Abstract
Merkel cell carcinoma (MCC) is a rare and aggressive form of skin cancer. In at least 80% of all MCC, Merkel cell polyomavirus (MCPyV) DNA has undergone clonal integration into the host cell genome, and most tumors express the MCPyV large and small T antigens. In all cases of MCC reported to date, the integrated MCPyV genome has undergone mutations in the large T antigen. These mutations result in expression of a truncated large T antigen that retains the Rb binding or LXCXE motif but deletes the DNA binding and helicase domains. However, the transforming functions of full-length and truncated MCPyV large T antigen are unknown. We compared the transforming activities of full-length, truncated, and alternatively spliced 57kT forms of MCPyV large T antigen. MCPyV large T antigen could bind to Rb but was unable to bind to p53. Furthermore, MCPyV-truncated large T antigen was more effective than full-length and 57kT large T antigen in promoting the growth of human and mouse fibroblasts. In contrast, expression of the MCPyV large T antigen C-terminal 100 residues could inhibit the growth of several different cell types. These data imply that the deletion of the C terminus of MCPyV large T antigen found in MCC serves not only to disrupt viral replication but also results in the loss of a distinct growth-inhibitory function intrinsic to this region.
INTRODUCTION
Merkel cell carcinoma (MCC) is an aggressive skin cancer with an annual incidence of 3 per million in the United States (1). Risk factors for developing MCC include advanced age, prolonged UV exposure, and immunosuppression due to HIV, hematologic malignancy, or solid-organ transplantation (2, 3). Recently, Merkel cell polyomavirus (MCPyV) was discovered to be clonally integrated in at least 80% of MCC, raising the possibility that this pathogen contributes to carcinogenesis (4, 5).
MCPyV is a typical polyomavirus with a circular, double-stranded DNA genome containing an early region that expresses large and small T antigens, a late region that encodes 3 viral coat proteins, VP1, VP2, and VP3, and a regulatory region that contains the origin of replication and a bidirectional promoter for the early and late genes. MCPyV was the fifth polyomavirus identified in humans, preceded by BKPyV, JCPyV, KIPyV, and WUPyV (6–9). Since then, 6 additional human polyomaviruses have been discovered, including HPyV6, HPyV7, TSPyV, HPyV9, MWPyV, and STLPyV (10–15). Although JCPyV and BKPyV have been detected in a variety of human cancers (16, 17), Merkel cell polyomavirus is the only polyomavirus DNA clonally integrated in human cancer. Expression of Merkel large and small T antigens can be detected in most MCC specimens (5, 18, 19).
The T antigens from several polyomaviruses have oncogenic activity. Notably, the simian virus 40 (SV40) large and small T antigens can transform a variety of rodent and human cells. Expression of SV40 large and small T antigens, together with human telomerase reverse transcriptase and an oncogenic form of H-RAS, can fully transform normal human fibroblasts (20, 21). At a minimum, the SV40 large T antigen transforming activity is dependent on binding to cellular tumor suppressor proteins, including p53 (TP53) and members of the Rb family (RB1, RBL1, and RBL2) (22). SV40 small T antigen binding to the serine/threonine phosphatase PP2A results in the perturbation in phosphorylation state of several host cell factors, including c-Myc (23, 24).
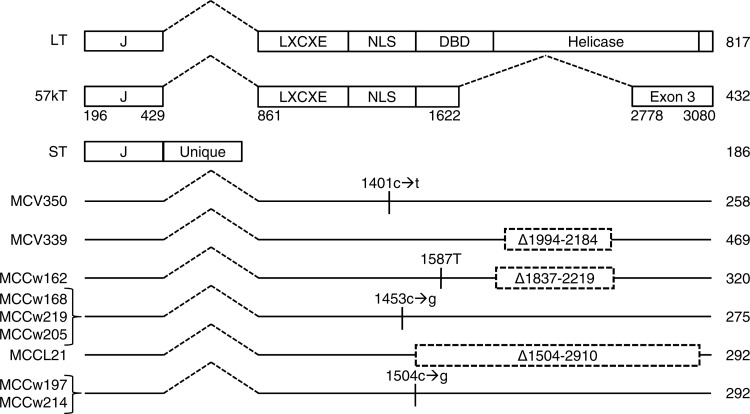
Similar to other polyomaviruses, the MCPyV large T antigen contains an N-terminal J domain, an LXCXE or Rb binding motif, a DNA binding domain, and a helicase domain (25, 26). MCPyV small T antigen contains the J domain and the unique region not shared with large T antigen (19). In addition, the MCPyV early region undergoes alternative splicing, resulting in expression of 57kT, which deletes most of the centrally located DNA binding and helicase domains of large T antigen (Fig. 1), but it retains the J domain and LXCXE motif in-frame with the C-terminal 100 residues expressed from Exon 3 (25, 27).
Fig 1.
Mutations truncate MCPyV large T antigen in MCC. Full-length (LT) and 57kT forms of large T antigen and small T antigen (ST) are shown. LT contains DnaJ (J), Rb-binding (LXCXE), nuclear localization signal (NLS), DNA binding (DBD), and helicase domains. MCV350 and MCV339 are from the original report of MCPyV in MCC (4). Predicted total numbers of residues are indicated on the right. Only mutations causing stop codons or deletions in large T antigen are shown.
Given the oncogenic properties of the canonical polyomavirus SV40 large and small T antigens, it is likely that the MCPyV T antigens also have transforming activities. MCPyV large T antigen has been shown to bind specifically to RB1 as well as VPS39 (vacuolar protein sorting 39 homolog, or Vam6) (25, 28). Furthermore, RNA interference (RNAi)-mediated knockdown of MCPyV large T and small T antigen results in decreased growth of MCC cell lines in vitro and as xenografts (19, 29). The LXCXE motif is required for MCPyV large T antigen for binding to Rb and for sustained growth of a Merkel cell carcinoma cell line (29). These results indicate that continued expression of MCPyV large and small T antigens is required for maintenance of the transformed phenotype in MCC cell lines.
