Abstract
Infection with human coronavirus 229E (HCoV-229E) is associated with the common cold and may result in pneumonia in immunocompromised patients. The viral spike (S) protein is incorporated into the viral envelope and mediates infectious entry of HCoV-229E into host cells, a process that depends on the activation of the S-protein by host cell proteases. However, the proteases responsible for HCoV-229E activation are incompletely defined. Here we show that the type II transmembrane serine proteases TMPRSS2 and HAT cleave the HCoV-229E S-protein (229E-S) and augment 229E-S-driven cell-cell fusion, suggesting that TMPRSS2 and HAT can activate 229E-S. Indeed, engineered expression of TMPRSS2 and HAT rendered 229E-S-driven virus-cell fusion insensitive to an inhibitor of cathepsin L, a protease previously shown to facilitate HCoV-229E infection. Inhibition of endogenous cathepsin L or TMPRSS2 demonstrated that both proteases can activate 229E-S for entry into cells that are naturally susceptible to infection. In addition, evidence was obtained that activation by TMPRSS2 rescues 229E-S-dependent cell entry from inhibition by IFITM proteins. Finally, immunohistochemistry revealed that TMPRSS2 is coexpressed with CD13, the HCoV-229E receptor, in human airway epithelial (HAE) cells, and that CD13+ TMPRSS2+ cells are preferentially targeted by HCoV-229E, suggesting that TMPRSS2 can activate HCoV-229E in infected humans. In sum, our results indicate that HCoV-229E can employ redundant proteolytic pathways to ensure its activation in host cells. In addition, our observations and previous work suggest that diverse human respiratory viruses are activated by TMPRSS2, which may constitute a target for antiviral intervention.
INTRODUCTION
The family Coronaviridae contains six human coronaviruses (HCoVs), namely, 229E, NL63, severe acute respiratory syndrome coronavirus (SARS-CoV), OC43, HKU1, and EMC, all of which target the respiratory tract. SARS-CoV and, potentially, HCoV-EMC were recently transmitted from animals to humans, and both viruses cause severe disease in infected patients (1–6). Thus, the SARS epidemic in 2002–2003 claimed more than 700 lives, mainly in East Asia, and the recent emergence of HCoV-EMC has so far been associated with 13 human infections, 7 of which had a fatal outcome (7–9). In contrast, the remaining four human coronaviruses, 229E, OC43, NL63, and HKU1, are believed to be adapted to spread in the human population and circulate worldwide. Infection by these viruses is associated with mild respiratory disease, mainly the common cold (10–13), although young children, the elderly, and immunocompromised patients might develop a more severe clinical presentation (12–17). For instance, HCoV-NL63 is associated with croup (14, 18), and a link between HCoV infection and lower respiratory tract disease has been suggested (16, 17, 19). In general, HCoV-229E, -OC43, -NL63, and -HKU1 cocirculate, exhibit seasonality, and are frequently involved in coinfections (15–17, 19–21).
HCoV-229E was one of the first isolated human coronavirus strains (22). The viral particle contains a single-stranded RNA genome of positive polarity which comprises about 27 kb. The viral envelope (E), membrane (M), and spike (S) proteins are incorporated into the viral envelope. The M and E proteins are important for assembly and budding of progeny particles, which proceed at the endoplasmic reticulum/Golgi intermediate compartment (ERGIC). The S-protein mediates host cell entry by binding to the cellular receptor CD13/aminopeptidase N (APN), which is expressed on the apical membranes of epithelial cells in the respiratory and enteric tracts, as well as on several other cell types (8, 23–26).
The domain organization of the spike protein of HCoV-229E (229E-S) resembles that of other viral envelope proteins termed class I membrane fusion proteins (27, 28). Thus, the 229E-S-protein contains an N-terminal surface unit, S1, which harbors the binding site for the cellular receptor, CD13, and a C-terminal transmembrane unit, S2, which encompasses the structural elements required for membrane fusion. The 229E-S-protein is synthesized as an inactive precursor in infected cells and depends on proteolytic processing by host cell proteases to enter an active state, another feature typical of class I membrane fusion proteins. Like SARS-S, the S-protein of HCoV-229E is activated by cathepsin L, a pH-dependent endosomal/lysosomal cysteine protease, upon viral uptake into target cells (29). However, recent studies indicate that the type II transmembrane serine proteases (TTSPs) HAT and TMPRSS2 can also activate SARS-S (30–32). Activation of SARS-S by TMPRSS2 renders viral entry independent from cathepsin L activity (30, 32, 33), which has important implications for therapeutic intervention. In addition, activation by TMPRSS2 protects SARS-S from inhibition by IFITMs (34), which are interferon-induced host cell proteins that inhibit cellular entry of several enveloped viruses (35). Whether TMPRSS2 and HAT activate 229E-S and are expressed in viral target cells is unknown.
