Abstract
Potato mop-top virus (PMTV) produces a defective RNA (D RNA) encompassing the 5′-terminal 479 nucleotides (nt) and 3′-terminal 372 nt of RNA-TGB (where TGB is triple gene block). The mechanism that controls D RNA biogenesis and the role of D RNA in virus accumulation was investigated by introducing deletions, insertions, and point mutations into the sequences of the open reading frames (ORFs) of TGB1 and the 8-kilodalton (8K) protein that were identified as required for efficient production of the D RNA. Transient expression of RNA-TGB in the absence of RNA-Rep (which encodes the replicase) did not result in accumulation of D RNA, indicating that its production is dependent on PMTV replication. The D RNA could be eliminated by disrupting a predicted minus-strand stem-loop structure comprising complementary sequences of the 5′ TGB1 ORF and the 3′ 8K ORF, suggesting intramolecular template switching during positive-strand synthesis as a mechanism for the D RNA biogenesis. Virus accumulation was reduced when the 8K ORF was disrupted but D RNA was produced. Conversely, the virus accumulated at higher titers when the 8K ORF was intact and D RNA production was blocked. These data demonstrate that the D RNA interferes with virus infection and therefore should be referred to as a defective interfering RNA (DI RNA). The 8K protein was shown to be a weak silencing suppressor. This study provides an example of the interplay between a pathogen and its molecular parasite where virus accumulation was differentially regulated by the 8K protein and DI RNA, indicating that they play antagonistic roles and suggesting a mechanism by which the virus can attenuate replication, decreasing viral load and thereby enhancing its efficiency as a parasite.
INTRODUCTION
The small genome size of RNA viruses means that most viral proteins are multifunctional and that viruses adopt several different strategies for gene expression including overlapping open reading frames (ORFs). In the course of infection many RNA viruses produce defective RNAs (D RNAs), which are deleted versions of the viral genome (see references 1 and 2 for a review). Because they lack essential genetic information, in plants D RNAs are totally dependent on the infectious helper virus for replication, encapsidation, and cell-to-cell and long-distance movement. Interference occurs when D RNA reduces symptom production by the parent virus or/and reduces multiplication of the helper virus; hence, the term defective interfering RNA (DI RNA) is used.
Production of D RNA molecules during replication cycles seems to be a common occurrence in many viruses. D and DI RNAs of plant viruses, including DNA and negative- and positive-stranded RNA viruses, have been described and characterized previously (1, 2). In some instances, for example, the criniviruses, DI RNAs consist of a mosaic of the multipartite parental viral genome (3). Multiple-deletion DI RNAs are typical for several members of genus Tombusvirus, and these DI RNAs have been extensively studied (4, 5). On the other hand, single-deletion D and DI RNAs have been reported in Potexvirus and Tobravirus genera as well as in several soilborne viruses of the genera Furo-, Peclu-, and Benyvirus (1).
Although the DI RNA could represent up to 60% of virus-specific RNA in the infected plants, as in case of tombusviruses, the percentage of encapsidated DI RNAs could be as low as 3 to 4%, and most DI RNAs are not transmitted by vectors (1, 2, 6). Moreover, while most DI RNAs can efficiently accumulate in inoculated tissue, they do not always move systemically, as exemplified by cucumber mosaic virus (CMV) D RNA, which moves long distances in tobacco species but not in tomato, zucchini squash, or muskmelon (1, 7, 8).
Three major mechanisms of interference by DI RNAs have been recognized (reviewed in references 2 and 5). These mechanisms are the following: (i) competition for viral and host resources, which impairs virus replication and attenuates the symptoms; (ii) DI RNA-triggered gene-silencing response; (iii) modulation of the functions of viral factors. In the first two cases, DI RNAs either compete with parental RNA for the virus replication machinery and are more efficient in recruiting the viral replicase, or they enhance posttranscriptional gene silencing against their parental virus; some DI RNAs are effective inducers but poor targets of RNA silencing (1, 2, 6). The third mechanism, modulation of function, for example, by interfering with viral gene products, is exemplified by the tomato bushy stunt virus (TBSV)/DI RNA disease complex. In this system, decreased accumulation of the TBSV p19 RNA silencing suppressor (RSS) and its subgenomic RNA was observed upon coinfection with TBSV and DI RNA compared to a less dramatic effect on the accumulation of RNA replication proteins (9, 10). Moreover, in the presence of DI RNAs, the generation of the virus-specific small interfering RNAs (siRNAs) was enhanced, leading to accumulation of larger amounts of free siRNAs that exceeded the capacity of the available p19 suppressor protein to bind to them. The excess siRNA enhanced gene silencing in newly invaded tissues, and virus infection was attenuated (9). In this way, the DI RNA interfered with the function of the p19 RSS.
The genus Pomovirus in the family Virgaviridae contains a number of important plant pathogens. Pomoviruses are widely distributed in Europe, the Far East, and the Americas and are transmitted by soilborne fungal vectors. Potato mop-top virus (PMTV) is of particular importance because it causes potato spraing disease, inducing brown arcs and circles in potato tubers, which make the tubers unmarketable. Most of the currently grown cultivars are susceptible to PMTV, but the virus accumulates at low, often hardly detectable, levels. While the reason for this is unknown, a D RNA has been shown to associate with PMTV infections. However, the impact of D RNA on virus accumulation has not been investigated.
PMTV has a genome that comprises three separate segments of single-stranded, positive-sense RNA. The genomic components have been recently renamed to RNA-Rep (former RNA 1), RNA-CP (where CP is capsid protein) (former RNA 2 or RNA 3 depending on the virus isolate) and RNA-TGB (where TGB is triple gene block) (former RNA 2 or RNA 3 depending on the virus isolate) to avoid further confusion with RNA segment nomenclature and to emphasize the function of each RNA component (11). Thus, RNA-TGB encodes a triple gene block of movement proteins and a small, cysteine-rich, 8-kilodalton (8K) zinc finger protein. A reverse genetics system is available (12), and full-length infectious cDNA clones have become a valuable tool to study cell-to-cell and long-distance movement of the virus (11–15). These studies revealed that the PMTV genome could tolerate quite extensive deletions in ORF1 and ORF4 of RNA-TGB, which encode the TGB1 movement protein and the 8K protein, respectively. A part of the N-terminal domain of TGB1 and the 8K protein appeared to be dispensable for the systemic movement of the virus in Nicotiana benthamiana (12, 14). Moreover, RNA-Rep and RNA-TGB can move long distances in the absence of RNA-CP (12, 13). So far, the only PMTV D RNA reported derives from RNA-TGB (16). This D RNA is formed from its cognate RNA and contains a large central deletion that encompasses 72% of RNA-TGB but retains the terminal sequences needed for replication (Fig. 1A). Whether the D RNA interferes with virus accumulation and/or symptom induction is not known, and that is why it was initially designated D RNA, not DI RNA.
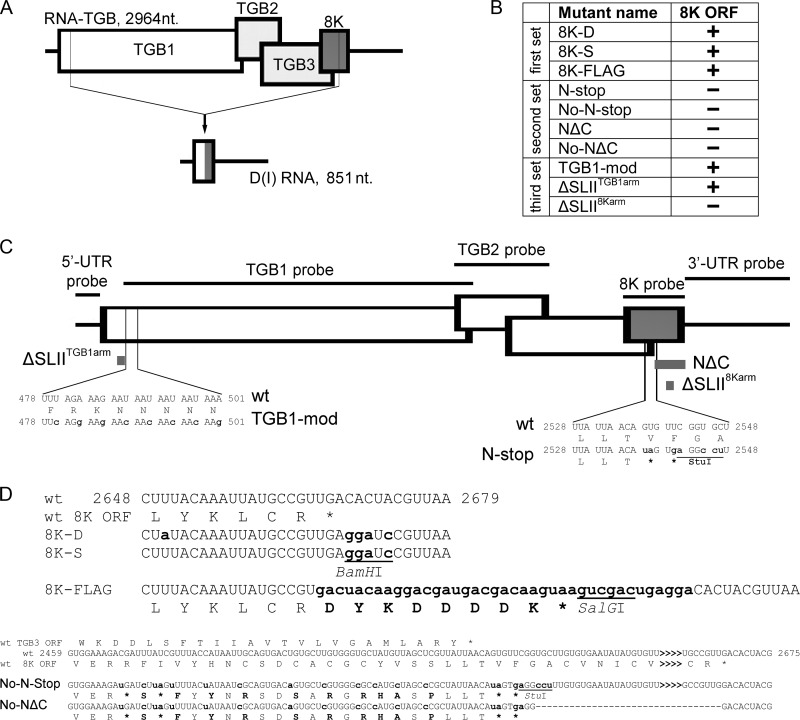
Fig 1.
Schematic representation of the RNA-TGB mutants engineered in this study or obtained by Savenkov et al. (12) and structure of D RNA. (A) Schematic of the D RNA formation through a single internal deletion in the parental RNA. Structure of the D RNA relative to the full-length RNA-TGB. The D RNA is composed of the fused ends of the viral RNA-TGB as described in the text. The length of the DI RNA (851 nt) is indicated. (B) Summary of the three sets of RNA-TGB mutants used in this study. (C) Graphical summary of RNA-TGB mutants. Small gray rectangles denote deletions engineered into infectious cDNA of RNA-TGB. Positions of the probes used for Northern blotting are indicated on the diagram. (D) Characteristic features of recombinants 8K-D, 8K-S, 8K-FLAG, No-N-Stop, and No-NΔC mutants compared to wt sequence. The sequences at the 3′ end of the 8K ORF (bottom sequence) and wt (top sequence) are shown. Translation of the ORFs is shown below the top and bottom sequences. Translation of the overlapping part of the TGB3 ORF is shown above the wt sequence. The mutated or inserted nucleotide residues in relation to the RNA-TGB wt sequence are shown in small script and in bold. The dashed line indicates the deletion of the sequence encompassing the 3′-proximal part of the 8K ORF, which does not overlap the TGB3 ORF. The sequence that is not shown is indicated with greater-than (>) symbols. Numbering refers to nucleotide positions in full-length RNA-TGB. Stop codons are indicated with asterisks. The restrictions sites used to facilitate cloning are underlined and refer to the cDNA (not RNA).
The aim of this study was to investigate the role of the D RNA in virus infection and gain some understanding about its biogenesis. We show that PMTV D RNA is a genuine DI RNA and interferes with virus accumulation and symptom induction. The 3′-proximal part of the 8K ORF was found to be essential for DI RNA production. In this work we report that the DI RNA and the 8K protein act antagonistically on virus accumulation: while DI RNA inhibits virus accumulation, the 8K protein appeared to enhance it.
MATERIALS AND METHODS
Construction of plasmids.
