Abstract
Coronaviruses possess a cap structure at the 5′ ends of viral genomic RNA and subgenomic RNAs, which is generated through consecutive methylations by virally encoded guanine-N7-methyltransferase (N7-MTase) and 2′-O-methyltransferase (2′-O-MTase). The coronaviral N7-MTase is unique for its physical linkage with an exoribonuclease (ExoN) harbored in nonstructural protein 14 (nsp14) of coronaviruses. In this study, the structure-function relationships of the N7-MTase were analyzed by deletion and site-directed mutagenesis of severe acute respiratory syndrome coronavirus (SARS-CoV) nsp14. The results showed that the ExoN domain is closely involved in the activity of the N7-MTase, suggesting that coronavirus N7-MTase is different from all other viral N7-MTases, which are separable from other structural domains located in the same polypeptide. Two of the 12 critical residues identified to be essential for the N7-MTase were located at the N terminus of the core ExoN domain, reinforcing a role of the ExoN domain in the N7-MTase activity of nsp14. The other 10 critical residues were distributed throughout the N7-MTase domain but localized mainly in the S-adenosyl-l-methionine (SAM)-binding pocket and key structural elements of the MTase fold of nsp14. The sequence motif DxGxPxA (amino acids [aa] 331 to 338) was identified as the key part of the SAM-binding site. These results provide insights into the structure and functional mechanisms of coronaviral nsp14 N7-MTase.
INTRODUCTION
Eukaryotic cellular mRNAs and many viral mRNAs contain a modified 5′-terminal “cap” structure that is essential for efficient splicing, nuclear export, translation, and stability of the mRNAs (1, 2). The cap structures are usually formed by three sequential enzymatic reactions: (i) the 5′-triphosphate end of the nascent mRNA is hydrolyzed to a diphosphate by RNA triphosphatase (TPase); (ii) a GMP residue derived from GTP is transferred to the diphosphate mRNA by RNA guanylyltransferase (GTase) via a two-step reaction; and (iii) the guanosine cap is methylated by guanine-N7-methyltransferase (N7-MTase) at the N7 position to generate a cap-0 structure (m7GpppN) in the presence of the methyl group donor S-adenosyl-l-methionine (SAM). Higher eukaryotes and some viruses further form cap-1 structures by methylating the cap-0 structure at the ribose 2′-O position by 2′-O-methyltransferase (2′-O-MTase) (1, 2). Although the final cap structures of cellular and viral mRNAs are similar, several major virus groups harbor capping enzymes with alternative molecular organization and biochemical mechanisms (3). For example, alphaviruses employ a noncanonical pathway for mRNA capping, in which the GTP molecule is N7 methylated before being transferred to the 5′-diphosphate end of the viral RNA (4), and rhabdoviruses use a unique RNA:GDP polyribonucleotidyltransferase, instead of the conventional capping enzyme GMP:RNA GTase, to transfer a monophosphorylated RNA onto GDP (5, 6).
Coronaviruses possess the largest and most complex genome (27 to 31 kb of single-stranded, positive-sense RNA) among RNA viruses (7). Many of them are identified as important pathogens of humans and animals, such as severe acute respiratory syndrome coronavirus (SARS-CoV) (8). Although it has been known that the genomic and subgenomic RNAs of coronaviruses carry a 5′ cap structure (9–11), the enzymes involved in coronavirus RNA capping were unknown for more than 2 decades. SARS-CoV nonstructural protein 13 (nsp13) has been shown to possess the TPase activity (12), but direct evidence for its involvement in RNA capping is lacking. Coronavirus nsp16 was predicted to be ribose 2′-O-MTase (13, 14), which was confirmed for nsp16 of feline coronavirus (FCoV) (15). However, recently we and others revealed structurally and biochemically that SARS-CoV nsp16 in complex with nsp10 acts as ribose 2′-O-MTase, with the latter as a stimulatory factor of nsp16 (16–18). Since bioinformatic analysis could not predict N7-MTase and GTase encoded by coronaviruses (13), we adopted a yeast genetic approach to functionally screen for SARS-CoV N7-MTase and GTase and revealed that SARS-CoV nsp14 possesses the N7-MTase activity (19). Interestingly, nsp14 was shown previously to be an exoribonuclease (20, 21). The coronavirus GTase is still unknown.
The N7-MTase and 2′-O-MTase identified in coronaviruses (16, 17, 19) belong to a large class of SAM-dependent methyltransferases (22, 23). One feature that distinguishes viral N7-MTases from cellular N7-MTases is the presence of RNA polymerase or other capping activities along with the MTase domain on the same polypeptide (18). For example, the N7-MTase of flaviviruses is combined with 2′-O-MTase and RNA polymerase, and that of alphaviruses is combined with GTase. Unexpectedly, we found that coronavirus N7-MTase is linked with a unique 3′-to-5′ exoribonuclease (ExoN) domain in nsp14 (19). The diversity of the capping apparatus makes viral RNA capping an attractive target for drug design and development.
Although we have previously shown that the core domains of MTase and ExoN are localized at the C- and N-terminal halves of SARS-CoV nsp14, respectively (19), the structure-function relationships of this protein are not known. In this study, we performed systematic mutagenesis of nsp14 by truncation, deletion, and alanine scanning and showed that the ExoN domain of nsp14 is also important for the N7-MTase activity, indicating that the functional domain of coronavirus N7-MTase is different from that of other viral MTases which can execute their MTase activity independently from other functional domains on the same protein. We further identified the critical amino acid residues essential for N7-MTase activity and defined the SAM-binding motif of N7-MTase in SARS-CoV nsp14.
MATERIALS AND METHODS
Construction of mutants for yeast screening.
The mutants of nsp14 of SARS-CoV were generated by overlap PCR with mutagenic primers from SARS-CoV strain WHU (24). The PCR fragments were cloned into yeast vector pMceK294A (2μm, TRP1) as described previously (19). The cloned mutants are fused in frame with an N-terminal leader peptide (MGSHHHHHHSGHMRI) and the C-terminal mouse capping enzyme Mce1 with an inactivating K294A mutation, and the expression is under the control of the yeast TPI1 promoter. All the clones and mutations were confirmed by DNA sequencing.
Yeast strains and assays of N7-MTase activity in vivo.
Yeast strain YBS40 carries a deletion at the chromosomal Abd1 locus encoding the yeast cap MTase (25), and its growth depends on the maintenance of plasmid p360-ABD1 (URA3). For analysis of N7-MTase activity of the SARS-CoV nsp14 mutants, yeast cells were transformed with expression plasmids carrying mutants of nsp14. Trp+ transformants were selected at 30°C on agar medium lacking tryptophan, and the cells were then streaked on agar medium containing 0.75 mg/ml of 5-fluoroorotic acid (5-FOA) to counterselect URA3 plasmids at 30°C. The plates were incubated for up to 5 days, and formation of FOA-resistant colonies indicated that the transformed mutants could replace or complement the endogenous cap N7-methyltransferase function residing in the URA3 plasmid (19).
Cloning, expression, and purification of recombinant proteins.
The mutants of nsp14 were generated as described for yeast screening plasmids and were inserted into plasmid pET30(a) (Novagen). All constructs were verified by DNA sequencing, and their expression gave rise to recombinant proteins with an N-terminal 6×His tag. Protein was induced with 0.4 mM isopropyl β-d-thiogalactopyranoside (IPTG) at 16°C for 12 to 16 h and was purified with nickel-nitrilotriacetic acid (Ni-NTA) resin (GenScript) as described previously (17, 19, 21).
Biochemical assays for MTase activity.
The capped but nonmethylated RNA substrate G*pppA-RNA (where the asterisk indicates that the following phosphate was 32P labeled) represented the 5′-terminal AGAUU- nucleotides of the SARS-CoV genome except that the second nucleotide was changed from U to G for enhanced efficiency of in vitro transcription. Purified mutant or truncated proteins (0.5 μg) and 2 ×103 cpm of 32P-labeled G*pppA-RNA substrates were added to an 8.5-μl reaction mixture (40 mM Tris-HCl [pH 8.0], 2 mM MgCl2, 2 mM dithiothreitol [DTT], 10 units RNase inhibitor, 0.2 mM SAM) and incubated at 37°C for 1.5 h. After the treatment of substrate RNAs with different recombinant proteins, the RNA cap structures (G*pppA- or m7G*pppA-) were digested with 5 μg of nuclease P1 (which cleaves capped RNAs into 3′-OH-terminated cap structures and 5′-pNOH) (Sigma) at 50°C for 30 min, and the resulting N7-methylated cap structures (G*pppA or m7G*pppA) were spotted onto polyethyleneimine cellulose-F plates (Merck) for thin-layer chromatography (TLC) and developed in 0.4 M ammonium sulfate. The methylated cap structure (m7G*pppA) could be readily separated from nonmethylated cap structure (G*pppA), and the amount of 32P-labeled cap was determined by scanning the chromatogram with a PhosphorImager (17, 19, 26).
