sigma; 1:2,000); mouse monoclonal and rabbit polyclonal MRE11, RAD50, and NBS1 antibodies (Abcam; 1:2,000); a mouse monoclonal WRN antibody against glutathione S-transferase (GST)-tagged WRN1072–1432 (Spring Valley Labs, Sykesville, MD; 1:1,000); and a mouse monoclonal BRCA1 antibody (Santa Cruz Biotechnology; 1:1,000). For FANCJ mapping studies, plasmids encoding Myc-FANCJ fragments were used to transfect 293T cells as described previously (19). Using these transfected 293T cells, immunoprecipitation assays were performed according to the kit protocol (mammalian c-Myc Tag IP/Co-IP kit; Pierce), with an additional 150 mM NaCl in the wash buffer. Proteins were separated using SDS-PAGE and electrotransferred to nitrocellulose membranes. Membranes were blocked in 5% milk–phosphate-buffered saline (PBS)–Tween and incubated with a primary antibody against the Myc epitope (9e10; Santa Cruz) (1:500) or against RAD50 or MRE11 (Abcam; 1:2,000) for 1 h. Membranes were washed, incubated with horseradish peroxidase-linked secondary antibodies (Santa Cruz; 1:5,000), and detected by use of an ECL-prime kit (Amersham).
Gel filtration analysis.
Nuclear extracts were prepared according to a previously described protocol (29) and then applied to a 24-ml Superdex 200 gel filtration column (GE Healthcare) on an Akta fast-performance liquid chromatography (FPLC) system (GE Healthcare). The column was equilibrated and protein eluted with 25 mM Tris-HCl, pH 7.5, 10% glycerol, 0.15 M NaCl, 1 mM EDTA, and 0.5 mM dithiothreitol (DTT), at a rate of 0.1 ml/min. Fractions (0.5 ml) were collected and analyzed by SDS-PAGE and Western blot detection. The column was calibrated using standard molecular mass markers containing blue dextran (2,000 kDa), thyroglobulin (670 kDa), alcohol dehydrogenase (150 kDa), albumin (66 kDa), carbonic anhydrase (29 kDa), and aprotinin (6.5 kDa) (Sigma).
Clonogenic survival assays.
Cells were trypsinized 12 h after the second siRNA transfection with various siRNAs (control, FANCJ, MRE11, or FANCJ plus MRE11). They were then seeded in equal numbers (500 cells per well) in 6-well plates in triplicate (untreated or treated with IR at specific doses) and incubated for 10 days. Cells were stained with crystal violet, and colonies were counted.
Laser irradiation and confocal microscopy.
Immunofluorescence localization of specific proteins at laser-activated psoralen-ICLs (Pso-ICLs) or laser-induced DSBs in vivo was performed as described previously (30). Briefly, a Nikon Eclipse 2000E spinning-disk confocal microscope with five laser imaging modules and a charge-coupled device (CCD) camera (Hamamatsu) was employed. The setup integrated a Stanford Research Systems (SRS) NL100 nitrogen laser with a Micropoint ablation system (Photonics Instruments). Site-specific DNA damage was induced using the SRS NL100 nitrogen laser adjusted to emit at 365 nm. Positions internal to the nuclei of either live untransfected cells or cells transfected with GFP- or mCherry-tagged MRE11 plasmids (GeneCopoeia) were targeted using a 60× oil objective lens. Cells were targeted at 5.5% laser intensity to induce DSBs or 1.7% laser intensity to create Pso-ICLs, and images were captured at various time points and analyzed using Volocity, version 5.0, build 6 (Improvision). The exposure to the laser at the intensities employed in our experiments did not affect cell viability assayed 24 h after treatment. Untransfected cells were stained with specific antibodies, and images were captured. Experiments were performed using an environmental chamber attached to the microscope to maintain experimental conditions (i.e., 37°C, 5% CO2, and 80% humidity). At the indicated time intervals, cells from different areas of the dish were targeted with the laser to generate a time course on a single plate. After the final time point, cells were fixed immediately in freshly prepared 4% formaldehyde in PBS for 10 min at room temperature.
Immunofluorescence staining.
Fixed cells were permeabilized with 0.5% Triton X-100, 1% bovine serum albumin, 100 mM glycine, and 0.2 mg/ml EDTA in PBS on ice for 10 min. Cells were subsequently digested with RNase A in a PBS-EDTA (5 mM) solution for 30 min at 37°C. Cells were blocked in 10% goat serum in PBS for 1 h at 37°C. For immunofluorescence staining, cells were incubated with an appropriate primary antibody diluted in blocking solution for 1 h at 37°C. After three 10-min washes using 0.05% Tween 20 in PBS, cells were incubated with a corresponding fluorescence-tagged secondary antibody (Alexa Fluor–goat anti-mouse, Alexa Fluor–goat anti-rabbit, or Alexa Fluor–donkey anti-goat antibody; Invitrogen). After another three 10-min washes, cells were mounted with ProLong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (Invitrogen). Primary antibodies used were a rabbit polyclonal FANCJ antibody (Sigma; 1:200); mouse monoclonal and rabbit polyclonal MRE11, RAD50, and NBS1 antibodies (Abcam; 1:200); a rabbit polyclonal FANCD2 antibody (Abcam; 1:100); a mouse monoclonal γH2AX antibody (Millipore; 1:400); a rabbit polyclonal γH2AX antibody (Santa Cruz Biotechnology; 1:500); a rabbit polyclonal XPB antibody (Santa Cruz Biotechnology; 1:100); a rabbit polyclonal RAD51 antibody (Santa Cruz Biotechnology; 1:500); the pS2056 DNA-dependent protein kinase catalytic subunit (DNA-PKcs) antibody (Abcam; 1:500); a rabbit polyclonal CtIP antibody (Abcam; 1:200); and a BRCA1 antibody (Santa Cruz Biotechnology; 1:500). Stained cells were visualized and imaged using a Hamamatsu EM-CCD digital camera attached to a Nikon Eclipse TE2000 confocal microscope. Cells (n = 20 to 25) were examined for each experimental point. In each experiment, images of cells at various time points were acquired using the same exposure, gain, sensitivity, and contrast settings.
Analysis of metaphase chromosomes.
Cells were treated with 0.1 μg/ml colcemid for the last 2 h before harvest. Cells were then trypsinized, incubated in 0.075 M KCl solution for 20 min, and subsequently fixed in methanol and acetic acid (3:1 ratio) for 30 min. Cells were dropped onto 50% ethanol-wetted slides and dried. They were then stained using 3% Giemsa solution (pH 6.8) for 10 min.
I-SceI-induced repair assays.
To analyze I-SceI-induced GFP+ frequencies in the indicated nonhomologous end-joining (NHEJ) or HR reporter cells depleted of FANCJ, MRE11, or both FANCJ and MRE11, we transfected I-SceI expression plasmids, as described previously, to induce a break that could be scored for repair by NHEJ (31) or HR (32).
DNA substrates.
The 44- and 66-bp blunt duplex DNA substrates used for this study were constructed as described previously (33, 34). PAGE-purified oligonucleotides used for the preparation of DNA substrates were purchased from Loftstrand Labs (Gaithersburg, MD).
Exonuclease assays.
Mre11/MRN exonuclease reactions were performed in 25 mM MOPS (morpholinepropanesulfonic acid; pH 7.0), 60 mM KCl, 2 mM dithiothreitol, and 5 mM MnCl2, with 1 nM DNA substrate. ExoIII exonuclease assays were performed in 66 mM Tris-HCl (pH 8.0), 0.66 mM MgCl2, with 1 nM DNA substrate. Reaction mixtures were incubated for the indicated times at 37°C, followed by treatment with 1 mg proteinase K for 15 min at 37°C. Twelve microliters of formamide loading buffer was added to the reaction mixtures, which were heated at 95°C for 5 min and then loaded onto 8% polyacrylamide denaturing gels. Gels were electrophoresed at 55 W for approximately 80 min and then exposed in a phosphorimager cassette or on film. The data represent percentages of intact DNA substrate in the reaction mixtures divided by the percentage of intact DNA substrate in the no-enzyme control, as determined from the nondigested/digested DNA ratios, for at least three independent experiments.
RESULTS
FANCJ is associated with the MRN complex.