Standard recombinant DNA procedures were performed by use of a combination of PCR, site-directed mutagenesis, and swapping of restriction fragments. The plasmid pPMTV-3 (12) bearing a full-length cDNA of RNA-TGB driven by a T7 promoter was used to generate PMTV mutants and constructs described in this study. 8K-D, 8K-S, NΔC, N-stop, No-N-stop, and 8K-FLAG were described previously (12, 17). In brief, to obtain 8K-D and 8K-S, the sequence immediately downstream of the 8K ORF stop codon (2607CACTAC2612 referred to as the full-length infectious cDNA clone of PMTV) was converted into a BamHI restriction site (GGATCC on the cDNA) by introducing four point mutations (mutated residues are in boldface). Thus, compared to the wild-type (wt) protein, 8K-S contains the BamHI restriction site and so differs by four point mutations immediately after the 8K ORF. 8K-FLAG had a 39-nt FLAG epitope sequence cloned at the 3′ terminus of the 8K ORF for expression of an in-frame fusion protein (17). The mutant, referred to as NΔC, has a 134-nt deletion (nt 2536 to 2667) of the 3′-proximal part of the 8K ORF, which does not overlap the TGB3 ORF. N-stop contains seven point mutations, which introduce two stop codons in the middle of the 8K ORF and an StuI restriction site to facilitate cloning. No-NΔC is a derivative of NΔC, in which 13 point mutations in addition to a 3′-proximal deletion of NΔC were introduced into the 5′-proximal part of the 8K cistron (nt 2462 to 2526) to block translation from the 8K ORF but not to change the sequence of the protein translated from the TGB3 ORF. No-N-stop is a derivative of N-stop, in which 20 point mutations (13 in addition to 7 of the N-stop) were introduced (nt 2462 to 2547) to block translation but to preserve the nucleotide sequence of the 3′-proximal part of the 8K ORF.
The sequences of the oligonucleotides will be provided upon request. To obtain TGB1-mod, a fragment of the TGB1 gene was amplified using primers P1-modif-DI-pos and P1-Xba-neg. The resulting amplification product was digested with MluI and KpnI and ligated into MluI- and KpnI-digested pPMTV-3. To engineer ΔSLIITGB1arm (where SLII is stem-loop two and TGB1 arm is nt 450 to 479) two fragments of RNA-TGB were amplified using two pairs of primers, Del451-471fw/P1Xba-neg and Del451-471rev/T7RNA-TGB. The amplification products were reamplified by overlap PCR using primers P1-Xba-neg and T7RNA-TGB. The resulting PCR product was digested with XbaI/KpnI and ligated into similarly digested pPMTV-3. To obtain ΔSLII8Karm (where 8K arm is nt 2593 to 2622), two fragments of pPMTV-3 were amplified using two pairs of primers, Del2593-2623rev/Del249Nco-minus-fw and Del2593-2623fw/M13fw. The amplification products were reamplified by overlap PCR using primers Del249Nco-minus-fw/M13fw. The resulting amplification product was digested with SpeI/KpnI and ligated into SpeI/KpnI-digested pPMTV-3. To engineer No-CSLIII (where CSLIII is a complementary stem-loop structure), a fragment of RNA-TGB was amplified using the primers PMTV-CSL3-Mlu-F and TGB-Kpn-rev. The resulting PCR product was digested with MluI/KpnI and ligated into similarly digested pPMTV-3. To obtain CSLIII-restore, a fragment of RNA-TGB was amplified using the primers CSL3-COMPENS-F and PMTV-Spe-rev. The obtained PCR product was reamplified by nested PCR with the primers PMTV-Stu-F and PMTV-Spe-rev. The amplification product was digested with StuI/SpeI and ligated into similarly digested pPMTV-3. To engineer No-SLII, a fragment of RNA-TGB was amplified using the primers PMTV-point-SL2-Mlu-R1 and T7RNA-TGB. The resulting PCR product was digested with MluI/SacI and ligated into similarly digested pPMTV-3. To engineer SLII-restore, two fragments of RNA-TGB were amplified using two pairs of primers, PMTV-comp-SLII-AvrII-R1/TGB-Kpn-F and TGB-Kpn-rev/T7RNA-TGB. The resulting PCR products were digested with AvrII/KpnI and MluI/KpnI, respectively, and ligated into pPMTV-3 digested with AvrII/MluI.
For expressing constructs under the control of 35S promoter, the 8K and TGB1 genes were cloned to the destination vectors pGWB14 and pGWB15 (18) using Gateway technology (Invitrogen). All the constructs were verified by sequencing.
Plant inoculations and analyses.
Inoculation of N. benthamiana plants with recombinant viruses and enzyme-linked immunosorbent assays (ELISAs) were carried out as described previously (12, 19). To detect PMTV virions, anti-CP monoclonal preparations were used to coat microtiter plates for ELISA, and the anti-CP monoclonal alkaline phosphatase conjugate was used for detection essentially as described previously (12, 13, 19).
RNA analysis.
Total RNA was extracted from leaf discs using a Spectrum Plant Total RNA kit (Sigma) according to the manufacturer's recommendations. For Northern blot analysis, 5 μg of total RNA preparation was separated by formaldehyde gel electrophoresis, transferred to a nylon membrane (Hybond-N; Amersham Biosciences AB), cross-linked with UV light, prehybridized, hybridized at 70°C, and washed in hybridization tubes. Antisense [α-32P]UTP-labeled gene-specific RNA probes were synthesized with T7 RNA polymerase (Promega) from XbaI-digested PMTV gene constructs according to the manufacturer's protocol. After a final washing, the blots were wrapped in polyethylene plastic and exposed to an exposure cassette (Molecular Dynamics) for 1 to 48 h. The cassette was then scanned with a Molecular Imager FX and processed with Quantity One software (Bio-Rad).
Agroinfiltration.
Agrobacterium tumefaciens was incubated overnight at 28°C in the presence of carbenicillin, rifampin, and kanamycin (or spectinomycin, depending on the construct) in LB medium supplemented with acetosyringone (20 μM final concentration) and morpholineethanesulfonic acid (MES; 10 mM final concentration). When the cultures reached an optical density at 600 nm (OD600) of ∼0.5, cells were pelleted by centrifugation at 3,500 × g for 5 min, resuspended in an infiltration buffer (10 mM MES, 10 mM MgCl2, and 150 μM acetosyringone), and incubated for 2 h at room temperature. Five- to seven-week-old N. benthamiana plants were infiltrated using a syringe without a needle. A. tumefaciens strains delivering a construct of interest were adjusted to an OD600 of 0.6 each. Turnip crinkle virus (TCV) expressing a green fluorescent protein (TCV-sGFP) and TCVΔ92-sGFP were used in complementation assays. The complementation assays and PZP-TCV-sGFP coinfiltration assays were performed as described previously (20, 21). Both TCV-sGFP and TCVΔ92-sGFP were linearized with XbaI prior to in vitro transcription. The obtained RNA transcripts were rub-inoculated on N. benthamiana leaves preinfiltrated with A. tumefaciens strains carrying 8K, TGB1, helper component-proteinase (HC-Pro) constructs, or an empty plasmid control 3 days before the inoculation. Fluorometric analysis of samples was performed as described in Zamyatnin et al. (22).
Reverse transcription-PCR (RT-PCR).
Total RNA was extracted from leaves using a Spectrum Plant Total RNA kit (Sigma) and On-Column DNase I Digest Set (Sigma) according to the manufacturer's instructions. One-microgram aliquots of total RNA samples were used for oligo(dT) and PMTV 3′-untranslated region (UTR)-primed cDNA synthesis with a RevertAid H Minus Fist Strand cDNA Synthesis Kit (MBI Fermentas). Obtained cDNAs were used as templates for a PCR with DreamTaq DNA Polymerase (MBI Fermentas) and PMTV-specific primers.
RESULTS
Experimental system designed to assess the significance of D RNA and the 8K protein in virus accumulation.
Previous studies demonstrated that a cysteine-rich protein encoded by the PMTV 8K ORF is not required for long-distance movement of the virus (12), but the role of the 8K protein in virus accumulation has not been addressed. In related hordeiviruses, a cysteine-rich protein called γb is a multifunctional protein which is needed for efficient virus accumulation, partially through suppression of the host antiviral RNA interference (RNAi) (19, 23). As previously stated, PMTV RNA-TGB that encodes 8K gives rise to a D RNA (16), and since this D RNA contains the 3′-proximal part of the 8K ORF, we reasoned that the sequence of this ORF might be involved in D RNA production. Furthermore, because of the overlap of the 8K ORF with the sequence that presumably controls D RNA production, it might be difficult to address the significance of the 8K protein and D RNA in the virus life cycle by straightforward mutagenesis. In this study, we wanted to separate the effect of D RNA on virus accumulation from the possible impact of 8K gene expression in the same process.
To address these questions, we developed an experimental system by constructing several deletion, insertion, and point-mutation mutants of PMTV in the 8K ORF and its downstream sequence (Fig. 1). The detailed description of the mutants is given in the Materials and Methods section and, schematically, in Fig. 1. The first set of mutants was designed to preserve the integrity of the 8K ORF (Fig. 1B to D). We used two PMTV isolates (Danish and Swedish) that differed in sequences of the 8K ORFs in the region thought to be important in D RNA formation (16). The first mutant, referred to as 8K-D, was constructed by replacing the 8K ORF and part of the TGB3 ORF (nt 1997 to 2606) of an infectious cDNA clone previously constructed using a Swedish isolate with that of a Danish isolate in order to change the nucleotide sequence of the 8K ORF (resulting in two nonsynonymous and one synonymous nucleotide substitutions) (Fig. 1D) and to compare the effect of the 8K protein from two different isolates (17). The second mutant, referred to as 8K-S, was constructed by replacing the 8K ORF and part of TGB3 ORF (nt 1997 to 2606) of 8K-D with that of the cognate sequence of the Swedish isolate (to introduce a BamHI restriction site). Thus, compared to the wt (Swedish isolate), 8K-S differs by four nucleotide mutations immediately after the 8K ORF (Fig. 1D). The third mutant, referred to as 8K-FLAG, had a FLAG epitope sequence cloned at the 3′ terminus of the 8K ORF for expression of an in-frame fusion protein (12) (Fig. 1D). The next set of mutants was designed to disrupt the 8K ORF and prevent 8K gene expression (Fig. 1B). Figures 1C and D show schematics of these mutants, referred to as N-stop, No-N-stop, NΔC, and No-NΔC. No-N-stop possesses 20 point mutations in the 5′-proximal part of 8K ORF, which overlaps the TGB3 ORF. N-stop has seven point mutations to create stop codons in the middle of the 8K ORF immediately after the TGB3 ORF stop codon. Consequently, N-stop, No-N-stop, NΔC, and No-NΔC will knock out the 8K ORF, while in 8K-D, 8K-S, and 8K-FLAG it remains intact (Fig. 1B). Next, we examined whether these mutants were competent to produce D RNA.
Mapping of the sequences involved in D RNA production.
The emergence of D RNA molecules from RNA-TGB was monitored by Northern blot hybridization using specific probes (Fig. 1C). In these and subsequent experiments, RNA-TGB transcripts from each of the mutants described above were prepared and inoculated onto N. benthamiana leaves along with transcripts of RNA-Rep and RNA-CP, and the type of inoculum used in each case was referred to by the name of the mutant. RNA extracted from upper noninoculated leaves of N. benthamiana plants systemically infected with PMTV and mutants for the 8K ORF were subjected to Northern blot analysis. Five identical Northern blots were prepared and hybridized independently with one of five probes (5′ UTR, TGB1, TGB2, 8K, and 3′ UTR) (Fig. 1C and 2). The faster-migrating molecule (presumably D RNA) could be detected by hybridization with 5′-UTR-, 8K-, and 3′-UTR-specific probes but not the TGB1 or TGB2 probes (Fig. 2, lane 2), suggesting that most of the TGB is deleted from this RNA. Low-molecular-weight cDNA fragments corresponding to the putative D RNA were cloned from five independent infections, and nine randomly selected clones for each infection were sequenced. The sequences confirmed that these faster-migrating RNA species were the authentic D RNA (Fig. 1A and data not shown). Moreover, its composition and junction point corresponded to the previously characterized D RNA from the Scottish T isolate (16) and included the complete 5′ UTR, a short sequence downstream of the TGB1 start codon, the 3′-proximal part of the 8K ORF, and the complete 3′ UTR. The joined ends of the RNA-TGB in the D RNA create a single continuous 186-nt ORF, with 110 nt coming from the TGB1 ORF and 76 nt coming from the 8K ORF, and the overall length of the D RNA is 851 nt (Fig. 1A).
