Abstract
APOBEC3 proteins mediate potent antiretroviral activity by hypermutating the retroviral genome during reverse transcription. To counteract APOBEC3 and gain a replicative advantage, lentiviruses such as human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus (SIV) have evolved the Vif protein, which targets APOBEC3 proteins for proteasomal degradation. However, the proteasome plays a critical role in the generation of T cell peptide epitopes. Whether Vif-mediated destruction of APOBEC3 proteins leads to the generation and presentation of APOBEC3-derived T cell epitopes on the surfaces of lentivirus-infected cells remains unknown. Here, using peptides derived from multiple Vif-sensitive APOBEC3 proteins, we identified APOBEC3-specific T cell responses in both HIV-1-infected patients and SIV-infected rhesus macaques. These results raise the possibility that these T cell responses may be part of the larger antiretroviral immune response.
INTRODUCTION
Viral sequence diversity is the Achilles' heel of traditional vaccine approaches, as exemplified by influenza vaccinology. The influenza vaccine must change yearly in anticipation of the predominant circulating viral strains, which differ from one another by only 1 to 2% (1, 2). In contrast, circulating human immunodeficiency virus type 1 (HIV-1) strains differ from each other by 20% in the more conserved proteins and up to 35% in the Env protein (1, 2). This enormous sequence diversity is a major stumbling block to the development of a conventional HIV-1 vaccine. Indeed, a neutralizing antibody-based vaccine has proven elusive, in part due to the extremely high sequence variation of the HIV-1 Env protein (3). While CD8+ T cell-based vaccines directed against more-conserved viral proteins have shown some ability to blunt viral replication (4, 5), they are also hindered by HIV-1 sequence variation (6). Even minor variations in sequence can have a dramatic effect on CD8+ T cell efficacy, as just one amino acid change can abrogate recognition and suppression of virus replication (7–9). Additionally, HIV-1 rapidly mutates to escape effective cytotoxic T lymphocyte (CTL) responses, thereby raising the possibility that any effective vaccine will only have a transient effect (6, 10, 11). On a population level, CD8+ T cells are driving HIV-1 evolution toward fixation of epitope escape variants that are more likely to evade the immune responses of the population in which they circulate (12, 13). Therefore, a successful HIV-1 vaccine must overcome the formidable challenge of HIV-1 sequence diversity. In this study, we sought to determine whether invariant self-antigens overexpressed within an HIV-1-infected cell could act as potential immune targets for vaccine development. Specifically, we examined HIV-infected patients and simian immunodeficiency virus (SIV)-infected macaques for the presence of APOBEC-specific T cell responses.
Mammalian host cells have developed intrinsic mechanisms to prevent lentiviral replication, as well as to maintain genomic stability by restricting the movement of retroelements. These mechanisms include postentry interference by tripartite motif-containing protein 5α (TRIM5α), transcriptional silencing through DNA methylation, posttranscriptional silencing via RNA interference, tetherin, and mutational inactivation of elements in the course of their retrotransposition cycle by cellular cytosine deaminases (14, 15). The apolipoprotein B mRNA-editing, enzyme-catalytic, polypeptide-like 3 (APOBEC3) family of cytidine deaminases consists of 7 members (APOBEC3A to APOBEC3H) in humans, all of which are encoded on the same gene cluster of chromosome 22. These enzymes are expressed in the majority of human cells and have been extensively studied since the discovery that APOBEC3G acts as a viral restriction factor in HIV-1 infection (16). Since that time, multiple additional APOBEC3s have been implicated in HIV restriction (17–19).
APOBEC3G is packaged into budding HIV-1 virions through an interaction with the nucleocapsid region of HIV-1 Gag, and its proclivity for binding single-stranded nucleic acids facilitates this process (20, 21). Following HIV-1 infection of a target cell, the viral RNA genome is uncoated, and reverse transcriptase generates a single strand of DNA complementary to the viral genome. During this transcription process, reverse transcriptase also degrades the viral RNA strand through its RNase H activity, leaving only the single strand of cDNA (20). APOBEC3G exerts its enzymatic activity on this single-stranded DNA, mutating cytosines to uracils. These extensive mutations can cause degradation of the viral genome or inhibit viral replication due to altered reading frames.
Alternatively, APOBEC3G has been shown to inhibit HIV-1 replication independent of its enzymatic function. First, it can impair reverse transcriptase activity by blocking the binding of tRNALys3 (the primer that initiates reverse transcription). Second, it can stifle the cleavage of tRNALys3 from the single-stranded DNA intermediate, leading to aberrant viral DNA ends (19). Finally, it can bind to HIV-1 integrase, a component of the viral preintegration complex, suggesting that it may obstruct nuclear homing of provirus (19).
HIV-1 expresses 6 accessory proteins that are integral to its replication and persistence. One of these accessory proteins is the viral infectivity factor (Vif) protein. The function of Vif was initially elucidated from experiments showing that Vif-deficient HIV could replicate in certain cells (permissive), but not in others (nonpermissive). Infection of a hybridoma of these two cell types with Vif-deficient HIV-1 yielded noninfectious virions, implying that a host cellular restriction factor was obstructing further infections.
Subsequent analysis of two highly related cell lines, one permissive and the other nonpermissive, using a cDNA subtraction strategy revealed that this cellular restriction factor was APOBEC3G (22). Since this discovery, Vif has been shown to ubiquitinate APOBEC3G, leading to degradation by the proteasome. Indeed, the levels of APOBEC3G are drastically reduced in virion-producing cells infected with HIV-1, and APOBEC3G is not packaged into progeny virions in the presence of Vif (23). Vif has been shown to inhibit APOBEC3G and APOBEC3F, but not APOBEC3B (19, 24). Thus, although the physiological role of APOBEC3 proteins is not fully understood, they represent an ancient form of antiviral or antiretroelement defense in mammals (19).
The proteasome is an important first step in the antigen presentation pathway for HLA class I peptide epitopes recognized by CD8+ T cells. We hypothesized that HIV-1 infection would give rise to CD8+ T cell responses against APOBEC3 proteins, due to increased Vif-triggered proteasomal processing and HLA class I presentation. In a cell not infected by HIV-1, there would be limited presentation of APOBEC3-derived peptide epitopes on the surface as part of the normal repertoire of self-antigens presented by major histocompatibility complex class I (MHC-I) molecules.
