Abstract
Phosphorylation of viral proteins plays important roles in the influenza A virus (IAV) life cycle. By using mass spectrometry, we identified tyrosine 132 (Y132) as a phosphorylation site of the matrix protein (M1) of the influenza virus A/WSN/1933(H1N1). Phosphorylation at this site is essential to the process of virus replication by controlling the nuclear import of M1. We further demonstrated that the phosphorylated tyrosine is crucial for the binding of M1 to the nuclear import factor importin-α1, since any substitutions at this site severely reduce this protein-protein interaction and damage the importin-α1-mediated nuclear import of M1. Additionally, the tyrosine phosphorylation which leads to the nuclear import of M1 is blocked by a Janus kinase inhibitor. The present study reveals a pivotal role of this tyrosine phosphorylation in the intracellular transportation of M1, which controls the process of viral replication.
INTRODUCTION
The genome of influenza A virus (IAV) consists of eight segments of negative-sense RNA coding for 14 viral proteins (1–4). With the exception of the newly found PB1 N40, PA-X, and M42 proteins, the remaining 11 viral proteins have been described as phosphorylated proteins either in virions or in infected cells. These remaining proteins include the RNA polymerase PA, PB1, and PB2 proteins, the nucleoprotein (NP), the two envelope proteins, i.e., hemagglutinin (HA) and neuraminidase (NA), nonstructural protein 1 (NS1), the nuclear export protein (NEP), the PB1-F2 protein, and the two matrix proteins, M1 and M2 (5–14). Phosphorylation of viral proteins plays an important role in the virus life cycle. Remarkably, phosphorylation of NP controls its intracellular distribution and then affects the transportation of viral RNPs (vRNPs) (15, 16). The phosphorylation of NS1 affects its double-stranded RNA (dsRNA)-binding capacity and has been shown to have critical roles in virus replication (13, 17).
M1, the most abundant protein in virions, also has multiple functions in the influenza A virus life cycle, including uncoating, transcription, nuclear export of vRNPs, assembly, and budding. Early studies indicated that M1 contains phosphoserine and phosphothreonine residues (5, 18) and has the potential to be phosphorylated by protein kinase C (PKC) and extracellular signal-regulated protein kinases (ERKs) (19). Recently, several phosphorylation sites on M1 (including a phosphotyrosine) were reported (14). However, the phosphorylated tyrosine of M1 has not yet been identified, and phosphorylated residues on M1 have not been confirmed functionally (20, 21). There is evidence that abnormal modification of M1 by hyperphosphorylation causes its retention in the nucleus (22), which further suggests that this important posttranslational modification of M1 is finely regulated for productive virus growth. In order to further understand the effect of phosphorylation on the functions of M1, we determined the status of M1 phosphorylation and identified tyrosine residue 132 as a phosphorylation site which is critical to controlling the nuclear import of M1 and virus replication.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic kidney 293T (HEK293T), A549, and MDCK cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) (both from Invitrogen) at 37°C and 5% CO2. The influenza virus used in this study was the A/WSN/1933(H1N1) strain and was rescued from cDNAs (23); A/chicken/Shandong/lx929/2007(H9N2) and A/Puerto Rico/8/1934(H1N1) were propagated in 10-day-old embryonated chicken eggs.
Reagents and antibodies.
Reagents and antibodies used in the study were obtained from the following sources: Phos-tag acrylamide was from Wako (Japan); U0126 and AG490 were from Beyotime (China); c-Myc antibody (C3956; produced in rabbits) and mouse anti-FLAG (M2) antibody were from Sigma-Aldrich; and dasatinib (sc-358114), β-actin (ac-1616-R), Myc (9E10), and p-Tyr (sc-508) antibodies were from Santa Cruz Biotechnology. A mouse anti-M1 monoclonal antibody and a rabbit anti-NP polyclonal antibody were generated as described previously (24). A rabbit anti-HA polyclonal antibody was kindly provided by George Fu Gao, while all secondary antibodies were obtained from Bai Hui Zhong Yuan Biotechnology (China).
Plasmid construction.
The full-length M1 [A/WSN/1933(H1N1)] sequence was cloned into the pCMV-Myc vector. Mutations in full-length M1 (Y132A, Y132F, or Y132D), either in pHH21 or in pCMV-Myc, were generated by use of a Newpep site-directed mutagenesis kit (China). The following primers were used: M1-Y132A-F, 5′-GGGCCTCATAGCCAACAGGATGGGGGCTGTGACCAC-3′; M1-Y132A-R, 5′-GCCCCCATCCTGTTGGCTATGAGGCCCATACAACTG-3′; M1-Y132F-F, 5′-GGGCCTCATATTCAACAGGATGGGGGCTGTGACCAC-3′; M1-Y132F-R, 5′-GCCCCCATCCTGTTGAATATGAGGCCCATACAACTG-3′; M1-Y132D-F, 5′-GGGCCTCATAGACAACAGGATGGGGGCTGTGACCAC-3′; and M1-Y132D-R, 5′-GCCCCCATCCTGTTGTCTATGAGGCCCATACAACTG-3′.
The cDNA clone of importin-α1 was obtained from OriGene Technologies. The full-length importin-α1 sequence was first inserted into pcDNA3-FLAG. Secondly, the full-length and ΔIBB-importin-α1 sequences were cloned into the pGEX-4T-2 vector to generate a glutathione S-transferase (GST)-fused protein.
Phosphate-affinity SDS-PAGE and preparation for nano-LC-MS/MS analysis.
IAV-infected 293T cells were lysed in lysis buffer (20 mM HEPES [pH 7.4], 1% Triton X-100, 150 mM NaCl, 10% glycerol, 1 mM EDTA) supplemented with Complete protease inhibitor cocktail (Roche Diagnostics) and a phosphatase inhibitor (5 mM Na3VO4; Sigma). The M1 protein was purified with protein G agarose beads prebound to a mouse anti-M1 monoclonal antibody for 3 h at 4°C. Proteins were separated by 15% Mn2+–Phos-tag SDS-PAGE, which was described previously (25, 26). Briefly, normal polyacrylamide gel electrophoresis was conducted according to the TaKaRa protocol, with an acrylamide-pendant phosphate-tagged (Phos-tag) ligand (50 μM) and 0.1 mM MnCl2 (Sigma) added to the separating gel before polymerization. The gel was silver stained, and the separated band was subjected to nano-liquid chromatography–tandem mass spectrometry (nano-LC-MS/MS) identification at the Technological Platform of the Institute of Zoology, Chinese Academy of Sciences (LCQ Deca XP Plus; Thermo).
Generation of recombinant influenza A viruses.
The wild-type (WT) A/WSN/1933(H1N1) virus and its M1 mutants were generated by using a 12-plasmid-based reverse genetic system (23). Briefly, 293T cells grown to 90% confluence in 60-mm dishes were transfected with 1 μg each of the 12 plasmids in the virus rescue system. Six hours later, the medium was replaced with DMEM plus 1 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin. The cells were further cultured for 72 h at 37°C in 5% CO2, and the supernatant containing the recombinant viruses was harvested and then centrifuged at 2,000 × g for 10 min to remove the cell debris.
Plaque assay.