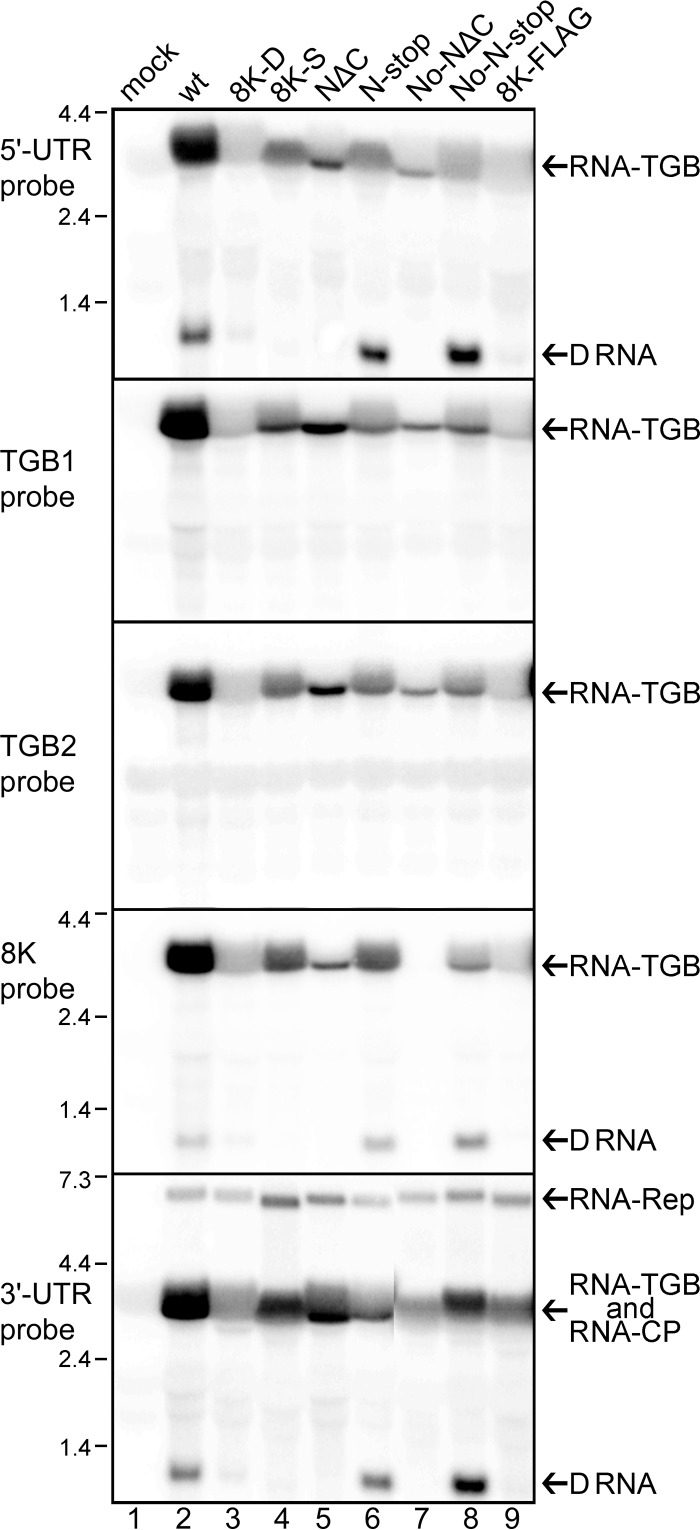
Fig 2.
Identification and location of RNA-TGB sequences within D RNA. Northern blot analyses depicting the accumulation of D RNA for N-stop, No-N-stop, and wt and the absence of D RNA accumulation for NΔC and No-NΔC. Note that 8K-D, 8K-S, and 8K-FLAG do not produce detectable D RNA. Five 32P-labeled RNA probes were used (shown schematically in Fig. 1). The identity of each probe and position of molecular size markers are given on the left. Note that the 8K probe does not detect No-NΔC because it completely lacks the 8K ORF.
Surprisingly, D RNA was detected in only two 8K knockout mutants (No-N-stop and N-stop) out of the seven constructs analyzed (Fig. 2). As described above, these two constructs possess either point mutations in the 5′-proximal part of the 8K ORF or point mutations in the middle of the 8K ORF immediately after the TGB3 ORF stop codon. These data suggest that the region of overlap between the TGB3 and 8K ORFs is not involved in D RNA formation. On the other hand, the mutants which failed to generate detectable amounts of D RNA (i.e., NΔC, No-NΔC, 8K-D, 8K-S, and 8K-FLAG) had either deletions in the 3′ end of the 8K ORF or point mutations or an insertion downstream of the 8K ORF stop codon, suggesting that this domain plays a role in D RNA production.
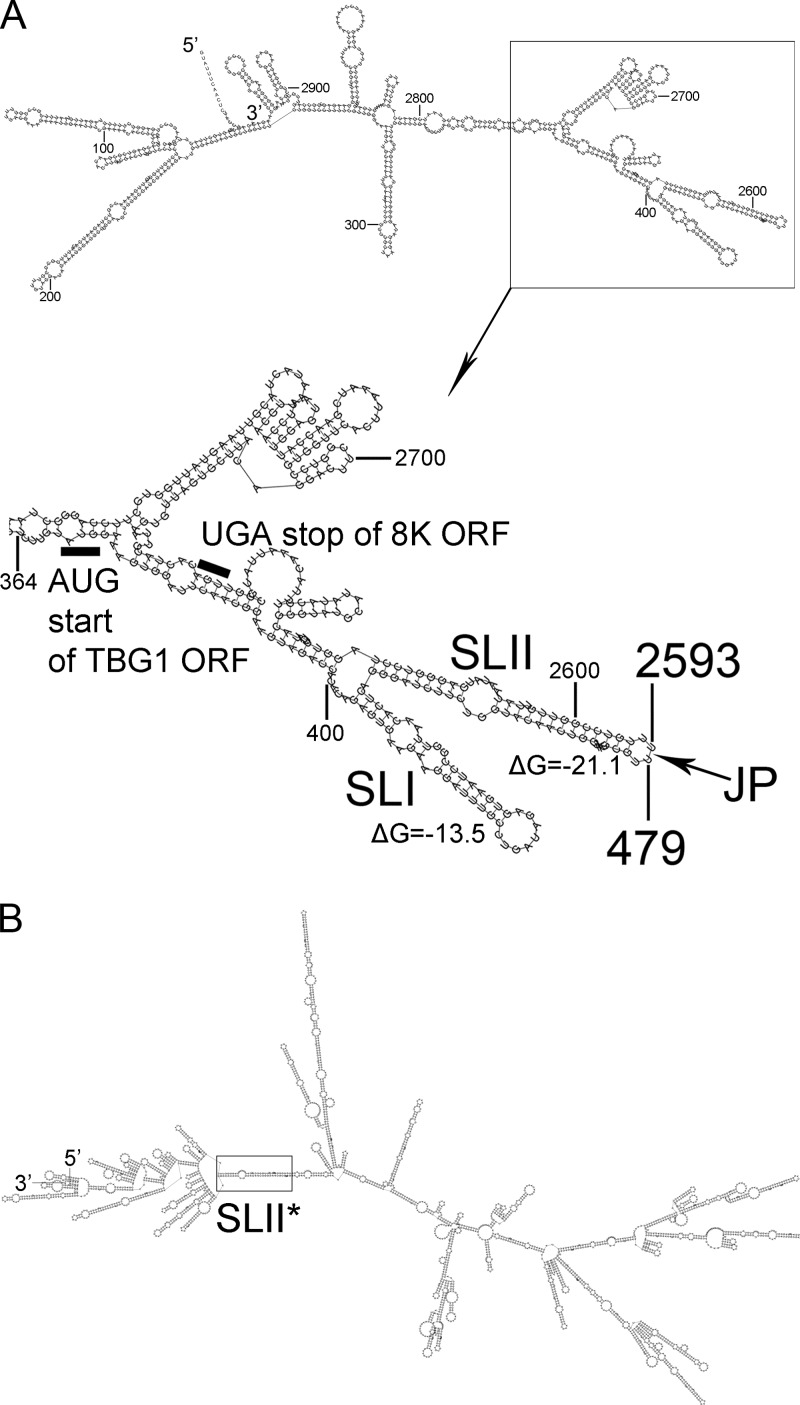
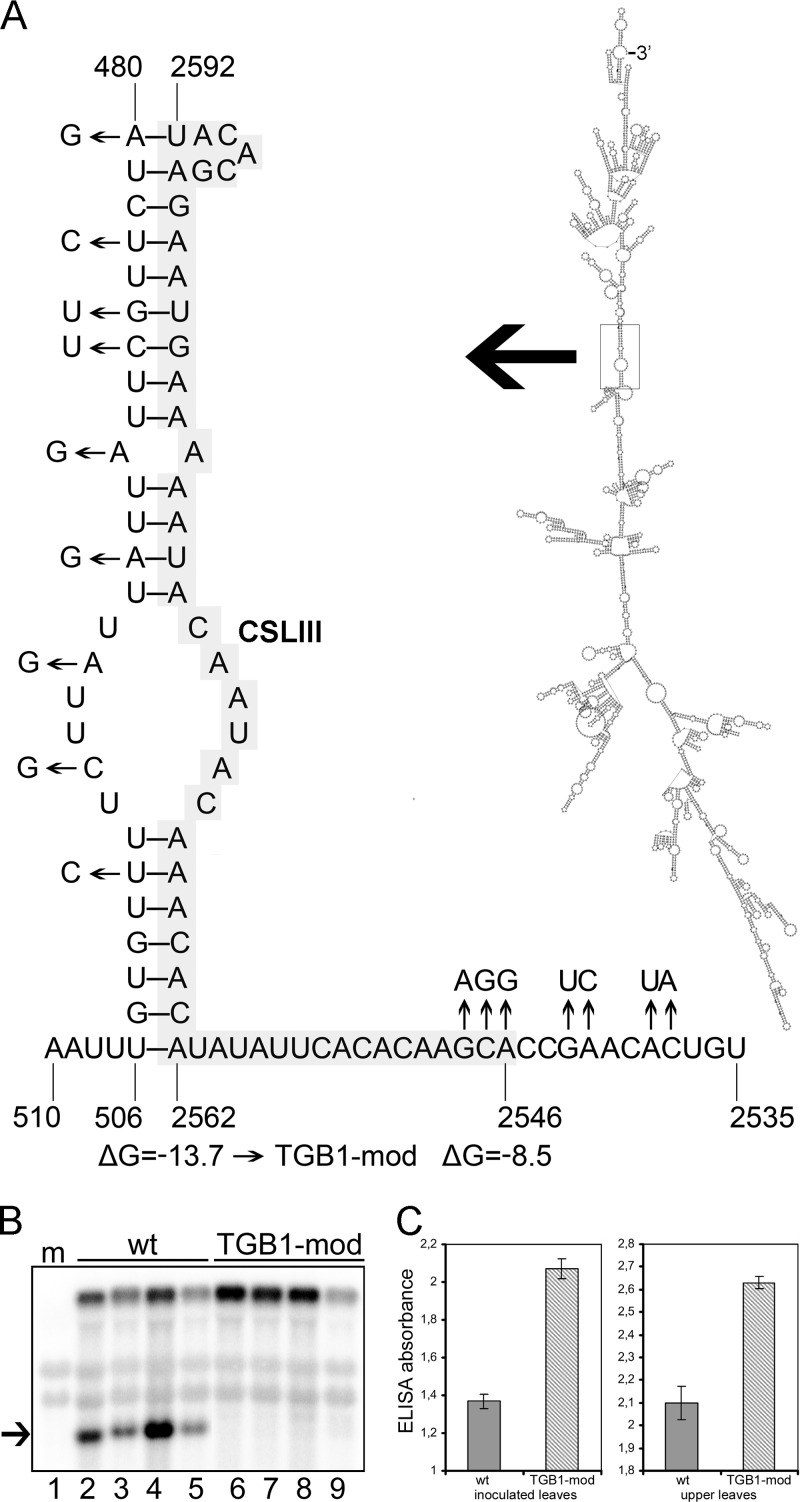
Indeed, the predicted secondary structure of D RNA supports our mutagenesis data (Fig. 3A). The figure shows two RNAfold-predicted, stem-loops, named SLI (nt 404 to 448) and SLII (TGB1 arm: nt 450 to 479; 8K arm: nt 2593 to 2622). Moreover, the structure similar to SLII designated SLII* (Fig. 3B), which spans the same sequence as SLII and has the same pattern of pairing between TGB1 and 8K arms, was predicted when the RNAfold algorithm was applied to the plus strand of the full-length RNA-TGB. In the mutants NΔC and No-NΔC, which did not give rise to D RNA, the cognate sequence that forms the SLII stem-loop was deleted or disrupted in the parental RNA. Point mutations or insertions downstream of the 8K ORF are predicted to destabilize the extensive stem-loop secondary structure (data not shown), resulting in suppression of D RNA production (Fig. 2, lanes 3, 4, and 9). Notably, the junction point of D RNA is located on the top of the SLII loop (Fig. 3A, JP), suggesting that formation of this secondary structure might be important during D RNA synthesis on the minus strand of RNA-TGB. To further explore this possibility, the RNAfold program was applied to the deleted part of the minus strand and to the full-length RNA-TGB. The predicted complementary stem-loop structure, named CSLIII, involved base pairing of the arm derived from the TGB1 gene(nt 480 to 506) with that derived from the 8K gene (nt 2562 to 2592) (Fig. 4A). Interestingly, two of the RNA-TGB mutants defective in D RNA production, namely, NΔC and No-NΔC, besides having a deleted SLII arm (Fig. 3) also had an extensive deletion which spans the complete CSLIII 8K-derived arm (Fig. 4A, gray shading). In order to address the direct role of CSLIII in D RNA production and validate our predictions, we introduced eight point mutations into nt 480 to nt 501 of the TGB1 ORF without changing the amino acid sequence of the TGB1 protein, and the obtained mutant was named TGB1-mod (Fig. 1C). Northern blot analysis of total RNA preparations recovered from the samples of leaves systemically infected with TGB-mod revealed the absence of D RNA accumulation in TGB1-mod-infected plants (Fig. 4B) and enhanced virus accumulation (Fig. 4C) compared to the plants infected with wt virus. Thus, we concluded that the point mutations in the sequence at the beginning of the TGB1 gene, adjacent to the sequence incorporated into D RNA, also blocks D RNA production. These data are in agreement with our computational predictions of CSLIII in the minus strand and previous predictions of template switching at nucleotide positions U480 and A2592 (16).
Fig 3.
Stem-loops in the D RNA and in the plus strand of full-length RNA-TGB as predicted by RNAfold. (A) Predicted secondary structure of the D RNA showing elements referred to in the text. The AUG and UGA codons are indicated by black bars. Numbering refers to nucleotide positions in the plus strand of full-length RNA-TGB. JP, junction point. (B) Predicted secondary structure of the plus strand of full-length RNA-TGB.
Fig 4.