We show here that TMPRSS2 and HAT cleave and activate 229E-S for cathepsin L-independent host cell entry. In addition, we demonstrate that TMPRSS2 is expressed in viral target cells in the human respiratory epithelium and might thus promote viral spread in humans.
MATERIALS AND METHODS
Cell culture.
293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen), and Caco-2 cells were cultured in DMEM-GlutaMAX (Invitrogen). All cell culture media were supplemented with 10% fetal bovine serum (FBS; Biochrom) and the antibiotics penicillin and streptomycin (Cytogen). Cells were maintained at 37°C under a 5% CO2 atmosphere. 293T cells stably expressing human CD13 (293T-CD13 cells) were generated by transient transfection of 293T cells with the plasmid pcDNA4.TO-hCD13, followed by zeocin (100 μg/ml; Invitrogen) selection. Human airway epithelial (HAE) cells were generated as previously described (36) and were maintained for 2 months.
Plasmids and antibodies.
Expression plasmids encoding the following glycoproteins were described previously: vesicular stomatitis virus glycoprotein (VSV-G), murine leukemia virus (MLV) envelope protein (Env), Lassa virus envelope glycoprotein (GPC), and 229E-S (37, 38). For generation of the 293T-CD13 stable cell line, the CD13 coding sequence from plasmid pcDNA-CD13 (39) was subcloned into pcDNA4.TO. Expression plasmids encoding the human proteases TMPRSS2, TMPRSS4, and HAT were published earlier (40, 41). Previously described expression plasmids for IFITM-1, -2, and -3 (35) were used as templates for amplification of the respective coding regions, using the oligonucleotides IFITM1-5N (GCGCGGCCGCACCATGCACAAGGAGGAACATGAG), IFITM2-3BE (CGAATTCCGGATCCCTATCGCTGGGCCTGGAC), IFITM2-5N (GCGCGGCCGCACCATGAACCACATTGTGCAAACC), IFITM3-5N (GCGCGGCCGCACCATGAATCACACTGTCCAAACC), and EMCV_IRES5b (GAAGACAGGGCCAGGTTTCC). The resulting PCR products were inserted into the plasmid pCAGGS, using NotI and EcoRI sites. A goat polyclonal serum directed against HCoV-229E was described previously (42). A mouse monoclonal antibody (MAb) against TMPRSS2 was obtained from Santa Cruz, a goat polyclonal serum reactive against the ZO-1 tight junction protein was purchased from Abcam, and a mouse anti-myc antibody was obtained from Biomol. A Cy3-conjugated anti-β-tubulin MAb and an anti-β-actin antibody were obtained from Sigma. Expression of IFITM1 was detected by using a MAb purchased from Proteintech Europe, while a rabbit serum obtained from the same provider was employed to detect IFITM2 and IFITM3. This serum was raised against IFITM2 and is cross-reactive with IFITM3. Secondary antibodies for Western blotting and immunofluorescence were purchased from Dianova and Jackson ImmunoResearch.
Western blot analysis of 229E-S cleavage by TMPRSS2 and HAT.
For the detection of 229E-S cleavage by TMPRSS2 and HAT, 293T cells were cotransfected with an expression plasmid for 229E-S (37) and either plasmids encoding the specified proteases or empty plasmid. The medium was replaced with fresh DMEM at 14 to 16 h posttransfection. At 48 h posttransfection, the cells were harvested in 1 ml phosphate-buffered saline (PBS), treated with PBS or 250 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin (Sigma) for 10 min at room temperature, and lysed in 2× sodium dodecyl sulfate (SDS) loading buffer. The lysates were separated by SDS gel electrophoresis and blotted onto nitrocellulose membranes, and 229E-S was detected by staining with a goat anti-229E coronavirus serum (42) at a dilution of 1:500, followed by incubation with a horseradish peroxidase (HRP)-coupled anti-goat secondary antibody (Dianova). As a loading control, expression of β-actin was detected by employing an anti-β-actin antibody (Sigma).
Real-time PCR.
Real-time PCR was performed essentially as described previously (40). In brief, total RNA (500 ng) was reverse transcribed using BioScript RNase H Low reverse transcriptase (Bioline), and aliquots of generated cDNA samples (25 ng total RNA equivalents) were used for real-time PCR in an ABI 7500 Fast real-time PCR system (Applied Biosystems). Specific amplification was ensured with TaqMan gene expression assays (Applied Biosystems) performed according to the manufacturer's recommendations. The average cycle threshold value (CT) for each individual assay was calculated from triplicate measurements by means of the instrument's software in “auto-CT” mode (model 7500 Fast system software v.1.3.0). Average CT values calculated for TMPRSS2 or HAT were normalized through subtraction from the CT values obtained for GUSB (a housekeeping reference).
