Abstract
Epstein-Barr virus (EBV) infects ∼95% of the adult population. The factors that confer protection in the remaining ∼5% remain unknown. In an exploratory study, we assessed immunogenetic factors and tonsillectomy in a cohort of 17 EBV-negative and 39 EBV-positive healthy individuals aged >60 years. Analyses of HLA genotypes revealed an association between EBV negativity and the presence of HLA-C-35T/T and/or HLA-Bw4 alleles. In addition, EBV-negative donors presented with a history of tonsillectomy more often than EBV-positive donors.
TEXT
For most, primary Epstein-Barr virus (EBV) infection occurs during childhood and is asymptomatic or causes an acute self-limiting lymphoproliferative disease (infectious mononucleosis). After acute infection, EBV enters life-long latency, which leaves the infected individual at risk for viral reactivation and, in rare cases, the development of EBV-associated malignancy (1). Why ∼5% of the adult population remain EBV-seronegative throughout their lives is not known, yet understanding natural resistance to EBV infection might provide fundamental insight into the host-pathogen interaction and pinpoint targets for novel preventive and/or therapeutic strategies.
Here, after Institutional Review Board (IRB) approval and written informed consent, 515 consecutive healthy blood donors aged >60 years who were routinely presenting at the Blood Transfusion Center Basel were serologically tested for EBV by multiplex microparticle technology (Luminex 200 Technology, Luminex, Austin, TX, USA). Seventeen of 515 donors were EBV seronegative (median age, 64 years; range, 62 to 70 years; 3 female, 14 male). The seropositive control cohort consisted of 39 individuals (median age, 64 years; range, 63 to 70 years; 5 female, 34 male). In the EBV-seronegative cohort, we (i) tested whether EBV seronegativity reflects the absence of viral genome and of EBV-specific cellular memory and (ii) searched for specific HLA-B and HLA-C polymorphisms associated with EBV negativity.
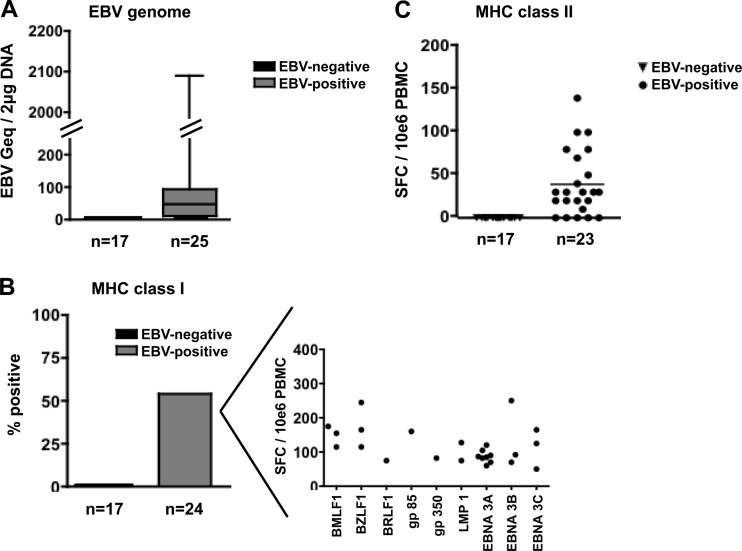
To relate serostatus and the presence of EBV DNA, a sensitive PCR was performed on DNA extracted from B cells (the primary target cells of EBV) (2), using a published real-time PCR protocol (3). The EBV genome was not detected in any of the 17 EBV-seronegative donors. In contrast, 22/25 EBV-seropositive donors tested positive [mean, 97 genome equivalents (geq)/(1 × 106) B cells; range, <3 to 1,072 geq/(1 × 106) B cells] (Fig. 1A). To assess whether a negative EBV serostatus also indicates the absence of EBV-specific cellular immunity, we applied a gamma interferon (IFN-γ) enzyme-linked immunosorbent spot (ELISpot) assay as previously described (4, 5; also data not shown). IFN-γ secretion in bulk peripheral blood mononuclear cells (PBMC) was measured in response to a pool of peptides consisting of 91 major histocompatibility complex (MHC) class I-restricted and 33 MHC class II-restricted optimal EBV epitopes, testing for CD8+- and CD4+-specific reactivity (4, 5). We did not detect responses to the peptide pools in any of the 17 EBV-seronegative individuals, whereas PBMC from 13/24 and 19/23 EBV-seropositive donors secreted IFN-γ in response to one or multiple MHC class I- and class II-restricted EBV peptide pools (Fig. 1B and C). Together, these data established that neither direct (EBV DNA) nor indirect (T cell reactivity) evidence for latent EBV infection was present in any of the 17 EBV-seronegative individuals. The expression of the EBV receptor CD21 and the coreceptor HLA-DR did not differ between the cohorts as assessed with fluorescence-activated cell sorter (FACS) staining. And, importantly, B cells from EBV-seronegative donors could readily be infected and transformed in vitro, excluding host resistance to EBV infection at the target cell level (data not shown).