Effects of CSLIII mutations on accumulation of D RNA and the virus progeny. (A) Predicted stem-loop structure in the minus strand of RNA-TGB. The predicted secondary structure of the minus strand of full-length RNA-TGB is shown on the right. Numbering refers to nucleotide positions in the plus strand of full-length RNA-TGB. Mutations used to test CSLIII requirements for D RNA production are shown on the left for TGB1-mod (nine point mutations), and a deletion and point mutations in NΔC and No-NΔC are highlighted in gray. The free energies refer to the stem-loops in the wt CSLIII and in the TGB1-mod. (B) Northern analysis to measure the accumulation of D RNA in total RNA preparations recovered from the leaves of N. benthamiana inoculated with wt and TGB1-mod. The 32P-labeled RNA probe specific for the 5′ UTR was used, and D RNA is indicated by an arrow. Lane m, mock. (C) Detection of PMTV by ELISA as indicated by absorbance values at 405 nm. Plant extracts were prepared from upper leaves at 14 dpi. The average absorbance of a healthy plant extract was equal to 0.18. ELISAs were conducted twice (n = 12).
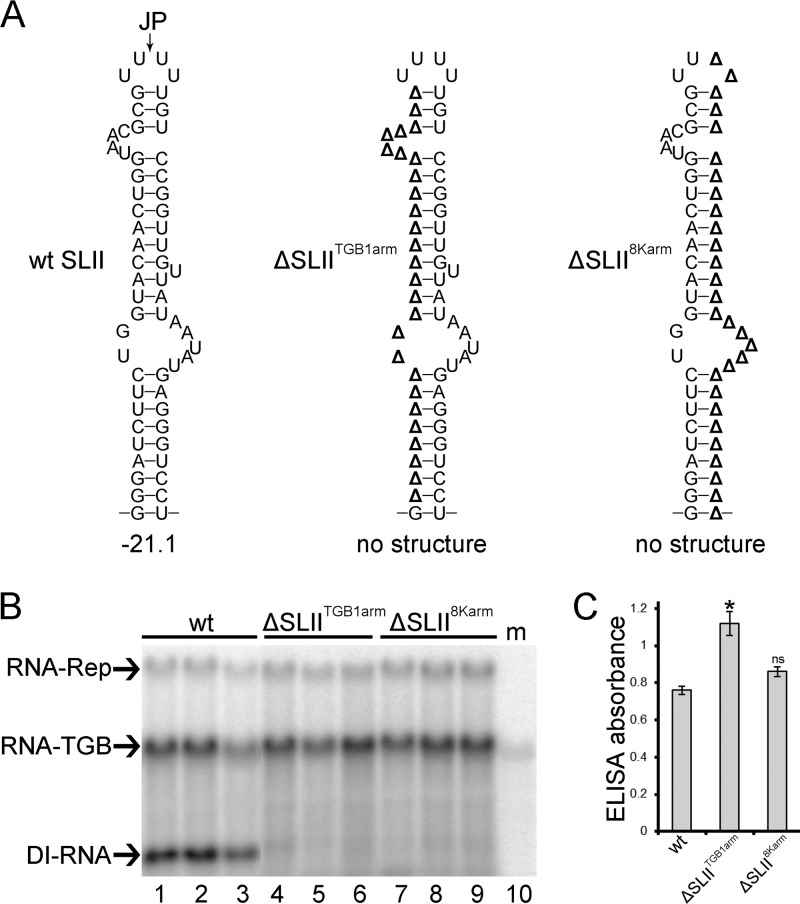
To specifically address the role of SLII in D RNA production, two mutants, ΔSLIITGB1arm and ΔSLII8Karm, were derived from RNA-TGB cDNA by mutagenesis. A 27-nt in-frame deletion was engineered into the TGB1 ORF (nt 451 to 477) to create the mutant ΔSLIITGB1arm, in which the TGB1-derived arm of SLII was removed (Fig. 1C and 5A). In ΔSLII8Karm, a 31-nt deletion was engineered into the 8K ORF (nt 2593 to 2622) to remove the 8K-derived arm of SLII (Fig. 1C and 5A). Consequently, both ΔSLIITGB1arm and ΔSLII8Karm were expected to be deficient in D RNA production; additionally ΔSLII8Karm would be a knockout for the 8K gene. Subsequently in vitro synthesized ΔSLIITGB1arm and ΔSLII8Karm transcripts were coinoculated with RNA-Rep and RNA-CP transcripts. Both inocula gave rise to viruses that were fully competent in cell-to-cell and long-distance movement since PMTV was detected in upper noninoculated leaves (Fig. 5B and C). These data are in agreement with previous observations that 5′-proximal deletions as long as 132 nt in the TGB1 cistron do not exert any significant effect on the cell-to-cell and long-distance movement of the virus in N. benthamiana (24). Northern blot analysis of RNA isolated from the samples of inoculated leaves and leaves systemically infected with either ΔSLIITGB1arm or ΔSLII8Karm revealed the absence of D RNA accumulation compared to the plants infected with wt virus (Fig. 5B). These results, therefore, indicate that SLII is required for D RNA production.
Fig 5.
Stem-loop II (SLII)-deficient mutants do not produce D RNA. (A) The RNAfold-predicted structures in the positive strand of D RNA and their calculated free energies in kcal/mole are shown. Deleted bases in ΔSLIITGB1arm and ΔSLII8Karm are represented by a delta (Δ). (B) Northern blot analysis to detect the DI in total RNA preparations recovered from the leaves of N. benthamiana inoculated with wt, ΔSLIITGB1arm, and ΔSLII8Karm inocula. The 32P-labeled RNA probe specific for the 3′ UTR was used. (C) Accumulation of PMTV was measured by ELISA as indicated by absorbance values at 405 nm. Plant extracts were prepared from upper leaves at 14 dpi. The average absorbance of a healthy plant extract was equal to 0.15. ELISAs were conducted twice (n = 8).
D RNA interferes with virus accumulation.
The significance of D RNA in the PMTV replication cycle is not known; however, a role for D RNA in symptom attenuation and virus accumulation/replication was proposed (16). Indeed, we found in this study that symptoms associated with inocula containing either RNA-Rep plus RNA-CP and TGB-mod (collectively referred as TGB-mod) or RNA-Rep plus RNA-CP and ΔSLIITGB1arm (collectively referred as ΔSLIITGB1arm) appeared 1 to 2 days earlier than with wt virus inoculation (data not shown). Moreover, quantitative analysis of progeny virus accumulation revealed that the ELISA absorbance values of TGB-mod and ΔSLIITGB1arm progeny were approximately 20 to 50% higher in upper leaves than those of the wt virus (Fig. 4C and 5C). These data demonstrate that the PMTV mutants with an unaffected 8K ORF, but which were completely defective in D RNA production, accumulated at higher levels than the wt virus in both inoculated and upper leaves. Collectively, these data confirmed that PMTV D RNA is a genuine DI RNA and therefore is referred to here as PMTV DI RNA.
8K protein and DI RNA act antagonistically on virus accumulation.