The immense sequence diversity of HIV-1 remains a major roadblock to a prophylactic HIV-1 vaccine. Targeting CD8+ T cells to HIV-1-infected cells by their higher expression of APOBEC3 antigens may be a novel way to circumvent the obstacle presented by viral sequence diversity. Here, we present the first data showing APOBEC3-specific T cell responses in both HIV-1-infected humans and SIV-infected rhesus macaques.
MATERIALS AND METHODS
Peptide antigens for humans.
We used in silico T cell immunogenicity prediction methods to identify peptide epitopes for the HLA-B*58, -A*2, and -B*7 superfamilies within APOBEC3F and APOBEC3G. Peptides were identified within the APOBEC3F and APOBEC3G proteins with NetCTL 1.2 software (http://www.cbs.dtu.dk/services/NetCTL/). Top scoring epitopes for the HLA-B*58, -A*2, and -B*7 supertypes were selected for peptide synthesis. We selected peptides on the basis of their NetCTL software affinity score and their capacity to be presented by several HLA supertypes and excluded one peptide similar to the activation-induced cytidine deaminase protein (AID). Priority was given to peptides shared by APOBEC3F and APOBEC3G proteins. A total of 12 peptides in total were synthesized and tested together in a peptide pool or individually in the assay. An APOBEC3A/B peptide pool consisting of the four top scoring epitopes for HLA-B*58, -A*2, and -B*7 was also used where specified.
Peptide antigens for rhesus macaques.
For the studies in macaques, we used the following two sets of peptides: predicted MHC-I-binding peptides for rhesus APOBEC3F and 15-mer peptides overlapping rhesus APOBEC3C, APOBEC3G, and APOBEC3H by 11 amino acids. For predicted binders, we used the MHCPathway Macaque algorithm (www.mamu.liai.org) to generate 36 CD8+ T cell epitopes derived from the rhesus APOBEC3F protein predicted to bind two common MHC-I molecules, Mamu-A1*001:01 and Mamu-A1*002:01. We then lumped these peptides into 3 Mamu-A1*001:01 pools and 6 Mamu-A1*002:01 pools, where each pool contained between 2 and 8 peptides. All peptides were synthesized by either Pepscan or Genscript and resuspended in 100% dimethyl sulfoxide (DMSO).
Study participants.
In this study, we measured T cell reactivity to APOBEC3F and APOBCEC3G peptides in the following groups: (i) low-risk healthy volunteers (n = 33); (ii) vertically exposed, but uninfected children (n = 7); (iii) vertically exposed, HIV-1-infected children (n = 73); (iv) long-term nonprogressors (LTNP), who were defined as having asymptomatic, untreated HIV-1 infection for at least 7 years with no consistent decline in peripheral blood CD4+ count and low or undetectable levels of plasma viremia (<10,000 copies/ml, bDNA) (n = 7); (v) HIV-1-infected adults with primary HIV-1 infection (n = 13); (vi) chronically infected adults not treated with an antiretroviral agent with viral loads below 5,000 copies RNA/ml (viral-load-controlled virus in the absence of therapy (“controllers”; n = 19); (vii) non-antiretroviral-treated individuals who had higher levels of viremia (“noncontrollers”; n = 21); and (viii) individuals treated with highly active antiretroviral therapy (HAART), defined by the treatment-mediated maintenance of an undetectable viral load (“HAART suppressed”; n = 20). HIV-1-negative controls were obtained from purchased buffy coats from low-risk donors attending the Stanford Blood Bank Center. Blood donors are asked about risk activity and are included or excluded as a blood donor based upon criteria established by blood banks. HIV-1-positive donors are from studies of adults with primary infections (Options cohort) and chronic HIV-1 infection (SCOPE cohort) at the University of California San Francisco (UCSF) and individuals with vertical infection under the care of clinicians at the Jacobi Hospital in the Bronx in New York, New York. All participants were enrolled in institutional review board (IRB)-approved studies, conducted under the directive of the Declaration of Helsinki.
Nonhuman primates.
The animals in this study were Indian rhesus macaques (Macaca mulatta) from either the Wisconsin or Oregon National Primate Research Center colony. The animals were infected with SIV for other unrelated studies and were cared for according to the regulations and guidelines of the Institutional Animal Care and Use Committee of the University of Wisconsin or the Oregon Health & Science University.
ELISPOT analysis.
Peptides were tested, as previously described, in a gamma interferon (IFN-γ) enzyme-linked immunosorbent spot assay (ELISPOT) using either fresh or cryopreserved peripheral blood mononuclear cells (PBMC) (25). Peptides were tested in duplicate wells at a concentration of 10 μg/ml or 10 μM (either individually or in pools) with 100,000 PBMC per well. The total numbers of spots for duplicate wells were averaged, and all the numbers of spots were normalized to the numbers of IFN-γ spot-forming cells (SFC) per 1 × 106 PBMC. The numbers of spots from control wells (no stimulation) were subtracted to determine the responses to each peptide. Responses were considered positive if they exceeded two times the standard deviation of the background count as previously described (8). We did not consider responses of <50 SFC per 1 × 106 PBMC as positive responses.
Flow cytometry.
Cryopreserved PBMC from chronically HIV-1-infected individuals or PBMC/bronchoalveolar lavage (BAL) cells were stimulated with or without 10 μg/ml APOBEC3F/G or APOBEC3A/B peptide pools for 6 h with 1 μg/ml costimulatory anti-CD28 and anti-CD49d antibodies (Biolegend) and in the presence of 5 μg/ml brefeldin A. An exogenous cytomegalovirus (CMV) pp65 peptide pool was used as a positive control. The cells were stained with fluorophore-conjugated antibodies to CD3 (clone UCHT1; Beckman Coulter), CD4 (clone RPA-T4; BD Pharmingen), CD8 (clone 3B5; Life Technologies), and IFN-γ (clone B27; BD Pharmingen) to determine phenotype and function, and an amine dye was used to discriminate between live and dead cells. Data were acquired with a BD LSR-II system. At least 100,000 events were collected and analyzed with FlowJo software (TreeStar, Inc.).
Generation of an APOBEC-specific cell line in vitro.