MDCK cell monolayers (5 × 106 cells at 100% confluence in a 12-well plate) were washed with phosphate-buffered saline (PBS) and infected with different dilutions of virus for 1 h at 37°C. The virus inoculums were removed by washing with PBS. Cell monolayers were then overlaid with agar overlay medium (DMEM supplemented with 1% low-melting-point agarose and 1 μg/ml TPCK-treated trypsin) and incubated at 37°C. Visible plaques were counted at 3 days postinfection, and the virus titers were determined. All data were expressed as means for three independent experiments.
Immunoprecipitation and Western blot analysis.
Cells were lysed in immunoprecipitation buffer (pH 7.4; 1% Triton X-100, 150 mM NaCl, 20 mM HEPES, 10% glycerol, 1 mM EDTA) further supplemented with Complete protease inhibitor cocktail and a phosphatase inhibitor (5 mM Na3VO4). After an incubation period of 15 min on ice, insoluble components were removed from lysates by centrifugation. Lysates were incubated with anti-FLAG M2 affinity gel (Sigma-Aldrich) for a minimum of 2 h. Following five washes in immunoprecipitation buffer, the precipitated proteins were separated by SDS-PAGE and then transferred onto Immobilon polyvinylidene difluoride (PVDF) membranes (Millipore Corporation, Billerica, MA). Membranes were probed with appropriate antibodies and then developed with a chemiluminescence detection reagent. All coimmunoprecipitation (co-IP) assays were performed with 293T cells.
IFAs.
Indirect immunofluorescence assays (IFAs) were performed with an Olympus FV500 confocal laser scanning microscope. The coverslips carrying cells were washed with PBS and then fixed with 4% paraformaldehyde. Cells were then blocked with 4% bovine serum albumin (BSA) (which was dissolved in PBS plus 1% Triton X-100) and stained with anti-NP, anti-M1, anti-FLAG, and anti-Myc antibodies. Secondary antibodies were fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG and tetramethyl rhodamine isocyanate (TRITC)-conjugated anti-rabbit IgG.
GST pulldown assay.
GST-fused importin-α1 was expressed in Escherichia coli strain BL21(DE3)pLysS and purified with Sepharose 4B-glutathione (GE Healthcare). Wild-type M1 and M1 mutants were expressed in 293T cells, and cells were stimulated by IAV infection before being lysed. An equal amount of either GST or GST-importin-α1 (100 μg) bound to Sepharose 4B-glutathione was mixed with cell lysate for 3 h at 4°C. Bound proteins were detected by Western blotting with the anti-M1 monoclonal antibody.
In vivo ubiquitination assays.
293T cells were transfected with expression constructs for Myc-tagged M1s and His-tagged ubiquitin. The subsequent procedures were performed as described previously (27). Briefly, at 40 h posttransfection (p.t.), the proteasome inhibitor MG-132 was added to the cells for 4 h, at a final concentration of 10 μM. Cell extracts were then immunoprecipitated with Ni2+-nitrilotriacetic acid (Ni2+-NTA) beads. The eluted proteins were analyzed by Western blotting using an anti-M1 antibody.
Cell membrane extraction.
The membrane fraction was extracted as described previously (28). Briefly, 293T cells were transfected with the 12 plasmids of the virus rescue system. After a 72-h transfection period, cells were washed with PBS and scraped into 250 μl of ice-cold TES buffer (20 mM Tris, pH 7.4; 100 mM NaCl; and 1 mM EDTA) supplemented with Complete protease inhibitor cocktail (Roche, Basel, Switzerland). Samples were kept on ice throughout the duration of the procedure. Cells were incubated on ice for 30 min, lysed by 30 strokes in a Dounce homogenizer, and then centrifuged for 10 min at 5,000 × g (at 4°C) to remove the nuclei and cell debris. The supernatants were then centrifuged for 1 h at 200,000 × g (at 4°C) to separate cellular membrane and cytoplasmic fractions.
Computer modeling and statistical analyses.
The three-dimensional crystal structure of the M1 protein (Protein Data Bank [PDB] accession no. 1EA3) was used to illustrate the location of Y132 and the nuclear localization signal (NLS). This modeling was performed with the program PyMOL (Schrödinger). Statistical analyses were performed using Prism5 software (GraphPad Software, San Diego, CA).
RESULTS
The conserved tyrosine 132 of M1 is a phosphorylation site.
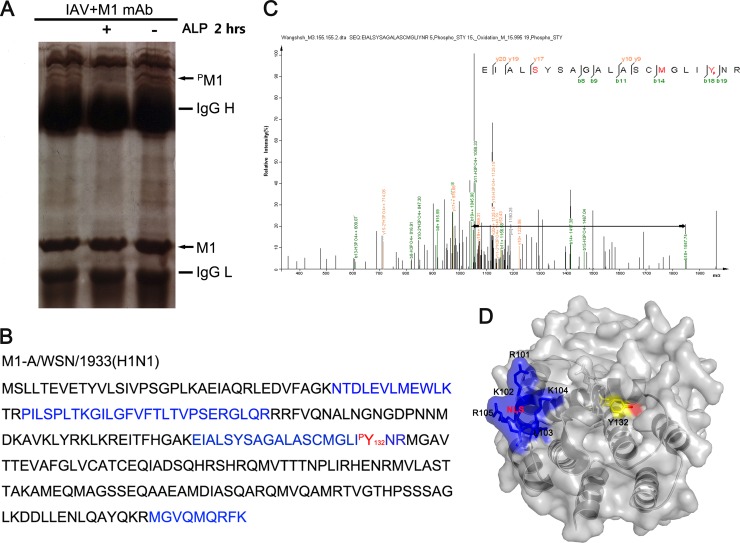
To identify the phosphorylated amino acid residues within the M1 protein, phosphate-affinity SDS-PAGE was performed to extract the phosphorylated M1 protein from virus-infected HEK293T cells (26, 29). A visible band exhibiting sensitivity to alkaline phosphatase (ALP) treatment and migrating much more slowly than wild-type M1 was considered to be phosphorylated M1 (Fig. 1A). This band was subjected to LC-MS/MS to confirm that it was in fact the matrix protein M1 [A/WSN/1933(H1N1)] (Fig. 1B). Further analysis of the spectra indicated that the peptide containing Y132 was phosphorylated at this position (Fig. 1C). We then analyzed the surface accessibility of this residue on the basis of its M1 crystal structure (30). As expected, Y132 and its hydroxyl group were exposed on the surface of M1, which implies that it is a functional phosphotyrosine (Fig. 1D).
Fig 1.
Tyrosine 132 is a phosphorylation site on the influenza A virus M1 protein. (A) Gel electrophoresis of phosphorylated M1 protein immunoprecipitated with a monoclonal anti-M1 antibody. The bands were digested in the gel and then identified by LC-MS/MS analysis. The locations of M1 and phosphorylated M1 (pM1) are indicated by arrows. ALP, alkaline phosphatase. (B) The protein was identified as the influenza A virus matrix protein M1 [A/WSN/1933(H1N1)]. Peptides (blue) detected by MS and the phosphorylated tyrosine site (red) are indicated. (C) Mass spectrometry analysis was performed on the purified M1 protein. The fragment ions b18 and b19, with correspondingly increased phosphoric acid, indicate that Y132 is a modification site. (D) Surface accessibility of Y132 as analyzed using PyMOL. Y132 (yellow) and its functional hydroxyl group (red) are visible, which indicates that they are on the surface of the M1 protein. The NLS of M1 (101RKLKR105; blue) (PDB accession no. 1EA3) is also shown.