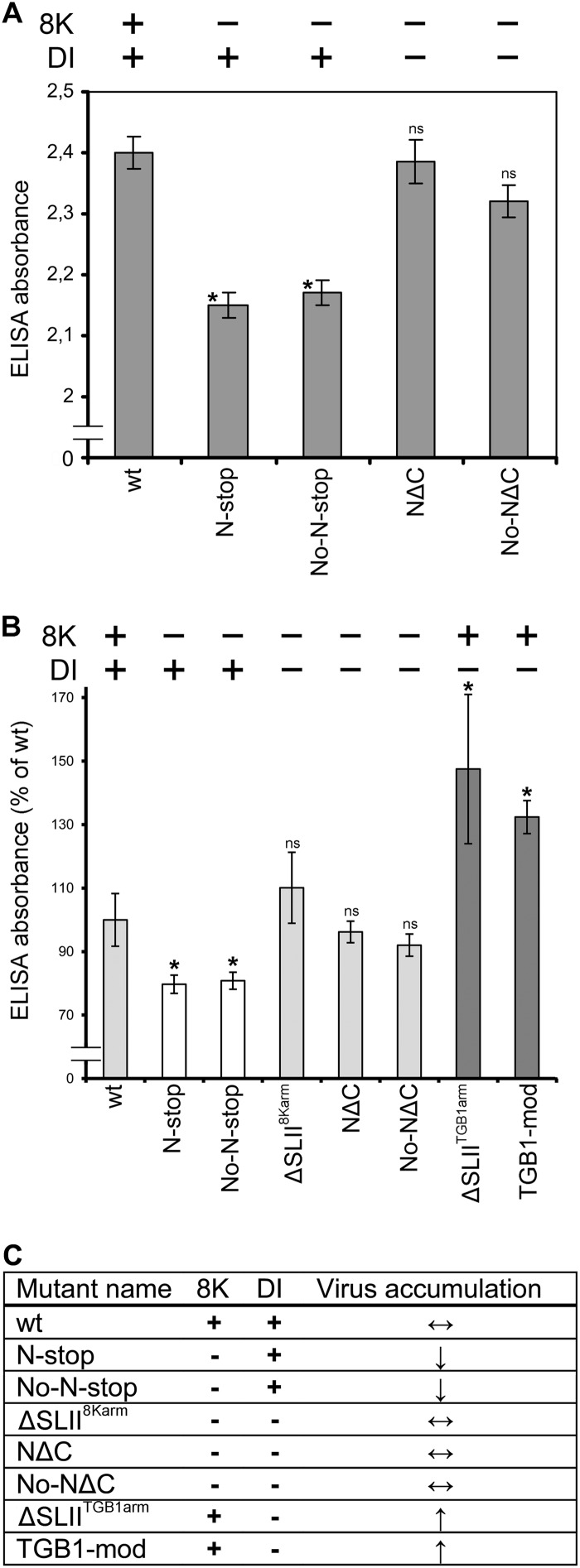
Virus accumulation in N. benthamiana was investigated following inoculation with RNA-TGB mutants. First, the 8K gene knockout mutants (N-stop and No-N-stop) were compared to the wt virus and to double mutants deficient in both 8K and DI RNA production (NΔC and No-NΔC). Comparative ELISAs of progeny virus accumulation revealed that the absorbance values (A405 nm) of NΔC and No-NΔC in systemically infected leaves were similar to the value for the wt virus (Fig. 6A). On the other hand, the absorbance values of N-stop and No-N-stop were approximately 15 to 20% lower than the value for the wt virus (Fig. 6A). These results are consistent with our previous observation that in the absence of DI RNA (TGB-mod and ΔSLIITGB1arm mutants), virus accumulated to a greater extent than in wt infection (Fig. 4C and 5C). The data are summarized in Fig. 6B and C and collectively demonstrate that 8K and DI RNA exert opposite effects on virus accumulation, with 8K enhancing the virus titer and DI RNA reducing it. Consistently, NΔC, No-NΔC, and ΔSLII8Karm, which lack both DI RNA and the 8K protein, accumulated to levels similar to the level of the wt virus, presumably because the positive effect on the virus accumulation by the absence of DI RNA was compensated for/balanced by the 8K gene knockout.
Fig 6.
Comparison of the absorbance values of the PMTV mutant deficient in 8K and DI RNA production. (A) Levels of PMTV virion accumulation detected by double-antibody sandwich ELISA as indicated by absorbance values at 405 nm. Plant extracts were prepared from upper leaves at 14 dpi. The average absorbance of a healthy plant extract was equal to 0.19. Asterisks indicate that the absorbance values were significantly different (P < 0.05, Student t test) from the wt. ELISAs were conducted three times (n = 18). (B) Effect of the lack/presence of 8K and DI on virus accumulation. Accumulation of wt progeny was considered to be 100%. The data were collected from three independent experiments (n = 18). Asterisks indicate that the absorbance values were significantly different (P < 0.05, Student t test) from the wt. The RNA-TGB mutants are also presented in the table (C). An upward arrow indicates statistically significant increase in virus accumulation compared to the wt. A downward arrow indicates statistically significant reduction in virus accumulation compared to the wt. Double-headed arrow, no change; ns, not significant (P > 0.05, Student t test).
Compensatory mutational analysis of SLII and CSLIII confirms their critical role in the DI RNA production.
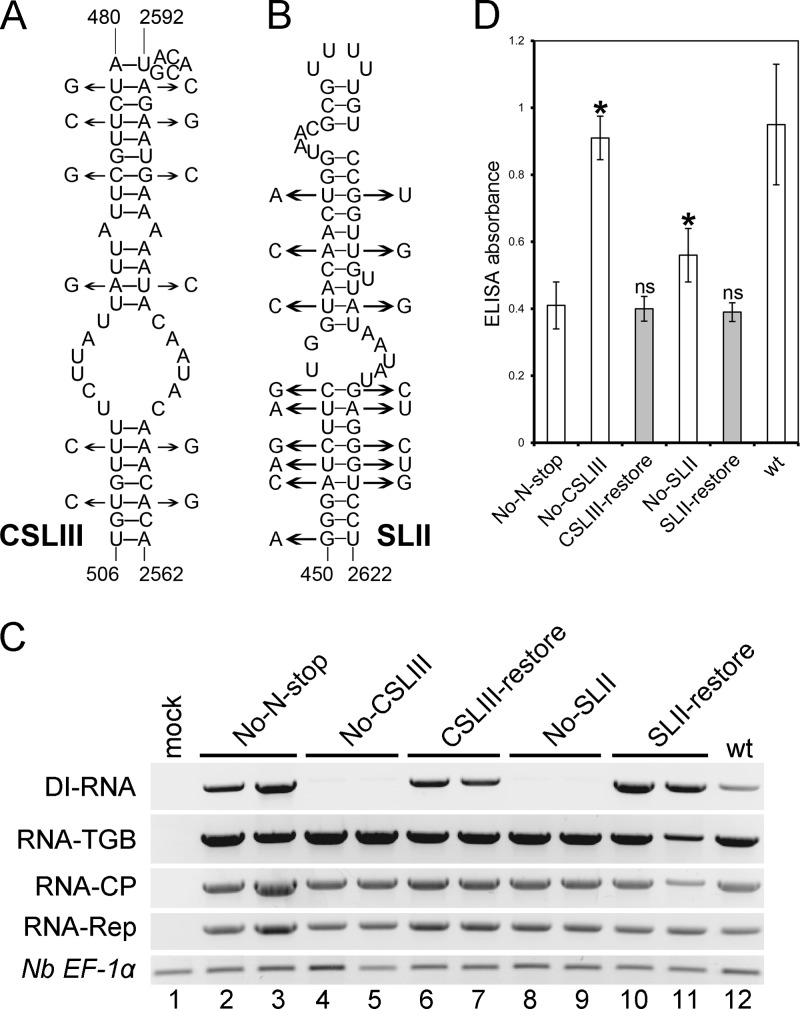
To specifically address the importance of SLII and CSLIII structures in DI RNA production, compensatory mutational analysis of the proposed critical stems was carried out using the No-N-stop mutant as a backbone for the engineering of additional mutants. In this mutant the 8K ORF is not expressed so that the corresponding compensatory substitutions in the 8K coding region will not be codon restricted. Two mutants, No-CSLIII and No-SLII, were derived from No-N-stop cDNA by mutagenesis. Six and nine point mutations were engineered into the wobble position or highly degenerate codons of the TGB1 ORF (nt 481 to 504 and nt 450 to 469) to create the mutants No-CSLIII and No-SLII, respectively (Fig. 7A, left side of each stem-loop). The next set of mutants was derived from No-CSLIII and No-SLII cDNA by mutagenesis. Six and eight compensatory mutations were engineered at nt 2563 to 2586 and nt 2601 to 2619 of No-CSLIII and No-SLII to restore the stem-loops and to create the mutants CSLIII-restore and SLII-restore, respectively (Fig. 7A). Consequently, both No-CSLIII and No-SLII were expected to be deficient in DI RNA production, whereas in the CSLIII-restore and SLII-restore mutants, DI RNA accumulation would be restored.
Fig 7.
Effects of CSLIII and SLII mutations on accumulation of PMTV DI RNA and the virions. (A and B) Mutations used to disrupt the stem-loops are shown on the left side of the secondary structures and refer to the No-CSLIII and No-SLII mutants, respectively. Mutations introduced into the No-CSLIII and No-SLII mutants to restore the stem-loops are shown on the right side of the secondary structures and refer to the CSLIII-restore and No-SLII-restore mutants, respectively. Numbering refers to nucleotide positions in the plus strand of full-length RNA-TGB. (C) RT-PCR of cDNA of N. benthamiana upper leaves to detect the accumulation of DI RNA and the virus genomic RNAs. The identity of each mutant is indicated above the gels. Twenty-four cycles were used for amplification. The constitutively expressed gene for N. benthamiana elongation factor 1α (Nb EF1α) served as a normalization control. The experiment was repeated twice with similar results. (D) Levels of PMTV virion accumulation detected by double-antibody sandwich ELISA as indicated by absorbance values at 405 nm. Plant extracts were prepared from upper leaves at 14 dpi. The average absorbance of a healthy plant extract was equal to 0.16. Asterisks indicate that the absorbance values were significantly different (P < 0.05, Student t test) from No-N-stop, which was used as a backbone to obtain all other constructs. ELISAs were conducted twice (n = 12).
Subsequently in vitro synthesized No-N-stop, No-CSLIII, CSLIII-restore, No-SLII, SLII-restore, and RNA-TGB transcripts were coinoculated with RNA-Rep and RNA-CP transcripts. All six inocula gave rise to viruses that were fully competent in cell-to-cell and long-distance movement since PMTV was detected in upper noninoculated leaves (Fig. 7C and D). RT-PCR analysis carried out on RNA isolated from the samples of leaves systemically infected either with No-CSLIII or No-SLII revealed significant reduction of DI RNA accumulation compared to the plants infected with No-N-stop or wt virus and restoration of DI RNA accumulation in leaves systemically infected with either CSLIII-restore or SLII-restore (Fig. 7C). These results, therefore, confirm the crucial role the CSLIII and SLII stem-loops in the DI RNA biogenesis.
Production of DI RNA is dependent on replication.