Fig 1.
EBV genome content of B cells and EBV-specific T cell reactivity. (A) The number of viral genome equivalents in 17 EBV-seronegative and 25 EBV-seropositive individuals was assessed by BALF5 (DNA polymerase catalytic subunit) reverse transcription (RT)-PCR using DNA extracted from 1 × 106 B cells (2 μg). For each donor, the RT-PCR was run twice from the same DNA stock. (B) Ex vivo CD8+ T cell reactivity to EBV-derived latent and lytic epitopes was assessed by IFN-γ ELISpot. Whereas 13/24 (54%) EBV-positive donors displayed IFN-γ secretion in response to at least one HLA-restricted EBV-derived epitope, no IFN-γ secretion could be detected in EBV-negative donors. The inset on the right shows the magnitude of CD8+ T cell responses of the 13 reactive study subjects to the individual peptide (some responding to more than one peptide). (C) IFN-γ secretion in response to MHC class II-restricted EBV peptides as measured by ELISpot. None of the 17 EBV-seronegative donors reacted in response to a pool of 33 MHC class II-restricted EBV peptides. In contrast, 19/23 EBV-seropositive donors displayed IFN-γ secretion when tested with the same peptide pool [mean, 39 spot-forming cells (SFC)/(1 × 106) PBMC; standard deviation, ±9 SFC/(1 × 106) PBMC; P < 0.001].
Control of established EBV infection depends on functional cytotoxic CD8+ T cells, and yet NK cells (i.e., innate immune cells) play an important role in shaping the clinical phenotype of EBV infection as well (6–9). NK cell functionality is largely determined by a family of receptors that interact with HLA I molecules, the killer cell immunoglobulin-like receptors (KIRs) (10–12). HLA and HLA-KIR compound genotypes have been shown to influence resistance to HIV infection among highly exposed seronegative individuals (13–16). In chronic HIV infection, HLA-Bw4 alleles, which interact with the inhibitory NK cell receptor KIR3DL1, have been collectively attributed a protective role, particularly the subset of Bw4 allotypes containing isoleucine at position 80 (Bw4 80Ile) as opposed to threonine (Bw4 80Thr) (17). Also of note is an allele variant 35 kb upstream from HLA-C (−35 C), known to be related to higher HLA-C mRNA expression levels, which has recently been associated with slower progression to AIDS (18). In EBV infection, no consistent roles for specific HLA class I and KIR variants have been reported.
Against this background, we specifically analyzed −35 C allele variants (by genotyping the HLA-C rs9264942 single-nucleotide polymorphism [SNP] using a commercial ABI TaqMan kit [Applied Biosystems, Branchburg, NJ, USA]) and HLA-Bw4 epitopes (by sequence-based genotyping [Histogenetics, New York, NY, USA]). HLA-B alleles were assigned to the Bw4 and HLA-Bw4 80Ile groups according to http://hla.alleles.org/wmda/index.html. KIR3DL1 and KIR3DS1 genotypes were determined by multiplex PCR, followed by a reverse sequence-specific oligonucleotide (rSSO) method according to the manufacturer's instructions (One Lambda, Inc., Canoga Park, CA, USA).