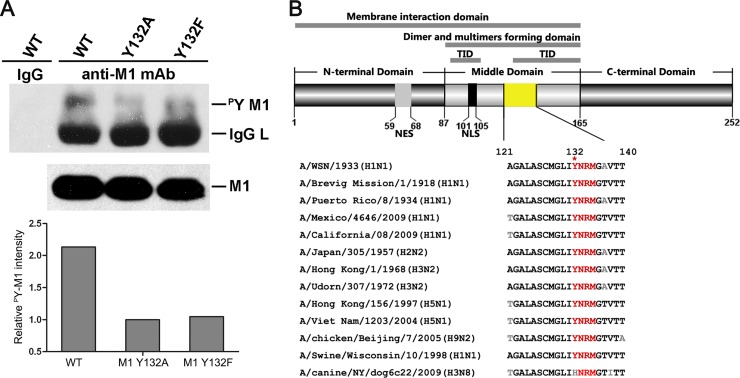
To determine whether Y132 is related to M1 tyrosine phosphorylation, we mutated Y132 to alanine (A) or phenylalanine (F) in vectors for virus rescue and purified the wild-type and mutated M1s from the 12-plasmid reverse genetic system by using an anti-M1 antibody. The levels of total M1 tyrosine phosphorylation were evaluated by use of an antiphosphotyrosine antibody (Fig. 2A). We found that conversion of Y132 to either A or F greatly reduced the level of M1 tyrosine phosphorylation compared to that of the wild-type protein. Quantified analysis of the relative intensity of the M1 phosphorylated tyrosine (pY) revealed that the loss of phosphorylation at Y132 resulted in a reduction of more than half the proportion of total M1 tyrosine phosphorylation. These results suggest that Y132 is a key site of phosphotyrosine on M1.
Fig 2.
Tyrosine 132 is a key site of phosphotyrosine on M1. (A) Phosphotyrosine-containing M1 was detected by specific anti-p-Tyr antibodies in a virus rescue system, and 1/10 the amount of whole loading protein was probed with anti-M1 antibody to show the levels of total M1. The relative pY M1 intensity was determined as the ratio of tyrosine-phosphorylated M1 to total M1. The loss of phosphorylation at Y132 drastically reduced the level of total M1 tyrosine phosphorylation (lanes 3 and 4). (B) Y132 and the putative SH2 binding motif (YRNM) are greatly conserved in M1 proteins among different subtypes of influenza A virus. Sequence alignment of the specific M1 domain (yellow) was performed by MegAlign. The schematic diagram of M1 shows the functional domains involving Y132 (upper panel). WT, wild type; TID, transcription inhibition domain; NES, nuclear export signal; NLS, nuclear localization signal.
Sequence alignment showed that the Y132 residue is almost 100% identical among different subtypes of the influenza A virus, with the exception of one strain of the H3N8 subtype (GenBank accession no. ADN86454.1), in which a histidine (H) was identified at this position (Fig. 2B). Notably, a putative SH2 (Src homology 2) binding motif (YNRM) containing Y132, which would be recognized by the SH2 domain through the phosphotyrosine to activate the downstream signal transduction pathway, was also greatly conserved (Fig. 2B). Additionally, Y132 is involved in many functional domains of M1, suggesting that it has important roles in virus replication.
Phosphorylated Y132 is essential for virus replication by controlling the cytoplasmic-nuclear translocation of M1.
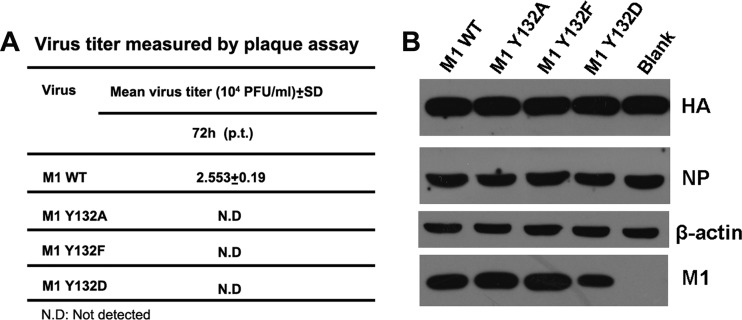
To directly assess the effect of Y132 on virus viability, we generated viruses possessing mutant M1 by way of a reverse genetic system. The rescued virus was titrated for 72 h p.t. by plaque assay. All mutations of Y132 (to A, F, or D) did not result in the recovery of infectious viruses (Fig. 3A). In the meantime, we also examined the viral proteins within transfected cells. The expression patterns of the major viral proteins (i.e., HA, NP, and M1) were similar in all of the wild-type and mutant M1-transfected cells, although the expression level of M1 Y132D was lower than that of wild-type M1 (Fig. 3B). These results provide evidence that Y132 of M1 is essential for virus replication, and the substitutions at Y132 impeded the key functions of the M1 protein rather than affecting the viral protein expression level.
Fig 3.
Phosphorylated Y132 of M1 is essential for virus rescue. A 12-plasmid reverse genetic system was used to rescue recombinant viruses with WT and mutated M1s (Y132A/F/D). (A) At 72 h p.t., the culture supernatants were harvested and subjected to plaque assays on MDCK cells. All recombinant viruses containing M1 mutants failed to be rescued. (B) At 72 h p.t., the transfected cells were lysed for Western blot analysis. HA, NP, and M1 were detected with respective antibodies. β-Actin was probed as a loading control. Blank, cells transfected without an added M1 plasmid.