MTase activity assays were carried out in 30-μl reaction mixtures (40 mM Tris-HCl [pH 8.0], 2 mM MgCl2, 2 mM DTT, 40 units RNase inhibitor, 0.01 mM SAM) with 1 μCi of S-adenosyl-[methyl-3H]methionine (67.3 Ci/mmol, 0.5 μCi/μl), 1 μg of mutant or truncated proteins, and 3 μg of GpppA-RNA substrates at 37°C for 1.5 h. The 3H-labeled product was isolated in small DEAE-Sephadex columns and quantitated by liquid scintillation (27). All the RNAs used here were produced as described previously (17, 19).
SAM binding assay.
Reaction mixtures (25 μl) (40 mM Tris-HCl [pH 7.5], 2 mM MgCl2, 2 mM DTT) containing 1 μg of mutant or truncated proteins and 1 μCi of S-adenosyl-[methyl-3H]methionine (67.3 Ci/mmol, 0.5 μCi/μl) were pipetted into wells of a microtiter plate. The reaction mixtures were incubated on ice and irradiated with 254-nm UV light in a Hoefer UVC500 cross-linking oven for 30 min. The samples were then analyzed by 12% SDS-PAGE. The gels were soaked in Enlightning solution (Perkin-Elmer) and used for fluorography (27).
Exoribonuclease assays.
Three micrograms of purified recombinant proteins and 300 cpm of 32P-labeled single-strand RNA of 58 nucleotides (nt), which was transcribed from pGEM-T vector, were added to a 10-μl reaction mixture (50 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 50 mM KCl, 50 mM NaCl, 2 mM DTT, 0.1 mg/ml bovine serum albumin [BSA], 10 U RNase inhibitor). The mixture was incubated at 37°C for 1.5 h and analyzed in a 10% urea-polyacrylamide gel (19, 21).
3D structure modeling.
The three-dimensional (3D) structure of the core MTase domain of SARS-CoV nsp14 (aa 291 to 527) was generated on the basis of the structures of Alfalfa caffeoyl 3-O-methyltransferase (Protein Data Bank [PDB] 1SUI) and Encephalitozoon cuniculi mRNA cap guanine-N7-methyltransferase (PDB 1RI4). The predicted structure was homology modeled with Swiss-PdbViewer v3.7. The bonds between nsp14 and SAM were calculated with Discovery studio2.1, and the final images were presented with PyMOL.
RESULTS
The global structure of nsp14 is essential for both N7-MTase and ExoN activity.
In our previous study, we showed that SARS-CoV nsp14 functions as the N7-MTase and that the core domain of the N7-MTase is located in the C-terminal half coupled with the ExoN domain at the N terminus of SARS-CoV nsp14 (19). In this study, we created a large set of SARS-CoV nsp14 mutants by serial truncations from both the N and C termini of nsp14 and various deletions inside the N-terminal ExoN domain (Table 1). We then analyzed the effects of the mutations on the N7-MTase activities of nsp14 in the yeast genetic system at different temperatures and with biochemical assays established previously (19). As shown in Table 1, at all tested temperatures, truncations of 78 amino acids (aa) from the N terminus (nsp14ΔN78) and 6 aa from C terminus (nsp14ΔC6) of nsp14 already attenuated the MTase activity dramatically, and truncations of 90 aa from the N terminus (nsp14ΔN90) and 17 aa from C terminus (nsp14ΔC17) completely abolished the MTase activity of nsp14. These results were consistent with our previous report (19) and suggest that the N-terminal ExoN domain is also important for the N7-MTase activity of nsp14.
Table 1.
Effects of truncations and deletions in nsp14 on the N7-MTase activity in yeast cells
| nsp14 | Position of deleted amino acids |
abd1Δ complementationa at: |
|||
|---|---|---|---|---|---|
| 20°C | 25°C | 30°C | 37°C | ||
| Wild type | +++ | +++ | +++ | +++ | |
| nsp14ΔN61 | 1–61 | +++ | +++ | +++ | ND |
| nsp14ΔN78 | 1–78 | + | + | + | ND |
| nsp14ΔN90 | 1–90 | − | − | − | ND |
| nsp14ΔC6 | 522–527 | + | + | + | ND |
| nsp14ΔC17 | 511–527 | − | − | − | ND |
| ΔA79 | 79 | − | − | − | − |
| ΔD90 | 90 | ++ | +++ | +++ | − |
| Δ90-92 | 90–92 | +++ | +++ | +++ | − |
| ΔE92 | 92 | +++ | +++ | +++ | − |
| Δ102-106 | 102–106 | +++ | +++ | +++ | +++ |
| Δ139-142 | 139–142 | ++ | +++ | +++ | + |
| Δ166-169 | 166–169 | − | − | − | − |
| ΔE191 | 191 | +++ | +++ | +++ | + |
| Δ191-193 | 191–193 | − | − | − | − |
| Δ198-200 | 198–200 | − | − | − | − |
| Δ238-243 | 238–243 | − | − | − | − |
| ΔD243 | 243 | + | + | + | − |
| Δ242-244 | 242–244 | − | − | − | − |
| Δ266-268 | 266–268 | +++ | +++ | +++ | +++ |
| Δ271-273 | 271–273 | +++ | +++ | +++ | − |
| ΔD273 | 273 | +++ | +++ | +++ | + |
| Δ277-280 | 277–280 | +++ | +++ | +++ | + |
| Δ297-299 | 297–299 | − | − | − | − |
In the yeast genetic system, the capability of nsp14 mutants for complementing yeast growth corresponds to the activity of nsp14 N7-MTase. +++, ++, +, and − indicate yeast cell growth in colony size and number as that of wild-type nsp14, slow growth with reduced colony size and number, slow growth with very few colonies, and no growth, respectively. ND, not determined.