Sequencing of the integrated MCPyV genome from several MCC tumors and cell lines revealed that while the small T antigen sequence remains intact, the large T antigen gene has undergone mutations that preserve the N-terminal J domain and LXCXE motif but delete most, if not all, of the DNA binding and helicase domains (25, 30). These truncated forms of MCPyV large T antigen were unable to bind to the viral origin of replication or initiate viral replication (25, 30, 31). This observation led to the model that truncation of large T antigen disrupts its ability to replicate the integrated viral genome. Although mutations within the MCPyV origin of replication or point substitutions within the DNA binding domain of large T antigen are predicted to disrupt viral replication, all reports of MCC tumors to date indicate that large T antigen has undergone truncating mutations (25).
Given the frequent presence of MCPyV in MCC, an important question remains whether the MCPyV large and small T antigens can transform normal cells. Furthermore, since MCC tumors contain truncations of the large T antigen, the relevant transforming activity of MCPyV must reside within the N-terminal part of large T antigen and the intact small T antigen. While loss of the DNA-binding domain and helicase domain of large T antigen can disrupt viral replication, it is not clear if any transforming functions of large T antigen are also lost by these truncations (25). For example, if the MCPyV large T antigen C-terminal helicase domain binds to p53, then these truncations would be predicted to lose a significant transforming activity. To investigate this, we examined the transforming potential of MCPyV full-length and 57kT large T antigen as well as truncated large T antigen derived from MCC tumors.
MATERIALS AND METHODS
Merkel cell carcinoma tumor specimens.
Total DNA was extracted from MCC tumor samples MCCw162, MCCw168, and MCCL21 using a DNeasy Blood & Tissue kit (catalog no. 69504; Qiagen) and used as the template to PCR amplify the MCPyV early region. The forward and reverse primers included sequences for attB sites to facilitate Gateway cloning (Invitrogen): forward primer, ggggacaagtttgtacaaaaaagcaggcATGGATTTAGTCCTAAA; reverse primer, ggggaccactttgtacaagaaagctgggtcTTATTGAGAAAAAGTACCAGAATCT, where capital letters correspond to the open reading frame of the MCPyV early region and lowercase letters correspond to Gateway sequences. PCR products were cloned into Gateway pDonor221 donor vector and propagated for sequencing.
DNA samples isolated from tumor specimens MCCw219, MCCw205, MCCw197, and MCCw214 were PCR amplified using the following sets of overlapping primers: 1161L (5′ AGC AGA GAG GAG ACC ACC AA 3′) and MCV1871R (5′ TGT AAG GGG GCT TGC ATA AA 3′), as well as MCV1712L (5′ GCA TGC CTG TGA ATT AGG ATG 3′) and MCV2158R (5′ GCT TGT TGG CAA ATG GTT TT 3′). PCR products were sequenced (Functional Biosciences, Inc.). Sequences for MCCw162 (KC426953), MCCw168 (KC426954), MCCw219 (KC512237), MCCw205 (KC512235), MCCL21 (KC426955), MCCw197 (KC512234), and MCCw214 (KC512236) are available from GenBank (32).
Plasmids.
The pQ-puro and pQ-neo retroviral expression vectors were modified from pQCXIP and pQCXIN, respectively (Clontech, Mountain View, CA), by replacing the cytomegalovirus (CMV) promoter between XbaI and NotI restriction sites with EF-1α (EEF1A1) promoter, which was PCR amplified from pEF6/V5-His A (Invitrogen). The full-length and 57kT cDNA versions of MCPyV large T antigen were generated by the QuikChange Lightning kit (Agilent Technologies) and were cloned between NotI and PacI restriction sites of the pQ-puro vector. MCPyV small T antigen was cloned between NotI and PacI restriction sites of the pQ-neo vector. MCPyV large T antigen Exon 3, corresponding to nucleotides 2778 to 3077 and encoding the C-terminal 100 residues, was inserted into the NotI and BamHI cloning sites of the pQ-neo vector to generate pQ-neo-V5-MCV-Exon 3-HA retroviral plasmid. pWZL-GFP-BSD was obtained from Addgene (plasmid 12269), and MCV Exon 3 was inserted between the BamHI and SalI sites to generate pWZL-V5-Exon 3-HA-BSD.
SV40 large T cDNA, MCPyV large T cDNA, and MCPyV early regions were cloned into pcDNA3.1/nV5-DEST (NT-V5) and pcDNA3.2/cV5-DEST (CT-V5) Gateway vectors (Invitrogen). Untagged SV40 large T antigen cDNA was cloned into BamHI sites of pBabe-puro retroviral vector (20). Full-length MCPyV and SV40 large T antigen cDNA was cloned into MSCV-N-Flag-HA-IRES-PURO by Gateway cloning (33). All constructs were sequence confirmed.
Cells.
BJ fibroblasts stably expressing human telomerase reverse transcriptase (hTERT) were a gift from William Hahn (Dana-Farber Cancer Institute) (20, 21). BJ-hTERT cells stably expressing MCPyV T antigens were produced using retroviruses generated in 293T cells transiently transfected with pQ-puro-HA-MCPyV-large T antigen or pQ-neo-HA-MCPyV-small T antigen, Gag-pol, and vesicular stomatitis virus G protein (VSV-G) plasmids. Infected BJ-hTERT cells were selected with 2 μg/ml puromycin (Sigma, St. Louis, MO) and 500 μg/ml G418 (Invitrogen). U-2OS cells were obtained from ATCC. MKL-1 cells were a gift from Masahiro Shuda, Yuan Chang, and Patrick Moore (University of Pittsburgh) (25). Mouse embryonic fibroblasts (MEF) were prepared from C57BL/6 E13.5 embryos (34).
All cells were cultured in Dulbecco's modified Eagle medium (DMEM) (Cellgro) supplemented with 10% fetal bovine serum (Omega), 100 U/ml penicillin, 100 μg/ml streptomycin (Gibco), and Glutamax (Invitrogen).
Immunoprecipitation, immunoblotting, and antibodies.
Confluent cultures of cells were washed with ice-cold phosphate-buffered saline (PBS) once, scraped from cell culture dishes, washed with cold PBS, and resuspended in EBC lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.5% NP-40, 1:10,000 β-mercaptoethanol, 0.5 mM EDTA) for 30 min on ice and then centrifuged. Clarified lysates were incubated overnight with agarose-immobilized V5 antibody (Bethyl) or anti-hemagglutinin (HA) affinity matrix (Roche Diagnostics). The beads were washed with high-salt EBC buffer (50 mM Tris, pH 8.0, 300 mM NaCl, 0.5% NP-40, 0.5 mM EDTA) 5 times and boiled in Laemmli sample buffer prior to SDS polyacrylamide gel electrophoresis.