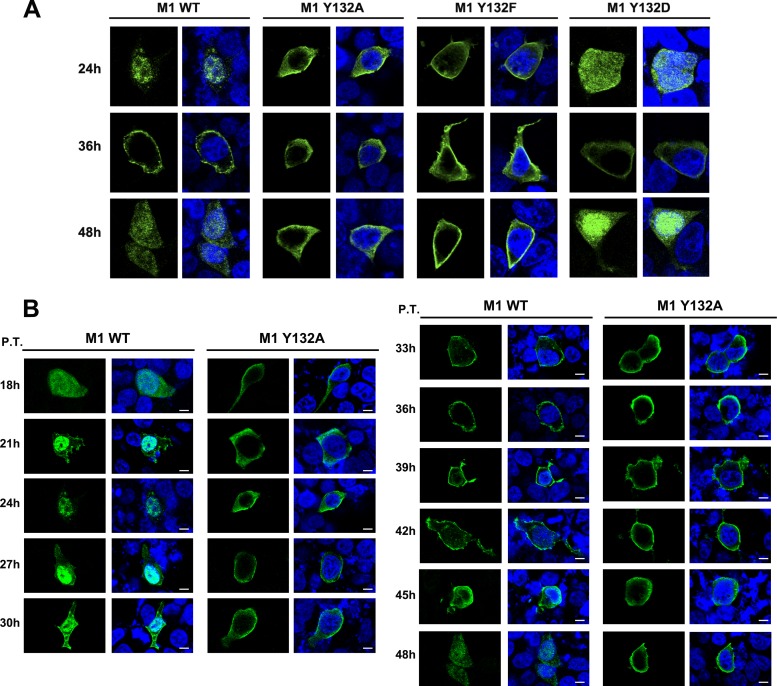
To further characterize the impact of mutant M1s on virus rescue, we examined the intracellular distribution of mutant M1s in cells transfected with 12 plasmids for reverse genetics. The cells were fixed at 24, 36, and 48 h p.t. and then incubated with anti-M1 antibody. The culture medium was collected at the corresponding time points and measured by plaque assay to evaluate harvested infectious virus particles with the wild-type M1 plasmid (date not shown). At each of these three time points, wild-type M1 localized to both the nuclear and cytoplasmic compartments, and at 36 h p.t., M1 was more concentrated on the cell membrane (Fig. 4A, left panels). Surprisingly, Y132A and Y132F mutant M1s were detected only in the cytoplasmic compartment and did not show nuclear localization at any time point (Fig. 4A, middle panels). To further confirm the effect of this tyrosine on the localization of M1 during viral replication, we continued to observe the localization of the M1 protein once every 3 h after transfection, beginning at 18 h p.t. (with observations not collected prior to this time due to minimal detection of M1 expression). Mutated M1 was not observed within the nuclear compartment at any of these time points, while wild-type M1 showed a normal intracellular distribution (Fig. 4B). In contrast, the replacement of Y132 with the phosphomimetic amino acid aspartic acid (D) showed that the nuclear distribution of M1 Y132D was similar to that of wild-type M1 (Fig. 4A, right panels), which suggests that the effect of Y132 on the nuclear import of M1 is dependent on the negative charge of the phosphate group. However, even the introduction of Y132D mutant M1 into the reverse genetic system could not recover the production of infectious virus.
Fig 4.
Phosphorylated Y132 of M1 is essential for nuclear import of M1. (A) The localization of WT and mutated M1s (Y132A/F/D) was determined in a 12-plasmid reverse genetic system. At 24, 36, and 48 h p.t., transfected cells were fixed and detected by anti-M1 antibody (green). (B) M1s (WT and Y132A mutant) were detected by an anti-M1 monoclonal antibody (green) at different time points during virus recuse. Bars, 10 μm. The nucleus was stained with DAPI (4′,6-diamidino-2-phenylindole; blue).
Since nuclear M1 has key functions in transcription inhibition and export of vRNPs during the virus life cycle (31, 32), the loss of phosphorylation impedes the translocation of M1 into the nucleus and finally results in the failure of virus recovery.
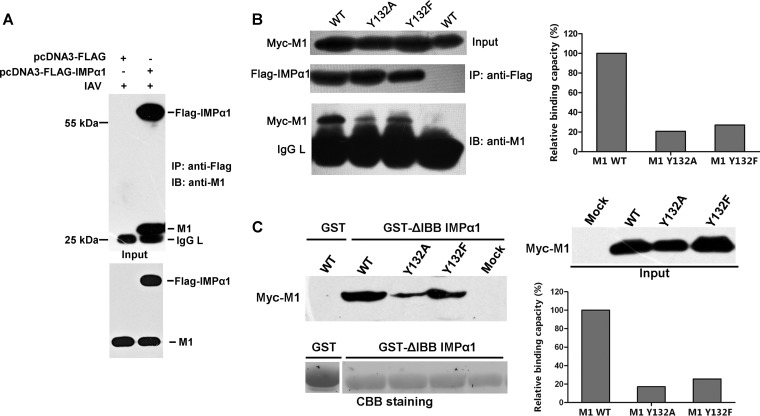
Phosphorylated Y132 is crucial for the association of M1 with the nuclear import factor importin-α1.
A major mechanism of protein nuclear import is the recognition of a classical, basic residue-rich NLS on cargo proteins by importin-α receptors. These receptors heterodimerize with the importin-β receptor and then perform nuclear translocation (33–36). The NLS sequence of the M1 protein [A/WSN/1933(H1N1)] was identified previously as a classical NLS sequence, i.e., 101RKLKR105 (37). Therefore, the nuclear import of M1 was considered to occur through the importin-dependent pathway (38), though we did not find any experimental evidence to prove it. In this case, the most likely explanation for the obstruction of the nuclear import of M1 was a dissociation of M1 from importins, which would have been caused by the loss of phosphorylation of Y132. To prove this hypothesis, we first demonstrated the binding of M1 to importin-α. Considering the monopartite NLS of M1, we chose importin-α1 to represent the importin-α family for the following work. We transfected importin-α1 into 293T cells and infected them with IAV; the co-IP results clearly show the binding of these two proteins (Fig. 5A). Since the introduction of mutated M1s by reverse genetics was shown to be unsuccessful in virus rescue assays, we transfected both Myc-M1 (wild type and mutants) and FLAG-importin-α1 into 293T cells and then performed the co-IP assay. The loss of phosphorylation of Y132 (M1 Y132A and M1 Y132F mutants) greatly reduced the binding of M1 and importin-α1 (Fig. 5B). To exclude other interferences in cells, we prepared a GST-fused importin-α1 without the IBB (importin-β binding) domain and incubated it with the M1-expressing cell lysates. Similar to the above result, GST-importin-α1 bound less mutated M1 than wild-type M1 (Fig. 5C). By quantifying the binding capacities of these two proteins, we found that the loss of phosphorylation reduced the association compared to that of wild-type M1 and importin-α1 by about 80%.
Fig 5.
Phosphorylated Y132 is crucial for binding of M1 to the nuclear import factor importin-α1. (A) The binding of M1 and importin-α1 (IMPα1) during virus infection was determined by coimmunoprecipitation. (B) Myc-M1 WT and Y132A and Y132F mutant proteins were coexpressed with FLAG-IMPα1 in 293T cells. FLAG-IMPα1 was immunoprecipitated with anti-FLAG agarose, and the associated Myc-M1 proteins were detected with the anti-M1 antibody. (C) Myc-M1 WT and Y132A and Y132F mutant proteins were expressed in 293T cells, and the cell lysates were incubated with GST-fused IMPα1 (ΔIBB [amino acids 60 to 529]). GST-ΔIBB IMPα1-bound M1s were blotted with the anti-M1 antibody. The relative binding capacities of M1s and IMPα1 were calculated by quantifying the ratio of IMPα1-bound Myc-M1s to input M1s. CBB, Coomassie brilliant blue; IAV, influenza A virus; IB, immunoblotting; IMP, importin; IgG L, IgG light chain; GST, glutathione S-transferase.