CD8+ T cell lines were produced as described previously (26). Briefly, PBMC were isolated using Ficoll-Paque Plus (Amersham Biosciences) density centrifugation. CD8+ cells were isolated using CD8 microbeads and columns purchased from Miltenyi Biotec and used according to the manufacturer's instructions. Isolated CD8+ cells were repeatedly stimulated with autologous, irradiated B lymphoblastoid cells pulsed with the peptide of interest. Cells were cultured in R15-100 (RPMI 1640 medium containing 15% fetal calf serum [FCS] and 100 U/ml interleukin 2 [IL-2]).
Restricting allele assay.
Transient expression of cloned MHC class I cDNA was achieved by electroporation of plasmid DNA into the MHC class I-deficient human B cell line 721.221. Briefly, 5 μg of plasmid DNA encoding the MHC-I allele of interest was added to 5 × 106 721.221 cells in 100 μl of Nucleofector solution C and electroporated using program G-16 on a Nucleofector I device (Amaxa, Köln, Germany). Maximum cell surface expression occurred 4 days postelectroporation. MHC class I surface expression on stable and transient MHC class I transfectants was measured by W6/32 antibody surface staining. Staining was also performed on 721.221 cells as a negative control and on immortalized macaque B cell lines as a positive control. These cells were incubated with the peptide of interest for 1.5 h at 37°C and then washed three times with warm phosphate-buffered saline (PBS) to remove excess peptide. The freshly washed peptide-pulsed cells were then incubated with CD8+ T cell lines specific for the peptide of interest for intracellular staining (ICS) or ELISPOT as described above to determine the restricting MHC-I molecule.
MHC-I tetramer production and staining.
MHC class I tetramers were constructed with minor modifications as previously described (8, 27). Ex vivo MHC class 1 tetramer stains were performed on CD8+ T cell lines or cryopreserved PBMC as previously described (8). Briefly, 5 × 105 cells were stained with 5 μl of 0.1-mg/ml tetramer stocks, 3 μl of CD3 conjugated to fluorescein isothiocyanate (FITC) (clone SP34-2; BD Biosciences), and 5 μl of CD8 conjugated to peridinin chlorophyll protein (PerCP) (clone SK1; BD Biosciences) in approximately 100 μl of R10 (RPMI 1640 with 10% FCS). After the cells were fixed in 1% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA), approximately 0.5 × 105 to 1 × 105 lymphocyte-gated events were acquired on a FACSCalibur (BD Biosciences) and analyzed using FlowJo (TreeStar, Inc.).
RESULTS
APOBEC3-specific responses in HIV-1-infected patients.
APOBEC3F and APOBEC3G are both potent inhibitors of HIV-1 replication and sensitive to HIV-1 Vif-induced proteasomal degradation (16, 19, 22). Therefore, we focused on these two proteins in our initial studies. Using the NetCTL MHC-I binding algorithm (www.cbs.dtu.dk/services/NetCTL), we generated peptide epitopes from APOBEC3F and APOBEC3G predicted to bind tightly to members of the common HLA-B2*8, -A*2, and -B*7 supertypes. We selected the top two predicted epitopes for each HLA allele derived from each APOBEC3 protein, resulting in 12 predicted APOBEC3 peptides. We then combined these peptides into a single APOBEC3 peptide pool and screened a cohort of healthy adults not infected with HIV-1, chronically HIV-1-infected adults, exposed uninfected children, and chronically HIV-1-infected children using IFN-γ ELISPOT (Table 1). We observed little or no response in the healthy, non-HIV-1-infected adults and exposed uninfected children. In contrast, we observed weak, yet detectable responses in the chronically HIV-1-infected adults. Intriguingly, we observed the strongest APOBEC3-specific responses in the long-term nonprogressors (5/7), with a mean of 486 SFC/106 PBMC (Table 1). In primary HIV-1-infected subjects, 5/13 had responses, albeit at lower magnitude, with a mean of 84 SFU/106 PBMC. HIV-1 noncontrollers had the lowest mean T cell response to the APOBEC3 pool (34 SFC/106 PBMC), although there was no substantial difference compared to the responses of the HAART-suppressed group (54 SFC/106 PBMC) or the controllers (45 SFC/106 PBMC). There were 13/73 responders in the group of HIV-1-infected children (88 SFC/106 PBMC) (Table 1).
Table 1.
T cell responses against APOBEC3 peptides
| HIV status and group | No. of subjects | Mean CD4 count (/mm3) | Mean viral load (no. of copies/ml) | No. of responders against APOBEC3 pool | Mean APOBEC3 pool response (no. of SFU/106 PBMC)a | Range of APOBEC3 pool responses (SFU/106 PBMC)a | % of responders |
|---|---|---|---|---|---|---|---|
| HIV infected | |||||||
| Chronic infection | |||||||
| LTNPb | 7 | 828 | 117 | 5 | 486 | 0–2,480 | 71 |
| Natural controllers | 19 | 643 | 393 | 6 | 45 | 0–285 | 32 |
| HAART suppressed | 20 | 626 | 51 | 8 | 54 | 0–250 | 40 |
| Viremic | 21 | 282 | 52,656 | 3 | 34 | 0–290 | 14 |
| Primary infection | 13 | 673 | 13,435 | 6 | 84 | 0–305 | 46 |
| Children with chronic infection | 73 | 836 | 18,488 | 13 | 88 | 0–1,780 | 18 |
| HIV-exposed children but uninfected | 7 | 0 | 5 | 0–35 | 0 | ||
| Nonexposed, healthy HIV-negative adults | 33 | 2 | 18 | 0–210 | 6 |
SFU, spot-forming units. The background SFU have been subtracted from the values shown.
LTNP, long-term nonprogressors.
APOBEC3-specific responses in HIV-1-infected patients are CD8+ T cell mediated.