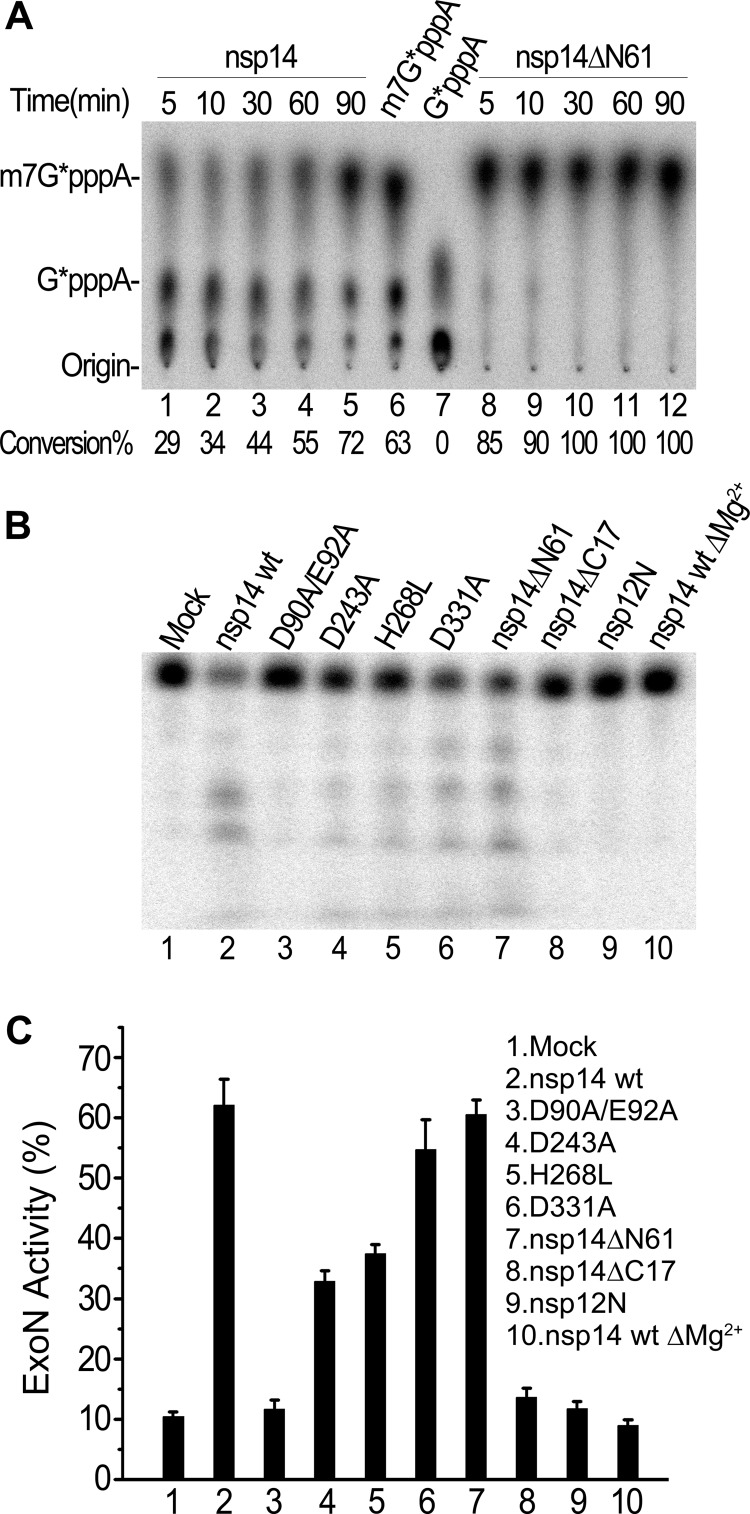
Interestingly, the truncation of the N-terminal 61 aa, which are outside the core domain of ExoN, did not impair the N7-MTase function in cells but instead increased the enzymatic activity in biochemical assays compared with that of wild-type nsp14 (Fig. 1A). As shown in Fig. 1A, 72% of the 32P-labeled RNA substrate (G*pppA-RNA) was converted to m7G*pppA-RNA (cap-0 structure) by wild-type nsp14 after a 90-min incubation (Fig. 1A, lane 5), while 85% of the RNA substrate was methylated by nsp14ΔN61 within 5 min (Fig. 1A, lane 8), and incubation for 30 min yielded entirely methylated RNA substrate (Fig. 1A, lane 10). This indicates that the N7-MTase activity of nsp14ΔN61 is remarkably higher than that of the wild-type nsp14.
Fig 1.
Biochemical assays of truncated nsp14 and mutants on ExoN motif. (A) TLC analysis of nuclease P1-resistant cap structures released from 32P-labeled G*pppA-RNA methylated by wild-type nsp14 (lanes 1 to 5) and truncated nsp14ΔN61 (lanes 8 to 12) at different time points. The markers m7G*pppA (lane 6) and G*pppA (lane 7) were prepared with commercial vaccinia virus capping enzymes. The positions of origin and migration of m7G*pppA and G*pppA are indicated on the left. (B) Analysis of ExoN activity of wild-type nsp14, nsp14 mutants, and truncations in 10% urea-PAGE using single-stranded RNA substrates that were 32P labeled at the 5′ end. Mock treatment and treatment with the N terminus of SARS-CoV nsp12 (nsp12N) were used as negative controls. (C) Quantitation of ExoN activity by analyzing the band intensity shown in panel B with ImageQuant software. The values on the y axis indicate the ratios of digestion products to input RNA within each lane. All products migrating below full-length substrate were quantitated together as digestion products. Input RNA represents digestion products plus undigested full-length substrate (n = 3; mean values ± standard deviations [SD] are shown).
We further generated a set of internal deletion mutants of nsp14 by removing 1- to 5-aa stretches inside the N-terminal ExoN domain. These deletion mutants were analyzed in the yeast genetic system at 20°C, 25°C, 30°C, and 37°C. As shown in Table 1, deletions all over the ExoN domain, including ΔA79, Δ166-169, Δ191-193, Δ198-200, Δ238-243, Δ242-244, and Δ297-299, completely disabled the N7-MTase activity of nsp14. Deletions of individual amino acids at the conserved D90-E92-D243-h268-D273 active-site motif of ExoN did not abolish the MTase activity of nsp14; however, 8- to 28-aa deletions at the same sites disabled the N7-MTase (data not shown), probably by destroying the structure of the ExoN domain. These results suggest that the global structure rather than the specific amino acids at the ExoN active site is important for the MTase activity. Interestingly, many of the yeast strains harboring the small deletion mutants were viable at 30°C but inviable at 37°C, suggesting that small deletions easily yielded temperature-sensitive proteins. These internal deletion analyses further strengthened the proposal that the N-terminal ExoN domain is essential to the enzymatic activity of the C-terminal MTase of nsp14.
We further analyzed the domain structure of nsp14 required for the ExoN activity. For this, terminal truncations (ΔN61 and ΔC17) and active-site mutations of both ExoN (D90A/E92A, D243A, and H268L) and MTase (D331A) were tested (Fig. 1B and C). In the biochemical ExoN assays, the 32P-labeled single-stranded RNA was used as the substrate and the magnesium-dependent ExoN activity was measured. As shown in Fig. 1B, nsp14ΔN61 (lane 7) and D331A (lane 6) mutations did not influence the ExoN activity, whereas mutations at the D-E-D-h motif (Fig. 1B, lanes 3 to 5) either disrupted or attenuated the ExoN activity. Surprisingly, the C-terminal truncation mutation nsp14ΔC17 abolished the ExoN activity (Fig. 1B, lane 8). The ratios of digestion products to input RNA within each lane were quantitated by analyzing the band intensity shown in Fig. 1B (Fig. 1C). These results show that the C-terminal MTase domain also contributes to the ExoN activity of nsp14. Taking the findings together, the global structure of aa 62 to 527 of nsp14 is essential for both N7-MTase and ExoN activities.
Alanine scanning of SARS-CoV nsp14 reveals critical residues affecting cap N7-MTase activity.
To define the exact active sites of N7-MTase of SARS-CoV nsp14, 60 mutations mainly with alanine substitutions for 57 amino acid residues from aa 61 to aa 523 of SARS-CoV nsp14 were generated (Table 2). We chose the residues for mutation based on the following rules: (i) the most conserved residues revealed in the primary sequence alignment of coronaviral nsp14, which might be active sites or RNA-binding sites of nsp14 (20, 28, 29); (ii) residues in the putative SAM-binding site motif predicted by structure-based sequence alignment of mRNA cap N7-MTases from Encephalitozoon cuniculi (Ecm1, PDB entry 1RI5), Homo sapiens (Hcm1, PDB entry 3BGV), and vaccinia virus (Vacci-D1) with the MTase domain of coronaviruses (transmissible gastroenteritis coronavirus [TGEV] nsp14 [amino acids 293 to 525] and SARS-CoV nsp14 [amino acids 291 to 527]) (19); (iii) residues corresponding to those which were identified in temperature-sensitive (ts) or virulence-attenuating isolates of the coronavirus mouse hepatitis virus (MHV) (30, 31); and (iv) corresponding conserved residues based on the mutagenic studies of vaccinia virus N7-MTase (32–34) and flavivirus cap MTase (35, 36).
Table 2.