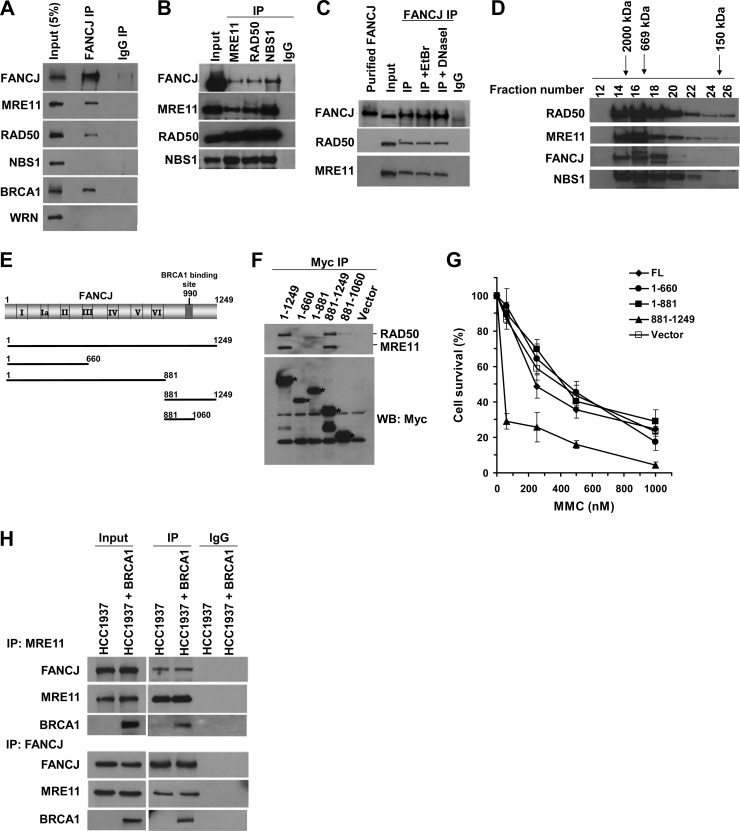
To identify novel proteins associated with FANCJ, we performed anti-FANCJ coimmunoprecipitation experiments followed by SDS-polyacrylamide gel analysis. Unique bands from FANCJ immunoprecipitates that were not observed in the precipitated control IgG incubations were sequenced by nano-liquid chromatography (nano-LC)–mass spectrometry. A band corresponding to an approximate molecular mass of 150 kDa was consistently identified as RAD50 (Table 1). To further explore if FANCJ is indeed associated with RAD50 and its partners (MRE11 and NBS1), we performed coimmunoprecipitation experiments and analyzed the precipitated samples by Western blotting. The FANCJ antibody precipitated MRE11 and RAD50, as well as BRCA1, from HeLa cell nuclear extracts, whereas neither protein was precipitated with a control IgG antibody (Fig. 1A). As a negative control, the WRN protein was not coprecipitated with FANCJ by the FANCJ antibody. Although NBS1 was not coprecipitated with FANCJ by the anti-FANCJ antibody, FANCJ was coimmunoprecipitated with NBS1 by the anti-NBS1 antibody (Fig. 1B). FANCJ was also coimmunoprecipitated with MRE11 and RAD50 by the anti-MRE11 and anti-RAD50 antibodies, respectively. Since FANCJ and MRN both interact with DNA, we tested if their association was DNA dependent. HeLa cell nuclear extracts were pretreated with either DNase I or ethidium bromide (EtBr) and subsequently incubated with anti-FANCJ antibody. Western blot analysis demonstrated that RAD50 and MRE11 were coimmunoprecipitated with FANCJ (Fig. 1C), suggesting that the interaction was DNA independent.
Table 1.
Identified RAD50 peptide sequences from FANCJ immunoprecipitate
| Peptide mass (Da) | Peptide sequence |
|---|---|
| 1,209.67 | LKEIEHNLSK |
| 1,532.82 | FQQEKEELINKK |
| 1,242.66 | LEENIDNIKR |
| 1,029.54 | SELLVEQGR |
| 1,423.66 | YELQQLEGSSDR |
| 1,212.65 | DVNGELIAVQR |
| 2,213.02 | VFQGTDEQLNDLYHNHQR |
| 1,764.86 | NVKYELQQLEGSSDR |
| 1,234.62 | GQDIEYIEIR |
| 1,212.70 | ILELDQELIK |
| 2,063.04 | VFQTEAELQEVISDLQSK |
| 2,072.17 | QIITFFSPLTILVGPNGAGK |
Fig 1.
FANCJ is associated with the MRN complex. (A) Coimmunoprecipitation (IP) was performed using the FANCJ antibody and HeLa nuclear extract (NE). The blot was probed with antibodies against the indicated proteins. Lane 1 represents 5% of the input. (B) Antibodies against MRE11, RAD50, or NBS1 were used to coimmunoprecipitate FANCJ with MRE11, RAD50, or NBS1. An IgG antibody was used as a negative control. (C) The FANCJ antibody coimmunoprecipitated FANCJ, RAD50, and MRE11 from HeLa NE in the presence of ethidium bromide (10 μg/ml) or DNase I (2 μg/ml). The blot was probed with MRE11, RAD50, and FANCJ antibodies. Note that the recombinant FANCJ protein migrated slightly higher than endogenous FANCJ. (D) Elution samples from HeLa NE fractionated by a Sephadex 200 gel filtration column were separated in an SDS gel, and the blot was probed with the indicated antibodies. Proteins with known molecular masses were used to calibrate the column, and their elution positions are shown above the figure. (E) Various Myc-tagged FANCJ constructs were expressed in 293T cells and tested for binding to endogenous MRE11/RAD50. (F) Myc IP experiments were performed with 293T cells that were transfected with vector alone, full-length FANCJ, or the specified FANCJ construct. IP products were analyzed by Western blotting (WB) with antibodies against Myc, MRE11, and RAD50. Asterisks denote the migration of the different Myc-tagged FANCJ species. (G) HeLa cells transfected with vector or the indicated FANCJ constructs were treated with the indicated concentrations of MMC. Cell proliferation was measured using the WST-1 assay (Roche, Indianapolis, IN) after 48 h. Experimental data are averages for three independent experiments done in triplicate, and standard deviations (SD) are indicated by error bars. (H) Immunoprecipitates from NE of HCC1937 (BRCA1 mutant) cells or BRCA1-corrected cells obtained using anti-FANCJ or anti-MRE11 antibody were analyzed by Western blotting with anti-FANCJ, anti-MRE11, and anti-BRCA1 antibodies.
Since FANCJ was found to coimmunoprecipitate with each member of the MRN complex, we wanted to establish if FANCJ could be found associated with all three proteins of the MRN complex. Gel filtration analysis of HeLa cell nuclear extracts demonstrated that FANCJ eluted in a large-molecular-weight fraction with all three MRN components (Fig. 1D). In addition, we observed that RAD50 and MRE11 as well as NBS1 and FANCJ, upon longer exposure of the Western blot membrane to X-ray film (our unpublished data), could be found in a lower-molecular-weight range that would be expected for their monomeric forms.
To define the FANCJ domain required for binding of RAD50/MRE11, several FANCJ-Myc fusion proteins of various lengths (Fig. 1E) were expressed in 293T cells, and the corresponding FANCJ protein fragments were precipitated from the cell lysates with c-Myc antibody-tagged agarose beads. Full-length Myc-tagged FANCJ precipitated endogenous RAD50/MRE11, but FANCJ1-660 or FANCJ1-881 did not. Thus, the FANCJ N-terminal region or helicase core domain is not required for binding to RAD50/MRE11 (Fig. 1F). Expression of FANCJ881-1249 was sufficient to coprecipitate RAD50/MRE11, suggesting that the C-terminal noncatalytic domain of FANCJ mediates the interaction with the RAD50-MRE11 complex. A very residual amount of RAD50/MRE11 was detected using Myc-FANCJ881-1060, suggesting that this region of FANCJ, which also mediates the interaction with BRCA1, may enhance the association of FANCJ with RAD50/MRE11. Since RAD50- or MRE11-deficient cells, like FANCJ mutant cells, are sensitive to ICL agents, we asked if 293T cells expressing the RAD50/MRE11-interacting Myc-FANCJ881-1249 construct were sensitive to MMC. Indeed, they were sensitive, whereas cells expressing Myc-FANCJ1-660 or Myc-FANCJ1-881 displayed MMC sensitivity comparable to that of cells transfected with empty vector expressing just the Myc epitope or Myc-FANCJ (full length) (Fig. 1G). These results suggest that expression of a FANCJ C-terminal fragment which interacts with RAD50/MRE11 exerts a dominant-negative effect on MMC resistance.
Since FANCJ was originally identified as a BRCA1-interacting protein (17), we wanted to assess if the FANCJ-MRN interaction was BRCA1 dependent. Coimmunoprecipitation experiments were performed using BRCA1 mutant and corrected HCC1937 cells. FANCJ and MRE11 were reciprocally coimmunoprecipitated from BRCA1 mutant or corrected cells (Fig. 1H), suggesting that the FANCJ-MRE11 interaction did not require BRCA1. BRCA1, which is also known to interact with the MRN complex (18, 35, 36), was coimmunoprecipitated by antibodies against FANCJ or MRE11 from the extracts of BRCA1-corrected HCC1937 cells (Fig. 1H).
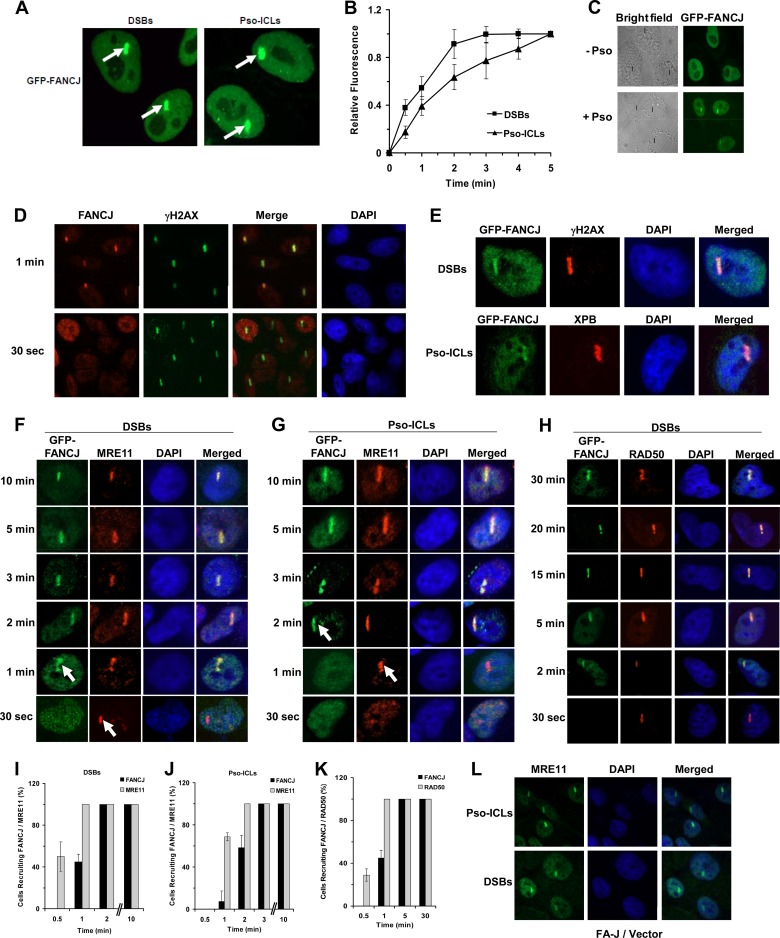
Recruitment kinetics of FANCJ, MRE11, and RAD50 to laser-induced DSBs or psoralen cross-links.