Western blotting was performed with antibodies to V5 (R96025; Life Technologies), p53 (DO-1; Thermo Scientific/Lab Vision), RB1 (G3-245; BD Biosciences), and HA (HA.11; Covance). Mouse monoclonal antibodies Ab3 and Ab5 were generated against MCPyV large T antigen residues 1 to 260 and produced as a glutathione S-transferase (GST) fusion protein in bacteria (5).
Proliferation assay.
Cells (20,000) were cultured in 60-mm-diameter plates in the presence of 10% serum and then trypsinized and counted with a hemocytometer at the indicated days. For cell growth at low serum concentration, cells were plated in the presence of 10% serum, washed, and changed to 1% serum 1 day later.
Soft-agar assay.
Adherent cells (5,000) were trypsinized and mixed with 0.3% Noble Agar (BD) and plated in 0.6% base agar in one well of a 6-well plate. Growth medium (0.2 ml) was used to cover the surface of agar and supplemented twice per week. After 4 weeks, colonies formed in soft agar were stained with 0.001% crystal violet (Sigma, St. Louis, MO) overnight at 37°C degrees and 10% CO2.
Use of human subjects and animals in research.
Research performed in this study complied with all relevant federal guidelines and institutional policies.
RESULTS
MCC contains truncated large T antigen and wild-type small T antigen.
DNA was extracted from 7 MCC tumors (MCCw162, MCCw168, MCCw197, MCCw205, MCCw214, MCCw219, and MCCL21). For three specimens, the MCPyV genome was amplified by PCR with primers covering the entire early region of MCPyV encoding large and small T antigens. For the remaining 4 specimens, primers were used to amplify and sequence the large T antigen unique region. In all 7 cases, sequencing revealed that the open reading frame for large T antigen contained point substitution mutations or deletions predicted to yield truncated proteins (Fig. 1). Notably, 3 independent tumors (MCCw168, MCCw219, and MCCw205) encoded a truncated large T antigen with a predicted length of 275 residues. Two additional tumors (MCCw197 and MCCw214) contained mutations predicted to yield a truncated large T antigen of 292 residues. All truncated large T antigens retained an intact N terminus, containing the J domain and Rb-binding LXCXE motif (residues 134 to 138; nucleotides 1261 to 1275) (22). Detection of truncated large T antigen in MCC is consistent with prior reports (4, 30, 35, 36).
MCPyV large T antigen does not bind to p53.
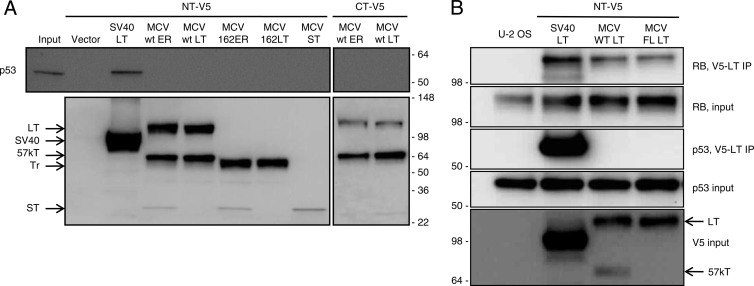
The polyomavirus T antigens transform cells by binding to key cellular proteins, such as the Rb and p53 tumor suppressor proteins. The large T antigens from SV40, BKPyV, JCPyV, and MCPyV can bind to Rb (25, 29). In addition, SV40, BKPyV, and JCPyV large T antigen can bind to and increase expression levels of p53. The SV40 large T antigen C-terminal helicase domain binds specifically to p53 (37). In contrast, mouse polyomavirus (MPyV) large T antigen is unable to bind to p53. To determine if MCPyV large T antigen could bind to p53, we generated expression vectors, containing the early region, encoding wild-type large and small T antigens or cDNAs for large T antigen and small T antigen. In addition, we generated MCPyV early region and large T antigen cDNA constructs that expressed a truncated LT with a premature stop codon after residue 320 identical to that isolated from tumor MCCw162 (Fig. 1).
U-2OS cells containing wild-type p53 were transiently transfected with V5 epitope-tagged SV40 large T antigen, MCPyV large T antigen, or MCPyV early region expression plasmids. Lysates were prepared and immunoprecipitated with a V5 antibody and immunoblotted for V5 and p53. As shown in Fig. 2A, the MCPyV large T antigen constructs were expressed at levels similar to those of SV40 large T antigen. The MCPyV early region and large T antigen constructs expressed the full length, as well as a smaller form of large T likely corresponding to 57kT (25). Notably, epitope tags on either the N or C terminus of MCPyV early region or large T antigen cDNA revealed similar-sized proteins, indicating that residues encoded by the alternative Exon 3 in 57kT are in frame with the first and second exons of large T antigen (Fig. 1). In addition, small T antigen was expressed from the MCPyV early region and the small T antigen cDNA constructs but not from the large T antigen cDNA constructs.
Fig 2.
MCPyV T antigens are unable to bind to p53. Transient transfection of U-2 OS cells with SV40 or MCPyV (MCV) large T (LT) antigen cDNA, small T (ST) antigen cDNA, or early region (ER) corresponding to wild-type (wt) or MCCw162 (Tr) tumor-derived sequence. N-terminal (NT) or C-terminal (CT) V5 epitope-tagged constructs were used. (A) Cell lysates (input) were immunoprecipitated with V5 antibody and blotted with p53 (top) or V5 (bottom) antibodies. (B) U-2 OS cells were transiently transfected with NT-V5 constructs as shown. Lysates were immunoprecipitated with V5 antibody and blotted with the indicated antibodies. Rb and p53 inputs are shown. Numbers indicate relative molecular mass. Arrows indicate LT, 57kT, Tr, and ST.
We compared SV40 and MCPyV large T antigens for binding to p53. As shown in Fig. 2A (top), immunoprecipitation for SV40 large T antigen coprecipitated p53. In contrast, immunoprecipitation for wild-type or truncated MCPyV large T antigen did not coprecipitate p53. We repeated this experiment using N-terminal V5 epitope-tagged cDNA constructs for SV40 and MCPyV large T antigens, as well as a modified form of MCPyV that expresses full-length large T antigen and not the 57kT form (see Fig. 4A). As shown in Fig. 2B, all T antigens were capable of binding to Rb, but only SV40 large T antigen bound to p53.