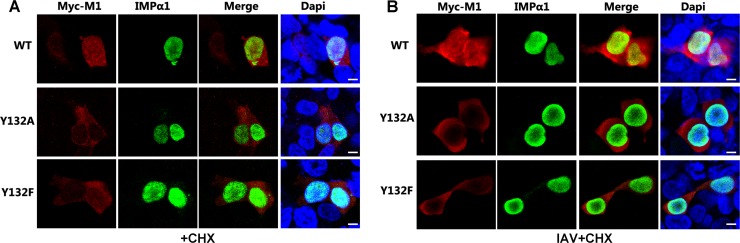
To validate the influence of the association of M1 with importin-α1 on the localization of M1, we performed an immunofluorescence assay to visualize the intracellular distributions of these two proteins. The transfected wild-type Myc-M1 protein was localized predominantly in the cytoplasm, as described previously, as it bears both nuclear export signal (NES) and NLS peptides (39). Nevertheless, a nuclear localization was obvious when the protein was coexpressed with importin-α1; in this case, the overexpressed importin-α1 would break the balance of nuclear export and import, thus greatly enhancing the nuclear import ability of M1 (Fig. 6A). When cells were stimulated by IAV infection, the colocalization of wild-type M1 and importin-α1 in the nucleus was observed to be much stronger (Fig. 6B). However, the large quantity of exogenously expressed importin-α1 could not promote translocation of either of the mutated M1s (Y132A and Y132F) from the cytoplasm to the nucleus as seen with the wild-type M1 protein. These results strongly proved that the dissociation of M1 from importin-α1 caused by the loss of phosphorylation of Y132 actually prevented the nuclear import of M1.
Fig 6.
Phosphorylated Y132 is essential for the importin-α1-mediated nuclear import of M1. (A and B) The association of M1 and IMPα1 was determined by immunofluorescence assay. Myc-M1s and FLAG-IMPα1 were cotransfected into 293T cells and stained with anti-Myc (red) and anti-FLAG (green) antibodies, respectively. The nucleus was stained with DAPI (blue). Cells were incubated with (B) or without (A) IAV (multiplicity of infection [MOI] = 5) for 1 h and then treated with cycloheximide (CHX; 50 μg/ml) for 3 h before fixation. “Input” represents 1/10 the total M1 proteins in each binding reaction mix. Bars, 10 μm.
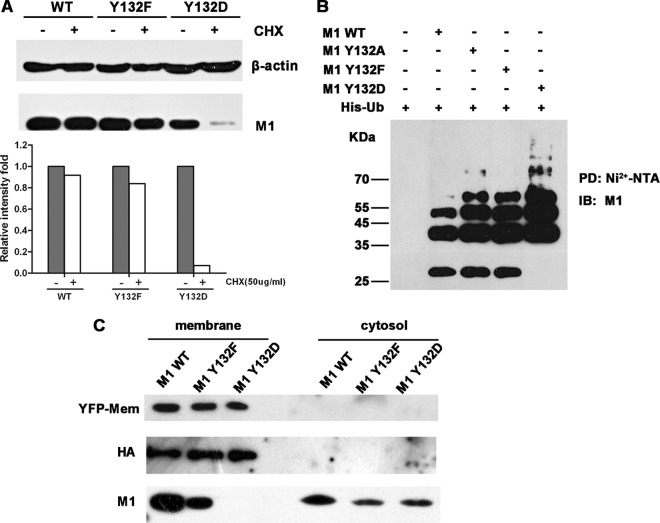
As for the phosphorylation mimic mutant M1 Y132D, which was imported into the nucleus (Fig. 4A), we performed additional analyses to ascertain why M1 Y132D did not support virus replication. We performed a degradation assay to confirm that M1 Y132D degraded quickly compared to other M1 proteins (Fig. 7A), and previous work in our lab showed that the degradation of M1 was actually mediated through the proteasome-dependent pathway (27). We then performed an in vivo ubiquitination assay of wild-type M1 and mutant M1s. The results showed that the level of polyubiquitinated M1 Y132D was greatly enhanced compared to those of other M1 proteins (Fig. 7B), and this phenomenon suggested that the Y-to-D substitution severely accelerated the ubiquitination of the M1 protein. On the other hand, it has been reported that M1 has the ability to associate with the cell membrane (which is the virus assembly and budding region) and that Y132 is located in one of the membrane binding domains (40, 41). Thus, we extracted the membrane fraction of cells transfected with the 12 plasmids from the virus rescue system and were able to detect the M1 protein. We used an expression construct for a membrane marker (pEYFP-Mem) as a control for fractionation. All the M1 proteins (WT, Y132F, and Y132D) were observed to appear in the cytosol, and while WT M1 and M1 Y132F were also observed in the membrane fraction, M1 Y132D was not (Fig. 7C). This result suggested that the M1 Y132D protein was not able to associate with the cell membrane.
Fig 7.
Y-to-D substitution at site 132 impairs the intracellular stability and membrane association of M1. (A) Twenty-four hours after transfection with constructs for WT, Y132F, or Y132D M1, 293T cells were treated with or without CHX (50 μg/ml) for 4 h. Cells were lysed and then detected with the anti-M1 antibody; β-actin was used as an internal control. The relative intensities were calculated by quantifying the results shown in the upper panel. (B) An in vivo ubiquitination assay suggested that mutation of Y to D increased the ubiquitination of the M1 protein. 293T cells were transfected with expression constructs for Myc-tagged M1s and His-tagged ubiquitin (His-Ub). At 40 hours posttransfection, the proteasome inhibitor MG-132 (10 μM) was added to the cells for 4 h. Cell extracts were then immunoprecipitated with Ni2+-NTA beads. The eluted proteins were analyzed by Western blotting using an anti-M1 antibody. PD, pulldown. (C) 293T cells were transfected with 12 plasmids from the virus rescue system and with an expression construct for a membrane marker, pEYFP-Mem (BD). After a 72-h transfection period, samples of cellular membrane and cytoplasmic fractions were separated and detected by anti-yellow fluorescent protein (anti-YFP), anti-HA, and anti-M1 antibodies.
A Janus kinase inhibitor blocks nuclear import of M1 during viral replication.
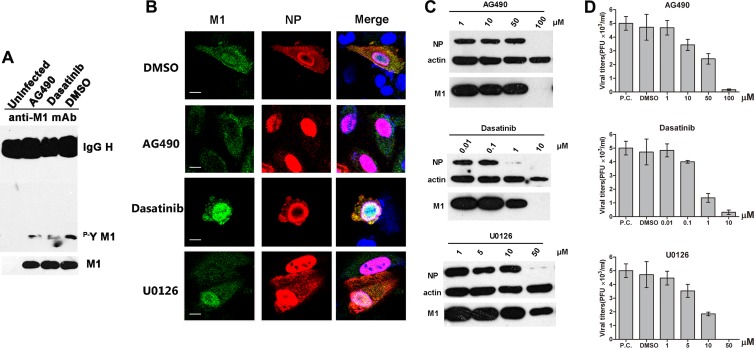
To verify the modulation of the subcellular localization of M1 by certain tyrosine kinases, we treated IAV-infected cells with several protein kinase inhibitors. These inhibitors included AG490 (JAK inhibitor) and dasatinib (Src kinase inhibitor), which are tyrosine kinase inhibitors, and U0126 (MEK1/2 inhibitor), which is a serine/threonine kinase inhibitor. First, we evaluated the effects of these inhibitors on the phosphorylation level of M1. We observed that either AG490 or dasatinib decreased the tyrosine phosphorylation of M1 (Fig. 8A). We then determined the influences of these inhibitors on the cellular localization of M1 during virus replication. After a 6-h treatment, the cells were fixed and detected with anti-M1 and anti-NP antibodies. Compared to the control (dimethyl sulfoxide [DMSO]-treated) cells, the distribution of M1 in U0126-treated cells was almost the same, which confirmed the previously reported results (42). Notably, in AG490-treated cells, most of the M1 was localized in the cytoplasm (Fig. 8B). Nevertheless, dasatinib treatment did not change the intracellular distribution of M1, thus indicating that the Src family kinases might be irrelevant to the M1 nuclear import induced by tyrosine phosphorylation. All tested inhibitors impaired viral protein expression and virus propagation at specific concentrations (Fig. 8C and D): while only AG490 seemed to inhibit nuclear import of M1, the other inhibitors must inhibit influenza virus replication by other means. These results confirmed that the intracellular localization of M1 was actually controlled by the protein modification of tyrosine phosphorylation and that Janus kinases were involved in the process.