The involvement of FANCJ and MRN in recombinational DNA repair led us to examine their recruitment kinetics to laser-induced DSBs or Pso-ICLs introduced at localized areas within the nuclei of actively dividing transformed HeLa cells. The kinetics of GFP-FANCJ recruitment to the target area was monitored by live-cell imaging. Representative cells with GFP-FANCJ recruitment to DSBs or Pso-ICLs are shown in Fig. 2A, and quantification is shown in Fig. 2B. GFP-FANCJ was recruited to the laser-irradiated sites of cells incubated with psoralen but not to those of cells incubated in the absence of psoralen (Fig. 2C). Similar to GFP-FANCJ, endogenous FANCJ was recruited to laser-induced DSBs within 1 min in untransfected HeLa cells, as shown by staining with anti-FANCJ antibody (Fig. 2D). γH2AX was used as a marker for DSBs (Fig. 2E). The TFIIH component XPB, required for nucleotide excision repair, a pathway necessary for removal of the cross-link remnant during ICL repair (37), is known to be recruited rapidly to psoralen photoadducts (30) and served as a positive control for the presence of Pso-ICLs (Fig. 2E).
Fig 2.
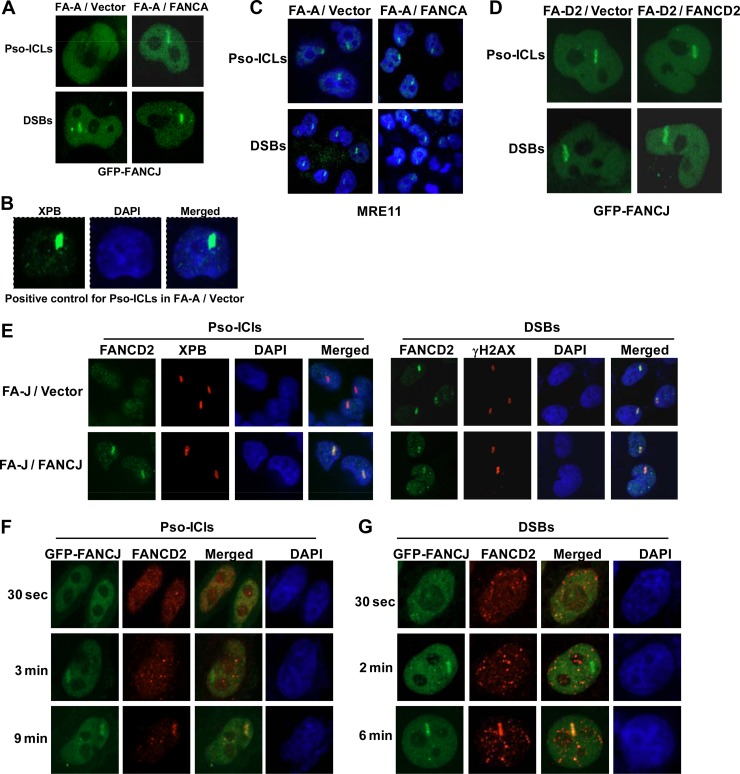
Recruitment kinetics of FANCJ and MRE11 to laser-induced DSBs or psoralen cross-links. (A) GFP-FANCJ-transfected HeLa cells were treated with a laser (5.5%) at defined regions to induce localized DSBs or incubated with psoralen followed by laser treatment (1.8%) to induce Pso-ICLs. GFP-FANCJ recruitment to Pso-ICLs or DSBs was observed by live-cell imaging using confocal immunofluorescence microscopy. Arrows indicate FANCJ recruitment to laser-induced DSBs or Pso-ICLs, as specified. (B) Graphical representation of GFP-FANCJ recruitment to sites of DSBs or Pso-ICLs from 0 to 5 min after laser-induced damage, as detected by live-cell imaging. Relative fluorescence values expressed as a function of time are means for three independent experiments, with SD indicated by error bars. (C) Live-cell imaging of GFP-FANCJ recruitment to laser-irradiated GFP-FANCJ-transfected HeLa cells in either the presence or absence of psoralen. Black lines in the bright-field images indicate the defined areas irradiated with the laser. (D) Laser-targeted untransfected HeLa cells were stained with anti-FANCJ antibody and anti-γH2AX antibodies as described in Materials and Methods. (E) Laser-targeted GFP-FANCJ-transfected HeLa cells were stained with anti-γH2AX and anti-XPB antibodies as positive controls for DSBs and Pso-ICLs, respectively. (F and G) GFP-FANCJ-transfected HeLa cells were targeted with a laser at specific time points to induce DSBs (F) or Pso-ICLs (G), and cells were fixed and stained with anti-MRE11 antibody. (H) GFP-FANCJ-transfected HeLa cells were targeted with a laser at specific time points to induce DSBs, and cells were fixed and stained with anti-RAD50 antibody. (I and J) Graphical representation of GFP-FANCJ and MRE11 recruitment to sites of DSBs (I) or Pso-ICLs (J) from 0.5 to 10 min after laser-induced damage. (K) Graphical representation of GFP-FANCJ and RAD50 recruitment to sites of DSBs from 0.5 to 30 min after laser-induced damage. (L) FA-J cells were targeted with a laser to create either DSBs or Pso-ICLs and then were fixed 2 min after laser irradiation, followed by staining with anti-MRE11 antibody. For each experimental point, 20 to 25 cells were examined.
To determine the temporal relationship between FANCJ and MRE11 for their recruitment to sites of DNA damage, laser-induced DSBs or ICLs were created at specific time intervals in HeLa cells transfected with GFP-FANCJ, and cells were fixed and stained with MRE11 antibody. MRE11 was recruited to DSBs within 30 s, whereas GFP-FANCJ was recruited within 30 to 60 s (Fig. 2F and I). The signals for both FANCJ and MRE11 persisted for at least 30 min (our unpublished data). For Pso-ICLs, MRE11 was recruited within 1 min, whereas FANCJ was recruited within 1 to 2 min (Fig. 2G and J). In either case, MRE11 and FANCJ strongly colocalized after treatments that induced Pso-ICLs or DSBs. Similar results were found for RAD50 and FANCJ recruitment kinetics to DSBs (Fig. 2H and K) and Pso-ICLs (unpublished data). These experiments demonstrated that both MRE11 and RAD50 colocalized with FANCJ at DSBs or Pso-ICLs; however, FANCJ consistently localized to the site of damage 30 to 60 s after MRE11 or RAD50. Consistent with these observations, MRE11 recruitment to DSBs or Pso-ICLs was not dependent on FANCJ, as evidenced by normal recruitment in FA-J cells (Fig. 2L).
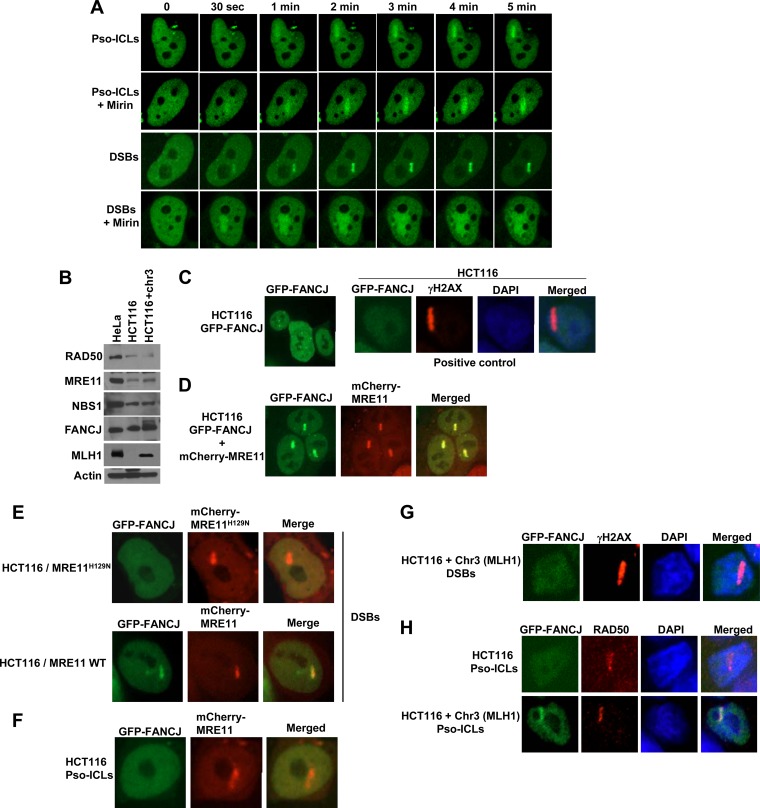
FANCJ recruitment to DSBs (but not Pso-ICLs) is dependent on MRE11 nuclease.
Since FANCJ is recruited to DSBs and Pso-ICLs after the appearance of MRE11, we wanted to determine if inhibition of MRE11's enzymatic function would exert any effect on FANCJ recruitment. We studied FANCJ recruitment in HeLa cells treated with the small-molecule MRE11 exonuclease inhibitor mirin [Z-5-(4-hydroxybenzylidene)-2-imino-1,3-thiazolidin-4-one]. Mirin was previously shown to inhibit MRE11 exonuclease activity without affecting the ability of MRE11 to interact with Rad50, NBS1, or damaged DNA (38). In the presence of mirin, FANCJ recruitment to DSBs was adversely affected, as evidenced by the diffuse staining pattern, whereas no detectable effect on FANCJ recruitment to Pso-ICLs was observed (Fig. 3A). Diffuse staining of FANCJ to laser-induced DSBs suggested that FANCJ failed to be localized properly or stably retained at the site of the targeted breaks when MRE11 exonuclease activity was inhibited in vivo by the pharmacological agent mirin.
Fig 3.