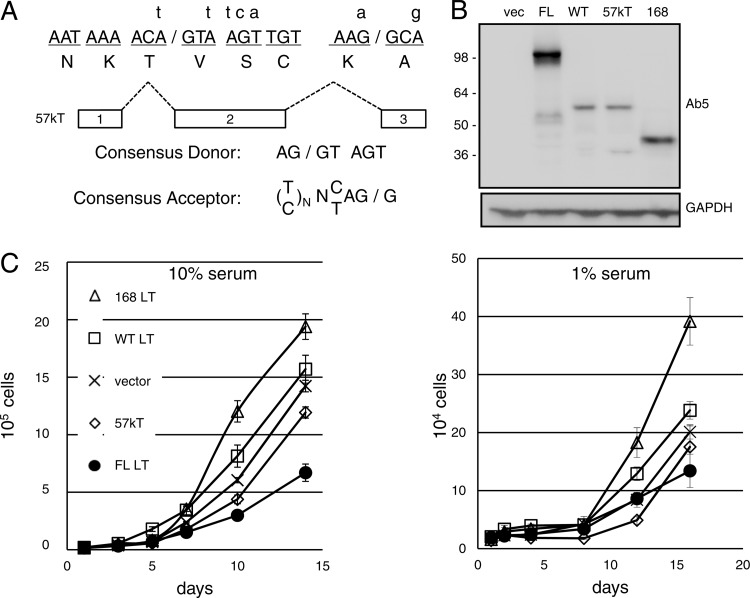
Fig 4.
Comparison of full-length, 57kT, and truncated MCPyV large T antigens. (A) Silent mutations (small letters) of the wild type (capital letters) to disable splice donor (1622) and acceptor (2778) sites of the second intron to generate full-length (FL) MCPyV large T antigen. (B) Extracts of BJ-hTERT cells stably expressing FL, wild-type (WT), 57kT, or truncated (168) cDNA of MCPyV large T antigen were blotted with antibody against MCPyV LT (Ab5) or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for a loading control. vec, vector. (C) BJ-hTERT cells were cultured in 10% (left) or 1% (right) serum and counted at the indicated days. The experiment was performed in triplicate and results averaged, with error bars indicating standard deviations. Similar results were obtained from at least three independent experiments.
Effects on cell growth by MCPyV large T antigen.
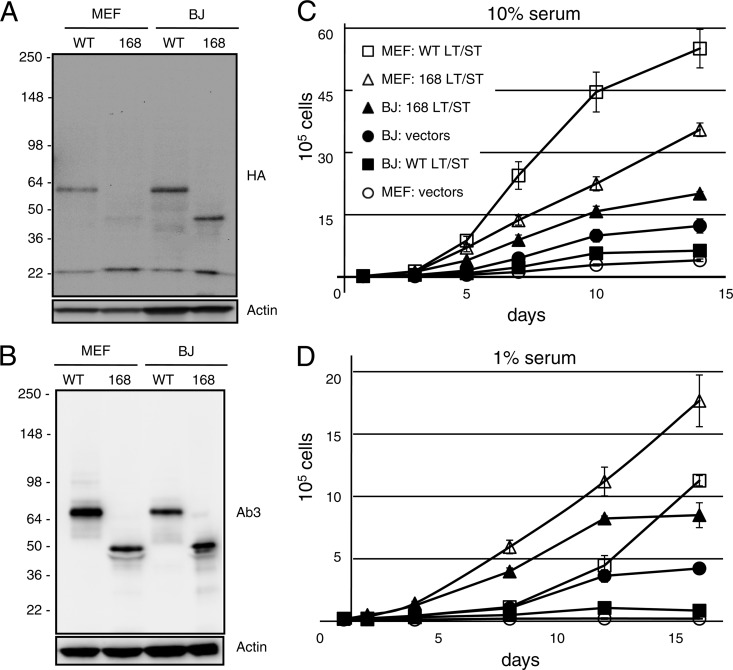
To study the transforming potential of MCPyV large and small T antigens, we generated stable derivatives of primary mouse and human fibroblasts with a retroviral vector, pQ-puro, expressing N-terminal, HA-tagged, wild-type, or truncated large T antigen cDNA corresponding to MCCw168 together with a separate vector, pQ-neo, expressing N-terminal, HA-tagged, wild-type small T antigen. As shown in Fig. 3A, the MCPyV 57kT form was preferentially expressed in the MEFs and BJ-hTERT fibroblasts containing the wild-type MCPyV large T antigen cDNA vector. Similar results, with the predominant expression of the 57kT form of large T antigen, were observed when cell lysates were subjected to Western blotting with Ab3 (Fig. 3B), a mouse monoclonal antibody specific for MCPyV large T antigen (5). Small T antigen was expressed equally well in MEFs and BJ-hTERT cells (Fig. 3A).
Fig 3.
Comparison of wild-type and truncated MCPyV T antigen growth-promoting activities. N-terminal HA-tagged wild-type or MCCw168 tumor-derived MCPyV early region was stably expressed in mouse embryo fibroblasts (MEF) or hTERT-immortalized BJ fibroblasts (BJ). Cell extracts were blotted with HA (A) or Ab3 (B) and actin for a loading control. Proliferation assay of cells was performed in triplicate in 10% serum (C) and 1% serum (D) as described in Materials and Methods. The results of a typical experiment are presented and represent the averages of triplicate samples. Similar results were obtained in at least three independent experiments. The error bars indicate standard deviations.
We tested the growth-promoting potential of the MCPyV T antigens by performing proliferation assays of these stable cell lines in the presence of 10% (Fig. 3C) or 1% (Fig. 3D) serum. MEFs expressing truncated or wild-type large T antigen grew faster than control MEFs containing both empty vectors when cultured in 10% serum. Of note, the truncated large T antigen expressing MEFs grew faster than cells expressing wild-type large T antigen in 1% serum. Similarly, BJ-hTERT cells expressing small T antigen and truncated large T antigen grew more rapidly than control cells containing vector only in media with 10% or 1% serum. In contrast, BJ-hTERT expressing wild-type large T antigen grew more slowly than control cells and cells expressing the truncated large T antigen.