Effects of point mutations on the N7-MTase activity in yeast cells and in vitro
| nsp14 | Position |
abd1Δ complementationa at: |
Methylation in vitro (%) | |||
|---|---|---|---|---|---|---|
| 20°C | 25°C | 30°C | 37°C | |||
| Wild type | +++ | +++ | +++ | +++ | 100 | |
| K61A | 61 | +++ | +++ | +++ | ++ | 97 |
| G68A | 68 | +++ | +++ | +++ | ++ | ND |
| F73A | 73 | ++ | ++ | ++ | ++ | 91 |
| T75A | 75 | +++ | +++ | +++ | ++ | ND |
| V83A | 83 | +++ | ++ | ++ | − | ND |
| R84A | 84 | − | − | − | − | 6 |
| W86A | 86 | − | − | − | − | 0 |
| G88A | 88 | +++ | +++ | +++ | ++ | ND |
| D90A/E92A | 90/92 | +++ | +++ | +++ | +++ | 93 |
| P106A | 106 | +++ | +++ | +++ | ++ | ND |
| K139A | 139 | +++ | +++ | +++ | ++ | ND |
| P141A | 141 | +++ | +++ | +++ | ++ | ND |
| P142A | 142 | +++ | +++ | +++ | ++ | ND |
| R163A | 163 | +++ | +++ | +++ | ++ | ND |
| E191A | 191 | +++ | +++ | +++ | ++ | ND |
| K200A | 200 | +++ | +++ | +++ | ++ | ND |
| D243A | 243 | +++ | +++ | +++ | ++ | 79 |
| N256A | 256 | +++ | +++ | +++ | +++ | 97 |
| H268L | 268 | +++ | +++ | +++ | +++ | 85 |
| D273A | 273 | +++ | +++ | +++ | +++ | 97 |
| R278A | 278 | +++ | +++ | +++ | ++ | ND |
| P297A | 297 | +++ | +++ | +++ | ++ | ND |
| R310A | 310 | − | − | − | − | 0 |
| Q313A | 313 | ++ | +++ | +++ | − | ND |
| D331A | 331 | − | − | − | − | 0 |
| D331E | 331 | + | + | ++ | + | 70 |
| D331Y | 331 | − | − | − | − | 0 |
| I332A | 332 | +++ | +++ | ++ | − | 83 |
| G333A | 333 | − | − | − | − | 0 |
| N334A | 334 | +++ | +++ | +++ | +++ | 89 |
| P335A | 335 | ++ | ++ | +++ | − | 61 |
| K336A | 336 | ++ | ++ | ++ | − | 50 |
| Y351A | 351 | +++ | +++ | +++ | ++ | ND |
| D352A | 352 | − | − | − | − | 47 |
| P355A | 355 | +++ | +++ | +++ | ++ | ND |
| L366A | 366 | +++ | +++ | +++ | ++ | ND |
| Y368A | 368 | − | − | − | − | 0 |
| D379A | 379 | +++ | +++ | +++ | +++ | ND |
| C382Y | 382 | +++ | +++ | ++ | + | 72 |
| N388A | 388 | − | − | − | − | 83 |
| Y392A | 392 | +++ | +++ | +++ | +++ | ND |
| P393A | 393 | +++ | +++ | +++ | +++ | ND |
| D402A | 402 | +++ | +++ | +++ | +++ | ND |
| C414R | 414 | +++ | +++ | ++ | − | 0 |
| G416R | 416 | +++ | +++ | − | − | 45 |
| L419A | 419 | ++ | + | − | − | 0 |
| Y420A | 420 | − | − | − | − | 5 |
| Y420H | 420 | − | − | − | − | ND |
| T428A | 428 | ++ | + | − | − | 27 |
| L439A | 439 | +++ | +++ | +++ | ++ | ND |
| P443A | 443 | +++ | +++ | +++ | ++ | ND |
| F445A | 445 | +++ | +++ | +++ | ++ | ND |
| W509A | 509 | +++ | +++ | +++ | ++ | ND |
| F514A | 514 | +++ | +++ | +++ | ++ | ND |
| D515A | 515 | +++ | +++ | +++ | ++ | ND |
| Y517A | 517 | +++ | +++ | +++ | ++ | ND |
| L519A | 519 | +++ | +++ | +++ | ++ | ND |
| W520A | 520 | +++ | +++ | +++ | ++ | ND |
| F523A | 523 | +++ | +++ | +++ | ++ | ND |
See Table 1, footnote a.
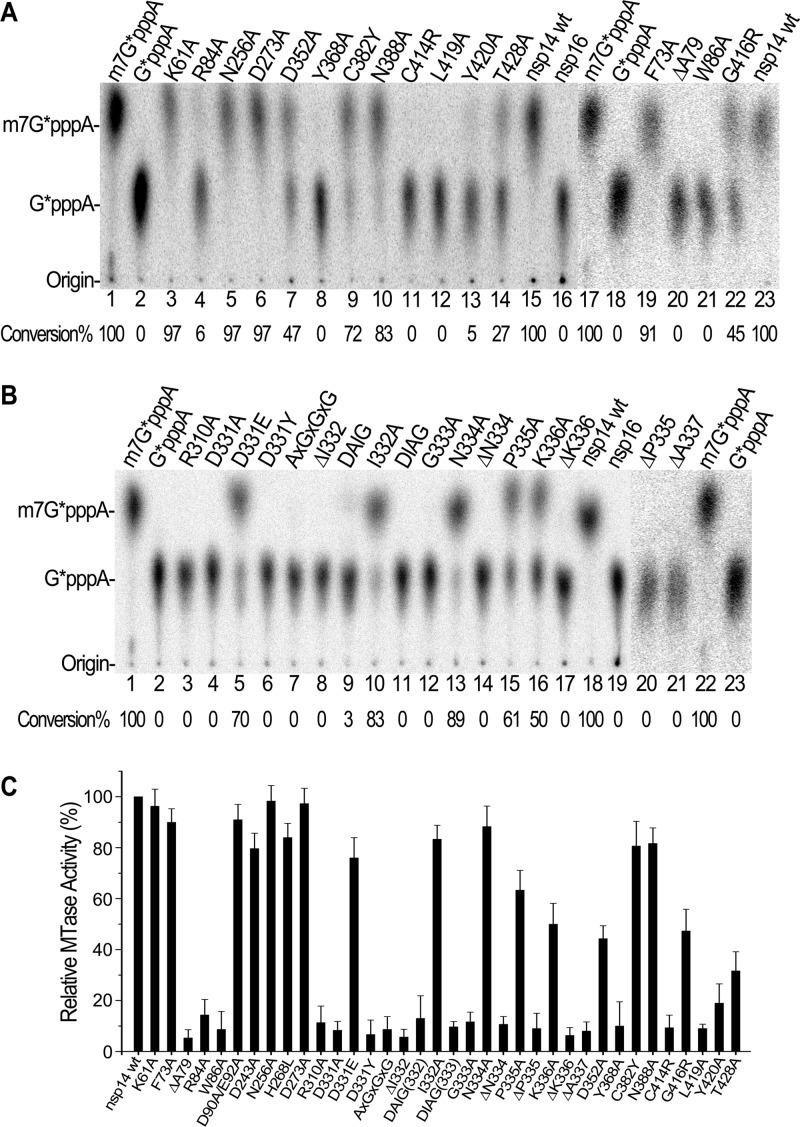
The effects of the amino acid substitutions on N7-MTase activity were tested in a powerful yeast genetic system as described previously (19). In this system, the growth status of yeast cells on the selection medium is correlated with the N7-MTase activities of nsp14 and its mutants (19, 37). The results are summarized in Table 2. Twelve lethal mutations were found, among which 10 (R310, D331, G333, D352, Y368, N388, G416, L419, Y420, and T428) are in the C-terminal MTase domain of nsp14 while two (R84 and W86) are located in the N-terminal ExoN domain of nsp14 (Table 2). All other mutations, including the active-site mutations of ExoN, did not inactivate the N7-MTase activity of nsp14 in yeast cells.
In addition, there were seven mutations that attenuated the N7-MTase activity of nsp14 by forming small and/or fewer colonies or obviously reducing the speed of yeast cell growth. Two of the seven residues (F73 and V83) are located in the N-terminal half of nsp14, close to the two critical residues R84 and W86 described above. Three sites (I332, P335, and K336) are likely at the putative SAM-binding site motif, and two (C382 and C414) were previously identified from the MHV ts strains (30).
In summary, critical residues for N7-MTase activity were identified mainly in two regions in nsp14. The first region, ranging from aa 310 to 428, overlaps the core domain of N7-MTase. Mutations at 15 residues in this area led to complete loss or remarkable attenuation of the N7-MTase activity of nsp14. The critical residues are clustered mainly at aa 331 to 336 and aa 414 to 428. The second region, ranging from aa 73 to 86, is located at the N terminus of the ExoN domain. The identification of critical residues of N7-MTase from the ExoN domain reinforces a role of the ExoN domain in the N7-MTase enzymatic function as revealed by the truncation and deletion analysis in this study.
Biochemical identification of residues essential to cap N7-methylation activity.