FANCJ recruitment to DSBs (but not Pso-ICLs) is dependent on MRE11. (A) GFP-FANCJ-transfected HeLa cells were either left untreated or exposed to mirin (100 μM) for 1 h and then monitored for FANCJ recruitment to DSBs or Pso-ICLs, as indicated. (B) Lysates of HeLa cells, HCT116 cells, and HCT116 cells complemented with chromosome 3 were immunoblotted with antibodies against the indicated proteins. β-Actin served as a loading control. (C to E) GFP-FANCJ recruitment to laser-induced DSBs was visualized using the human colon carcinoma cell line HCT116 transfected with GFP-FANCJ (C), GFP-FANCJ and mCherry-MRE11 (D), or GFP-FANCJ and mCherry-MRE11H129N (E). γH2AX served as a marker for DSBs in HCT116 cells. (F) GFP-FANCJ was not recruited to Pso-ICLs in HCT116 cells transfected with mCherry-MRE11. (G and H) Chromosome 3 (which harbors the MLH1 gene)-complemented HCT116 cells were transfected with GFP-FANCJ and examined for FANCJ recruitment to DSBs (G) or Pso-ICLs (H). HCT116 cells or HCT116 cells complemented with chromosome 3 were targeted with a laser to induce DSBs or Pso-ICLs and were stained with the indicated antibodies. For each experimental point, 20 to 25 GFP-FANCJ-transfected cells were examined and showed similar responses to laser treatment.
To further investigate the importance of MRE11 nuclease activity for FANCJ recruitment to DSBs, we performed live-cell imaging experiments with an MRE11 exonuclease-deficient human colon carcinoma cell line, HCT116 (39). MRE11, RAD50, NBS1, FANCJ, and MLH1 protein levels in HCT116 cells and HCT116 cells complemented with chromosome 3 in comparison to HeLa cells were analyzed by Western blotting (Fig. 3B). The level of MRE11 protein was confirmed to be reduced in HCT116 cells. Confocal analysis of HCT116 cells revealed that FANCJ failed to be recruited to DSBs (Fig. 3C) or Pso-ICLs (Fig. 3H). For positive controls in HCT116 cells, γH2AX formed at DSBs (Fig. 3C) and RAD50 was recruited to Pso-ICLs (Fig. 3H). Cotransfection of mCherry-MRE11 and GFP-tagged FANCJ into HCT116 cells restored FANCJ recruitment to DSBs (Fig. 3D). In contrast, HCT116 cells transfected with mCherry-MRE11H129N, encoding a nuclease-defective MRE11 protein (40), failed to rescue GFP-FANCJ recruitment to the laser-induced DSBs (Fig. 3E). In contrast to the case with laser-induced DSBs, MRE11 expression in HCT116 cells did not restore GFP-FANCJ recruitment to Pso-ICLs (Fig. 3F).
In addition to defective MRE11, HCT116 cells are also deficient in the mismatch repair protein MLH1 (41). It was reported that the physical association of FANCJ with MLH1 is required for normal cross-link resistance (19); therefore, the inability of FANCJ to be recruited to Pso-ICLs in HCT116 cells or HCT116 cells exogenously expressing nuclease-active MRE11 may reflect the MLH1 deficiency. To address this possibility, HCT116 cells complemented with chromosome 3, which carries the wild-type MLH1 gene, were tested for FANCJ recruitment to both DSBs and Pso-ICLs. These experiments demonstrated that FANCJ was recruited, with a modest delay (∼5 min instead of 2 min), to Pso-ICLs in HCT116 cells, but very poorly, if at all, to DSBs (Fig. 3G and H), suggesting that the presence of MLH1 is required for FANCJ recruitment to cross-links but not to DSBs. γH2AX served as a positive control for DSBs in HCT116 cells complemented with chromosome 3 (Fig. 3G). These results, together with the mirin studies, suggest that MRE11 exonuclease activity is necessary for FANCJ recruitment to DSBs. MLH1, on the other hand, is important for FANCJ recruitment to Pso-ICLs.
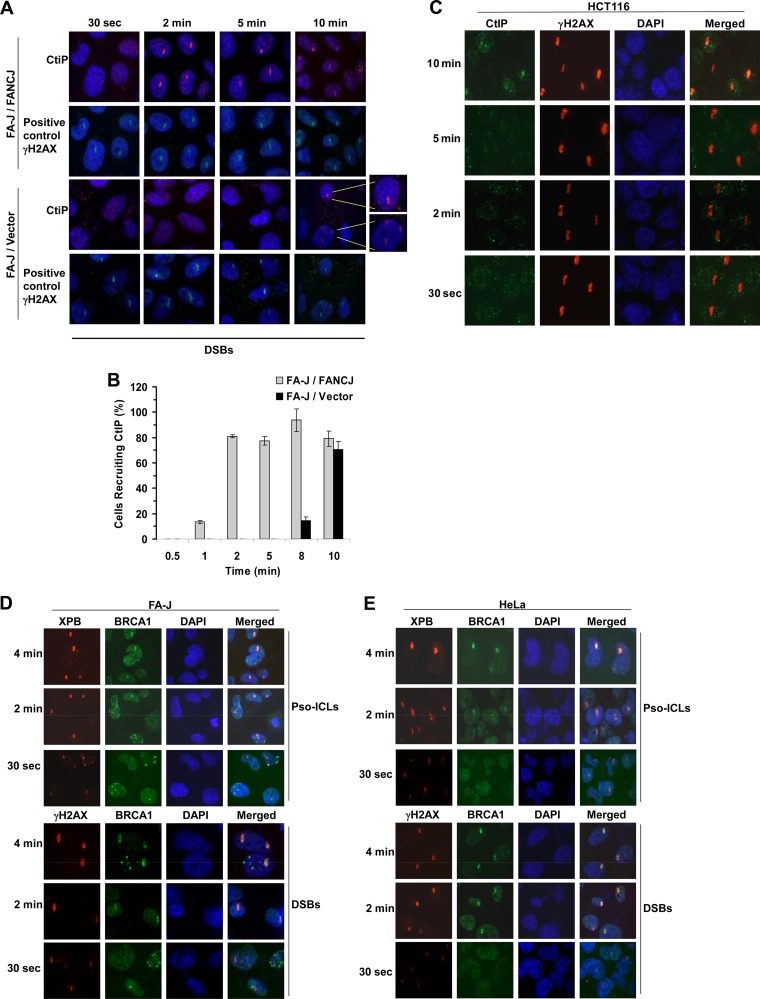
Effect of FANCJ on recruitment of CtIP to DSBs.
The evidence presented thus far suggested that FANCJ recruitment to DSBs is dependent on MRE11 exonuclease; therefore, we asked what influence FANCJ status had on the recruitment of CtIP, an accessory factor for the strand resection nuclease MRE11 that is implicated in downstream events of DSB repair (23). CtIP recruitment to DSBs was delayed by 8 to 10 min in FA-J cells compared with FA-J cells complemented with wild-type FANCJ (Fig. 4A and B), consistent with our observations that CtIP is recruited after FANCJ to DSBs in FANCJ-proficient cells (unpublished data). These results suggest that the absence of FANCJ perturbs normal recruitment of CtIP to DSBs. Accordingly, CtIP recruitment to DSBs was delayed in MRE11 exonuclease-deficient HCT116 cells (Fig. 4C), which were also shown to be defective in recruiting FANCJ to laser-induced DSBs (Fig. 3C). MRE11-deficient cells are also defective in the initial localization of BRCA1 to IR-induced DNA damage (42). In contrast to what was observed for CtIP, BRCA1 was recruited normally to laser-induced DSBs or Pso-ICLs in FA-J mutant cells (Fig. 4E), comparable to what was observed in HeLa cells (Fig. 4D).
Fig 4.
Effect of FANCJ on recruitment of DNA repair factors to DSBs. (A) An isogenic pair of FA-J null and corrected cells was analyzed for CtIP recruitment to laser-induced DSBs. γH2AX served as a positive control. (B) Graphical representation of CtIP recruitment to sites of DSBs from 0.5 to 10 min after laser-induced damage in FA-J null and corrected cells. (C) HCT116 cells were targeted with a laser at specific times to create DSBs. Cells were fixed and stained with antibodies against CtIP or γH2AX. (D and E) HeLa (D) or FA-J (E) cells were targeted with a laser at specific times to create DSBs or Pso-ICLs. Cells were fixed and stained with antibodies against BRCA1 or XPB.
FANCJ recruitment to cross-linked DNA but not DSBs is dependent on FA core complex.
Our results presented in Fig. 3 demonstrate that FANCJ recruitment to laser-induced DSBs but not Pso-ICLs is dependent on the MRE11 nuclease. To address the specificity of the effect of MRE11 deficiency on FANCJ localization to DNA damage, we wanted to evaluate if FANCJ recruitment to laser-induced Pso-ICLs displayed an expected genetic dependence on an upstream player (FANCA) of the ICL repair pathway. To add further specificity, we hypothesized that FANCJ would not be dependent on FANCD2 for recruitment to Pso-ICLs because, in contrast to members of the FA core complex, FANCJ is not required for FANCD2 monoubiquitination (13), a key activation event in the FA pathway. Therefore, it has been proposed that FANCJ operates downstream in the pathway to help mediate recombinational repair. We examined DNA damage-induced FANCJ localization in an isogenic pair of FA-A or FA-D2 mutant and corrected cells. FANCJ was recruited normally to DSBs but not Pso-ICLs in FA-A cells. FA-A corrected cells exhibited normal FANCJ localization to Pso-ICLs (Fig. 5A). XPB served as a positive control for the presence of Pso-ICLs in FA-A cells (Fig. 5B). Although FANCJ was not recruited to Pso-ICLs in FA-A-deficient cells, MRE11 was recruited normally (Fig. 5C), suggesting a distinction between FANCJ and MRE11 in their dependence on FANCA for recruitment to cross-links.
Fig 5.