Given the predominant expression of the 57kT form of MCPyV large T antigen from the wild-type vector in MEFs and BJ-hTERT cells, we made several modifications to the large T antigen gene. We generated a cDNA that encodes only the 57kT form of large T antigen by eliminating the nucleotides corresponding to the first and second introns but retaining the C-terminal 100 residues encoded by Exon 3 in frame with the N terminus (Fig. 1 and 4A), and we cloned it into the pQ-puro retroviral expression vector. We generated a full-length (FL) MCPyV large T antigen cDNA by introducing silent mutations that disrupt the splice donor and acceptor sites for intron 2 while retaining the wild-type coding sequence for large T antigen to generate a cDNA predicted to express only FL large T antigen (Fig. 4A), and then we cloned it into MSCV-N-Flag-HA-IRES-Puro vector.
BJ-hTERT cells were infected with retroviruses encoding cDNAs corresponding to wild-type, FL, 57kT, and truncated (MCV168) MCPyV large T antigens. Expression was confirmed by immunoblotting with an antibody specific for MCPyV T antigen (Fig. 4B). Consistent with the results shown in Fig. 3A and B, the wild-type large T antigen cDNA construct predominantly expressed a 57-kDa form that comigrated with the 57kT cDNA construct in BJ-hTERT cells. In contrast, the MCPyV FL large T antigen construct migrated at the expected size of approximately 100 kDa. To compare the growth-promoting ability of these forms of MCPyV large T antigen, replica plates of cells were cultured in medium containing 10 or 1% serum and counted over several days. As shown in Fig. 4C, BJ-hTERT fibroblasts expressing truncated MCPyV large T antigen grew faster than cells containing the empty vector. In contrast, cells expressing FL or 57kT MCPyV large T antigen grew at a rate slower than that of the vector control cells. Similar results were observed in medium containing 10 or 1% serum.
Merkel cell polyomavirus large T antigen Exon 3 inhibits cell growth.
BJ-hTERT cells expressing truncated MCPyV large T antigen appeared to have a growth advantage over cells expressing wild-type, FL, and 57kT large T antigens (Fig. 4C). Since wild-type, FL, and 57kT large T antigens share the C-terminal 100 residues that are not present in the MCC tumor-specific truncated large T antigen, we suspected that the Exon 3 region contained a growth-inhibitory function. To test this possibility, we generated a retroviral construct that expresses Exon 3 only.
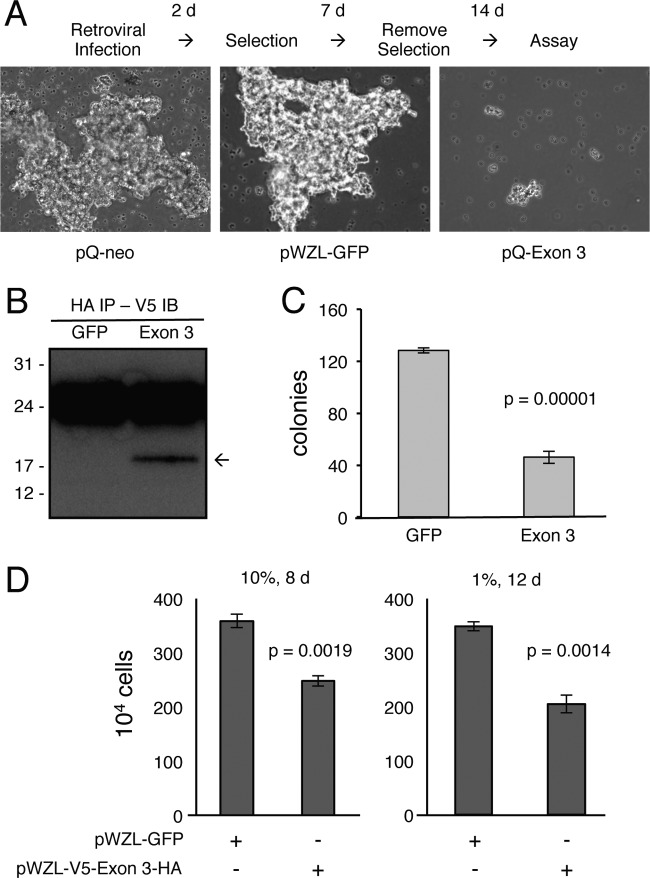
We transduced the MCC cell line MKL-1 with retroviral constructs containing the MCPyV large T antigen Exon 3, green fluorescent protein (GFP), or empty vector. The MKL-1 cells normally grow in large aggregates. The control vectors readily established stable cell lines that grew as clumps, but cells containing the Exon 3 construct failed to propagate and formed tiny clumps containing very few cells (Fig. 5A). To test if MCPyV Exon 3 could inhibit growth of cells transformed with SV40, we transduced the Exon 3 or GFP control vector in BJ-hTERT cells that stably expressed H-Ras and SV40 large and small T antigens (20). We were able to establish a stable line with detectable expression of the Exon 3 construct (Fig. 5B). However, the SV40-transformed BJ cells expressing the C-terminal 100 residues of MCPyV large T antigen had a significantly reduced ability to grow in an anchorage-independent manner compared to cells expressing GFP (Fig. 5C). We also observed that Exon 3 significantly reduced the growth rate of the SV40-transformed BJ-hTERT cells (Fig. 5D).
Fig 5.
C terminus of MCPyV large T antigen inhibits cell growth. (A) MCC cell line MKL-1 was infected with pQ-neo, pWZL-GFP-BSD, or pQ-neo-V5-Exon 3-HA retrovirus for 2 days, selected with antibiotics for 7 days, and maintained in culture medium without antibiotics for an additional 14 days. Representative images of cultures at completion of the assay are shown (100× magnification). (B) BJ-hTERT cells stably expressing SV40 large T antigen, small T antigen, and H-Ras cells were stably infected with pWZL-GFP-BSD or pWZL-V5-Exon 3-HA-BSD retrovirus. Equal amounts of protein lysates were immunoprecipitated with HA antibody, followed by immunoblotting with V5 antibody. The arrow points to the position of V5-Exon 3-HA. (C) Cells described for panel B were plated in soft agar and counted at 1 day after plating. Colonies were counted after 14 days of culture. The experiment was performed in triplicate, and results were averaged, with error bars indicating standard deviations. (D) Cells described for panel B were cultured in 10% (left) or 1% (right) serum for the indicated number of days. The experiment was performed in triplicate, and results were averaged, with error bars indicating standard deviations.