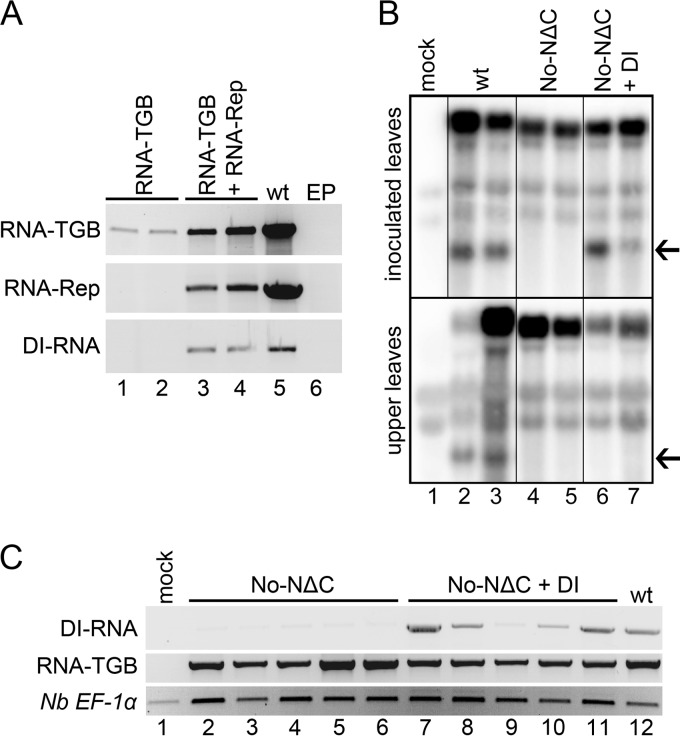
The data show that DI RNA accumulated in significant amounts, was readily detectable by Northern blotting, and interfered with virus accumulation, which implies that production of DI RNA might be replication dependent. To verify this possibility, we engineered transfer DNA (T-DNA) constructs of PMTV genomic RNAs. The RNA-TGB agro-construct was infiltrated alone into N. benthamiana leaves, and, in parallel, infiltrations were performed with a mixture of inocula containing wt plasmids of PMTV RNA-TGB and RNA-Rep (which provides a replication function). RT-PCR analysis of DNase I-treated total RNA preparations isolated from infiltrated leaf samples are shown in Fig. 8A. As a positive control, RT-PCR analysis was done for RNA preparations isolated from PMTV-infected N. benthamiana plants. In the absence of replication, DI RNA was not detected (Fig. 8A, lower panel, lanes 1 and 2), despite the high levels of RNA-TGB accumulation upon transcription from the strong constitutive 35S promoter of cauliflower mosaic virus in infiltrated patches (Fig. 8A, upper panel, lanes 1 and 2). These data confirmed that DI RNA production is dependent on replication.
Fig 8.
Dependence of DI RNA production on replication and inefficient DI RNA movement into the upper leaves. (A) Electrophoresis of RT-PCR-amplified DNA fragments corresponding to RNA-TGB, RNA-Rep, and DI. N. benthamiana leaves were infiltrated autonomously with the RNA-TGB genomic component (lanes 1 and 2), or agro-transformants of RNA-TGB and RNA-Rep were coexpressed (lanes 3 and 4). Control infiltration was performed with an Agrobacterium strain transformed with an empty plasmid (EP). (B) Northern blot analysis of RNA extracted from inoculated and upper leaves of the N. benthamiana plants inoculated with wt (RNA-Rep plus RNA-CP plus RNA-TGB; lanes 2 and 3), No-NΔC (RNA-Rep plus RNA-CP plus No-NΔC; lanes 4 and 5), or No-NΔC inoculum supplemented with T7 RNA polymerase-generated transcripts of DI (lanes 6 and 7). The experiment was repeated three times with similar results. DI RNA is indicated by an arrow. (C) RT-PCR on cDNA of N. benthamiana upper leaves to detect accumulation of the DI RNA and the virus genomic RNAs at 21 dpi. The identity of each type of inoculum is indicated above the gels. Twenty-four cycles were used for amplification. The constitutively expressed gene for N. benthamiana elongation factor 1α (Nb EF1α) served as a normalization control. The data are from two independent experiments.
DI RNA moves long distances inefficiently and is produced de novo during virus replication cycles.
To address the question of whether DI RNA can move from the inoculated to the upper leaves in the presence of helper virus, experiments were designed using the RNA-TGB mutant No-NΔC that lacks the 8K ORF and does not produce DI RNA (Fig. 1D and 2). Two different inocula were assembled comprising RNA-Rep plus RNA-CP and No-NΔC (collectively referred to as No-NΔC) and RNA-Rep plus RNA-CP and No-NΔC and DI (referred to as No-NΔC plus DI). An additional inoculum was assembled to serve as a wt control. Comparison between these inocula would allow us to determine whether DI RNA is capable of long-distance movement to upper noninoculated leaves. Fourteen days postinoculation (dpi) of N. benthamiana, total RNA was extracted from both inoculated and upper noninoculated leaves and subjected to analyses by Northern blot hybridization (Fig. 8B). While DI RNA was readily detectable in the inoculated leaves of the No-NΔC plus DI inoculum due to its efficient replication and cell-to-cell movement, it was not detected in upper leaves (Fig. 8B). On the other, hand DI RNA was readily detectable in both inoculated and upper leaves infected with wt virus. This implies that in wt virus the DI RNA is regenerated from RNA-TGB following virus replication and systemic movement. RT-PCR analysis carried out at late stages of virus infection (21 dpi) on RNA isolated from the samples of upper noninoculated leaves of the plants inoculated either with No-NΔC plus DI or No-NΔC alone revealed that DI RNA did move long distances to upper noninoculated leaves, albeit inefficiently (Fig. 8C, lanes 7 to 11). Regardless of the fact that equal amounts of the RNA transcript were used for plant inoculation in two independent experiments, the presence of the DI RNA, if any, in some upper leaves was either very low or below detection limits even by RT-PCR (e.g., Fig. 8C, lanes 9 and 10). The results further reinforce the idea that DI RNA moves long distances inefficiently and was mostly produced de novo during virus replication.
Knockout of potential coding ORF within the DI RNA does not inhibit its effect on virus accumulation.
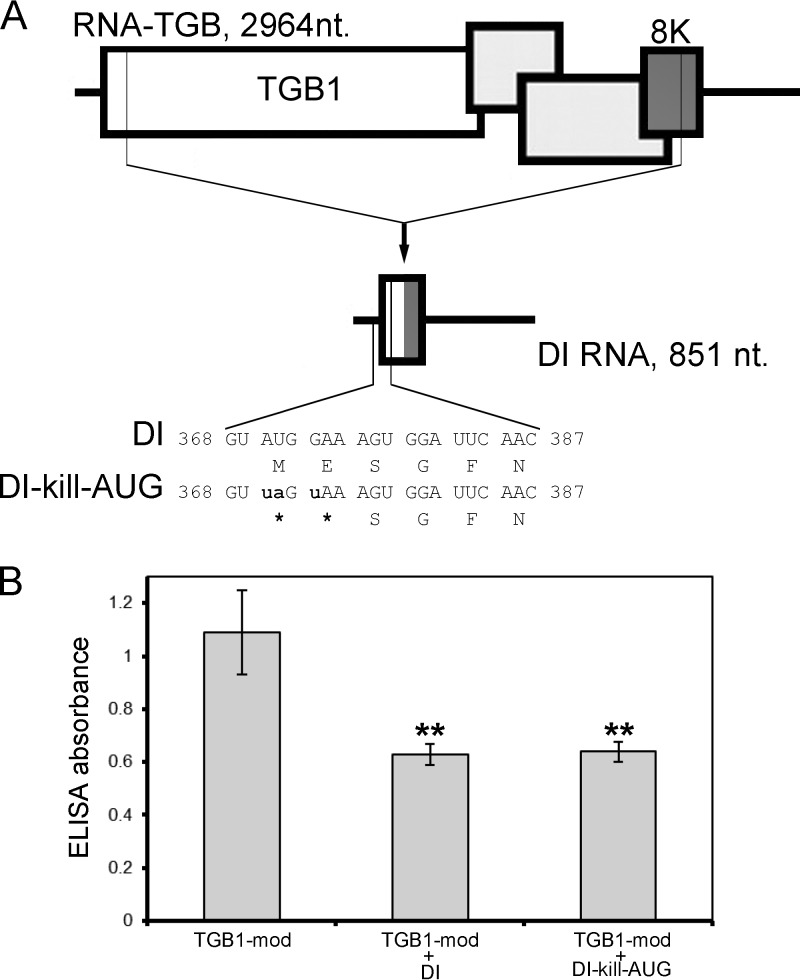
As the DI RNA harbors a single continuous 186-nt ORF (Fig. 1A) that potentially could encode a 7-kDa chimeric fusion protein (7K) consisting of the TGB1 N terminus and the 8K C terminus, it is possible that the putative 7K may have a role in interference by DI RNA on virus accumulation. To address this question, we knocked out the putative 7K ORF by introducing two nonsense mutations in the beginning of the ORF (Fig. 9A), and the engineered DI RNA mutant was named DI-kill-AUG (Fig. 9A). Three different inocula were assembled comprising RNA-Rep plus RNA-CP and TGB1-mod (collectively referred to as TGB1-mod), RNA-Rep plus RNA-CP and TGB1-mod and DI (referred to as TGB1-mod plus DI), and RNA-Rep plus RNA-CP and TGB1-mod and DI-kill-AUG (referred to as TGB1-mod plus DI-kill-AUG). The TGB1-mod (that does not produce DI RNA) inoculum served as a control. Comparison between these inocula would allow us to determine whether a coding ORF within the DI RNA is needed for DI interference with virus accumulation. Comparative ELISAs of progeny virus accumulation revealed that the absorbance values (A405 nm) of TGB1-mod plus DI and TGB1-mod plus DI-kill-AUG in inoculated leaves were similar to each other (P > 0.05; Student t test) (Fig. 9B) but were significantly lower (approximately 45%; P < 0.005; Student t test) than the absorbance value of the TGB1-mod control (Fig. 8B). These data explicitly demonstrated that the RNA DI ORF is dispensable for the interference with virus accumulation since its knockout did not result in significant change of the virus yield compared with the inoculum supplemented with DI RNA unaffected for the ORF. Moreover, sequencing of 16 randomly selected clones obtained by cloning the full-length DI-kill-AUG cDNA generated by RT-PCR did not reveal evidence for reversion to wt (data not shown).
Fig 9.
Knockout of the putative coding ORF within DI RNA does not affect its capability to interfere with virus accumulation. (A) Schematic representation of the DI mutant engineered to prevent translation from the DI-encoding ORF. The mutated nucleotide residues in relation to the DI RNA sequence are shown in small script and in bold. (B) Levels of PMTV CP antigen detected by double-antibody sandwich ELISA as indicated by absorbance values at 405 nm. Plant extracts were prepared from inoculated leaves at 14 dpi. The average absorbance of a healthy plant extract was equal to 0.18. Asterisks indicate that the absorbance values were significantly different (P < 0.0005, Student t test) from the control inoculation with TGB1-mod. The data were collected from two independent experiments (n = 8).
Analysis of RNA silencing suppressor activity of the 8K protein.