Fig 8.
The nuclear import of M1 is impaired by inhibiting Janus kinases. (A) A549 cells were infected with IAV (MOI = 0.1) for 12 h and then treated with the tyrosine kinase inhibitors AG490 (50 μM) and dasatinib (1 μM), with DMSO as a control (the inhibitors were dissolved in DMSO). After a 6-h treatment, cells were lysed and assayed for phosphotyrosine-containing M1. One-tenth the amount of whole loading protein was probed with anti-M1 antibody to show the levels of total M1. (B) A549 cells were infected with IAV (MOI = 2); after 1 h at 4°C for virus absorption, cells were washed with PBS and treated with several protein kinase inhibitors, including AG490 (100 μM), dasatinib (10 μM), and U0126 (50 μM), with DMSO as a control. Cells were fixed after 6 h of treatment and stained with anti-M1 (green) and anti-NP (red) antibodies. The nucleus was stained with DAPI (blue). Views of single cells with viral protein expression are shown. Bars, 10 μm. (C) A549 cells were infected with IAV (MOI = 2) for 1 h at 4°C, and then the medium was changed and cells were treated with kinase inhibitors at different concentrations. After 6 h of incubation at 37°C, cells were lysed and detected with anti-M1 and anti-NP antibodies. β-Actin was used as a loading control. (D) Virus titers of the supernatants in panel C were measured by plaque assay on MDCK cells. The positive control (P.C.) was IAV-infected cells without any additions (DMSO or inhibitors).
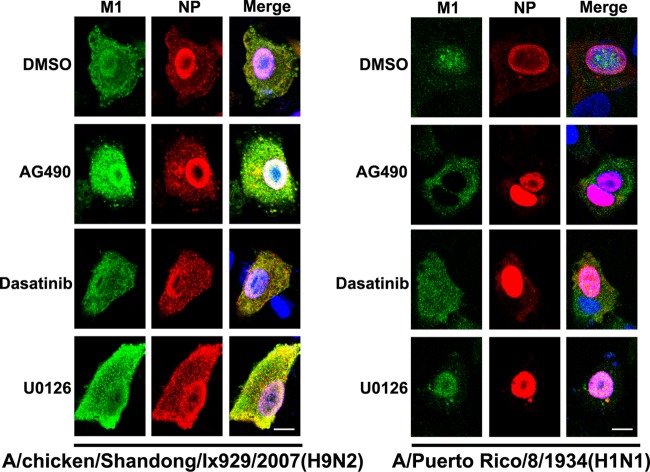
To further strengthen the suggestion that phosphotyrosine control of M1 is a conserved mechanism for M1 nuclear import, we performed the kinase inhibitor assay on two other strains, both of which have Y132: A/chicken/Shandong/lx929/2007(H9N2) and A/Puerto Rico/8/1934(H1N1). All the procedures we performed were the same as those used with the A/WSN/1933(H1N1) strain. The results were interesting. The two strains of influenza A virus had different responses to treatment with the same kinase inhibitors (Fig. 9). Remarkably, the nuclear import of A/Puerto Rico/8/1934 M1 was blocked by treatment with AG490, similar to the case with A/WSN/1933, while that of the H9N2 M1 protein was not blocked. The antibodies we used in this experiment were qualified to detect M1 and NP from these two strains (data not shown). These results suggest that the mechanism of phosphotyrosine control of nuclear import of M1 is different in different subtypes and strains and might be conserved in the H1N1 subtype.
Fig 9.
A Janus kinase inhibitor has different effects on nuclear localization of influenza A virus M1 proteins from different strains. A549 cells were infected with A/chicken/Shandong/lx929/2007(H9N2) or A/Puerto Rico/8/1934(H1N1) (MOI = 2); after 1 h at 4°C for virus absorption, cells were washed with PBS and then treated with AG490 (100 μM), dasatinib (10 μM), or U0126 (50 μM), with DMSO as a control. Cells were fixed after 6 h of treatment and then stained with anti-M1 (green) and anti-NP (red) antibodies. The nucleus was stained with DAPI (blue). Following staining, coverslips were analyzed using a model FV500 laser scanning confocal microscope. Views of single cells with viral protein expression are shown. Bars, 10 μm.
DISCUSSION
Although phosphorylation of M1 during virus infection has been reported previously (5, 14, 18), the precise modification site was not functionally proven. According to previous results, M1 contained phosphoserine and phosphothreonine but lacked phosphotyrosine; as such, the resulting effect of this important posttranslational modification of M1 on viral replication was still unknown. In our study, we detected the phosphotyrosine in M1 along with phosphoserine and phosphothreonine (data not shown), and we further confirmed that the specific phosphotyrosine is essential for virus replication. We determined that a loss of phosphorylation at Y132 impairs the binding of M1 to importin-α1, prevents its nuclear import, and, consequently, impairs virus replication. For the first time, we provide evidence of the existence of a phosphotyrosine in M1, while simultaneously uncovering the crucial role of phosphorylated Y132 in virus replication.
Phosphorylated proteins play significant roles in biological processes by changing protein structure, enzymatic activity, ligand associations, and subcellular localization (20). It was previously reported that phosphorylation regulates nucleocytoplasmic trafficking of several proteins (43–45). One of these general patterns involves phosphorylation upregulating the nuclear localization of substrates by enhancing the recognition of transport signals (NLS) on cargos to importin receptors, such as the simian virus 40 (SV40) large T antigen. Phosphorylation of serine residues upstream from the classical SV40 NLS enhances the nuclear import of the SV40 large T antigen (44). The effect of phosphorylation of M1 may follow the same pattern as that for the SV40 large T antigen. The NLS-neighboring phosphorylated Y132 residue increases the binding capacity of M1 and importin-α1 to make sure that there is adequate M1 in the nucleus. According to the M1 structure, we tried to figure out the molecular mechanism for the interaction of Y132 and the NLS peptide. When the Y132 residue is phosphorylated, it can have a negatively charged phosphate group. The surrounding residues (R134, R76, R77, and R78) are positively charged. Furthermore, the neighboring NLS peptide is also rich in positively charged residues. Thus, we speculate that the phosphorylation of Y132 probably changes the charge network and induces a large conformational change in the NLS region. This putative structural arrangement may enhance the binding of M1 to importin-α.