DISCUSSION
The discovery of the human polyomavirus MCPyV in MCC immediately led to the question of whether this virus and the virus-encoded T antigens contribute to cancer development (4). In support of this model, most MCC tumors express the MCPyV large and small T antigens (5, 19). In addition, expression of MCPyV large T and small T antigen is required to maintain growth of MCPyV-positive MCC cell lines (19, 29). However, in every case of MCC reported to date, the MCPyV early region has undergone mutations that truncate large T antigen while keeping small T antigen intact. Therefore, if MCPyV contributes to cancer development, then its transforming activity must reside within small T antigen and the N-terminal fragment of large T antigen.
Consistent with prior studies, we sequenced MCPyV DNA isolated from 7 unique MCC tumors and found that each one contained mutations in the MCPyV genome predicted to encode truncated forms of large T antigen and an intact small T antigen. We went on to evaluate the transforming potential of full-length, 57kT, and tumor-specific truncated versions of MCPyV large T antigen. We determined that truncated MCPyV large T antigen has growth-promoting activities that were more effective than full-length or 57kT versions of large T antigen in primary mouse and human fibroblasts. In contrast, we observed that expression of the C-terminal 100 residues of MCPyV large T antigen inhibited the growth of several cell types, including an MCC cell line. These results indicate that the MCC-specific truncations of large T antigen results in the loss of the C-terminal growth-inhibitory function present in both the full-length and 57kT versions. Notably, of all the large T antigen sequences reported from MCC tumors, only MCV339 is predicted to encode an intact 57kT large T antigen that retains Exon 3, since the deleted nucleotides are spliced out of the second intron (4, 25). It is not known if the MCV339 tumor expressed 57kT.
The polyomavirus T antigens transform cells by binding to key cellular proteins. In particular, the large T antigen from SV40, as well as JCPyV and BKPyV, can bind to the Rb and p53 tumor suppressor proteins (38–41). We confirmed previous reports that MCPyV large T antigen can bind specifically to Rb (25, 29). In contrast, we were unable to demonstrate that MCPyV large T antigen binds to p53. Our data indicate that MCPyV full-length, 57kT, and truncated large T antigen or small T antigen were unable to bind to p53. We observed a similar result in a large-scale study of human polyomaviruses, where the large T antigens from JCPyV, WUPyV, HPyV6, HPyV7, TSPyV, and MCPyV could bind to Rb and all except for MCPyV could bind to p53 (42). These results indicate that the MCC-specific truncations of MCPyV large T antigen do not result in loss of p53 binding activity. It should be noted that most MCC tumors contain wild-type p53, although it is not known if p53 function is perturbed in MCC containing wild-type p53 (5, 43, 44). Although MCPyV T antigens do not bind to p53, they may serve to indirectly inactivate p53 function.
Shuda and colleagues proposed that mutations found in MCPyV large T antigen isolated from MCC tumors resulted from the negative selection of cells that contained wild-type large T antigen (25). It was proposed that wild-type MCPyV large T antigen would promote unregulated replication of an integrated MCPyV origin, resulting in collisions between cellular and virus-initiated replication forks. To date, all of the MCPyV large T antigens isolated from tumors are truncated due to nonsense mutations or deletions. At least to date, missense mutations that render the full-length large T antigen deficient in DNA binding or helicase activity have not been identified (25, 45). Similarly, mutations in the MCPyV viral origin of replication predicted to disrupt the binding of wild-type MCPyV large T antigen DNA binding domain have not been identified.
Our studies suggest that in addition to loss of the replication function of large T antigen, the truncating mutations in MCPyV large T antigen also serve to eliminate a growth-inhibitory property contained within the C terminus. Deletion of the large T antigen C terminus eliminates this potential growth-inhibitory function while retaining Rb binding by the LXCXE motif in the N terminus. Therefore, truncation of large T antigen disables the viral DNA replication function as well as the growth-suppressing potential of the large T antigen C terminus. To date, there has been only one exception reported that does not fit with this model. The MCV339 tumor contains a deletion entirely within the second intron of MCPyV large T antigen and is predicted to express the 57kT form of large T antigen with the C-terminal 100 residues (Fig. 1) (4, 25). However, it is not known if the MCV339 tumor expressed the 57kT form of large T antigen that retains the C-terminal 100 residues.
We observed that a wild-type cDNA construct of MCPyV large T antigen preferentially expressed the 57kT form of large T antigen when stably expressed in human and mouse fibroblasts. Similarly, the 57kT form was preferentially expressed compared to the full-length form when wild-type or tumor-derived MCV339 was transiently expressed in 293 cells (25). The normal function of the 57kT form of MCPyV large T antigen is not known, although it does retain the capability of binding to Rb (25). Other human polyomaviruses, including JCPyV, express alternatively spliced large T antigens that delete the C terminus. These spliced versions also retain the Rb binding motif and can promote cellular growth (40, 46). However, MCPyV large T antigen appears to be unique, since the C-terminal 100 residues of the full-length version are retained in the 57kT form.
It is not known how the MCPyV large T antigen C-terminal 100 residues encoded by Exon 3 can inhibit cellular growth. This region is unlikely to affect the Rb pathway, since the N terminus of MCPyV large T antigen can bind to Rb and should be sufficient to inactivate this growth-suppressing pathway. Similarly, Exon 3 is unlikely to activate the p53 growth-inhibitory pathway, since expression was able to reduce the growth rate of SV40 large T antigen-transformed cells that have fully inactivated p53. The C-terminal 100 residues of MCPyV large T antigen contain some residues corresponding to the helicase domain. However, this fragment, corresponding to Exon 3, was unable to bind to full-length or truncated MCPyV large T antigen and is unlikely to function in a dominant-negative manner (data not shown). We suspect that Exon 3, as well as full-length and 57kT forms of MCPyV large T antigen, bind specifically to cellular proteins involved in growth regulation. If the C terminus of MCPyV large T antigen binds to a cellular protein, then it appears to interfere with an essential factor required for cellular proliferation of normal and cancer cells. Identification of host cell proteins that bind to the C-terminal domain of MCPyV large T antigen may bring insight into an important cellular growth pathway.