FANCJ recruitment to cross-linked DNA but not DSBs is dependent on the FA core complex. (A) FANCJ recruitment to Pso-ICLs or DSBs was examined in an isogenic pair of FA-A and corrected cells transfected with GFP-FANCJ. (B) FA-A cells were stained for endogenous XPB with an anti-XPB antibody as a positive control for Pso-ICLs. (C) An isogenic pair of FA-A mutant and corrected cells was analyzed for MRE11 recruitment to laser-induced Pso-ICLs and DSBs. (D) An isogenic pair of FA-D2 mutant and corrected cells transfected with GFP-FANCJ was analyzed for GFP-FANCJ recruitment to laser-induced Pso-ICLs and DSBs. (E) An isogenic pair of FA-J mutant and corrected cells was analyzed for FANCD2 recruitment to laser-induced Pso-ICLs and DSBs. XPB and γH2AX served as positive controls for Pso-ICLs and DSBs, respectively. (F and G) HeLa cells transfected with GFP-FANCJ were analyzed for recruitment of FANCJ and FANCD2 to Pso-ICLs (F) or DSBs (G) at the indicated time points. For each experimental time point, 20 to 25 cells were examined, and a typical response is shown.
In contrast to what was observed in FA-A cells, FANCJ recruitment to Pso-ICLs was normal in FA-D2 cells (Fig. 5D). These results are consistent with a recent study by the Andreassen lab in which it was found that the FANCD2 mutational status had no effect on FANCJ foci following cellular exposure to the DNA cross-linking agent mitomycin C (16). FANCJ recruitment to DSBs was also normal in FA-D2 cells (Fig. 5D). From these results, we conclude that FANCJ recruitment to Pso-ICLs but not DSBs is dependent on the FA core complex, whereas FANCJ recruitment to Pso-ICLs as well as DSBs is independent of FANCD2.
Although FANCJ recruitment to laser-induced DSBs or Pso-ICLs was not found to be dependent on FANCD2 status, we wanted to determine if the converse was true, i.e., was FANCD2 recruitment dependent on FANCJ? This analysis demonstrated that FANCD2 recruitment to Pso-ICLs was dependent on FANCJ (Fig. 5E). On the other hand, FANCD2 was recruited to DSBs similarly in both FA-J null and corrected cells (Fig. 5E). These results led us to test the recruitment kinetics of FANCJ and FANCD2. We observed that FANCJ was recruited to Pso-ICLs in HeLa cells in ∼2 min, whereas FANCD2 was recruited later, in ∼9 min (Fig. 5F). FANCD2 was also recruited later than FANCJ to DSBs (Fig. 5G).
FANCJ binds directly to MRE11 and inhibits MRE11 exonuclease activity.
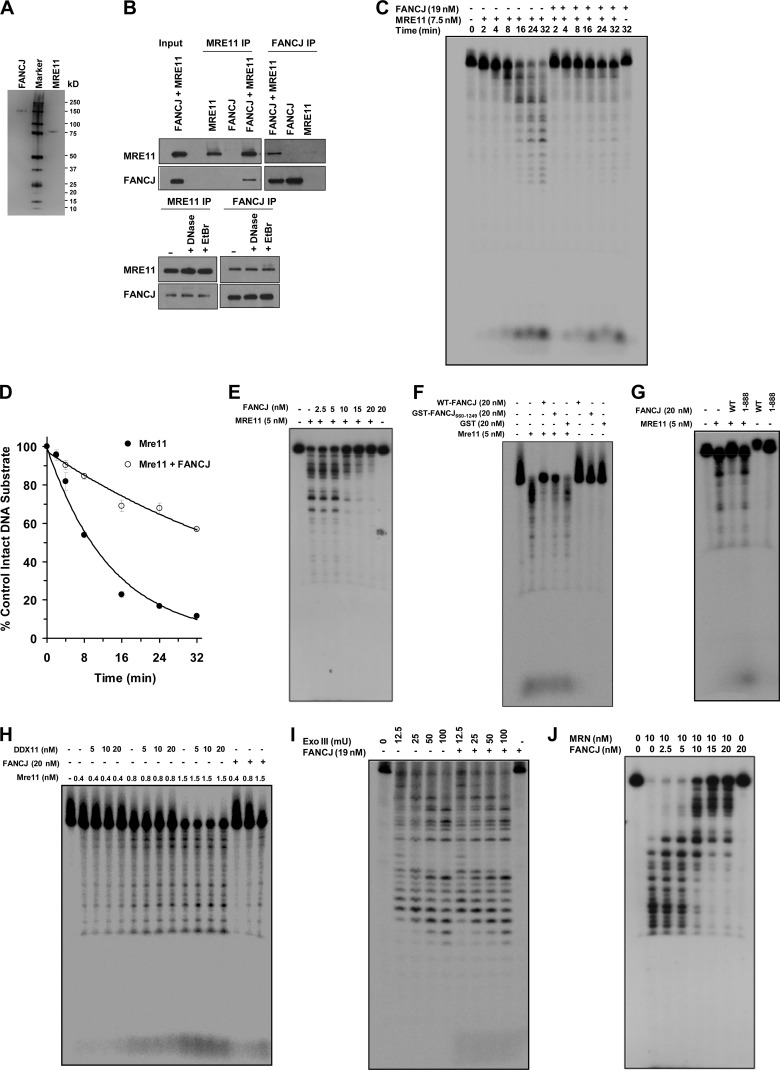
The fact that FANCJ recruitment to laser-induced DSBs was dependent on MRE11 but not the FA core complex or FANCD2 prompted us to test for a direct physical and functional interaction between FANCJ and MRE11. Purified recombinant FANCJ and MRE11 proteins, shown by silver staining in Fig. 6A, were reciprocally coimmunoprecipitated with each other (Fig. 6B). In control experiments, FANCJ failed to be precipitated by the anti-MRE11 antibody when MRE11 was omitted from the binding incubation mixture. FANCJ and MRE11 were reciprocally coimmunoprecipitated with each other in the presence of DNase I or EtBr, suggesting a DNA-independent protein interaction.
Fig 6.
FANCJ binds directly to MRE11 and inhibits MRE11 exonuclease activity in a specific manner. (A) Silver-stained polyacrylamide gel showing purified FANCJ (100 ng) and MRE11 (100 ng) recombinant proteins. (B) Purified FANCJ and MRE11 proteins were reciprocally coimmunoprecipitated from a mixture of the two proteins by using antibodies against FANCJ and MRE11. The blots were probed with anti-MRE11 (top) and anti-FANCJ (bottom) antibodies. (C) Representative gel showing products from reaction mixtures containing MRE11 (7.5 nM) incubated in MRE11 exonuclease reaction buffer with 1 nM 5′-32P-end-labeled, 44-bp blunt duplex DNA substrate in the presence or absence of FANCJ (19 nM) for the indicated times, as described in Materials and Methods. (D) Quantitation of Mre11 exonuclease activity in the absence (closed circles) or presence (open circles) of FANCJ (19 nM) as shown in panel C. The ratio of nondigested DNA to the total DNA in each reaction was determined using ImageQuant TL software (GE Healthcare). Results are averages for three independent experiments, and standard deviations are indicated by error bars. (E) Representative gel showing products from reaction mixtures containing MRE11 (5 nM) and the indicated concentrations of FANCJ and incubated with 1 nM 44-bp blunt duplex DNA substrate in MRE11 exonuclease reaction buffer for 30 min as described in Materials and Methods. (F) Representative gel showing the MRE11 exonuclease assay products from reaction mixtures containing MRE11 and purified GST (20 nM) or GST-FANCJ660–1249. (G) Representative gel showing products from reaction mixtures containing MRE11 (5 nM) and 19 nM FANCJ-WT or FANCJ1–888 incubated with 1 nM 44-bp blunt duplex DNA substrate in MRE11 exonuclease reaction buffer for 30 min. (H) Representative gel showing products from reaction mixtures containing the indicated concentrations of DDX11 incubated with 1 nM 44-bp blunt duplex DNA substrate in the presence or absence of the indicated concentrations of MRE11 for 30 min under MRE11 reaction conditions, as described in Materials and Methods. (I) Representative gel showing products from reaction mixtures containing the indicated amounts of ExoIII incubated with 1 nM 44-bp blunt duplex DNA substrate in the presence or absence of FANCJ (19 nM) for 30 min under ExoIII reaction conditions, as described in Materials and Methods. (J) FANCJ inhibits MRN 3′-to-5′ exonuclease activity. The image shows a representative gel showing products from reaction mixtures containing MRN (10 nM) and the indicated concentrations of FANCJ incubated with 1 nM 66-bp blunt duplex DNA substrate for 30 min in MRN exonuclease reaction buffer as described in Materials and Methods.
We next asked if FANCJ and MRE11 interact functionally. Using a forked duplex DNA substrate that FANCJ actively unwinds, we detected no effect of MRE11 or the MRN complex on FANCJ helicase activity (our unpublished data). However, FANCJ inhibited MRE11 3′-to-5′ exonuclease activity on a blunt duplex DNA substrate, as demonstrated by rate analysis experiments (Fig. 6C). In the absence of FANCJ, MRE11 (7.5 nM) digested approximately 80% of the blunt duplex substrate in 16 min; however, in the presence of FANCJ (19 nM), MRE11 digested only 30% of the substrate. Linear regression analysis of the first 16 min of the reaction demonstrated that MRE11 exonuclease activity was inhibited 2.5-fold by FANCJ (Fig. 6D). Moreover, FANCJ inhibited MRE11 3′-to-5′ exonuclease activity in a protein concentration-dependent manner (Fig. 6E). A recombinant GST-tagged FANCJ660–1249 fragment retained the ability to inhibit MRE11 exonuclease activity (Fig. 6F). In contrast, an N-terminal region of FANCJ (FANCJ1–888) that lacks the interacting domain for MRE11 did not inhibit MRE11 exonuclease activity (Fig. 6G). Moreover, the sequence-related Fe-S DNA helicase DDX11, which is mutated in Warsaw breakage syndrome (43), did not inhibit MRE11 exonuclease activity (Fig. 6H), suggesting that the effect of FANCJ was specific. FANCJ did not affect ExoIII 3′-to-5′ exonuclease activity (Fig. 6I). Consistent with these observations, FANCJ binds poorly to blunt duplex DNA (8), suggesting that the inhibitory effect of FANCJ on MRE11 exonuclease activity was specific and mediated by a direct physical interaction between the proteins.