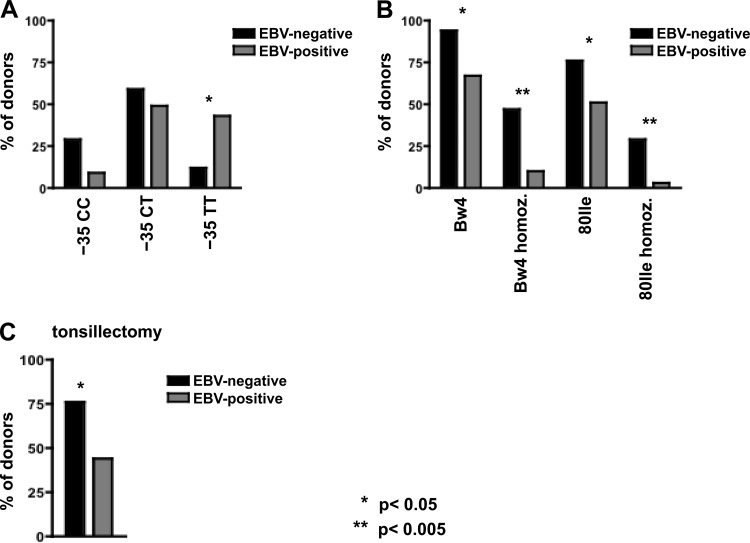
The HLA-C variant with TT at position −35, which is associated with lower HLA-C levels, was significantly underrepresented in the EBV-negative cohort (15/35 EBV-positive donors were homozygous for the −35 T allele, compared to only 2/17 EBV-negative donors [P = 0.03]) (Fig. 2A). In contrast, the frequency of HLA-Bw4 epitopes was significantly higher among the EBV-negative group, with 16/17 donors carrying at least one HLA-Bw4 epitope, versus 26/39 among the EBV-positive control cohort (P = 0.04) (Fig. 2B). Eight of 17 EBV-negative donors were homozygous for HLA-Bw4, compared to only 4/39 EBV-positive donors (P = 0.004), and individuals homozygous for HLA-Bw4 80Ile were significantly overrepresented among the EBV-negative cohort (P = 0.003) (Fig. 2B). In chronic HIV infection, the protective effect of HLA-Bw4 is thought to be mediated through interaction with KIR3DL1 and/or KIR3DS1 (19, 20). In our hands, the combined genotype of HLA-Bw4 homozygous and KIR3DL1 homozygous (KIR3DL1 homozygous equals the absence of KIR3DS1) was present significantly more often in the EBV-negative cohort (4/17 [24%]) than in the EBV-positive cohort (2/38 [5%]; P = 0.045). No significant association was found between HLA-Bw4 alleles and KIR3DS1 (data not shown).
Fig 2.
HLA I genotype and tonsillectomy in long-term EBV-protected individuals and EBV-positive controls. (A) Individuals (n = 52) were grouped according to −35 genotype into −35 CC, CT, and TT HLA allele variants. Within each group, EBV-negative individuals (n = 17 total, filled bars) were compared to EBV-positive individuals (n = 35 total, gray bars). A significant difference was found within the group of −35 T homozygous donors, with only 2/17 (12%) within the EBV-negative and 15/35 (43%) within the EBV-positive group. (B) HLA-Bw4-positive, HLA-Bw4-homozygous, HLA-Bw4 80Ile-positive, and HLA-Bw4 80Ile-homozygous donors were compared according to EBV status. In EBV-negative individuals, HLA-Bw4 and HLA-Bw4 80Ile alleles were overrepresented compared to their frequency in EBV-positive controls. (C) A significant difference was found in tonsillectomy rates between EBV-negative individuals (filled bars) and EBV-positive individuals (gray bars). *, P < 0.05; **, P < 0.005.
The oropharyngeal lymphatic tissue represents the entry site for EBV (1). Reduction of oropharyngeal lymphoid tissue, thereby removing substrate that can be infected by EBV, may affect an individual's risk to become infected. In addition to the genetic makeup, we therefore assessed how tonsillectomy, ethnicity, and socioeconomic status influence susceptibility to infection. No differences were found in ethnicity and socioeconomic status (data not shown). Intriguingly, 17/39 (44%) EBV-positive versus 13/17 (76%) EBV-negative individuals had a history of tonsillectomy (P = 0.023) (Fig. 2C). No significant difference in median age at tonsillectomy was found between the two groups (EBV negative, age 7 at tonsillectomy, versus EBV positive, age 9.5; P = 0.75). Of note, tonsillectomy and protective genetic variants in HLA-B alleles did not cocluster among long-term EBV-negative individuals (data not shown).
In summary, our preliminary study for the first time provides a genetic/anatomic signature capturing long-term protection from EBV. Genetic studies suggest a model whereby inhibition of NK cells is weaker by specific KIR-HLA combinations than by others, with weaker inhibition resulting in a more pronounced activation of NK cells and, therefore, better control of viral infections (21). However, more analyses of KIR/HLA combined genotypes are required to interpret the between-group differences in HLA-Bw4 and HLA-Bw4 80Ile-homozygous genotypes in the context of EBV infection observed here.
ACKNOWLEDGMENTS
We thank Hojjat Nozad Charoude, Department of Biomedicine, University Hospital Basel, for performing the HLA-C rs9264942 SNP PCRs. We thank Alexis Dumoulin, Department of Biomedicine, Institute for Medical Microbiology, University of Basel, for performing the BALF5 (DNA polymerase catalytic subunit) reverse transcription-PCRs.
The authors declare no competing financial interests that are relevant for the study.
B.D. is supported by a grant from the Swiss National Science Foundation (SNSF) (grant 323500-119221). O.G. is supported by a grant from the University of Basel. C.H. is supported by grants from the SNSF (grant 31003A_135677/1) and the Basel Cancer League (grant 03-2009).