To confirm the essential roles of the critical residues identified in the genetic system, we further tested all the key mutants of nsp14 in biochemical MTase assays. For this, recombinant proteins with an N-terminal 6×His tag for 27 SARS-CoV nsp14 mutants were expressed in Escherichia coli cells and purified by nickel-nitrilotriacetic acid affinity chromatography. The results are shown in Fig. 2 and are also summarized in Table 2 for direct comparison with those of genetic assays. In general, remarkable consistency of genetic and biochemical assays for N7-MTase activity was demonstrated. As shown in the thin-layer chromatographic (TLC) analysis in Fig. 2, treatment of G*pppA-RNAs (where the asterisk indicates that the following phosphate is 32P labeled) with nsp14 mutants K61A, F73A, N256A, and D273A (Table 2) in the presence of SAM resulted in nuclease P1 cleavage products (Fig. 2A, lanes 3, 19, 5, and 6) that comigrated with m7G*pppA (Fig. 2A, lanes 1 and 17), indicating that these four mutations did not significantly affect the N7-MTase activity of nsp14. The conversion ratios were quantified based on the dot intensity (Table 2; Fig. 2A and B). In contrast, treatment of G*pppA-RNAs with nsp14 mutants R84A, W86A, R310A, D331A, D331Y, G333A, Y368A, C414R, L419A, and Y420A (Fig. 2A, lanes 4 and 21; Fig. 2B, lanes 3, 4, 6, and 12; Fig. 2A, lanes 8 and 11 to 13) did not generate a similar product corresponding to the position of m7G*pppA, revealing the nine functionally critical residues for N7-MTase. This observation conforms to the lethal phenotype of the same mutations in the yeast genetic system. The N7-MTase activities of 10 nsp14 mutants (D331E, I332A, N334A, P335A, K336A, D352A, C382Y, N388A, G416R, and T482A) were attenuated, causing incomplete conversion from G*pppA-RNA to m7G*pppA-RNA (Fig. 2B, lanes 5, 10, 13, 15, and 16; Fig. 2A, lanes 7, 9, 10, 22, and 14). All the results described above are strictly consistent with the results of genetic assays. However, a discrepancy for two nsp14 mutants was observed in biochemical and genetic assays. The nsp14 mutants D352A and N388A were lethal in the yeast genetic system (Table 2) but still retained 47% and 83% of N7-MTase activity in biochemical assays, respectively (Fig. 2A, lanes 7 and 10). This discrepancy could be explained by the high concentrations of SAM and GpppA-RNA included in the cap methylation reaction mixtures in the in vitro assays, which were in excess of the Km values for the N7-MTase (33, 38), or by changes of protein stability or localization in yeast cells.
Fig 2.
Mutational effects on cap N7-MTase activity of nsp14 in vitro. (A and B) TLC analysis of nuclease P1-resistant cap structures released from 32P-labeled G*pppA-RNA methylated by nsp14 mutants. The markers m7G*pppA and G*pppA were prepared with commercial vaccinia virus capping enzymes. The positions of origin and migration of m7G*pppA and G*pppA are indicated on the left. The ratios of conversion of G*pppA to m7G*pppA are show at the bottom. (C) GpppA-RNA was used to test the methylation activities of nsp14 mutants (n = 3; mean values ± SD are shown).
Furthermore, we also tested the nsp14 mutants in another biochemical system, where 3H-labeled methyl donor [3H]SAM was used and thereby the radiolabeled methyl group was transferred to the unlabeled GpppA-RNA. The reaction products were purified and quantified by liquid scintillation. The MTase activities of the mutants were normalized to that of wild-type nsp14 (Fig. 2C). In this set of experiments, we also included the ExoN active-site D-E-D-h motif mutants (D90A/E92A, D243A, and H268L), which did not affect the activity of N7-MTase of nsp14 (19). As shown in Fig. 2C, the results are in accord with those obtained in TLC analysis. Taken together, the two sets of biochemical assays of the nsp14 mutants consolidated the findings in the genetic assays.
Mapping of the SAM-binding site motif of N7-methyltransferase in coronavirus nsp14.
The SAM-binding motif (D/E/YxGxGxG, where x means any amino acid) is the best-conserved motif of MTases, including cap MTases. However, the SAM-binding and catalytic residues still vary considerably in different MTases or in different species (22, 23). In our previous study (19), we proposed the DxG motif (aa 331 to 333) in the core MTase domain of nsp14 as the putative SAM-binding motif of nsp14 N7-MTase, but experimental evidence is still lacking.
To investigate which residues are involved in the SAM binding, 3H-labeled SAM was incubated with the nsp14 mutants, and after UV cross-linking, the enzyme-[3H]SAM complex was analyzed by SDS-PAGE and autoradiography. As shown in Fig. 3, many of the mutations attenuated the SAM-binding affinity of N7-MTase, and six mutations at five positions (W86A, R310A, D331A, D331Y, G333A, and Y368A) entirely abolished the SAM-binding activity of nsp14 (Fig. 3A, lane 5; Fig. 3B, lanes 18, 4, 3, and 10; Fig. 3A, lane 12) as well as the cap N7-MTase activity (Table 2; Fig. 2). While glutamate could replace the conserved aspartate (D331E) and enhanced the SAM-binding activity of nsp14 (Fig. 3B, lane 2), it attenuated the enzymatic activity of the N7-MTase and reduced the viability of yeast cells (Table 2; Fig. 2B, lane 5; Fig. 2C). In contrast, in the same biochemical assay system, we observed that four nsp14 mutants (R84A, C414R, L419A, and Y420A) which abolished the N7-MTase activity in the in vitro assays (Fig. 2) still retained partial SAM-binding ability (Fig. 3A, lanes 4, 15, 17, and 18), indicating that these mutations also affected other properties of the N7-MTase besides SAM binding. In particular, mutant T428A showed the same SAM-binding capacity as wild-type nsp14 (Fig. 3A, lane 19) but was lethal in yeast cells at 30°C and 37°C (Table 2), suggesting that this site is not directly involved in SAM binding but is essential for other functions of N7-MTase of nsp14. In addition, four nsp14 mutants (F73A, D243A, H268L, and C382Y) attenuated the SAM-binding affinity (Fig. 3A, lanes 2, 7, 9, and 13) but sustained high enzymatic activities both in cells and in the in vitro assays (Table 2; Fig. 2C), indicating that partial SAM-binding ability was sufficient for the activity of N7-MTase. Collectively, these results clearly show that the DxG motif at aa 331 to 333 of nsp14 is involved in the SAM binding of coronavirus nsp14. In addition, the residues W86, R310, and Y368 are also essential for SAM binding of nsp14.
Fig 3.
Mutational effects on the SAM binding affinity of cap N7-MTase of nsp14 in vitro, showing 12% SDS-PAGE analysis of SAM UV cross-linking with nsp14 mutants.
In comparison with the conventional SAM-binding motif D/E/YxGxGxG, the corresponding conserved motif in nsp14 of coronaviruses is DxGxPxG/A (at the last position, alanine is found in SARS-CoV, whereas glycine is conserved in all other coronaviruses). We then tested the effects of mutations at all other positions in this motif on the SAM-binding and enzymatic activities of nsp14 N7-MTase. The mutant P335A possessed weak SAM-binding capacity (Fig. 3) but retained relatively high enzymatic activity both in vivo and in vitro (Table 2; Fig. 2). Mutations at the nonconserved residues (I332A, N334A, and K336A) in the DxGxPxG/A motif (marked by “x”) did not disrupt either the SAM-binding capacities of nsp14 (Fig. 3B, lanes 8, 11, and 15) or the enzymatic activities of N7-MTase (Table 2; Fig. 2). However, deletion of any amino acids within the motif (ΔI332, ΔN334, ΔP335, ΔK336, and ΔA337) or insertion into the DxG site (DAIG, DIAG) abolished the SAM-binding ability (Fig. 3B) and inactivated the N7-MTase activity (Table 3; Fig. 2). These results demonstrate that the structural patterning of the SAM-binding motif or distance between the critical residues is more important than the properties of the specific residues (except D331 and G333) for the functions of this motif.
Table 3.