We next tested the effects of increasing concentrations of FANCJ on the MRN complex 3′-to-5′ exonuclease activity (Fig. 6J). We observed that FANCJ concentrations as low as 2.5 nM inhibited MRN complex exonuclease activity. Inhibition of MRN exonuclease activity was dependent on the FANCJ concentration.
Effects of FANCJ and MRE11 depletion on IR sensitivity, chromosomal instability, and recombinational repair.
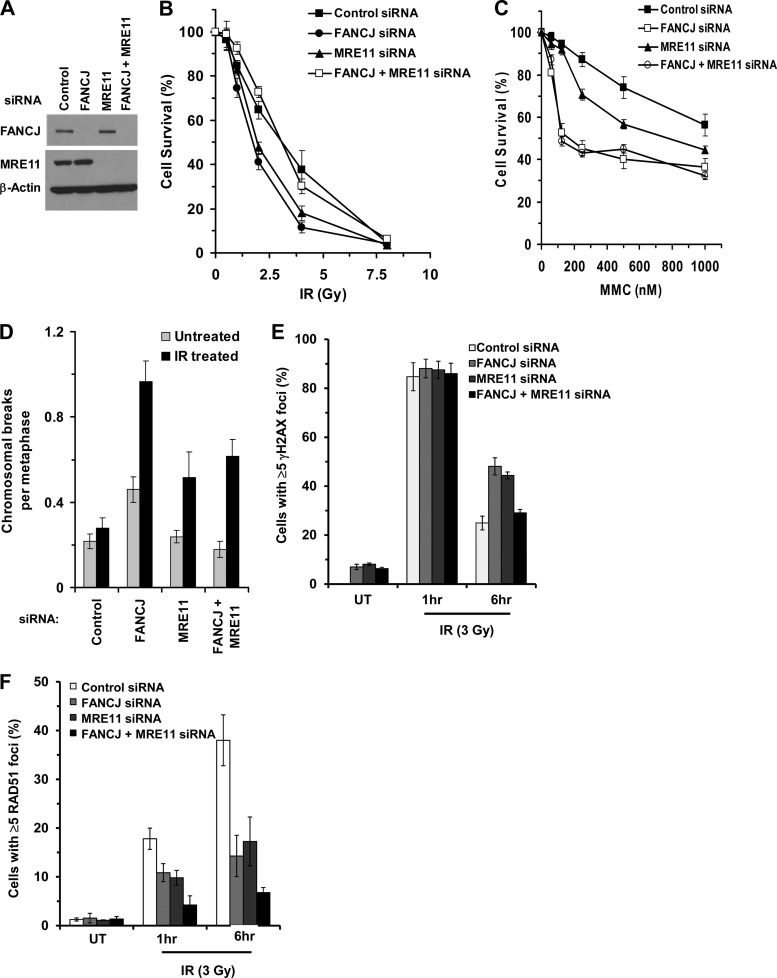
Having demonstrated that FANCJ and MRE11 interact and that the MRE11 status affects FANCJ localization at DSBs, we sought to determine if FANCJ and MRE11 behave epistatically to each other in terms of DSB resistance as measured by cell survival. Therefore, we depleted FANCJ, MRE11, or both FANCJ and MRE11 by RNA interference (Fig. 7A), and we examined IR-treated cells for the ability to form colonies (Fig. 7B). Untreated cells transfected with siRNA (control, FANCJ, MRE11, or combined) displayed only a modest reduction (≤10%) in cell growth. Depletion of FANCJ or MRE1 sensitized cells to IR compared to control cells transfected with a scrambled siRNA. However, codepletion of both FANCJ and MRE11 restored IR resistance to a level similar to that of control siRNA-treated cells. Since codepletion of FANCJ and MRE11 restored IR resistance, we assessed what the outcome of FANCJ-MRE11 codepletion would be for MMC resistance. In this case, we observed that cells depleted of both FANCJ and MRE11 were as sensitive to MMC as cells depleted only of FANCJ (Fig. 7C); however, codepletion did not restore MMC resistance as it did for IR treatment.
Fig 7.
Effects of FANCJ and MRE11 depletion on IR-induced double-strand breaks. (A) HeLa cells were transfected twice with siRNAs against FANCJ and/or MRE11 or with scrambled siRNA, as described in Materials and Methods, and were evaluated by Western blotting for FANCJ or MRE11, with β-actin serving as a loading control. (B) One day after the second siRNA transfection, cells were exposed to the indicated doses of IR, and cell survival was measured using a colony formation assay. Experimental data are means for three independent experiments done in triplicate, and SD are indicated by error bars. (C) One day after the second siRNA transfection, cells were exposed to the indicated concentrations of MMC, and a cell proliferation assay (WST-1 assay) was performed after 48 h. Experimental data are means for three independent experiments done in triplicate, and SD are indicated by error bars. (D) Metaphase spreads from cells transfected as in panel A and either left untreated or exposed to IR (2 Gy) were analyzed for chromosomal aberrations (chromatid breaks and radials). The graphical data represent analyses of a total of 50 metaphase spreads for each sample with a P value of <0.001. (E and F) The specified control siRNA-, FANCJ siRNA-, and/or MRE11 siRNA-transfected cells were either left untreated (UT) or exposed to IR (3 Gy). At the indicated time after IR exposure, cells were fixed and stained with γH2AX (E) or RAD51 (F) antibodies. The cells with >5 γH2AX or RAD51 foci were counted and expressed as a percentage. The graphs show averages for three independent experiments, and SD are indicated by error bars. More than 100 cells were counted for each sample in each experiment with a P value of <0.001.
We also examined spontaneous chromosomal instability in FANCJ-, MRE11-, or FANCJ-plus-MRE11-depleted cells. MRE11 depletion of ∼90% showed a low level of chromosomal abnormalities, similar to that of control siRNA-treated cells (Fig. 7D), and consistent with earlier studies (40). FANCJ depletion (∼90%) in HeLa cells resulted in a 2-fold increase in the number of spontaneous chromosomal abnormalities per metaphase, consistent with an earlier observation (12). Codepletion of both FANCJ and MRE11 restored the level of chromosomal anomalies to that of the control siRNA-treated cells. These results indicate that the additional depletion of MRE11 in FANCJ-depleted cells suppressed the spontaneous chromosomal instability.
Similarly, we examined the effects of FANCJ and/or MRE11 depletion on chromosomal stability in cells exposed to IR (2 Gy) (Fig. 7D). FANCJ-depleted cells showed a 4-fold increase in the number of chromosomal breaks per metaphase, whereas MRE11-depleted cells displayed a 2-fold increase. Cells that were codepleted of both FANCJ and MRE11 exhibited a level of chromosomal abnormalities similar to that of MRE11-depleted cells and one-third less than that of FANCJ-depleted cells.
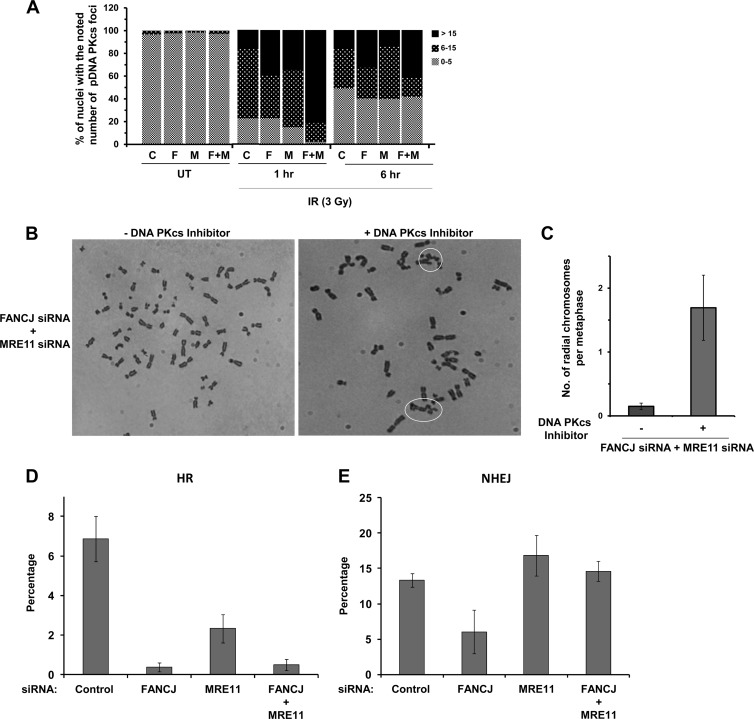
Since double depletion of FANCJ and MRE11 restored IR resistance as measured by colony formation, we asked what the effect would be on DSB accumulation as measured by γH2AX foci. After 6 h of recovery from IR exposure, cells depleted of both FANCJ and MRE11 showed fewer γ-H2AX foci than cells depleted of either FANCJ or MRE11, with a very comparable number to that in the control siRNA-treated cells (Fig. 7E). Since RAD51 foci are formed during HR after strand resection at sites of DSBs, we examined the effect of FANCJ/MRE11 depletion on RAD51 foci. We observed that in control siRNA-treated cells, approximately 40% of cells showed ≥5 RAD51 foci 6 h after IR exposure, whereas FANCJ- or MRE11-depleted cells displayed only 15 to 20% of cells with ≥5 RAD51 foci (Fig. 7F). Depletion of both FANCJ and MRE11 resulted in a further reduction of the percentage of cells with ≥5 RAD51 foci (7%). Diminished RAD51 staining in FANCJ- and/or MRE11-depleted cells suggested a reduction in HR, raising the possibility that NHEJ would be elevated to deal with the accumulation of DSBs. As evident at the 1-h or 6-h post-IR time point, the percentage of nuclei with ≥15 DNA-PKcs foci was increased in cells depleted of both FANCJ and MRE11 compared to FANCJ- or MRE11-depleted cells (Fig. 8A). Collectively, these results demonstrated that cells deficient in FANCJ and MRE11 showed increased IR resistance, reduced numbers of γH2AX and RAD51 foci, and elevated numbers of DNA-PKcs foci, suggesting that NHEJ was elicited to help cells cope with IR-induced strand breaks when both FANCJ and MRE11 were deficient. Consistent with this hypothesis, when HeLa cells depleted of FANCJ and MRE11 were treated with a DNA-PKcs inhibitor (NU-7441) and exposed to IR (2 Gy), there was a 4-fold increase in the number of radial chromosomes per metaphase (Fig. 8B and C).