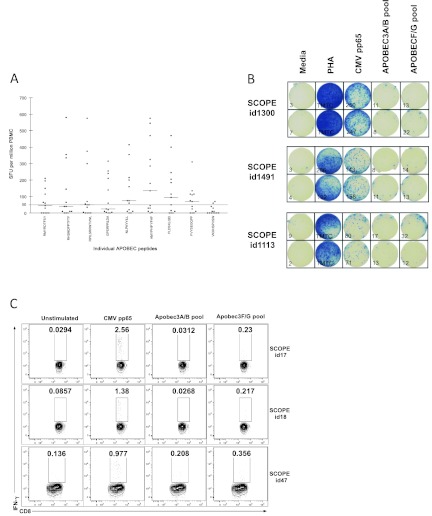
To further investigate these APOBEC3-specific T cell responses, we selected 13 chronically HIV-1-infected children with responses against the APOBEC3F/G peptide pool and tested them against individual APOBEC3 peptides using IFN-γ ELISPOT (Fig. 1A). Figure 1B illustrates typical results of a IFN-γ ELISPOT of three chronically infected adults. We then tested PBMC IFN-γ production by flow cytometry in three chronically infected adults comparing APOBEC3F/G-specific CD8+ T cell responses to those generated by a known ubiquitous T cell immunogen, CMV pp65 (Fig. 1C). In addition, a pool consisting of four predicted peptides derived from Vif-insensitive APOBEC3A/B proteins was included as a negative control. IFN-γ-producing T cells were shown to be CD8+, and there was little to no response to APOBEC3A/B peptide pools, suggesting that Vif-induced proteasomal destruction of APOBEC3F/G but not APOBEC3A/B, may occur in HIV-1-infected cells (Fig. 1C). Together with the responses observed in LTNP, these data suggest that APOBEC3-specific T cell responses may expand during HIV-1 infection. No CD4+ T cell responses were identified using flow cytometry from these human studies, although we cannot rule out the possibility that some APOBEC3-specific IFN-γ production in the ELISPOT assay was produced by CD4+ T cells.
Fig 1.
APOBEC3-specific T cell responses in HIV-1-infected children. (A) PBMC from 13 chronically HIV-1-infected children were screened against 9 of the 12 APOBEC3 peptides present in the APOBEC3 peptide pool. Results are shown as the mean numbers of spot-forming units (SFU) per million PBMC of duplicate wells in the IFN-γ ELISPOT, and background SFUs have been subtracted from the values. The sequences of the individual peptide epitopes are shown on the x axis. (B) Examples of wells from an IFN-γ ELISPOT assay. One hundred thousand PBMC from three chronically infected adults were left unstimulated (Media) or stimulated with phytohemagglutinin (PHA), CMV pp65 peptide pool, APOBEC3A/B peptide pool, or APOBEC3F/G peptide pool. The number of spot-forming units/105 cells are indicated in the lower left corner for each well. (C) IFN-γ production by the CD8+ lymphocyte population from PBMC isolated from three chronically HIV-1-infected patients from the SCOPE cohort. The percentage of cells within the gate is shown in each plot: unstimulated, stimulated with a pool of CMV pp65 peptides, stimulated with a pool of APOBEC3A/B peptides, and stimulated with APOBEC3F/G pool.
Identification of APOBEC3-specific T cell responses in SIV-infected rhesus macaques.
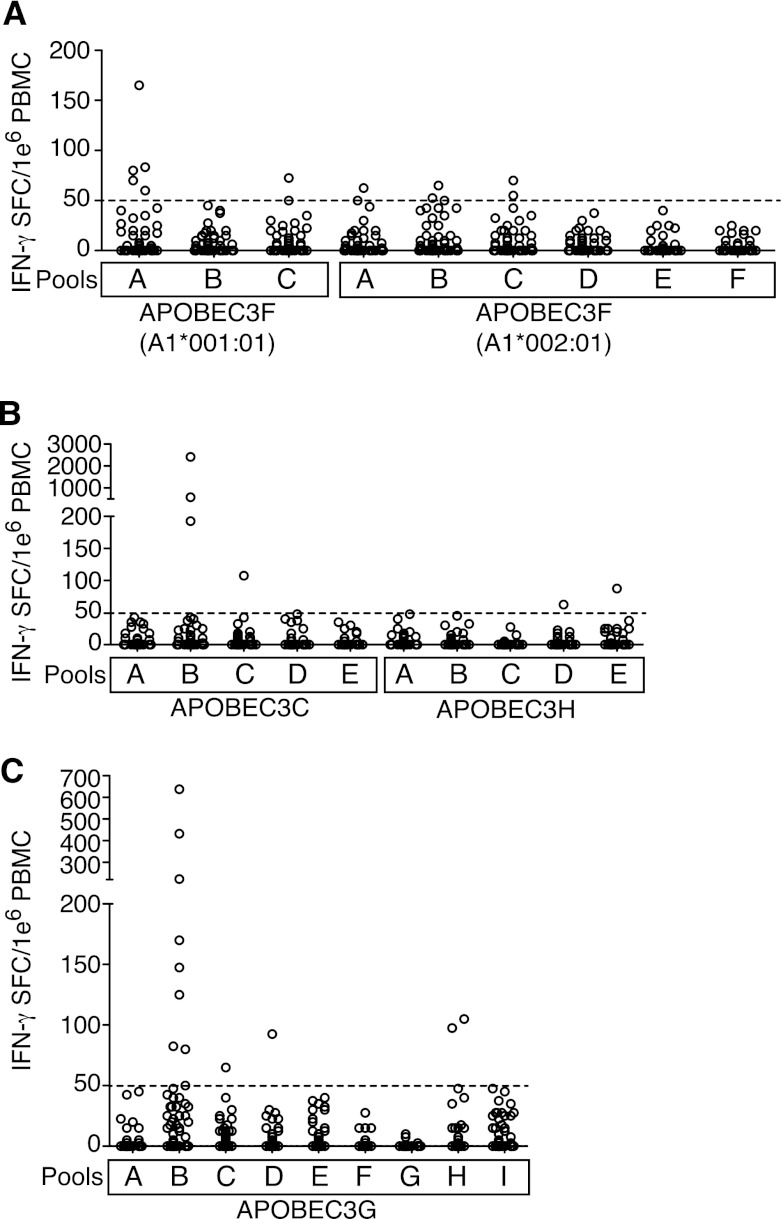
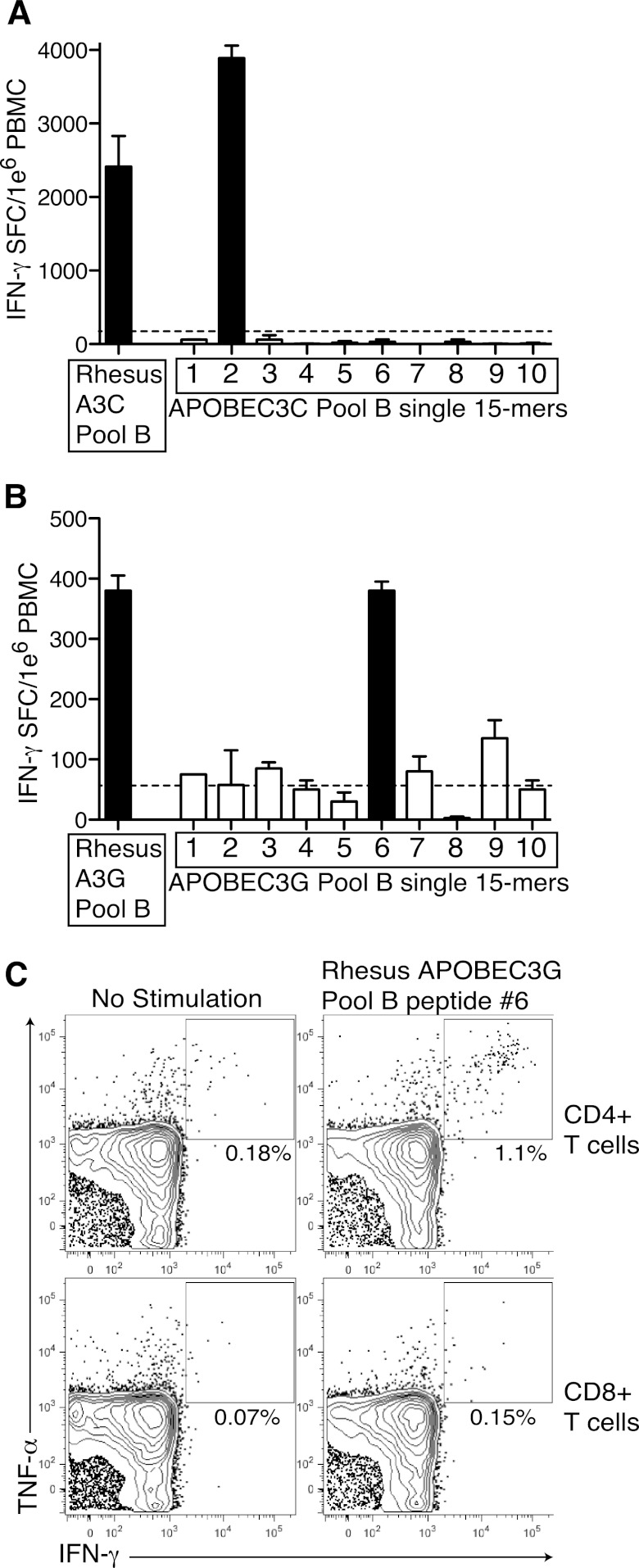