Footnotes
Published ahead of print 27 March 2013
REFERENCES
- 1. Young LS, Rickinson AB. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757–768 [DOI] [PubMed] [Google Scholar]
- 2. Nemerow GR, Mold C, Schwend VK, Tollefson V, Cooper NR. 1987. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. J. Virol. 61:1416–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kimura H, Morita M, Yabuta Y, Kuzushima K, Kato K, Kojima S, Matsuyama T, Morishima T. 1999. Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay. J. Clin. Microbiol. 37:132–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gasser O, Bihl FK, Wolbers M, Loggi E, Steffen I, Hirsch HH, Gunthard HF, Walker BD, Brander C, Battegay M, Hess C. 2007. HIV patients developing primary CNS lymphoma lack EBV-specific CD4+ T cell function irrespective of absolute CD4+ T cell counts. PLoS Med. 4:e96 doi:10.1371/journal.pmed.0040096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Woodberry T, Suscovich TJ, Henry LM, Davis JK, Frahm N, Walker BD, Scadden DT, Wang F, Brander C. 2005. Differential targeting and shifts in the immunodominance of Epstein-Barr virus-specific CD8 and CD4 T cell responses during acute and persistent infection. J. Infect. Dis. 192:1513–1524 [DOI] [PubMed] [Google Scholar]
- 6. Biron CA, Byron KS, Sullivan JL. 1989. Severe herpesvirus infections in an adolescent without natural killer cells. N. Engl. J. Med. 320:1731–1735 [DOI] [PubMed] [Google Scholar]
- 7. Lotz M, Tsoukas CD, Fong S, Carson DA, Vaughan JH. 1985. Regulation of Epstein-Barr virus infection by recombinant interferons. Selected sensitivity to interferon-gamma. Eur. J. Immunol. 15:520–525 [DOI] [PubMed] [Google Scholar]
- 8. Strowig T, Brilot F, Arrey F, Bougras G, Thomas D, Muller WA, Munz C. 2008. Tonsilar NK cells restrict B cell transformation by the Epstein-Barr virus via IFN-gamma. PLoS Pathog. 4:e27 doi:10.1371/journal.ppat.0040027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Williams H, McAulay K, Macsween KF, Gallacher NJ, Higgins CD, Harrison N, Swerdlow AJ, Crawford DH. 2005. The immune response to primary EBV infection: a role for natural killer cells. Br. J. Haematol. 129:266–274 [DOI] [PubMed] [Google Scholar]
- 10. Moretta L, Moretta A. 2004. Unravelling natural killer cell function: triggering and inhibitory human NK receptors. EMBO J. 23:255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parham P. 2003. Innate immunity: the unsung heroes. Nature 423:20. [DOI] [PubMed] [Google Scholar]
- 12. Trinchieri G. 1989. Biology of natural killer cells. Adv. Immunol. 47:187–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jennes W, Verheyden S, Demanet C, Adje-Toure CA, Vuylsteke B, Nkengasong JN, Kestens L. 2006. Cutting edge: resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. J. Immunol. 177:6588–6592 [DOI] [PubMed] [Google Scholar]
- 14. Paximadis M, Minevich G, Winchester R, Schramm DB, Gray GE, Sherman GG, Coovadia AH, Kuhn L, Tiemessen CT. 2011. KIR-HLA and maternal-infant HIV-1 transmission in sub-Saharan Africa. PLoS One 6:e16541 doi:10.1371/journal.pone.0016541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boulet S, Kleyman M, Kim JY, Kamya P, Sharafi S, Simic N, Bruneau J, Routy JP, Tsoukas CM, Bernard NF. 2008. A combined genotype of KIR3DL1 high expressing alleles and HLA-B*57 is associated with a reduced risk of HIV infection. AIDS 22:1487–1491 [DOI] [PubMed] [Google Scholar]
- 16. Boulet S, Sharafi S, Simic N, Bruneau J, Routy JP, Tsoukas CM, Bernard NF. 2008. Increased proportion of KIR3DS1 homozygotes in HIV-exposed uninfected individuals. AIDS 22:595–599 [DOI] [PubMed] [Google Scholar]
- 17. Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O'Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, Carrington M. 2007. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 39:733–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thomas R, Apps R, Qi Y, Gao X, Male V, O'HUigin C, O'Connor G, Ge D, Fellay J, Martin JN, Margolick J, Goedert JJ, Buchbinder S, Kirk GD, Martin MP, Telenti A, Deeks SG, Walker BD, Goldstein D, McVicar DW, Moffett A, Carrington M. 2009. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat. Genet. 41:1290–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carrington M, Martin MP, van Bergen J. 2008. KIR-HLA intercourse in HIV disease. Trends Microbiol. 16:620–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. 2006. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J. Exp. Med. 203:633–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parham P. 2005. MHC class I molecules and KIRs in human history, health and survival. Nat. Rev. Immunol. 5:201–214 [DOI] [PubMed] [Google Scholar]