Fig 8.
Effects of FANCJ and MRE11 depletion on chromosomal instability and DNA repair. (A) The specified control siRNA-, FANCJ siRNA-, and/or MRE11 siRNA-transfected cells were either left untreated or exposed to IR (3 Gy). At the indicated time after IR exposure, cells were fixed and stained with the DNA-PKcs pS2056 antibody. Cells with the noted numbers of pDNA-PKcs foci were counted and expressed as percentages. The graph shows the averages for three independent experiments. More than 100 cells were counted for each sample in each experiment with a P value of <0.001. (B) Metaphase spreads of HeLa cells treated with siRNAs against FANCJ and MRE11 or with control siRNA were treated with IR (2 Gy) in the presence or absence of 10 μM NU-7441 (DNA-PKcs inhibitor) and then analyzed for radial chromosomes. Radial chromosomes are circled. (C) Quantitation of three independent experiments performed as described in panel B. (D and E) I-SceI-induced HR in U2OS cells (D) or NHEJ in MCF7 cells (E) depleted of FANCJ, MRE11, or FANCJ plus MRE11 by siRNA. Percentages of I-SceI-induced GFP+ cells were measured. Graphs show the mean values for at least three independent experiments. Error bars indicate SD values.
Taken together, the IR survival and chromosome instability data, along with protein interaction results, suggest the cooperative involvement of FANCJ and MRE11 in DNA repair of strand breaks. To assess the roles of FANCJ and MRE11 in DSB repair, we performed I-SceI-induced DSB repair in cells depleted of FANCJ, MRE11, or both FANCJ and MRE11, using a GFP reporter construct, to evaluate HR (32) or NHEJ (31). The results demonstrated that HR repair of the I-SceI DSB was significantly reduced in either FANCJ- or MRE11-depleted cells, with a greater effect observed for FANCJ-depleted cells (Fig. 8D). Depletion of both FANCJ and MRE11 resulted in an HR-dependent DSB repair defect comparable to that of FANCJ-depleted cells. The results from the NHEJ I-SceI DSB repair assay showed that FANCJ depletion, but not MRE11 depletion, reduced NHEJ, although not to the extent observed for HR repair (Fig. 8E). Codepletion of FANCJ and MRE11 restored NHEJ to a level comparable to that of the control siRNA-treated cells. These results suggest that FANCJ and MRE11 play a prominent role in HR repair of a DSB and that the adverse effect of FANCJ deficiency on NHEJ can be suppressed by MRE11 deficiency.
DISCUSSION
Although FANCJ has been implicated in the DNA damage response, its molecular functions in DNA repair are not well understood. For example, FANCJ is believed to participate in DSB repair through a BRCA1 protein interaction (17); however, it is still unclear at what step and precisely how FANCJ participates in the metabolism of DNA broken ends or a processed DNA cross-link substrate intermediate. Presumably, FANCJ DNA unwinding activity and protein partnerships are critical for its role during HR repair (13, 17, 44). Here we showed that FANCJ exists in a protein complex with RAD50 and MRE11 and that the FANCJ-MRE11 interaction is not dependent on BRCA1. This subcomplex may be distinct from the previously identified BARD1 complex, which contains BRCA1, FANCJ, TopBP1, and MLH1 (18). FANCJ interacts directly with MRE11, and the interaction is mediated by a C-terminal noncatalytic domain of FANCJ. Since BRCA1 also binds to the FANCJ C-terminal domain, it is conceivable that MRE11 may compete with BRCA1 for binding to FANCJ and that FANCJ may exist in distinct complexes with either MRE11 or BRCA1.
To better understand the dynamics of FANCJ protein interactions in a cellular context, we examined the localization of FANCJ and MRE11 after DNA damage. Using laser-targeted DNA damage and confocal microscopy, we found that FANCJ is recruited to Pso-ICLs or DSBs soon after the very early arrival of MRE11. Based on our finding that FANCJ interacts with MRE11, the results from the DNA damage localization studies suggest that after the rapid recruitment of MRE11 to break sites, FANCJ arrives at the lesion either as a complex with MRE11 or independently, without MRE11 bound. Furthermore, FANCJ recruitment to DSBs but not ICLs is dependent on MRE11, suggesting that the MRE11-FANCJ protein interaction is tailored for specific functions that are dependent on the type of DNA damage being repaired. Further analysis demonstrated that inhibition of MRE11 nuclease activity or expression of nuclease-defective MRE11 perturbed normal localization of FANCJ at the DSB. The dependence of FANCJ recruitment to DSBs on MRE11 nuclease activity is unique and contrasts with the recent observation that the human heterogeneous ribonucleoprotein U-like (hnRNPUL) proteins important for the DSB response require the presence of the MRN complex but not MRN nuclease activity for recruitment to sites of laser-induced DNA damage (45).
Although FANCJ did not require MRE11 for recruitment to Pso-ICLs, other proteins were necessary, in a lesion-specific manner. FANCJ recruitment to Pso-ICLs (but not DSBs) required the interacting partner MLH1. The dependence of FANCJ on MLH1 for recruitment to Pso-ICLs is consistent with a recent study showing that the physical interaction between FANCJ and MLH1 is required for a normal cellular response to the cross-link-inducing agent MMC (19). FANCJ recruitment to Pso-ICLs was also dependent on FANCA, a member of the FA core complex. This is logical, because FANCA (46, 47), unlike FANCJ (13), is required for FANCD2 monoubiquitination, suggesting that FANCJ operates downstream of the FA core complex. FANCD2, on the other hand, was not required for FANCJ recruitment to Pso-ICLs, consistent with an earlier report that FANCJ foci were able to form proficiently in FA-D2 cells exposed to MMC (16). However, it is likely that FANCD2 collaborates with FANCJ and other HR proteins to orchestrate the necessary steps for repair of the processed cross-link. The implication is that the FA pathway and MLH1 regulate FANCJ's involvement in cross-link repair, whereas the MRN complex controls FANCJ's participation in repair of direct DSBs.
Since MRN is implicated in early events of DSB repair, it can be reasoned that FANCJ and MRN collaborate with other proteins during the early end resection phase to ensure that HR can occur in an efficient and timely manner. Our results from cellular assays that measure repair of an I-SceI-induced DSB are consistent with a role of FANCJ (and MRE11) in HR repair. Furthermore, the auxiliary factor CtIP and BLM helicase, both of which are implicated in DNA end processing, are dependent on FANCJ for their timely recruitment to DSBs (Fig. 4 and our unpublished data). Recent evidence implicates BLM helicase in two pathways of strand resection after the early events of MRN sensing of the DSB and initiation of DSB processing (25). In the current study, biochemical assays with purified recombinant proteins demonstrated that FANCJ interacts directly with MRE11 and inhibits its 3′-to-5′ exonuclease activity on a blunt duplex DNA substrate in a specific manner. The interaction of FANCJ with MRE11 suggests that FANCJ may regulate MRE11 exonuclease activity to prevent excessive or inappropriate end resection. Inhibition of MRE11 exonuclease activity would presumably prevent the generation of overly long 5′ single-stranded DNA (ssDNA) tails. Rather, processive strand resection involving the BLM helicase and the EXO-1 or DNA2 nuclease with other proteins takes over to allow the creation of long 3′ ssDNA tailed molecules for strand invasion into the recipient duplex by RAD51 recombinase.
The processing and resection of DNA breaks by specific protein complexes are highly dependent on cell cycle stages. Although HR repair is known to occur during the S or G2 phase when sister chromatids are available, our observation that the majority of cycling cells display fairly rapid recruitment of FANCJ to DSBs or ICLs suggests that FANCJ, like other FA proteins (FANCD2 and FANCA) (48), participates in a pan-cell cycle DNA damage response. Presumably, FANCJ participates in multiple pathways of the DNA damage response in addition to its role in HR repair of replicated DNA molecules. Consistent with this proposal, a recent study provided evidence that FANCJ participates in HR repair and nonhomologous end joining (49), a pathway that can occur in all phases of the cell cycle. It will be interesting to determine if (and how) the FANCJ helicase takes part in long-strand resection in addition to its proposed role in the regulation of early DNA processing events through its interaction with the MRN nuclease. One possible scenario is that the FANCJ and BLM helicases collaborate to unwind the processed DSB break in a processive manner by simultaneously translocating on opposite strands of the duplex as they catalytically unwind the double-stranded DNA end. A coordinate action of BLM (and possibly FANCJ) with the structure-specific nuclease EXO-1 or Dna2 would serve to mediate degradation of the same strand to which FANCJ translocates in a 5′-to-3′ direction. Further studies are required to test this model. The ability of MRN to stimulate resection by either BLM-DNA2-RPA or EXO-1 (25) and the interaction of MRN with FANCJ (this study) suggest that FANCJ may have early and late roles in DSB end resection.
Coordination between FANCJ and MRN in the metabolism of DSBs is borne out by our cellular studies. When both FANCJ and MRE11 are deficient, human cells show improved IR resistance and reduced DSB accumulation, suggesting that other proteins are elicited to compensate for their absence. Cells deficient in both FANCJ and MRE11 showed a significant reduction in IR-induced RAD51 foci and increased numbers of DNA-PKcs foci, suggesting diminished HR and a concomitant elevation of NHEJ to deal with the accumulation of DSBs. A collaborative role of FANCJ and MRE11 in DSB repair through NHEJ and HR repair was further suggested by the I-SceI-induced DSB repair assay results. Although NHEJ lacks fidelity, the pathway serves to join DSBs incurred by IR and to improve overall survival. Redundancy in DNA damage response pathways is likely to contribute to resistance of tumors to anticancer DNA-damaging treatments. The balance between HR and NHEJ is regulated by protein interactions between DNA repair enzymes that are important for the correction of DNA cross-links and DSBs which threaten genomic stability.