Given that HIV-1-infected patients, LTNP in particular, mounted APOBEC3G- and APOBEC3F-specific CD8+ T cell responses, we next investigated whether SIV-infected Indian rhesus macaques might also target SIVmac239 Vif-sensitive APOBEC3 protein sequences (28). To this end, we screened PBMC from a total of 52 chronically SIV-infected Indian rhesus macaques for T cell responses against rhesus APOBEC3F, APOBEC3C, APOBEC3H, and APOBEC3G by IFN-γ ELISPOT (Fig. 2A, B, and C). The majority of animals did not respond to any of the APOBEC3 peptides, but similar to what we had observed in HIV-1-infected humans, we detected the presence of low-magnitude responses, particularly against APOBEC3G (Fig. 2C). Because the strongest APOBEC3-specific responses in humans were observed in LTNP, we focused subsequent analysis predominantly on the small number of LTNP Indian rhesus macaques in our cohort, which have spontaneously controlled SIV replication to low levels. Interestingly, we observed the highest frequency response against APOBEC3C pool B in an animal that had previously undergone experimental depletion of CD8+ cells (Fig. 3A). To confirm this high frequency APOBEC3C T cell response, we next screened PBMC from this animal against the individual 15-mer peptides in APOBEC3C pool B and identified a single 15-mer peptide eliciting a high frequency IFN-γ T cell response (Fig. 3A). We also confirmed a high frequency T cell response against APOBEC3G in a second LTNP macaque and identified a single 15-mer peptide triggering this response (Fig. 3B). To further characterize that APOBEC3G-specific T cell response, we performed cytokine flow cytometry. Surprisingly, we identified that this APOBEC3G-specific response was CD4+ T cell mediated (Fig. 3C). Thus, we determined that both CD4+ and CD8+ T cells target APOBEC3 proteins.
Fig 2.
APOBEC3 responses in chronically SIV-infected rhesus macaques. (A to C) PBMC from SIV-infected rhesus macaques were screened against peptides derived from rhesus APOBEC3F and predicted to bind Mamu-A1*001:01 or Mamu-A1*002:01 (A), against overlapping 15-mer peptides spanning rhesus APOBEC3C and APOBEC3H (B), or against overlapping 15-mer peptides spanning rhesus APOBEC3G (C). The mean of each response minus the background from each animal is plotted. The broken line indicates the threshold for positivity.
Fig 3.
Confirmation of APOBEC3-specific responses in SIV-infected rhesus macaques. (A) PBMC from LTNP rhesus macaques were screened against the APOBEC3C pool B and each individual 15-mer peptide within that pool. Results are shown as means plus standard deviations (error bars) of duplicate wells. (B) PBMC from LTNP rhesus macaques were screened against the APOBEC3G pool B and each individual 15-mer peptide within that pool. Black bars indicate positive responses. Results are shown as means plus standard deviations of duplicate wells. The broken line represents the threshold for positivity. (C) Bronchoalveolar lavage cells were stimulated with media (no stimulation) or the rhesus APOBEC3G pool B single 15-mer peptide identified in panel B. The percentages of cells positive for IFN-γ and tumor necrosis factor alpha (TNF-α) are shown.