Effects of insertions and deletions in the SAM-binding/active-site motif on the N7-MTase activity
| nsp14 | Position |
abd1Δ complementationa at: |
Methylation in vitro (%) | |||
|---|---|---|---|---|---|---|
| 20°C | 25°C | 30°C | 37°C | |||
| Wild type | +++ | +++ | +++ | +++ | 100 | |
| AxGxGxG | 331–337 | − | − | − | − | 0 |
| DIG→DAIG(332) | 332 | − | − | − | − | 3 |
| ΔI332 | 332 | − | − | − | − | 0 |
| DIG→DIAG(333) | 333 | − | − | − | − | 0 |
| ΔN334 | 334 | − | − | − | − | 0 |
| ΔP335 | 335 | − | − | − | − | 0 |
| ΔK336 | 336 | − | − | − | − | 0 |
| ΔA337 | 337 | − | − | − | − | 0 |
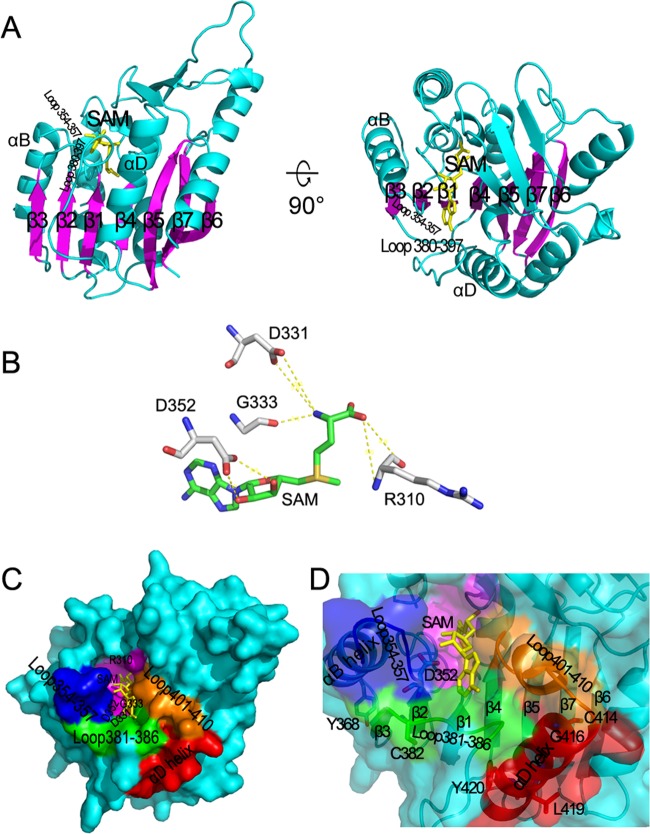
To reveal more structural information on the SAM-binding site of coronavirus nsp14, the 3D structure of the core MTase domain (aa 291 to 527) of nsp14 was modeled by homologous modeling on the basis of the structure of a cellular mRNA cap N7-MTase in complex with SAM (PDB entry 1RI4). In secondary structure-based sequence alignment, the SARS-CoV nsp14 N7-MTase domain is more similar to Alfalfa caffeoyl 3-O-methyltransferase (PDB entry 1SUI), and therefore the latter was used as a guide to refine the modeled structure of nsp14 N7-MTase (Fig. 4). As shown in Fig. 4A, the ligand SAM lies at the C termini of strands β1 and β2, similar to that of most SAM-dependent MTases, and the experimentally confirmed critical residues within 5 Å from SAM are shown in Fig. 4B. The ribose hydroxyl groups of the adenosine ribose of SAM are stabilized by the side chain carboxyl group of D352 via two hydrogen bonds. The amino group of the methionine moiety of SAM is maintained by three polar contacts. An N atom forms a hydrogen bond with main chain carbonyl group of G333 and two polar contacts with the side chain carboxyl group of D331. The carbonyl O atom of the methionine moiety of SAM has two polar contacts with the N atom and carbonyl O atom at the main chain of R310, respectively. Based on the model, these residues are located at the bottom of the SAM-binding pocket (Fig. 4C). Taken together, the structure modeling analysis corroborated the observation in genetic and biochemical assays that the residues such as D331 and G333 in the DxG motif are critical for SAM-binding and enzymatic activities of nsp14.
Fig 4.
Structure-function insight into cap N7-MTase of nsp14 (aa 291 to 527). (A) Overall structure of the predicted 3D model of the core N7-MTase. The proteins are shown as ribbons. The 7 main strands are colored in magenta, and the other structures are colored in cyan. SAM is depicted as a stick model and colored in yellow. (B) Experimentally confirmed residues of nsp14 interacting with SAM. Amino acids are colored by atoms (C, white; O, red; N, blue). SAM is colored by atoms (C, green; O, red; N, blue; S, yellow). Interactions between residues and SAM are shown as yellow dotted lines. (C and D) The SAM-binding pocket of N7-MTase is shown as surface (C) and surface with 40% transparency (D). nsp14 is colored in cyan. The αD helix is colored in red, loop 401-410 is colored in orange, loop 381-386 is colored in green, loop 354-357 is colored in blue, the bottom of the SAM-binding pocket is colored in magenta, and SAM is colored in yellow. Residues are colored by atoms (O, red; N, blue; S, yellow).
In addition to the most conserved DxG motif, the SAM-binding pocket consists of the flexible loop 401-410 as the right flanking wall, which is supported by a rigid αD helix (aa 411 to 424), the loop 381-386 as one part of the long flexible region 380-397 between strands β3 and β4 (Fig. 4A), which supports SAM at the entrance and bottom of the pocket, and the loop 354-357 as the left flanking wall, which was strongly sustained by the strand β2 and αB helix (Fig. 4A and C). It is conceivable that the conformation and stability of the pocket may affect the SAM-binding affinity. Thus, the model suggests why mutations in these loops (such as C382Y in loop 381-386) or sustaining strands and helix (Y368A in αB and C414R, G416R, L419A, Y420H, and Y420A in αD) (Fig. 4D) disrupted or attenuated the activity of nsp14 N7-MTase.
DISCUSSION
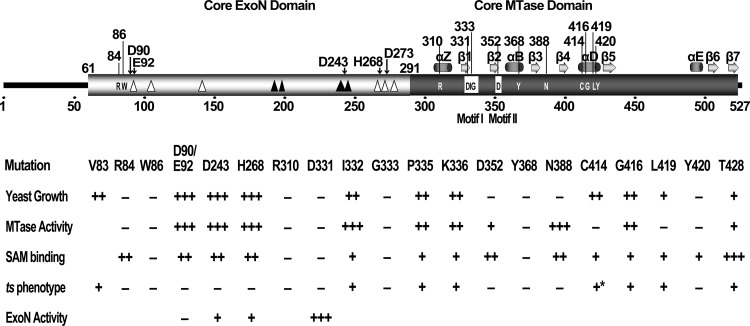
Coronaviruses represent the largest RNA viruses and employ a complicated replication and transcription complex (RTC) for genome replication and expression. The RTC consists of at least a dozen viral nonstructural proteins, including several RNA-processing enzymes such as exoribonuclease and endonuclease that are usually not found in small RNA viruses (13). In previous studies, SARS-CoV nsp14 was shown to possess 3′-to-5′ exoribonuclease activity (20, 21, 39) and N7-MTase activity (19). The combination of N7-MTase and ExoN activities in one protein is unique for coronaviruses. We previously proposed that the two domains for N7-MTase (aa 291 to 527) at the C terminus and ExoN (aa 62 to 290) at the N terminus of nsp14 are structurally interdigitated and functionally inseparable (19), and in this study we dissected the complicated structure-function relationships of nsp14 N7-MTase in biochemical and genetic systems (summarized in Fig. 5).
Fig 5.
Summary of the predicted domain structure of SARS-CoV nsp14 and the mutational effects of critical residues in different assays. The core domains of ExoN and MTase are indicated with open and solid black boxes, respectively. The predicted secondary structure of MTase is shown above the MTase domain, where α indicates α-helix and β represents β-sheet. The critical residues for MTase activity are shown inside the domain boxes of nsp14 (with residue positions above the boxes). The boxed residues indicate the conserved motif I (SAM-binding site) and motif II of the MTase. The open triangles indicate the internal deletions of 2 to 5 amino acid residues which did not affect the MTase activity, and the solid black triangles indicate the lethal internal deletions which inactivated the MTase. The conserved residues and positions of the DEDDh motif of ExoN are indicted by black arrows above the structure box. The arabic numbers indicate the positions or scales of the residues. The effects and phenotypes of the mutations of the critical residues are listed in the lower panel. All amino acids are mutated to alanine except H268L, C414R, and G416R. Results of five different assays are shown: yeast growth, the assays in yeast complementation system; MTase activity, in vitro biochemical assays; SAM binding, in vitro binding assay; ts phenotype, temperature-sensitive tests in yeast system (the asterisk indicates the ts phenotype identified in virus replication); and ExoN activity, in vitro exonuclease assay. The values corresponding to +++, ++, +, and − represent >60%, >20%, <20%, and < 5% of the activity of the wild type (or wild-type growth), except for the ts phenotype, in which + and − indicate the ts phenotype and lethality observed in yeast cells.