ACKNOWLEDGMENTS
This work was supported in part by Public Health Service grants R01CA162522 and K24-CA139052 to P.N. and P01CA050661, RO1CA93804, and R01CA63113 to J.A.D. We gratefully acknowledge the support of the Claudia Adams Barr Program in Innovative Basic Cancer Research at the Dana-Farber Cancer Institute to J.A.D.
Footnotes
Published ahead of print 20 March 2013
REFERENCES
- 1. Miller RW, Rabkin CS. 1999. Merkel cell carcinoma and melanoma: etiological similarities and differences. Cancer Epidemiol. Biomarkers Prev. 8:153–158 [PubMed] [Google Scholar]
- 2. Bichakjian CK, Lowe L, Lao CD, Sandler HM, Bradford CR, Johnson TM, Wong SL. 2007. Merkel cell carcinoma: critical review with guidelines for multidisciplinary management. Cancer 110:1–12 [DOI] [PubMed] [Google Scholar]
- 3. Heath M, Jaimes N, Lemos B, Mostaghimi A, Wang LC, Penas PF, Nghiem P. 2008. Clinical characteristics of Merkel cell carcinoma at diagnosis in 195 patients: the AEIOU features. J. Am. Acad. Dermatol. 58:375–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Feng H, Shuda M, Chang Y, Moore PS. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319:1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rodig SJ, Cheng J, Wardzala J, Dorosario A, Scanlon JJ, Laga AC, Martinez-Fernandez A, Barletta JA, Bellizzi AM, Sadasivam S, Holloway DT, Cooper DJ, Kupper TS, Wang LC, Decaprio JA. 2012. Improved detection suggests all Merkel cell carcinomas harbor Merkel polyomavirus. J. Clin. Investig. 122:4645–4653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH. 1971. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet i:1257–1260 [DOI] [PubMed] [Google Scholar]
- 7. Gardner SD, Field AM, Coleman DV, Hulme B. 1971. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet i:1253–1257 [DOI] [PubMed] [Google Scholar]
- 8. Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MA, Dalianis T, Ramqvist T, Andersson B. 2007. Identification of a third human polyomavirus. J. Virol. 81:4130–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, Brennan DC, Storch GA, Sloots TP, Wang D. 2007. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 3:e64 doi:10.1371/journal.ppat.0030064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. 2010. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 7:509–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van der Meijden E, Janssens RW, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MC. 2010. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLoS Pathog. 6:e1001024 doi:10.1371/journal.ppat.1001024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sauvage V, Foulongne V, Cheval J, Ar Gouilh M, Pariente K, Dereure O, Manuguerra JC, Richardson J, Lecuit M, Burguiere A, Caro V, Eloit M. 2011. Human polyomavirus related to African green monkey lymphotropic polyomavirus. Emerg. Infect. Dis. 17:1364–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Scuda N, Hofmann J, Calvignac-Spencer S, Ruprecht K, Liman P, Kuhn J, Hengel H, Ehlers B. 2011. A novel human polyomavirus closely related to the African green monkey-derived lymphotropic polyomavirus. J. Virol. 85:4586–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lim ES, Reyes A, Antonio M, Saha D, Ikumapayi UN, Adeyemi M, Stine OC, Skelton R, Brennan DC, Mkakosya RS, Manary MJ, Gordon JI, Wang D. 2012. Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology 436:295–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DeCaprio JA, Garcea RL. 2013. A cornucopia of human polyomaviruses. Nat. Rev. Microbiol. 11:1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maginnis MS, Atwood WJ. 2009. JC virus: an oncogenic virus in animals and humans? Semin. Cancer Biol. 19:261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abend JR, Jiang M, Imperiale MJ. 2009. BK virus and human cancer: innocent until proven guilty. Semin. Cancer Biol. 19:252–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shuda M, Arora R, Kwun HJ, Feng H, Sarid R, Fernandez-Figueras MT, Tolstov Y, Gjoerup O, Mansukhani MM, Swerdlow SH, Chaudhary PM, Kirkwood JM, Nalesnik MA, Kant JA, Weiss LM, Moore PS, Chang Y. 2009. Human Merkel cell polyomavirus infection I. MCV T antigen expression in Merkel cell carcinoma, lymphoid tissues and lymphoid tumors. Int. J. Cancer 125:1243–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shuda M, Kwun HJ, Feng H, Chang Y, Moore PS. 2011. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J. Clin. Investig. 121:3623–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hahn WC, Dessain SK, Brooks MW, King JE, Elenbaas B, Sabatini DM, DeCaprio JA, Weinberg RA. 2002. Enumeration of the simian virus 40 early region elements necessary for human cell transformation. Mol. Cell. Biol. 22:2111–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. 1999. Creation of human tumour cells with defined genetic elements. Nature 400:464–468 [DOI] [PubMed] [Google Scholar]
- 22. Cheng J, DeCaprio JA, Fluck MM, Schaffhausen BS. 2009. Cellular transformation by simian virus 40 and murine polyoma virus T antigens. Semin. Cancer Biol. 19:218–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, Hahn WC, Stukenberg PT, Shenolikar S, Uchida T, Counter CM, Nevins JR, Means AR, Sears R. 2004. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat. Cell Biol. 6:308–318 [DOI] [PubMed] [Google Scholar]