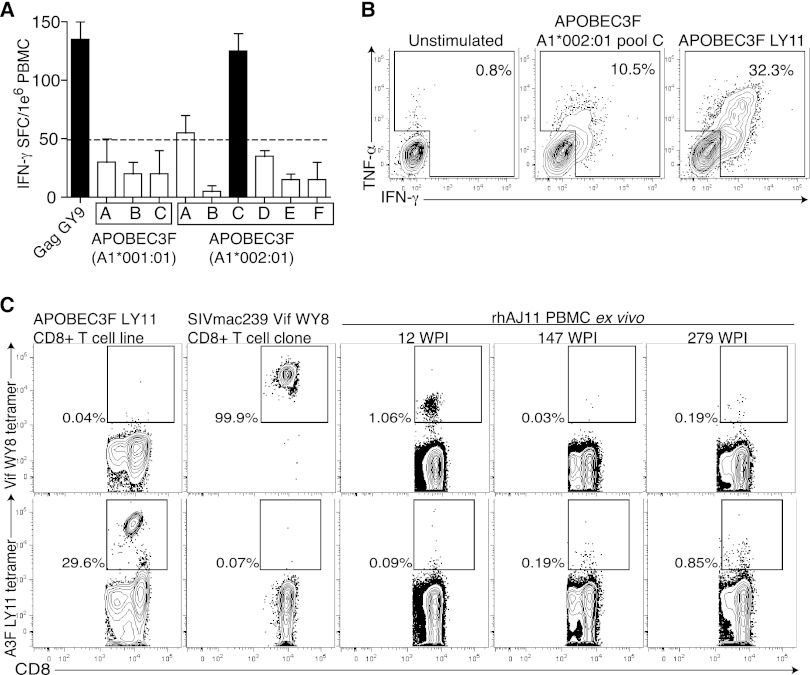
Finally, we identified a LTNP macaque with no detectable viral load that mounted a T cell response against a pool of predicted Mamu-A1*002:01-binding, APOBEC3F-derived peptides (Fig. 4A). Although this response was not robust, the magnitude of this response was similar to the magnitude of the Mamu-A1*002:01-restricted immunodominant SIVmac239 Gag71-79 GY9 response in this animal (Fig. 4A). Next, we stimulated PBMC from this animal with the predicted APOBEC3F Mamu-A1*002:01 pool to generate an APOBEC3-specific cell line in vitro. Using this cell line, we mapped the APOBEC3 pool response to a single predicted CD8+ T cell epitope, APOBEC3F115-125 LY11 (Fig. 4B). Using single MHC-I-expressing transfectants, we then confirmed the restricting allele of APOBEC3F115-125 LY11-specific CD8+ T cells to be Mamu-A1*002:01 as predicted (data not shown). As we had identified both the minimal optimal peptide epitope and its restricting allele, we generated a CD8+ T cell tetramer that specifically stained our in vitro cell line (Fig. 4C). We next used a Vif WY8 tetramer and our APOBEC3 LY11 tetramer reagent to longitudinally examine the immune response in this LTNP macaque to both APOBEC3F and the SIVmac239 Vif protein. As expected, tetramers for APOBEC3F115-125 LY11 stained only cells specific for that epitope and not Vif97-104 WY8. We noted that while CD8+ T cells specific for both Vif and APOBEC3F were detected at low frequencies, they expanded and contracted asynchronously with respect to each other (Fig. 4C). Furthermore, consistent with our other observations, APOBEC3-specific CD8+ T cells appeared to be in the peripheral blood compartment.
Fig 4.
A LTNP macaque targets APOBEC3F. (A) PBMC from a LTNP (with no detectable plasma virus) were screened against APOBEC3F pools predicted to bind Mamu-A1*001:01 or Mamu-A1*002:01, and the SIVmac239 Gag71-79 GY9 epitope in an IFN-γ ELISPOT. Results are shown as means plus standard deviations of duplicate wells. (B) A CD8+ T cell line was generated by multiple rounds of stimulation using the APOBEC3F Mamu-A1*002:01 pool C. The cell line reacts against the autologous B lymphocyte cell line (BLCL) presenting both the pool and the single epitope APOBEC3F115-125 LY11. Graphs were generated by gating on CD8+ cells. (C) Tetramers specific for either APOBEC3F115-125 LY11 or SIVmac239 Vif97-104 WY8 were used to stain cryopreserved PBMC, an in vitro CD8+ T cell line specific for Mamu-A1*002:01-restricted APOBEC3F115-125 LY11, and an in vitro CD8+ T cell clone specific for Mamu-A1*002:01-restricted SIVmac239 Vif97-104 WY8. Dot plots were generated by gating on CD3+ or CD8+ cells. The percentages of tetramer-positive events are shown. rhAJ11, recombinant human AJ11; WPI, weeks post-SIVmac239 infection.
DISCUSSION
We show here for the first time that APOBEC3-specific CD8+ T cells exist in humans infected with HIV-1 and in macaques infected with SIV. It is intriguing that the long-term nonprogressors had the highest magnitude responses to the APOBEC3 peptides, suggesting that they may be involved in helping control HIV-1 replication. Within the remaining HIV-1-infected subjects, APOBEC3-specific T cell responses were detected at lower magnitude and there was a trend toward higher responses within the controller and HAART-suppressed groups.
The data suggest that immune responses to APOBEC3 proteins may be triggered by lentivirus infection. Indeed, we found APOBEC3-specific T cell responses in healthy individuals at a very low frequency. Since APOBEC3-derived peptides will stabilize nascent HLA molecules and be transported to the surface of any nucleated cell at low levels as part of the repertoire of self-peptides processed within the cell, we hypothesize that APOBEC3-specific T cells are present normally at low frequency as part of the normal low-level self-peptide T cell repertoire. However, we also hypothesize that in the inflammatory context of HIV-1 or SIV infection, Vif-induced degradation of APOBEC3 will result in a marked increase in the amount of APOBEC3-derived peptides on the surfaces of virus-infected cells. Thus, while APOBEC3 peptides will be present at low levels on the surfaces of healthy cells, they should be present at much higher levels in virus-infected cells due to Vif-induced destruction of APOBEC3. Alternatively, the generalized immune deregulation in the context of HIV-1 infection might provide cytokine-driven expansion of self-antigen responses.
Recognition of APOBEC3-derived peptides is theoretically independent of the sequence of the infecting viral sequence. This could be used to circumvent the obstacle of virus sequence diversity by targeting a stable target induced by the viral replication cycle. To escape APOBEC3-specific T cell immune responses, HIV-1 would need to mutate Vif such that it would no longer interact with and degrade APOBEC proteins. Such a mutation would render the virus vulnerable to APOBEC3-induced hypermutation.