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute on Aging, and by the Fanconi Anemia Research Fund (R.M.B.). J.-Y.M. is an FRSQ senior investigator and is supported by the Canadian Institute of Health Research.
We thank Marina Bellani and Dan McNeill (NIA, NIH) for critically reading the manuscript and Al May for technical advice on using the laser confocal microscope.
We declare that there are no conflicts of interest.
Footnotes
Published ahead of print 25 March 2013
REFERENCES
- 1. Kee Y, D'Andrea AD. 2010. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 24:1680–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kitao H, Takata M. 2011. Fanconi anemia: a disorder defective in the DNA damage response. Int. J. Hematol. 93:417–424 [DOI] [PubMed] [Google Scholar]
- 3. Joenje H. 2011. Metabolism: alcohol, DNA and disease. Nature 475:45–46 [DOI] [PubMed] [Google Scholar]
- 4. Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. 2011. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 475:53–58 [DOI] [PubMed] [Google Scholar]
- 5. Cybulski KE, Howlett NG. 2011. FANCP/SLX4: a Swiss Army knife of DNA interstrand crosslink repair. Cell Cycle 10:1757–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ali AM, Singh TR, Meetei AR. 2009. FANCM-FAAP24 and FANCJ: FA proteins that metabolize DNA. Mutat. Res. 668:20–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cantor S, Drapkin R, Zhang F, Lin Y, Han J, Pamidi S, Livingston DM. 2004. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc. Natl. Acad. Sci. U. S. A. 101:2357–2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gupta R, Sharma S, Sommers JA, Jin Z, Cantor SB, Brosh RM., Jr 2005. Analysis of the DNA substrate specificity of the human BACH1 helicase associated with breast cancer. J. Biol. Chem. 280:25450–25460 [DOI] [PubMed] [Google Scholar]
- 9. London TB, Barber LJ, Mosedale G, Kelly GP, Balasubramanian S, Hickson ID, Boulton SJ, Hiom K. 2008. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 283:36132–36139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu Y, Shin-Ya K, Brosh RM., Jr 2008. FANCJ helicase defective in Fanconi anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 28:4116–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu Y, Brosh RM., Jr 2009. FANCJ helicase operates in the Fanconi anemia DNA repair pathway and the response to replicational stress. Curr. Mol. Med. 9:470–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bridge WL, Vandenberg CJ, Franklin RJ, Hiom K. 2005. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat. Genet. 37:953–957 [DOI] [PubMed] [Google Scholar]
- 13. Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, Andreassen PR, Cantor SB. 2005. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell 8:255–265 [DOI] [PubMed] [Google Scholar]
- 14. Suhasini AN, Rawtani NA, Wu Y, Sommers JA, Sharma S, Mosedale G, North PS, Cantor SB, Hickson ID, Brosh RM., Jr 2011. Interaction between the helicases genetically linked to Fanconi anemia group J and Bloom's syndrome. EMBO J. 30:692–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shen X, Do H, Li Y, Chung WH, Tomasz M, De Winter JP, Xia B, Elledge SJ, Wang W, Li L. 2009. Recruitment of Fanconi anemia and breast cancer proteins to DNA damage sites is differentially governed by replication. Mol. Cell 35:716–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang F, Fan Q, Ren K, Auerbach AD, Andreassen PR. 2010. FANCJ/BRIP1 recruitment and regulation of FANCD2 in DNA damage responses. Chromosoma 119:637–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, Livingston DM. 2001. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 105:149–160 [DOI] [PubMed] [Google Scholar]
- 18. Greenberg RA, Sobhian B, Pathania S, Cantor SB, Nakatani Y, Livingston DM. 2006. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 20:34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peng M, Litman R, Xie J, Sharma S, Brosh RM, Jr, Cantor SB. 2007. The FANCJ/MutLalpha interaction is required for correction of the cross-link response in FA-J cells. EMBO J. 26:3238–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14:927–939 [PMC free article] [PubMed] [Google Scholar]
- 21. Meetei AR, Sechi S, Wallisch M, Yang D, Young MK, Joenje H, Hoatlin ME, Wang W. 2003. A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol. Cell. Biol. 23:3417–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Deans AJ, West SC. 2009. FANCM connects the genome instability disorders Bloom's syndrome and Fanconi anemia. Mol. Cell 36:943–953 [DOI] [PubMed] [Google Scholar]
- 23. Chapman JR, Taylor MR, Boulton SJ. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47:497–510 [DOI] [PubMed] [Google Scholar]
- 24. Symington LS, Gautier J. 2011. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45:247–271 [DOI] [PubMed] [Google Scholar]
- 25. Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. 2011. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 25:350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Boisvert FM, Dery U, Masson JY, Richard S. 2005. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 19:671–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yu Z, Vogel G, Coulombe Y, Dubeau D, Spehalski E, Hebert J, Ferguson DO, Masson JY, Richard S. 2012. The MRE11 GAR motif regulates DNA double-strand break processing and ATR activation. Cell Res. 22:305–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu Y, Sommers JA, Khan I, De Winter JP, Brosh RM., Jr 2012. Biochemical characterization of Warsaw breakage syndrome helicase. J. Biol. Chem. 287:1007–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dignam JD, Lebovitz RM, Roeder RG. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Muniandy PA, Thapa D, Thazhathveetil AK, Liu ST, Seidman MM. 2009. Repair of laser-localized DNA interstrand cross-links in G1 phase mammalian cells. J. Biol. Chem. 284:27908–27917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xie A, Kwok A, Scully R. 2009. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat. Struct. Mol. Biol. 16:814–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bennardo N, Cheng A, Huang N, Stark JM. 2008. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 4:e1000110 doi:10.1371/journal.pgen.1000110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gupta R, Sharma S, Doherty KM, Sommers JA, Cantor SB, Brosh RM., Jr 2006. Inhibition of BACH1 (FANCJ) helicase by backbone discontinuity is overcome by increased motor ATPase or length of loading strand. Nucleic Acids Res. 34:6673–6683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gupta R, Sharma S, Sommers JA, Kenny MK, Cantor SB, Brosh RM., Jr 2007. FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood 110:2390–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu X, Rathbun G, Lane WS, Weaver DT, Livingston DM. 2000. Interactions of the Nijmegen breakage syndrome protein with ATM and BRCA1. Cold Spring Harb. Symp. Quant. Biol. 65:535–545 [DOI] [PubMed] [Google Scholar]
- 36. Zhong Q, Chen CF, Li S, Chen Y, Wang CC, Xiao J, Chen PL, Sharp ZD, Lee WH. 1999. Association of BRCA1 with the hRad50-hMre11-p95 complex and the DNA damage response. Science 285:747–750 [DOI] [PubMed] [Google Scholar]
- 37. Wood RD. 2010. Mammalian nucleotide excision repair proteins and interstrand crosslink repair. Environ. Mol. Mutagen. 51:520–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee JH, Nicolette ML, Kopelovich L, Jasin M, Baer R, Paull TT, Gautier J. 2008. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat. Chem. Biol. 4:119–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wen Q, Scorah J, Phear G, Rodgers G, Rodgers S, Meuth M. 2008. A mutant allele of MRE11 found in mismatch repair-deficient tumor cells suppresses the cellular response to DNA replication fork stress in a dominant negative manner. Mol. Biol. Cell 19:1693–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. 2008. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 135:85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yanamadala S, Ljungman M. 2003. Potential role of MLH1 in the induction of p53 and apoptosis by blocking transcription on damaged DNA templates. Mol. Cancer Res. 1:747–754 [PubMed] [Google Scholar]
- 42. Yuan J, Chen J. 2010. MRE11-RAD50-NBS1 complex dictates DNA repair independent of H2AX. J. Biol. Chem. 285:1097–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van der Lelij P, Chrzanowska KH, Godthelp BC, Rooimans MA, Oostra AB, Stumm M, Zdzienicka MZ, Joenje H, De Winter JP. 2010. Warsaw breakage syndrome, a cohesinopathy associated with mutations in the XPD helicase family member DDX11/ChlR1. Am. J. Hum. Genet. 86:262–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xie J, Litman R, Wang S, Peng M, Guillemette S, Rooney T, Cantor SB. 2010. Targeting the FANCJ-BRCA1 interaction promotes a switch from recombination to poleta-dependent bypass. Oncogene 29:2499–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Polo SE, Blackford AN, Chapman JR, Baskcomb L, Gravel S, Rusch A, Thomas A, Blundred R, Smith P, Kzhyshkowska J, Dobner T, Taylor AM, Turnell AS, Stewart GS, Grand RJ, Jackson SP. 2012. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair. Mol. Cell 45:505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D'Andrea AD. 2001. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7:249–262 [DOI] [PubMed] [Google Scholar]
- 47. Yang YG, Herceg Z, Nakanishi K, Demuth I, Piccoli C, Michelon J, Hildebrand G, Jasin M, Digweed M, Wang ZQ. 2005. The Fanconi anemia group A protein modulates homologous repair of DNA double-strand breaks in mammalian cells. Carcinogenesis 26:1731–1740 [DOI] [PubMed] [Google Scholar]
- 48. Yan Z, Guo R, Paramasivam M, Shen W, Ling C, Fox D, III, Wang Y, Oostra AB, Kuehl J, Lee DY, Takata M, Hoatlin ME, Schindler D, Joenje H, De Winter JP, Li L, Seidman MM, Wang W. 2012. A ubiquitin-binding protein, FAAP20, links RNF8-mediated ubiquitination to the Fanconi anemia DNA repair network. Mol. Cell 47:61–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dohrn L, Salles D, Siehler SY, Kaufmann J, Wiesmuller L. 2012. BRCA1-mediated repression of mutagenic end-joining of DNA double-strand breaks requires complex formation with BACH1. Biochem. J. 441:919–926 [DOI] [PubMed] [Google Scholar]