When we made a Y-to-D replacement to mimic constitutive phosphorylation, the M1 mutant with D132 relocalized to the nucleus just like WT M1 did (Fig. 4A). This was consistent with the finding that the nuclear import of protein was upregulated by its phosphorylation. However, the Y132D mutant M1 protein was still lethal to virus rescue, despite its normal intracellular localization. Previous reporting on the nuclear retention of hyperphosphorylated M1 provides a clue regarding the abnormal function of M1 Y132D. In addition, in considering the multiple functions of M1 in the virus life cycle, the status of protein modification must be modulated precisely (such as with phosphorylation and dephosphorylation). In this case, the constitutively phosphorylated protein might lose other functions and be recognized as a mistaken protein that quickly undergoes degradation. Accordingly, we observed that the ubiquitin-dependent degradation of M1 Y132D was greatly accelerated compared to that of WT M1 and the other M1 mutants (Fig. 7A and B). Additionally, M1 Y132D was not able to associate with the cell membrane (Fig. 7C), which is the assembly and budding site of influenza virus, suggesting that the Y-to-D substitution impaired the virus life cycle at these stages. Otherwise, the Y132-containing SH2 binding motif of M1 (132YNRM135) would be recognized by some important proteins having the SH2 domain, and downstream signal transduction would then be induced. This process may also be essential for influenza virus replication but cannot be compensated by the Y-to-D substitution.
The enzymes responsible for the addition of a phosphate moiety to tyrosine are protein tyrosine kinases (PTKs). The identification of kinases is greatly helpful for studies of the functions of substrate proteins. We used tyrosine kinase inhibitors to seek for the putative kinase which can phosphorylate Y132. It turned out that AG490 (JAK inhibitor) but not dasatinib (Src kinase inhibitor) prevented the nuclear import of M1 during virus replication, like the effect of the loss of phosphorylation of Y132 (as has been proven). Previous studies showed that AG490 inhibits both JAK1 and JAK2 activities (46). Since A549 cells did not have JAK1 kinase activity, it was indicated that JAK2 might be responsible for the phosphorylation of Y132. So far, a number of genomewide screens to identify host proteins required for influenza virus replication have been published (2, 47–49), and JAK2 was reported to have a potential interaction with M1 and thus was considered necessary for virus replication (47). All findings in this field provide evidence for the putative roles of JAK2 in the phosphorylation of M1. Nevertheless, whether JAK2 itself (or other tyrosine kinases activated by JAK2) can phosphorylate M1 remains unknown. We need to conduct further investigations to confirm the role of the kinase which functions in phosphorylating Y132 and the effects of proteins which can recognize the phosphorylated M1 protein and have specific roles in the downstream pathway. Blocking of these interactions will destroy the bond between viruses and host cells, subsequently killing the viruses. This point becomes even more important with our observation that the phosphorylation site of Y132 is conserved in almost all strains of influenza A virus (WSN and PR8 strains were functionally verified in this study), including the highly pathogenic H5 subtype, providing a potential target for beating this virus.
In summary, we report a tyrosine phosphorylation site on the influenza A virus M1 protein, which was found to be essential in controlling the binding of M1 to the host factor importin-α1. These findings provide a new mechanism for the phosphorylation-regulated nuclear translocation of the M1 protein, uncovering a new target on the viral protein for antiviral drug development.
ACKNOWLEDGMENTS
We thank Yi Shi for help with computer modeling, Yuanming Luo for technological help with MS analysis, and Jing Li and Xiaojuan Jia for continuous support.
This work was supported by the National Basic Research Program (973) of China (grants 2012CB518900 and 2012CB955501) and the National Natural Science Foundation of China (grants 81101253 and 31101830). Wenjun Liu is the principal investigator of the Innovative Research Group of the National Natural Science Foundation of China (grant 81021003).
Footnotes
Published ahead of print 27 March 2013
REFERENCES
- 1. Samji T. 2009. Influenza A: understanding the viral life cycle. Yale J. Biol. Med. 82:153–159 [PMC free article] [PubMed] [Google Scholar]
- 2. Watanabe T, Watanabe S, Kawaoka Y. 2010. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe 7:427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wise HM, Hutchinson EC, Jagger BW, Stuart AD, Kang ZH, Robb N, Schwartzman LM, Kash JC, Fodor E, Firth AE, Gog JR, Taubenberger JK, Digard P. 2012. Identification of a novel splice variant form of the influenza A virus M2 ion channel with an antigenically distinct ectodomain. PLoS Pathog. 8:e1002998 doi:10.1371/journal.ppat.1002998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gregoriades A, Guzman GG, Paoletti E. 1990. The phosphorylation of the integral membrane (M1) protein of influenza virus. Virus Res. 16:27–41 [DOI] [PubMed] [Google Scholar]
- 6. Arrese M, Portela A. 1996. Serine 3 is critical for phosphorylation at the N-terminal end of the nucleoprotein of influenza virus A/Victoria/3/75. J. Virol. 70:3385–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mahmoudian S, Auerochs S, Grone M, Marschall M. 2009. Influenza A virus proteins PB1 and NS1 are subject to functionally important phosphorylation by protein kinase C. J. Gen. Virol. 90:1392–1397 [DOI] [PubMed] [Google Scholar]
- 8. Mitzner D, Dudek SE, Studtrucker N, Anhlan D, Mazur I, Wissing J, Jansch L, Wixler L, Bruns K, Sharma A, Wray V, Henklein P, Ludwig S, Schubert U. 2009. Phosphorylation of the influenza A virus protein PB1-F2 by PKC is crucial for apoptosis promoting functions in monocytes. Cell. Microbiol. 11:1502–1516 [DOI] [PubMed] [Google Scholar]
- 9. Petri T, Dimmock NJ. 1981. Phosphorylation of influenza virus nucleoprotein in vivo. J. Gen. Virol. 57:185–190 [DOI] [PubMed] [Google Scholar]
- 10. Richardson JC, Akkina RK. 1991. NS2 protein of influenza virus is found in purified virus and phosphorylated in infected cells. Arch. Virol. 116:69–80 [DOI] [PubMed] [Google Scholar]
- 11. Sanz-Ezquerro JJ, Santaren JF, Sierra T, Aragon T, Ortega J, Ortin J, Smith GL, Nieto A. 1998. The PA influenza virus polymerase subunit is a phosphorylated protein. J. Gen. Virol. 79:471–478 [DOI] [PubMed] [Google Scholar]
- 12. Holsinger LJ, Shaughnessy MA, Micko A, Pinto LH, Lamb RA. 1995. Analysis of the posttranslational modifications of the influenza virus M2 protein. J. Virol. 69:1219–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hsiang TY, Zhou L, Krug RM. 2012. Roles of the phosphorylation of specific serines and threonines in the NS1 protein of human influenza A viruses. J. Virol. 86:10370–10376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hutchinson EC, Denham EM, Thomas B, Trudgian DC, Hester SS, Ridlova G, York A, Turrell L, Fodor E. 2012. Mapping the phosphoproteome of influenza A and B viruses by mass spectrometry. PLoS Pathog. 8:e1002993 doi:10.1371/journal.ppat.1002993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Neumann G, Castrucci MR, Kawaoka Y. 1997. Nuclear import and export of influenza virus nucleoprotein. J. Virol. 71:9690–9700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bui M, Myers JE, Whittaker GR. 2002. Nucleo-cytoplasmic localization of influenza virus nucleoprotein depends on cell density and phosphorylation. Virus Res. 84:37–44 [DOI] [PubMed] [Google Scholar]