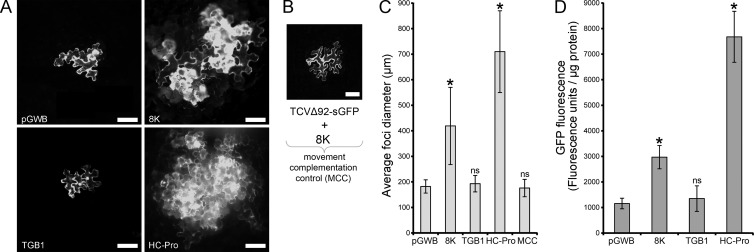
As expression of the 8K gene in the absence of DI RNA appeared to enhance virus accumulation, it is possible that the 8K protein may act an RNA silencing suppressor (RSS). Our previous studies that used a patch Agrobacterium infiltration assay did not reveal such a function (17). To further examine PMTV suppression of RNA silencing (and to avoid a nuclear-localized transgene reporter bias of a patch agro-infiltration assay), complementation of the movement of a turnip crinkle virus construct harboring a green fluorescent protein reporter gene (TCV-sGFP), but having a gene for RSS deleted (20, 21), was employed to test PMTV 8K protein and TGB1, the most obvious candidates, for RNA silencing suppressor activity. To this end the ORFs of 8K and TGB1 were cloned into binary vectors using Gateway technology. In the TCV-sGFP complementation assay, the 8K and TGB1 proteins were transiently expressed by infiltration of Agrobacterium cultures, followed 3 days later by rub inoculation of in vitro generated infectious TCV-sGFP transcripts. Infiltration of the empty binary vector or a plasmid expressing the HC-Pro silencing suppressor from potato virus A (PVA) (19) was used as a negative or positive control, respectively. As expected, in the absence of an RSS, TCV-sGFP movement was confined to foci of 1 to 4 cells at 3 dpi (Fig. 9A). A similar pattern of TCV-sGFP movement was observed in the presence of the TGB1 construct (fluorescent foci consisting of 1 to 4 cells). On the other hand, both PMTV 8K and PVA HC-Pro were able to complement the movement of RSS-deficient TCV-sGFP although PVA HC-Pro was more efficient, resulting in foci consisting of 4 to 60 cells compared with foci of 3 to 20 cells for 8K (Fig. 10A). Measurements of the diameter of 40 fluorescent foci confirmed these visual observations (Fig. 10C) that the 8K protein as an RSS was weaker. To rule out the possibility that the 8K protein assists in movement rather than being an RSS, the movement-deficient construct TCVΔ92-sGFP was employed. To this end in vitro generated infectious TCVΔ92-sGFP transcripts were inoculated onto N. benthamiana leaves preinfiltrated with the Agrobacterium strain carrying the 8K construct. Three days postinoculation, examination of 40 fluorescent foci revealed that GFP fluorescence was exclusively confined to single cells (Fig. 10B), demonstrating that the 8K protein cannot assist or complement the movement-deficient TCV.
Fig 10.
Analysis of local RNA silencing suppression activity in N. benthamiana plants. (A) Representative images of TCV-sGFP complementation assay with constructs indicated on the panels. Preinfiltrated Agrobacterium cultures carrying either an empty plasmid (pGWB) or TGB1 construct does not complement the movement of TCV-sGFP, while preinfiltrated 8K and PVA HC-Pro constructs complement the movement of TCV-sGFP at 3 dpi. Scale bar, 100 μm. (B) Representative image showing the lack of movement. Preinfiltrated Agrobacterium cultures carrying the 8K construct do not complement the movement of TCVΔ92-sGFP at 3 dpi. Scale bar, 100 μm. (C) Quantification of the movement of TCV-sGFP in the complementation assay. The diameter of 40 foci of infection were measured at 3 dpi for each construct indicated below the graph. An asterisk (*) indicates a statistically significant (P < 0.0005) difference in focus diameter compared to the empty plasmid control (pGWB). ns, not significant (P > 0.05). (C) GFP fluorometric analysis of the protein extracts from N. benthamiana leaves of the PZP-TCV-sGFP coinfiltration assay. Leaves were coinfiltrated with PZP-TCV-sGFP and various constructs, indicated below the graph. The baseline represents the level of fluorescence of the protein extract from the leaf expressing an empty T-DNA. Both PMTV 8K and PVA HC-Pro showed statistically significant increases in GFP fluorescence (P < 0.0005). ns, not significant (P > 0.05). Error bars denote standard deviation of the measurements.
Next, leaf discs coinfiltrated with PZP-TCV-sGFP and the different constructs were harvested at 5 dpi from the infiltrated leaves and analyzed for evidence of increased GFP accumulation, indicative of silencing suppression. GFP fluorescence was quantified by a fluorometric analysis as described by Zamyatnin et al. (22). As before, both PMTV 8K and PVA HC-Pro constructs showed statistically significant (P values of <0.0005) increases in GFP fluorescence compared to PZP-TCV-sGFP coinfiltration with an empty plasmid control (Fig. 10D). TGB1 was found to have no increase in GFP fluorescence at a significance level of 0.05 (P > 0.05) (Fig. 10D). Collectively, these data demonstrate that the 8K protein appears to be a genuine RSS, albeit a weak one, and enhances virus accumulation.
DISCUSSION
Helper virus-dependent D or DI RNA has been reported to associate with many plant viruses and is often observed during RNA virus infections of animal and human cell cultures. For some viruses, e.g., coronaviruses, DI RNAs have become a valuable tool in studies of virus replication and identification of cis replication elements because the large coronavirus genome still remains a technical challenge even with use of reverse genetics systems (24). Among plant viruses, DI RNAs associated with the genera Tombusvirus and Cucumovirus are the most extensively studied (2, 5). Despite the fact that PMTV DI was first described more than a decade ago, it has not been extensively characterized. In this work we took advantage of two previously reported unusual features of the PMTV genome to investigate PMTV DI RNA. First, the 8K protein was shown not to be required for virus replication or cell-to-cell and long-distance movement in N. benthamiana (12). Second, deletions as long as 132 nt could be engineered into the 5′-proximal part of the ORF encoding the TGB1 protein without significant effect on virus replication and cell-to-cell movement (14). These features allowed us to engineer point mutations and deletions into 8K- and TGB1-encoding regions of the viral genome and subsequently to test the effect of mutations on DI RNA formation, accumulation, and movement in planta.
This study has shown that the coding regions of TGB1 and 8K are involved in DI RNA biogenesis and that the regions of the TGB1 and 8K ORFs which are deleted from the DI RNA control DI RNA production. The mechanism of production is most likely through base pairing on the minus strand to form a stem-loop structure (CSLIII), and the DI RNA is formed during synthesis of positive-strand RNA by intramolecular template switching. Indeed, deletions or point mutations engineered into either the 8K-derived arm or the TGB1-derived arm of the minus strand involved in CSLIII formation blocked DI RNA production, suggesting that CSLIII brings the 5′ and 3′ ends of the RNA-TGB minus strand together such that it forms an omega-like structure (Ω) or “panhandle” with unpaired 5′ and 3′ ends. PMTV RNA-dependent RNA polymerase (RdRP) then binds to the 3′ end of the minus strand to initiate plus-strand RNA synthesis. The RNA synthesis continues until RdRP encounters CSLIII, which causes the RdRP to pause and jump from nt 480 to nt 2593. The stability of CSLIII (a minimum free energy of −13.7 kcal/mol) may explain the precision of intramolecular template switching and the invariability of the junction site within the DI RNA. Moreover, the suggested mechanism implies that PMTV DI RNA may arise as a result of a single internal deletion.
The results also suggest the existence of higher-order RNA structures identified as SLI and SLII in close proximity of the 8K coding region and within DI RNA that might be required for DI RNA production. This idea is supported by our mutagenesis data and RNAfold secondary structure predictions because point mutations and an insertion engineered into a pentanucleotide sequence (of which 4 nucleotides are involved in the secondary structure formation) immediately downstream of the 8K coding region suppressed DI RNA production (8K-D, 8K-S, and 8K-FLAG mutants). Furthermore, these results emphasize the impact of a pentanucleotide sequence on the stability of the stem-loop structure. Recently, it was shown that a 2-nucleotide sequence influenced the stability of a stem-loop structure and was involved in production of a redundant RNA5 of CMV (25). Collectively, these observations suggest that even very short sequences of 2 to 5 nucleotides might control DI RNA production. Moreover, SLII is a stable stem-loop (a minimum free energy of −21.1 kcal/mol) that shows considerable structural but not nucleotide sequence conservation among many isolates of PMTV (our unpublished data), regardless of the fact that the 8K gene is the most variable gene in the numerous PMTV isolates sequenced so far (17, 26; also our unpublished data). While direct biochemical evidence for intramolecular template switching is lacking, it is noteworthy that the predicted location of the junction point within the DI RNA is at the very top of the SLII and involves the arms derived from the TGB1 and 8K genes. Thus, from the discussion above, it is possible that formation of SLII in the plus strand of the DI RNA in addition to CSLIII formation of the minus strand promotes intramolecular template switching during plus-strand RNA synthesis and results in DI RNA production, probably through stabilization of higher-order RNA structures involving SLII. This suggestion is supported by our mutagenesis data (ΔSLIITGB1arm and ΔSLII8Karm mutants). Moreover, by being an abundant molecule, DI RNA was very often amplified, cloned, and sequenced during molecular characterization of the PMTV isolates from various geographic locations. Remarkably, the junction point within DI RNA was always the same (our unpublished data). These results contrast with a study of a DI RNA associated with cymbidium ringspot virus (CymRSV), which addressed the mechanism of formation of smaller DI RNA molecules from the longer DI RNA (27). While a highly base-paired structure in the CymRSV DI RNA directed a smaller DI RNA formation through intramolecular template switching, similar to our results but on the plus strand, the deletions (and the junction points, accordingly) within the newly formed smaller DI RNAs were highly heterogeneous depending on the inoculation, spanned a 90-nt region, and were distributed upstream and downstream of the stem-loop structure (27).
Compensatory mutagenesis first to disrupt and then to restore CSLIII or SLII confirmed the importance of both stem-loops for DI RNA production. The presence of CSLIII alone (e.g., in ΔSLIITGB1arm, ΔSLII8Karm, and No-SLII mutants) was insufficient for efficient DI RNA production and could not fully compensate for the absence of SLII. Likewise, the presence of SLII alone (e.g., in TGB1-mod and No-CSLIII mutants) was insufficient for DI RNA production and could not fully compensate for the absence of CSLIII. Therefore, the data suggest that CSLIII and SLII act in concert during DI RNA biogenesis, where CSLIII is likely needed for efficient intramolecular template switching and where SLII formation during production of DI RNA enhances the template switching and helps to shift the equilibrium between normal full-length plus-strand RNA synthesis and DI RNA production.
Notably, the DI RNA possess a short ORF; i.e., the region of the TGB1 ORF downstream of the deletion is in-frame with the 3′-proximal part of the 8K ORF. Translation of this chimeric ORF would yield a small fusion protein of 7 kDa consisting of the TGB1 N terminus and the 8K C terminus. Potentially, if expressed, the chimeric protein might act as a dominant negative protein (produced by a dominant negative mutant) to impair normal functioning of either TGB1 and/or 8K or some other TGB1 and 8K protein interactors which could, at least partially, explain the mechanism of interference by DI RNA with virus accumulation. However, the results showed that the DI RNA ORF does not interfere with virus accumulation. While sequencing of a number of clones did not reveal any reversion of the point mutations introduced to knock down the putative chimeric ORF in the DI RNA, one cannot exclude the possibility that initiation of the 7-kDa protein translation from the knockdown construct might occur at non-AUG codons (often referred to as CUG, AUA, and GUG codons). Scanning the 7K ORF sequence revealed two GUG codons located 33 nt (406GUG408) and 57 nt (430GUG432) downstream of the AUG, but both are surrounded by suboptimal Kozak consensus.