By the analysis of the truncation and deletion mutants of SARS-CoV nsp14, we demonstrated that a large portion of nsp14 (aa 62 to 527) is essential for full activity of both N7-MTase and ExoN. The N-terminal 61 aa of nsp14 are dispensable for the enzymes, and the removal of this part even enhanced the activity of N7-MTase. Deletions of even a few amino acid residues throughout the N-terminal ExoN domain could abolish the activity of N7-MTase (Table 1 and Fig. 5). This indicates that the function of the N7-MTase is also strongly dependent on the ExoN domain structure on the same polypeptide. Such properties of coronavirus N7-MTase are different from those of other viral N7-MTases that are commonly separable from other functional domains (1, 3, 36, 40, 41). As the domain size of coronaviral N7-MTase is similar to that of other viral N7-MTases, it is unclear why the enzymatic activity of coronaviral N7-MTase needs the assistance of the ExoN domain in the same protein. Our hypothesis is that the ExoN domain may be involved in substrate binding of the N7-MTase, as discussed below. Resolution of nsp14 crystal structure is needed to finally answer this question.
We generated 60 mutations at 57 positions of nsp14 and revealed 12 critical residues essential for N7-MTase enzymatic activity. Although the residues for mutation were carefully selected, as described in Results, it would be reasonable to assume that more critical residues could be identified if more mutations were analyzed. The critical residues found in this study could be divided into four groups according to their locations: (i) residues at the N-terminal half of nsp14 (R84 and W86), (ii) residues at the SAM-binding and active site motifs (R310, D331, G333, and D352), (iii) residues at the supporting structure of the SAM-binding pocket (Y368, C382, C414, G416, L419, and Y420), and (iv) a residue at the C-terminal half of nsp14 that did not affect SAM binding (T428).
Very interestingly, two critical residues (R84 and W86) were identified at the N terminus of the core ExoN domain (aa 62 to 297) but no critical residues were found in other parts of the ExoN domain (aa 90 to 297). Some internal deletions (1 to 5 aa) of the ExoN domain disabled the activity of N7-MTase, while deletions of individual residues of the conserved DEDDh active site motif maintained the N7-MTase activity (Table 1). In contrast, deletion of one amino acid (ΔA79) at the N terminus of the ExoN domain (aa 62 to 86), which harbors two critical residues (R84 and W86), abolished the N7-MTase activity of nsp14 (Table 1; Fig. 2A, lane 20; Fig. 2C; Fig. 3A, lane 3). Based on these observations, it is tempting to speculate that the conserved residues such as R84 and W86 at the N terminus of the ExoN domain may be directly involved in the enzymatic function of N7-MTase. W86 was shown to be essential for SAM binding of nsp14 (Fig. 3), but it is not known whether it has a direct contact with the SAM molecule. The basic residue R84 could plausibly play a crucial role in binding substrate RNA needed for both N7-MTase and ExoN activities. Such hypothesis is supported by our recent finding that the W86A mutation did not influence the ExoN activity, while the R84A mutation dramatically reduced the ExoN activity (data not shown).
Ten of the 12 critical residues identified are distributed throughout the core domain of N7-MTase, but unexpectedly, no critical residues were found in the C-terminal part of the N7-MTase domain (aa 439 to 527). Truncation analysis revealed that removal of 6 aa from the C terminus already severely attenuated the enzymatic activity of nsp14 N7-MTase. It is important to mention that the C-terminal 5 residues represent the conserved P5 to P1 residues of the cleavage site of coronaviral 3C-like proteinase (3CLpro) and are essential for the proteolysis of 3CLpro between nsp14-nsp15 and viral viability (42). Taking these findings together, this suggests that the C-terminal part of the N7-MTase domain not only contains the conserved cleavage site of 3CLpro but also might contribute to fold formation and substrate RNA binding. This is consistent with the case for other SAM-dependent MTases, in which the C-terminal part varies tremendously in sequence and structure and forms the site for binding different substrates (22, 23). The cap N7-MTase belongs to a large family of SAM-dependent MTases, which share little sequence identity but incorporate a highly conserved Rossmann-like structural fold in the MTase domain (22, 23). The Rossmann-like fold consists of alternating β-stranded and α-helical regions, with 7 strands (β1 to -7) forming a central relatively planar β-sheet and a varying number of helices filling two layers, one on each side of the plane. The order of the strands is generally 3, 2, 1, 4, 5, 7, 6, with strand 7 antiparallel to the other strands (22, 23). The modeled structure of the SARS-CoV nsp14 MTase domain shows an MTase fold similar to that of other MTases (Fig. 4A). However, the MTase domain of nsp14 alone is enzymatically inactive, as discussed above.
The sequence alignment of various SAM-dependent MTases based on structural superimposition shows that the only residues that are highly conserved in the SAM-binding region of the core fold are the glycine-rich sequence E/DxGxGxG (often referred to as motif I) between β1 and αA, which interacts with the amino acid portion of SAM, and an acidic loop between β2 and αB (motif II), which interacts with the ribose hydroxyls (22). At the corresponding position of coronavirus N7-MTase, the conserved sequence motif I could be recognized as DxGxPxG/A (aa 331 to 337), which lies between the putative β1 and αA, and motif II could be recognized as D352, located at the end of β2 (Fig. 5). However, in this study, only two residues (D331 and G333) in the motif were demonstrated to be essential to SAM-binding and N7-MTase activities. The residue R310, which is in the loop between the ExoN domain and the first alpha helix (αZ) of the N7-MTase domain, was also critical for SAM binding. Based on the experimental data and the model structure (Fig. 4), R310, D331, G333, and D352 are most likely involved in direct binding with the SAM molecule. Moreover, alanine substitutions of 6 other residues (Y368A, C382Y, C414R, G416R, L419A, Y420A, and Y420H) affected the SAM-binding affinity and enzymatic activity. Y368 and C382 are predicted to reside in the αB helix and one loop, respectively, and residues C414, G416, L419, and Y420 in the αD helix of the MTase fold (Fig. 4D). These residues may not be in direct contact with SAM but may play a structural role in stabilizing the SAM-binding pocket.
Critical roles have been previously described for some of the residues identified for nsp14 N7-MTase. In our previous study, we showed that mutation of the SAM-binding residue D331 severely impaired SARS-CoV genome replication (19). Sperry and colleagues showed that the mutation of tyrosine to histidine at position 414 (Y414H) of MHV (corresponding to Y420 in SARS-CoV) attenuated the viral viability and virulence in mice but did not affect replication in cell culture (31). In this study, we showed that the same mutations attenuated SAM binding of nsp14 and disrupted the N7-MTase activity (Table 2), which will affect the formation of cap-0 and further cap-1 structure of viral RNA (16, 17, 19). Recently, it was reported that the cap-1 structure of viral RNA provides a molecular signature for the distinction of self and nonself mRNA, and the formation of the viral RNA cap structure may help the virus to evade host innate immunity (43, 44). Altogether, these studies may suggest that the enzymatic activity of coronavirus N7-MTase may not be essential for viral replication in cells but may play a crucial role in viral infection and pathogenesis due to its contribution to viral RNA capping and methylation. In addition, MHV mutations C376Y and C408R (corresponding to SARS-CoV C382Y and C414R) led to a temperature-sensitive phenotype of the mutant viruses (30). In the current study, we showed that such mutants grew well at low temperature (20°C and 25°C) but attenuated or abolished the viability of yeast cells at higher temperature (30°C and 37°C), which may destabilize the structure of the MTase fold and consequently attenuate the activity of N7-MTase. The temperature sensitivity of the yeast strains suggests that the MTase activity is directly responsible for the temperature sensitivity of the virus mutants. Therefore, these mutants will be useful tools in dissecting the significance of the enzymatic activity during virus infection. As N7-MTase plays important roles in coronavirus infection and pathogenesis, the structure-function studies of N7-MTase will contribute to the development of antiviral drugs specifically targeting at the nsp14 enzymatic activities of coronaviruses, including SARS-CoV.
ACKNOWLEDGMENTS
Y.C. thankfully acknowledges the Institute of Biotechnology, University of Helsinki, and Biocenter Finland for providing research facilities and support during his visits.
This study was supported by the China “973” Basic Research Program (2010CB911800 and 2013CB911101), China NSFC grants (81130083, 30925003, 81271817, and 31221061), and the Fundamental Research Funds for the Central Universities (1101003).