- 17. Hale BG, Randall RE, Ortin J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:2359–2376 [DOI] [PubMed] [Google Scholar]
- 18. Gregoriades A, Christie T, Markarian K. 1984. The membrane (M1) protein of influenza virus occurs in two forms and is a phosphoprotein. J. Virol. 49:229–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reinhardt J, Wolff T. 2000. The influenza A virus M1 protein interacts with the cellular receptor of activated C kinase (RACK) 1 and can be phosphorylated by protein kinase C. Vet. Microbiol. 74:87–100 [DOI] [PubMed] [Google Scholar]
- 20. Tan CS. 2011. Sequence, structure, and network evolution of protein phosphorylation. Sci. Signal. 4:mr6 doi:10.1126/scisignal.2002093 [DOI] [PubMed] [Google Scholar]
- 21. Li L, Wu CG, Huang HM, Zhang KZ, Gan J, Li S. 2008. Prediction of phosphotyrosine signaling networks using a scoring matrix-assisted ligand identification approach. Nucleic Acids Res. 36:3263–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Whittaker G, Kemler I, Helenius A. 1995. Hyperphosphorylation of mutant influenza virus matrix protein, M1, causes its retention in the nucleus. J. Virol. 69:439–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. U. S. A. 96:9345–9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu XL, Sun L, Yu MR, Wang ZF, Xu CF, Xue QH, Zhang K, Ye X, Kitamura Y, Liu WJ. 2009. Cyclophilin A interacts with influenza A virus M1 protein and impairs the early stage of the viral replication. Cell. Microbiol. 11:730–741 [DOI] [PubMed] [Google Scholar]
- 25. Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. 2006. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol. Cell. Proteomics 5:749–757 [DOI] [PubMed] [Google Scholar]
- 26. Kinoshita-Kikuta E, Aoki Y, Kinoshita E, Koike T. 2007. Label-free kinase profiling using phosphate affinity polyacrylamide gel electrophoresis. Mol. Cell. Proteomics 6:356–366 [DOI] [PubMed] [Google Scholar]
- 27. Liu X, Zhao Z, Xu C, Sun L, Chen J, Zhang L, Liu W. 2012. Cyclophilin A restricts influenza A virus replication through degradation of the M1 protein. PLoS One 7:e31063 doi:10.1371/journal.pone.0031063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu MR, Liu XL, Sun L, Chen CW, Ma GP, Kitamura Y, Gao GF, Liu WJ. 2010. Subcellular localization and topology of porcine reproductive and respiratory syndrome virus E protein. Virus Res. 152:104–114 [DOI] [PubMed] [Google Scholar]
- 29. Kinoshita E, Kinoshita-Kikuta E, Koike T. 2009. Separation and detection of large phosphoproteins using Phos-tag SDS-PAGE. Nat. Protoc. 4:1513–1521 [DOI] [PubMed] [Google Scholar]
- 30. Arzt S, Baudin F, Barge A, Timmins P, Burmeister WP, Ruigrok R. 2001. Combined results from solution studies on intact influenza virus M1 protein and from a new crystal form of its N-terminal domain show that M1 is an elongated monomer. Virology 279:439–446 [DOI] [PubMed] [Google Scholar]
- 31. Watanabe K, Handa H, Mizumoto K, Nagata K. 1996. Mechanism for inhibition of influenza virus RNA polymerase activity by matrix protein. J. Virol. 70:241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bui M, Wills EG, Helenius A, Whittaker GR. 2000. Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins. J. Virol. 74:1781–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hodel MR, Corbett AH, Hodel AE. 2001. Dissection of a nuclear localization signal. J. Biol. Chem. 276:1317–1325 [DOI] [PubMed] [Google Scholar]
- 34. Marfori M, Lonhienne TG, Forwood JK, Kobe B. 2012. Structural basis of high-affinity nuclear localization signal interactions with importin-α. Traffic 13:532–548 [DOI] [PubMed] [Google Scholar]
- 35. Melen K, Fagerlund R, Franke J, Kohler M, Kinnunen L, Julkunen I. 2003. Importin alpha nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J. Biol. Chem. 278:28193–28200 [DOI] [PubMed] [Google Scholar]
- 36. Jans DA, Xiao CY, Lam M. 2000. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays 22:532–544 [DOI] [PubMed] [Google Scholar]
- 37. Ye Z, Robinson D, Wagner RR. 1995. Nucleus-targeting domain of the matrix protein (M1) of influenza virus. J. Virol. 69:1964–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boulo S, Akarsu H, Ruigrok R, Baudin F. 2007. Nuclear traffic of influenza virus proteins and ribonucleoprotein complexes. Virus Res. 124:12–21 [DOI] [PubMed] [Google Scholar]
- 39. Cao S, Liu XL, Yu MR, Li J, Jia XJ, Bi YH, Sun L, Gao GF, Liu WJ. 2012. A nuclear export signal in the matrix protein of influenza A virus is required for efficient virus replication. J. Virol. 86:4883–4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gregoriades A, Frangione B. 1981. Insertion of influenza M protein into the viral lipid bilayer and localization of site of insertion. J. Virol. 40:323–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gregoriades A. 1980. Interaction of influenza M protein with viral lipid and phosphatidylcholine vesicles. J. Virol. 36:470–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pleschka S, Wolff T, Ehrhardt C, Hobom G, Planz O, Rapp UR, Ludwig S. 2001. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat. Cell Biol. 3:301–305 [DOI] [PubMed] [Google Scholar]
- 43. Gallay P, Swingler S, Song J, Bushman F, Trono D. 1995. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell 83:569–576 [DOI] [PubMed] [Google Scholar]
- 44. Nardozzi JD, Lott K, Cingolani G. 2010. Phosphorylation meets nuclear import: a review. Cell Commun. Signal. 8:32 doi:10.1186/1478-811X-8-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. White WO, Seibenhener ML, Wooten MW. 2002. Phosphorylation of tyrosine 256 facilitates nuclear import of atypical protein kinase C. J. Cell. Biochem. 85:42–53 [PubMed] [Google Scholar]
- 46. Blaskovich MA, Sun J, Cantor A, Turkson J, Jove R, Sebti SM. 2003. Discovery of JSI-124 (cucurbitacin I), a selective Janus kinase/signal transducer and activator of transcription 3 signaling pathway inhibitor with potent antitumor activity against human and murine cancer cells in mice. Cancer Res. 63:1270–1279 [PubMed] [Google Scholar]
- 47. Konig R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, Gao Q, Andrews SE, Bandyopadhyay S, De Jesus P, Tu BP, Pache L, Shih C, Orth A, Bonamy G, Miraglia L, Ideker T, Garcia-Sastre A, Young JA, Palese P, Shaw ML, Chanda SK. 2010. Human host factors required for influenza virus replication. Nature 463:813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shaw ML. 2011. The host interactome of influenza virus presents new potential targets for antiviral drugs. Rev. Med. Virol. 21:358–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hirsch AJ. 2010. The use of RNAi-based screens to identify host proteins involved in viral replication. Future Microbiol. 5:303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]