APOBEC3 is a self-protein that is widely expressed (22, 29). Self-directed T cell responses raise the possibility of autoimmunity. However, as we have shown in our data, elite controllers of HIV-1 infection make anti-APOBEC3-specific T cell responses without apparent deleterious clinical autoimmune manifestations. The anti-APOBEC3 T cell responses observed do not appear to have led to any overt systemic autoimmunity in these patients or primates. The immune system exists with continuous surveillance and recognition of self-antigens, which in the absence of danger signals does not result in an immunopathogenic state (30, 31). Analogous to a cancer cell with aberrant protein expression, an HIV-1-infected cell will express APOBEC3-derived peptide epitopes at a higher frequency than uninfected cells. In the cancer field, many studies have approached cancer immunotherapy with self-antigens expressed in tumors (32, 33). In addition, the cancer therapeutic vaccination field has used self-antigens (e.g., melanoma-associated antigen [MAGE] peptides for melanoma) with good safety profiles and successful induction of immune responses (34). Further studies will be needed to investigate the potential of APOBEC3 epitopes as “safe” antigens in a vaccine against HIV-1. An important caveat for this proposed vaccination approach is that noninfected cells might also upregulate APOBEC3 during infections and could also possibly be marked for destruction.
On the basis of the observations presented here, we hypothesize that lentiviral infection generates APOBEC3-specific T cells, which are capable of recognizing and eliminating virally infected cells. Further, a vaccine immunogen using APOBEC3 sequences could generate APOBEC3-specific T cells, which might recognize and kill a cell infected with any variant of HIV-1. This novel approach targets infected cells based on their presentation of APOBEC3-derived peptides, not HIV-1 peptides. These studies are, therefore, the first steps toward a novel vaccine approach, which circumvents the obstacle of HIV-1 sequence diversity by targeting a surrogate marker of HIV-1 infection.
ACKNOWLEDGMENTS
We thank Emily Eriksson, Mark Peakman, and Mike Malim for helpful discussions.
This work was supported in part by the AIDS biology program of the AIDS Research Institute, UCSF, NIH grants AI093179 and P01 AI071713, and an Explorations grant from the Bill & Melinda Gates Foundation, grant 51950. This work was also supported in part by grant NIH OD 011092. Steven G. Deeks and Jeffrey N. Martin are supported in part by the National Institutes of Health (grants AI069994 and AI071713) and the UCSF Center for AIDS Research (grant AI027763). Rafael R. Almeida is a recipient of São Paulo State Research Funding Agency (FAPESP) fellowships.
Footnotes
Published ahead of print 27 March 2013
REFERENCES
- 1. Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, Novitsky V, Haynes B, Hahn BH, Bhattacharya T, Korber B. 2002. Diversity considerations in HIV-1 vaccine selection. Science 296:2354–2360 [DOI] [PubMed] [Google Scholar]
- 2. Korber B, Gaschen B, Yusim K, Thakallapally R, Kesmir C, Detours V. 2001. Evolutionary and immunological implications of contemporary HIV-1 variation. Br. Med. Bull. 58:19–42 [DOI] [PubMed] [Google Scholar]
- 3. Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233–236 [DOI] [PubMed] [Google Scholar]
- 4. Liu J, O'Brien KL, Lynch DM, Simmons NL, La Porte A, Riggs AM, Abbink P, Coffey RT, Grandpre LE, Seaman MS, Landucci G, Forthal DN, Montefiori DC, Carville A, Mansfield KG, Havenga MJ, Pau MG, Goudsmit J, Barouch DH. 2009. Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature 457:87–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wilson NA, Keele BF, Reed JS, Piaskowski SM, MacNair CE, Bett AJ, Liang X, Wang F, Thoryk E, Heidecker GJ, Citron MP, Huang L, Lin J, Vitelli S, Ahn CD, Kaizu M, Maness NJ, Reynolds MR, Friedrich TC, Loffredo JT, Rakasz EG, Erickson S, Allison DB, Piatak M, Jr, Lifson JD, Shiver JW, Casimiro DR, Shaw GM, Hahn BH, Watkins DI. 2009. Vaccine-induced cellular responses control simian immunodeficiency virus replication after heterologous challenge. J. Virol. 83:6508–6521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barouch DH, Kunstman J, Kuroda MJ, Schmitz JE, Santra S, Peyerl FW, Krivulka GR, Beaudry K, Lifton MA, Gorgone DA, Montefiori DC, Lewis MG, Wolinsky SM, Letvin NL. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335–339 [DOI] [PubMed] [Google Scholar]
- 7. Bennett MS, Ng HL, Ali A, Yang OO. 2008. Cross-clade detection of HIV-1-specific cytotoxic T lymphocytes does not reflect cross-clade antiviral activity. J. Infect. Dis. 197:390–397 [DOI] [PubMed] [Google Scholar]
- 8. Loffredo JT, Burwitz BJ, Rakasz EG, Spencer SP, Stephany JJ, Vela JP, Martin SR, Reed J, Piaskowski SM, Furlott J, Weisgrau KL, Rodrigues DS, Soma T, Napoe G, Friedrich TC, Wilson NA, Kallas EG, Watkins DI. 2007. The antiviral efficacy of simian immunodeficiency virus-specific CD8+ T cells is unrelated to epitope specificity and is abrogated by viral escape. J. Virol. 81:2624–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Valentine LE, Piaskowski SM, Rakasz EG, Henry NL, Wilson NA, Watkins DI. 2008. Recognition of escape variants in ELISPOT does not always predict CD8+ T-cell recognition of simian immunodeficiency virus-infected cells expressing the same variant sequences. J. Virol. 82:575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, Tang Y, Holmes EC, Allen T, Prado JG, Altfeld M, Brander C, Dixon C, Ramduth D, Jeena P, Thomas SA, St. John A, Roach TA, Kupfer B, Luzzi G, Edwards A, Taylor G, Lyall H, Tudor-Williams G, Novelli V, Martinez-Picado J, Kiepiela P, Walker BD, Goulder PJ. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282–289 [DOI] [PubMed] [Google Scholar]