- 24. Sablina AA, Hector M, Colpaert N, Hahn WC. 2010. Identification of PP2A complexes and pathways involved in cell transformation. Cancer Res. 70:10474–10484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shuda M, Feng H, Kwun HJ, Rosen ST, Gjoerup O, Moore PS, Chang Y. 2008. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc. Natl. Acad. Sci. U. S. A. 105:16272–16277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Garneski KM, DeCaprio JA, Nghiem P. 2008. Does a new polyomavirus contribute to Merkel cell carcinoma? Genome Biol. 9:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Feng H, Kwun HJ, Liu X, Gjoerup O, Stolz DB, Chang Y, Moore PS. 2011. Cellular and viral factors regulating Merkel cell polyomavirus replication. PLoS One 6:e22468 doi:10.1371/journal.pone.0022468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu X, Hein J, Richardson SC, Basse PH, Toptan T, Moore PS, Gjoerup OV, Chang Y. 2011. Merkel cell polyomavirus large T antigen disrupts lysosome clustering by translocating human Vam6p from the cytoplasm to the nucleus. J. Biol. Chem. 286:17079–17090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Houben R, Adam C, Baeurle A, Hesbacher S, Grimm J, Angermeyer S, Henzel K, Hauser S, Elling R, Brocker EB, Gaubatz S, Becker JC, Schrama D. 2012. An intact retinoblastoma protein-binding site in Merkel cell polyomavirus large T antigen is required for promoting growth of Merkel cell carcinoma cells. Int. J. Cancer 130:847–856 [DOI] [PubMed] [Google Scholar]
- 30. Schmitt M, Wieland U, Kreuter A, Pawlita M. 2012. C-terminal deletions of Merkel cell polyomavirus large T-antigen, a highly specific surrogate marker for virally induced malignancy. Int. J. Cancer 131:2863–2868 [DOI] [PubMed] [Google Scholar]
- 31. Duncavage EJ, Magrini V, Becker N, Armstrong JR, Demeter RT, Wylie T, Abel HJ, Pfeifer JD. 2011. Hybrid capture and next-generation sequencing identify viral integration sites from formalin-fixed, paraffin-embedded tissue. J. Mol. Diagn. 13:325–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paulson KG, Iyer JG, Tegeder AR, Thibodeau R, Schelter J, Koba S, Schrama D, Simonson WT, Lemos BD, Byrd DR, Koelle DM, Galloway DA, Leonard JH, Madeleine MM, Argenyi ZB, Disis ML, Becker JC, Cleary MA, Nghiem P. 2011. Transcriptome-wide studies of Merkel cell carcinoma and validation of intratumoral CD8+ lymphocyte invasion as an independent predictor of survival. J. Clin. Oncol. 29:1539–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sowa ME, Bennett EJ, Gygi SP, Harper JW. 2009. Defining the human deubiquitinating enzyme interaction landscape. Cell 138:389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zalvide J, DeCaprio JA. 1995. Role of pRb-related proteins in simian virus 40 large-T-antigen-mediated transformation. Mol. Cell. Biol. 15:5800–5810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Katano H, Ito H, Suzuki Y, Nakamura T, Sato Y, Tsuji T, Matsuo K, Nakagawa H, Sata T. 2009. Detection of Merkel cell polyomavirus in Merkel cell carcinoma and Kaposi's sarcoma. J. Med. Virol. 81:1951–1958 [DOI] [PubMed] [Google Scholar]
- 36. Laude HC, Jonchere B, Maubec E, Carlotti A, Marinho E, Couturaud B, Peter M, Sastre-Garau X, Avril MF, Dupin N, Rozenberg F. 2010. Distinct Merkel cell polyomavirus molecular features in tumour and nontumour specimens from patients with Merkel cell carcinoma. PLoS Pathog. 6:e1001076 doi:10.1371/journal.ppat.1001076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lilyestrom W, Klein MG, Zhang R, Joachimiak A, Chen XS. 2006. Crystal structure of SV40 large T-antigen bound to p53: interplay between a viral oncoprotein and a cellular tumor suppressor. Genes Dev. 20:2373–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Harris KF, Christensen JB, Imperiale MJ. 1996. BK virus large T antigen: interactions with the retinoblastoma family of tumor suppressor proteins and effects on cellular growth control. J. Virol. 70:2378–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shivakumar CV, Das GC. 1996. Interaction of human polyomavirus BK with the tumor-suppressor protein p53. Oncogene 13:323–332 [PubMed] [Google Scholar]
- 40. Bollag B, Prins C, Snyder EL, Frisque RJ. 2000. Purified JC virus T and T′ proteins differentially interact with the retinoblastoma family of tumor suppressor proteins. Virology 274:165–178 [DOI] [PubMed] [Google Scholar]
- 41. Poulin DL, Kung AL, DeCaprio JA. 2004. p53 targets simian virus 40 large T antigen for acetylation by CBP. J. Virol. 78:8245–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rozenblatt-Rosen O, Deo RC, Padi M, Adelmant G, Calderwood MA, Rolland T, Grace M, Dricot A, Askenazi M, Tavares M, Pevzner SJ, Abderazzaq F, Byrdsong D, Carvunis AR, Chen AA, Cheng J, Correll M, Duarte M, Fan C, Feltkamp MC, Ficarro SB, Franchi R, Garg BK, Gulbahce N, Hao T, Holthaus AM, James R, Korkhin A, Litovchick L, Mar JC, Pak TR, Rabello S, Rubio R, Shen Y, Singh S, Spangle JM, Tasan M, Wanamaker S, Webber JT, Roecklein-Canfield J, Johannsen E, Barabasi AL, Beroukhim R, Kieff E, Cusick ME, Hill DE, Munger K, Marto JA, Quackenbush J, Roth FP, DeCaprio JA, Vidal M. 2012. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 487:491–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lill C, Schneider S, Item CB, Loewe R, Houben R, Halbauer D, Heiduschka G, Brunner M, Thurnher D. 2011. P53 mutation is a rare event in Merkel cell carcinoma of the head and neck. Eur. Arch. Otorhinolaryngol. 268:1639–1646 [DOI] [PubMed] [Google Scholar]
- 44. Nardi V, Song Y, Santamaria-Barria JA, Cosper AK, Lam Q, Faber AC, Boland GM, Yeap BY, Bergethon K, Scialabba VL, Tsao H, Settleman J, Ryan DP, Borger DR, Bhan AK, Hoang MP, Iafrate AJ, Cusack JC, Engelman JA, Dias-Santagata D. 2012. Activation of PI3K signaling in Merkel cell carcinoma. Clin. Cancer Res. 18:1227–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Harrison CJ, Meinke G, Kwun HJ, Rogalin H, Phelan PJ, Bullock PA, Chang Y, Moore PS, Bohm A. 2011. Asymmetric assembly of Merkel cell polyomavirus large T-antigen origin binding domains at the viral origin. J. Mol. Biol. 409:529–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bollag B, Kilpatrick LH, Tyagarajan SK, Tevethia MJ, Frisque RJ. 2006. JC virus T'135, T'136, and T'165 proteins interact with cellular p107 and p130 in vivo and influence viral transformation potential. J. Neurovirol. 12:428–442 [DOI] [PubMed] [Google Scholar]