Footnotes
Published ahead of print 27 March 2013
REFERENCES
- 1. Furuichi Y, Shatkin AJ. 2000. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 55:135–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cougot N, van Dijk E, Babajko S, Seraphin B. 2004. ‘Cap-tabolism’. Trends Biochem. Sci. 29:436–444 [DOI] [PubMed] [Google Scholar]
- 3. Decroly E, Ferron F, Lescar J, Canard B. 2012. Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 10:51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ahola T, Kaariainen L. 1995. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. U. S. A. 92:507–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ogino T, Banerjee AK. 2007. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell 25:85–97 [DOI] [PubMed] [Google Scholar]
- 6. Li J, Wang JT, Whelan SP. 2006. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc. Natl. Acad. Sci. U. S. A. 103:8493–8498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 117:17–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguiere AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967–1976 [DOI] [PubMed] [Google Scholar]
- 9. Lai MM, Stohlman SA. 1981. Comparative analysis of RNA genomes of mouse hepatitis viruses. J. Virol. 38:661–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lai MM, Patton CD, Stohlman SA. 1982. Further characterization of mRNAs of mouse hepatitis virus: presence of common 5′-end nucleotides. J. Virol. 41:557–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Vliet AL, Smits SL, Rottier PJ, de Groot RJ. 2002. Discontinuous and non-discontinuous subgenomic RNA transcription in a nidovirus. EMBO J. 21:6571–6580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ivanov KA, Thiel V, Dobbe JC, van der Meer Y, Snijder EJ, Ziebuhr J. 2004. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 78:5619–5632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LL, Guan Y, Rozanov M, Spaan WJ, Gorbalenya AE. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. von Grotthuss M, Wyrwicz LS, Rychlewski L. 2003. mRNA cap-1 methyltransferase in the SARS genome. Cell 113:701–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Decroly E, Imbert I, Coutard B, Bouvet M, Selisko B, Alvarez K, Gorbalenya AE, Snijder EJ, Canard B. 2008. Coronavirus nonstructural protein 16 is a cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J. Virol. 82:8071–8084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bouvet M, Debarnot C, Imbert I, Selisko B, Snijder EJ, Canard B, Decroly E. 2010. In vitro reconstitution of SARS-coronavirus mRNA cap methylation. PLoS Pathog. 6:e1000863 doi:10.1371/journal.ppat.1000863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen Y, Su C, Ke M, Jin X, Xu L, Zhang Z, Wu A, Sun Y, Yang Z, Tien P, Ahola T, Liang Y, Liu X, Guo D. 2011. Biochemical and structural insights into the mechanisms of SARS coronavirus RNA ribose 29-O-methylation by nsp16/nsp10 protein complex. PLoS Pathog. 7:e1002294 doi:1002210.1001371/journal.ppat.1002294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Decroly E, Debarnot C, Ferron F, Bouvet M, Coutard B, Imbert I, Gluais L, Papageorgiou N, Sharff A, Bricogne G, Ortiz-Lombardia M, Lescar J, Canard B. 2011. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 7:e1002059 doi:10.1371/journal.ppat.1002059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen Y, Cai H, Pan J, Xiang N, Tien P, Ahola T, Guo D. 2009. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 106:3484–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Minskaia E, Hertzig T, Gorbalenya AE, Campanacci V, Cambillau C, Canard B, Ziebuhr J. 2006. Discovery of an RNA virus 3′->5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 103:5108–5113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen P, Jiang M, Hu T, Liu Q, Chen XS, Guo D. 2007. Biochemical characterization of exoribonuclease encoded by SARS coronavirus. J. Biochem. Mol. Biol. 40:649–655 [DOI] [PubMed] [Google Scholar]
- 22. Martin JL, McMillan FM. 2002. SAM (dependent) I AM: the S-adenosylmethionine-dependent methyltransferase fold. Curr. Opin. Struct. Biol. 12:783–793 [DOI] [PubMed] [Google Scholar]
- 23. Kozbial PZ, Mushegian AR. 2005. Natural history of S-adenosylmethionine-binding proteins. BMC Struct. Biol. 5:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hussain S, Pan J, Chen Y, Yang Y, Xu J, Peng Y, Wu Y, Li Z, Zhu Y, Tien P, Guo D. 2005. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J. Virol. 79:5288–5295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saha N, Schwer B, Shuman S. 1999. Characterization of human, Schizosaccharomyces pombe, and Candida albicans mRNA cap methyltransferases and complete replacement of the yeast capping apparatus by mammalian enzymes. J. Biol. Chem. 274:16553–16562 [DOI] [PubMed] [Google Scholar]
- 26. Ray D, Shah A, Tilgner M, Guo Y, Zhao Y, Dong H, Deas TS, Zhou Y, Li H, Shi PY. 2006. West Nile virus 5′-cap structure is formed by sequential guanine N-7 and ribose 2′-O methylations by nonstructural protein 5. J. Virol. 80:8362–8370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ahola T, Laakkonen P, Vihinen H, Kaariainen L. 1997. Critical residues of Semliki Forest virus RNA capping enzyme involved in methyltransferase and guanylyltransferase-like activities. J. Virol. 71:392–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zuo Y, Deutscher MP. 2002. Mechanism of action of RNase T. I. Identification of residues required for catalysis, substrate binding, and dimerization. J. Biol. Chem. 277:50155–50159 [DOI] [PubMed] [Google Scholar]
- 29. Zuo Y, Deutscher MP. 2002. Mechanism of action of RNase T. II. A structural and functional model of the enzyme. J. Biol. Chem. 277:50160–50164 [DOI] [PubMed] [Google Scholar]
- 30. Sawicki SG, Sawicki DL, Younker D, Meyer Y, Thiel V, Stokes H, Siddell SG. 2005. Functional and genetic analysis of coronavirus replicase-transcriptase proteins. PLoS Pathog. 1:e39 doi:10.1371/journal.ppat.0010039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sperry SM, Kazi L, Graham RL, Baric RS, Weiss SR, Denison MR. 2005. Single-amino-acid substitutions in open reading frame (ORF) 1b-nsp14 and ORF 2a proteins of the coronavirus mouse hepatitis virus are attenuating in mice. J. Virol. 79:3391–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. De la Pena M, Kyrieleis OJ, Cusack S. 2007. Structural insights into the mechanism and evolution of the vaccinia virus mRNA cap N7 methyl-transferase. EMBO J. 26:4913–4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zheng S, Shuman S. 2008. Structure-function analysis of vaccinia virus mRNA cap (guanine-N7) methyltransferase. RNA 14:696–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zheng S, Shuman S. 2008. Mutational analysis of vaccinia virus mRNA cap (guanine-N7) methyltransferase reveals essential contributions of the N-terminal peptide that closes over the active site. RNA 14:2297–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kroschewski H, Lim SP, Butcher RE, Yap TL, Lescar J, Wright PJ, Vasudevan SG, Davidson AD. 2008. Mutagenesis of the dengue virus type 2 NS5 methyltransferase domain. J. Biol. Chem. 283:19410–19421 [DOI] [PubMed] [Google Scholar]
- 36. Zhou Y, Ray D, Zhao Y, Dong H, Ren S, Li Z, Guo Y, Bernard KA, Shi PY, Li H. 2007. Structure and function of flavivirus NS5 methyltransferase. J. Virol. 81:3891–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saha N, Shuman S, Schwer B. 2003. Yeast-based genetic system for functional analysis of poxvirus mRNA cap methyltransferase. J. Virol. 77:7300–7307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schwer B, Shuman S. 2006. Genetic analysis of poxvirus mRNA cap methyltransferase: suppression of conditional mutations in the stimulatory D12 subunit by second-site mutations in the catalytic D1 subunit. Virology 352:145–156 [DOI] [PubMed] [Google Scholar]
- 39. Eckerle LD, Lu X, Sperry SM, Choi L, Denison MR. 2007. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 81:12135–12144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shuman S. 2002. What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell Biol. 3:619–625 [DOI] [PubMed] [Google Scholar]
- 41. Ferron F, Decroly E, Selisko B, Canard B. 2012. The viral RNA capping machinery as a target for antiviral drugs. Antiviral Res. 96:21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fang S, Shen H, Wang J, Tay FP, Liu DX. 2010. Functional and genetic studies of the substrate specificity of coronavirus infectious bronchitis virus 3C-like proteinase. J. Virol. 84:7325–7336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M, Jr, Shi PY, Diamond MS. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468:452–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, Siddell SG, Ludewig B, Thiel V. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 12:137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]