- 11. O'Connor DH, Allen TM, Vogel TU, Jing P, DeSouza IP, Dodds E, Dunphy EJ, Melsaether C, Mothe B, Yamamoto H, Horton H, Wilson N, Hughes AL, Watkins DI. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493–499 [DOI] [PubMed] [Google Scholar]
- 12. Dilernia DA, Jones L, Rodriguez S, Turk G, Rubio AE, Pampuro S, Gomez-Carrillo M, Bautista CT, Deluchi G, Benetucci J, Lasala MB, Lourtau L, Losso MH, Perez H, Cahn P, Salomon H. 2008. HLA-driven convergence of HIV-1 viral subtypes B and F toward the adaptation to immune responses in human populations. PLoS One 3:e3429 doi:10.1371/journal.pone.0003429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kawashima Y, Pfafferott K, Frater J, Matthews P, Payne R, Addo M, Gatanaga H, Fujiwara M, Hachiya A, Koizumi H, Kuse N, Oka S, Duda A, Prendergast A, Crawford H, Leslie A, Brumme Z, Brumme C, Allen T, Brander C, Kaslow R, Tang J, Hunter E, Allen S, Mulenga J, Branch S, Roach T, John M, Mallal S, Ogwu A, Shapiro R, Prado JG, Fidler S, Weber J, Pybus OG, Klenerman P, Ndung'u T, Phillips R, Heckerman D, Harrigan PR, Walker BD, Takiguchi M, Goulder P. 2009. Adaptation of HIV-1 to human leukocyte antigen class I. Nature 458:641–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rowe HM, Trono D. 2011. Dynamic control of endogenous retroviruses during development. Virology 411:273–287 [DOI] [PubMed] [Google Scholar]
- 15. Strebel K, Luban J, Jeang KT. 2009. Human cellular restriction factors that target HIV-1 replication. BMC Med. 7:48 doi:10.1186/1741-7015-7-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sheehy AM, Gaddis NC, Malim MH. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404–1407 [DOI] [PubMed] [Google Scholar]
- 17. Berger G, Durand S, Fargier G, Nguyen XN, Cordeil S, Bouaziz S, Muriaux D, Darlix JL, Cimarelli A. 2011. APOBEC3A is a specific inhibitor of the early phases of HIV-1 infection in myeloid cells. PLoS Pathog. 7:e1002221 doi:10.1371/journal.ppat.1002221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cullen BR. 2006. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 80:1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harris RS, Hultquist JF, Evans DT. 2012. The restriction factors of human immunodeficiency virus. J. Biol. Chem. 287:40875–40883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chiu YL, Greene WC. 2008. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 26:317–353 [DOI] [PubMed] [Google Scholar]
- 21. Luo K, Liu B, Xiao Z, Yu Y, Yu X, Gorelick R, Yu XF. 2004. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 78:11841–11852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sheehy AM, Gaddis NC, Choi JD, Malim MH. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650 [DOI] [PubMed] [Google Scholar]
- 23. Kao S, Khan MA, Miyagi E, Plishka R, Buckler-White A, Strebel K. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 77:11398–11407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Malim MH. 2009. APOBEC proteins and intrinsic resistance to HIV-1 infection. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:675–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meiklejohn DA, Karlsson RK, Karlsson AC, Chapman JM, Nixon DF, Schweighardt B. 2004. ELISPOT cell rescue. J. Immunol. Methods 288:135–147 [DOI] [PubMed] [Google Scholar]
- 26. Sacha JB, Chung C, Rakasz EG, Spencer SP, Jonas AK, Bean AT, Lee W, Burwitz BJ, Stephany JJ, Loffredo JT, Allison DB, Adnan S, Hoji A, Wilson NA, Friedrich TC, Lifson JD, Yang OO, Watkins DI. 2007. Gag-specific CD8+ T lymphocytes recognize infected cells before AIDS-virus integration and viral protein expression. J. Immunol. 178:2746–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hutchinson SL, Wooldridge L, Tafuro S, Laugel B, Glick M, Boulter JM, Jakobsen BK, Price DA, Sewell AK. 2003. The CD8 T cell coreceptor exhibits disproportionate biological activity at extremely low binding affinities. J. Biol. Chem. 278:24285–24293 [DOI] [PubMed] [Google Scholar]
- 28. Virgen CA, Hatziioannou T. 2007. Antiretroviral activity and Vif sensitivity of rhesus macaque APOBEC3 proteins. J. Virol. 81:13932–13937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N. 2002. An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22. Genomics 79:285–296 [DOI] [PubMed] [Google Scholar]
- 30. Matzinger P. 2007. Friendly and dangerous signals: is the tissue in control? Nat. Immunol. 8:11–13 [DOI] [PubMed] [Google Scholar]
- 31. Wing K, Sakaguchi S. 2010. Regulatory T cells exert checks and balances on self tolerance and autoimmunity. Nat. Immunol. 11:7–13 [DOI] [PubMed] [Google Scholar]
- 32. Jaini R, Kesaraju P, Johnson JM, Altuntas CZ, Jane-Wit D, Tuohy VK. 2010. An autoimmune-mediated strategy for prophylactic breast cancer vaccination. Nat. Med. 16:799–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parmiani G, De Filippo A, Novellino L, Castelli C. 2007. Unique human tumor antigens: immunobiology and use in clinical trials. J. Immunol. 178:1975–1979 [DOI] [PubMed] [Google Scholar]
- 34. Slingluff CL, Jr, Petroni GR, Yamshchikov GV, Barnd DL, Eastham S, Galavotti H, Patterson JW, Deacon DH, Hibbitts S, Teates D, Neese PY, Grosh WW, Chianese-Bullock KA, Woodson EM, Wiernasz CJ, Merrill P, Gibson J, Ross M, Engelhard VH. 2003. Clinical and immunologic results of a randomized phase II trial of vaccination using four melanoma peptides either administered in granulocyte-macrophage colony-stimulating factor in adjuvant or pulsed on dendritic cells. J. Clin. Oncol. 21:4016–4026 [DOI] [PubMed] [Google Scholar]
- 35. Guo F, Cen S, Niu M, Saadatmand J, Kleiman L. 2006. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 80:11710–11722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mbisa JL, Bu W, Pathak VK. 2010. APOBEC3F and APOBEC3G inhibit HIV-1 DNA integration by different mechanisms. J. Virol. 84:5250–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]