The results of this study established that PMTV DI is replication dependent and produced during virus replication cycles. However, DI RNA moved long distances inefficiently regardless of the efficient long-distance movement of the helper virus in N. benthamiana. DI RNA could be detected in the upper leaves by RT-PCR at later stages of virus infection (21 dpi). It was shown previously that DI RNA was encapsidated (16); however, it may be that the DI particles (or DI RNA) are unable to bind to the necessary virus or host factors required for long-distance movement (11, 13). Similar observations were also reported in other studies involving CMV and different hosts as CMV D RNA accumulated in upper leaves of Nicotiana species but did not move systemically in muskmelon, tomato, or zucchini squash (7, 8). Therefore, it is also possible that the capacity of DI RNA to move efficiently is host dependent, and the mechanism of inhibition of DI long-distance movement remains to be studied.
The symptoms associated with inocula deficient in DI RNA production consistently appeared 1 to 2 days earlier than with wt virus or inocula supplemented with DI RNA. Results of our experiments using various TGB1 and 8K mutants revealed that virus accumulation was strongly dependent on the presence of the 8K protein and the DI RNA. Accordingly, DI RNA had a strong influence on virus accumulation, and the mutants deficient in DI RNA production (e.g., TGB1-mod and ΔSLIITGB1arm), but expressing the 8K gene, accumulated more efficiently than the wt virus. Interestingly, our initial characterization of 8K-deficient mutants, namely, NΔC and No-NΔC, did not reveal any differences in their accumulation levels compared to the level of the wt virus (12). In this study, we clarified this situation by showing that NΔC and No-NΔC, besides being knockouts for the 8K gene, are also deficient in DI RNA production.
The 8K protein appeared to be recalcitrant to purification; i.e., attempts to express and purify it either from Escherichia coli, Saccharomyces cerevisiae, or plants failed. Nevertheless, when expressed from the PMTV genome, the FLAG-tagged 8K was detected in the extracts prepared from infected plants, demonstrating that the 8K gene is expressed in planta during virus infection (17). In this study, we demonstrated that the 8K gene is needed for efficient virus accumulation. Many plant viruses encode an RSS (28, 29), and a strong RSS would allow virus accumulation to be sustained and at a high level. Conversely, a low level of a virus accumulation could be attributed to the absence of or weak suppressor activity. While our previous studies did not reveal RNA silencing suppression activity for the 8K protein in a patch Agrobacterium infiltration assay (17), in this work using an assay based on complementation of movement of the RSS-deficient TCV-GFP (20, 21), we show that the 8K protein acts as a weak RSS. Notably, PMTV TGB1, the major movement protein of the virus, was not able to complement TCV-GFP movement, while a well-characterized RSS PVA HC-Pro complemented TCV-GFP movement much more efficiently (loci consisting of 4 to 60 cells). Therefore, it seems that the sequences of the genes required for efficient accumulation of the virus progeny (e.g., 8K RSS) are involved in DI RNA production, which in turn impairs the efficient accumulation of the virus progeny. Since wt virus infections always contain this DI RNA species, it is tempting to speculate that the D RNA is a crucial regulatory RNA rather than a DI RNA in the sense of a molecular parasite that results from errors during replication of genomic RNA. It might be that PMTV DI RNA is in a separate category from the traditional DI RNAs. The results support the contention that the interplay between RSSs and DI RNAs may be a more common occurrence in plant RNA viruses and is now found in at least in two groups of viruses, namely, tombusviruses (9) and pomoviruses.
ACKNOWLEDGMENTS
We thank Tim L. Sit and Steve A. Lommel (Department of Plant Pathology, North Carolina State University) for PZP-TCV-sGFP, TCV-sGFP, and TCVΔ92-sGFP constructs. We thank Jens Tilsner for providing agroconstructs of RNA-TGB and RNA-Rep and Andrey Solovyev for helpful discussions.
Financial support from the Swedish Research Council Formas (grants 2007-256, 2008-1047, and 2009-1979 to E.I.S.), the Carl Tryggers Foundation (E.I.S. and N.I.L.), and the Swedish Institute (postdoctoral fellowship to N.I.L.) is gratefully acknowledged. L.T. is funded by the Scottish Government Rural and Environmental Science and Analytical Services Division.
Footnotes
Published ahead of print 20 March 2013
REFERENCES
- 1. Graves MV, Pogany J, Romero J. 1996. Defective interfering RNAs and defective viruses associated with multipartite RNA viruses of plants. Semin. Virol. 7:399–408 [Google Scholar]
- 2. Simon AE, Roossinck MJ, Havelda Z. 2004. Plant virus satellite and defective interfering RNAs: new paradigms for a new century. Annu. Rev. Phytopathol. 42:415–437 [DOI] [PubMed] [Google Scholar]
- 3. Eliasco E, Livieratos IC, Müller G, Guzman M, Salazar LF, Coutts RH. 2006. Sequences of defective RNAs associated with potato yellow vein virus. Arch. Virol. 151:201–204 [DOI] [PubMed] [Google Scholar]
- 4. Havelda Z, Szittya G, Burgyán J. 1998. Characterization of the molecular mechanism of defective interfering RNA-mediated symptom attenuation in tombusvirus-infected plants. J. Virol. 72:6251–6256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pathak KB, Nagy PD. 2009. Defective interfering RNAs: foes of viruses and friends of virologists. Viruses 1:895–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Szittya G, Molnár A, Silhavy D, Hornyik C, Burgyán J. 2002. Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell 14:359–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Graves MV, Roossinck MJ. 1995. Characterization of defective RNAs derived from RNA 3 of the Fny strain of cucumber mosaic cucumovirus. J. Virol. 69:4746–4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kaplan IB, Lee KC, Canto T, Wong SM, Palukaitis P. 2004. Host-specific encapsidation of a defective RNA 3 of cucumber mosaic virus. J. Gen. Virol. 85:3757–3763 [DOI] [PubMed] [Google Scholar]
- 9. Havelda Z, Hornyik C, Válóczi A, Burgyán J. 2005. Defective interfering RNA hinders the activity of a tombusvirus-encoded posttranscriptional gene silencing suppressor. J. Virol. 79:450–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scholthof KB, Scholthof HB, Jackson AO. 1995. The effect of defective interfering RNAs on the accumulation of tomato bushy stunt virus proteins and implications for disease attenuation. Virology 211:324–328 [DOI] [PubMed] [Google Scholar]
- 11. Torrance L, Wright KM, Crutzen F, Cowan GH, Lukhovitskaya NI, Bragard C, Savenkov EI. 2011. Unusual features of pomoviral RNA movement. Front. Microbiol. 2:259 doi:10.3389/fmicb.2011.00259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Savenkov EI, Germundsson A, Zamyatnin AA, Jr, Sandgren M, Valkonen JPT. 2003. Potato mop-top virus: the coat protein-encoding RNA and the gene for cysteine-rich protein are dispensable for systemic virus movement in Nicotiana benthamiana. J. Gen. Virol. 84:1001–1005 [DOI] [PubMed] [Google Scholar]
- 13. Torrance L, Lukhovitskaya NI, Schepetilnikov MV, Cowan GH, Ziegler A, Savenkov EI. 2009. Unusual long-distance movement strategies of Potato mop-top virus RNAs in Nicotiana benthamiana. Mol. Plant Microbe Interact. 22:381–390 [DOI] [PubMed] [Google Scholar]
- 14. Wright KM, Cowan GH, Lukhovitskaya NI, Tilsner J, Roberts AG, Savenkov EI, Torrance L. 2010. The N-terminal domain of PMTV TGB1 movement protein is required for nucleolar localization, microtubule association, and long-distance movement. Mol. Plant Microbe Interact. 23:1486–1497 [DOI] [PubMed] [Google Scholar]
- 15. Zamyatnin AA, Jr, Solovyev AG, Savenkov EI, Germundsson A, Sandgren M, Valkonen JPT, Morozov SY. 2004. Transient co-expression of individual genes encoded by the triple gene block of Potato mop-top virus reveals requirements for TGBp1 trafficking. Mol. Plant Microbe Interact. 17:921–930 [DOI] [PubMed] [Google Scholar]
- 16. Torrance L, Cowan GH, Sokmen MA, Reavy B. 1999. A naturally occurring deleted form of RNA 2 of Potato mop-top virus. J. Gen. Virol. 80:2211–2215 [DOI] [PubMed] [Google Scholar]
- 17. Lukhovitskaya NI, Yelina NE, Zamyatnin AA, Jr, Schepetilnikov MV, Solovyev AG, Sandgren M, Morozov SY, Valkonen JP, Savenkov EI. 2005. Expression, localization and effects on virulence of the cysteine-rich 8 kDa protein of Potato mop-top virus. J. Gen. Virol. 86:2879–8289 [DOI] [PubMed] [Google Scholar]
- 18. Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T. 2007. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104:34–41 [DOI] [PubMed] [Google Scholar]
- 19. Yelina NE, Savenkov EI, Solovyev AG, Morozov SY, Valkonen JP. 2002. Long-distance movement, virulence, and RNA silencing suppression controlled by a single protein in hordei- and potyviruses: complementary functions between virus families. J. Virol. 76:12981–12991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Powers JG, Sit TL, Qu F, Morris TJ, Kim KH, Lommel SA. 2008. A versatile assay for the identification of RNA silencing suppressors based on complementation of viral movement. Mol. Plant Microbe Interact. 21:879–890 [DOI] [PubMed] [Google Scholar]
- 21. Powers JG, Sit TL, Heinsohn C, George CG, Kim KH, Lommel SA. 2008. The Red clover necrotic mosaic virus RNA-2 encoded movement protein is a second suppressor of RNA silencing. Virology. 381:277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zamyatnin AA, Jr, Solovyev AG, Bozhkov PV, Valkonen JP, Morozov SY, Savenkov EI. 2006. Assessment of integral membrane protein topology in living cells. Plant J. 46:145–154 [DOI] [PubMed] [Google Scholar]
- 23. Donald RG, Jackson AO. 1994. The barley stripe mosaic virus gamma b gene encodes a multifunctional cysteine-rich protein that affects pathogenesis. Plant Cell 6:1593–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Masters PS. 2006. The molecular biology of coronaviruses. Adv. Virus Res. 66:193–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Wispelaere M, Rao AL. 2009. Production of cucumber mosaic virus RNA5 and its role in recombination. Virology 384:179–191 [DOI] [PubMed] [Google Scholar]
- 26. Latvala-Kilby S, Aura JM, Pupola N, Hannukkala A, Valkonen JP. 2009. Detection of potato mop-top virus in potato tubers and sprouts: combinations of RNA2 and RNA3 variants and incidence of symptomless infections. Phytopathology 99:519–531 [DOI] [PubMed] [Google Scholar]
- 27. Havelda Z, Dalmay T, Burgyán J. 1997. Secondary structure-dependent evolution of Cymbidium ringspot virus defective interfering RNA. J. Gen. Virol. 78:1227–1234 [DOI] [PubMed] [Google Scholar]
- 28. Ding SW. 2010. RNA-based antiviral immunity. Nat. Rev. Immunol. 10:632–644 [DOI] [PubMed] [Google Scholar]
- 29. Burgyán J, Havelda Z. 2011. Viral suppressors of RNA silencing. Trends Plant Sci. 16:265–272 [DOI] [PubMed] [Google Scholar]