Cell-cell fusion assay.
The activation of 229E-S-driven cell-cell fusion by TMPRSS2 and HAT was assessed by employing a cell-cell fusion assay previously described by us (43). In brief, 293T effector cells seeded in 6-well plates at 2.4 × 105 cells/well were transfected with either the 229E-S expression plasmid or empty plasmid (pcDNA) in combination with a plasmid encoding the herpes simplex virus VP16 transactivator. In parallel, 293T target cells seeded in 48-well plates at 0.4 × 105 cells/well were cotransfected with a plasmid encoding human CD13 and a plasmid encoding TMPRSS2, TMPRSS4, or HAT or encoding no protease (empty plasmid), jointly with a plasmid carrying a luciferase gene under the control of a VP16-responsive promoter (44). Alternatively, target cells were transfected with a plasmid encoding either CD13 or protease. The culture medium was replaced with fresh DMEM at 8 h posttransfection. At 24 h posttransfection, effector cells were resuspended in fresh medium, diluted 1:2.5, and cocultured with target cells. Six to 8 h after the start of the cocultivation, medium containing TPCK-trypsin (Sigma; final concentration, 100 ng/ml) was added and then was replaced with fresh DMEM after an incubation period of 8 h. Finally, at 72 h posttransfection, the cells were lysed, and luciferase activities in cell lysates were measured by employing a commercially available kit (Promega).
Infection experiments with lentiviral pseudoparticles.
Lentiviral vectors pseudotyped with 229E-S (pseudotypes) were generated as described previously (37, 40). In brief, 293T cells were cotransfected with the HIV-1-derived vector pNL4-3 E-R-Luc (45) and an expression plasmid for 229E-S or VSV-G or empty plasmid (pcDNA). At 16 h posttransfection, the culture medium was replaced with fresh DMEM, and at 48 h posttransfection, the supernatants were harvested, passed through 0.45-μm-pore-size filters, aliquoted, and stored at −80°C. To analyze the impact of transiently expressed TMPRSS2 and HAT on 229E-S-driven host cell entry, either 293T-CD13 cells were transiently transfected with plasmids encoding proteases or 293T cells were transiently cotransfected with plasmids encoding human CD13 and the indicated proteases or empty plasmid. For the analysis of 229E-S activation by endogenous TMPRSS2, Caco-2 cells were used as targets, since these cells were previously shown to express TMPRSS2 (40). One day prior to infection, the target cells were seeded in 96-well plates at 30,000 cells/well. One hour prior to infection, the target cells were preincubated with dimethyl sulfoxide (DMSO) or the indicated concentration of MDL-28170 (Calbiochem), leupeptin (Sigma), or camostat mesylate (Tocris) at 37°C. Subsequently, pseudotypes normalized for infectivity were added and the cells incubated for 6 h at 37°C. Thereafter, the medium was replaced with fresh DMEM without inhibitor. Finally, the luciferase activities in cell lysates were determined at 72 h postinfection, using a commercially available kit (Promega).
Infection experiments with authentic HCoV-229E.
A recombinant HCoV-229E strain expressing luciferase upon genome replication (HCoV-REN) was described previously (46). 293T cells were seeded in 24-well plates at 4 × 105 cells/well and then transfected to express CD13 and the indicated protease or with empty plasmid (pcDNA) before incubation with MDL-28170 (20 μM) or DMSO for 1 h at 37°C. Thereafter, the cells were infected with HCoV-229E at a multiplicity of infection (MOI) of 1, incubated for 8 h at 33°C, and then harvested for determination of luciferase activity in cell extracts, as described above.
Analysis of TMPRSS2 expression in human airway epithelial cells by indirect immunofluorescence.
Analysis of the TMPRSS2 distribution in differentiated HAE cell cultures was performed as described elsewhere (47), using mock- and HCoV-229E-infected (MOI = 0.1) cultures. After 48 h, fully differentiated HAE cultures were fixed with 4% paraformaldehyde (PFA; FormaFix) for 30 min at room temperature, followed by rinsing of the apical and basolateral sides three times with PBS. Fixed HAE cultures were immunostained using a previously described procedure (48). Mouse monoclonal anti-229E N (1E7) (49), sheep polyclonal anti-CD13 (R&D Systems), and rabbit polyclonal anti-TMPRSS2 (Abcam) were applied as primary antibodies. Donkey-derived Dylight 488-labeled anti-mouse IgG(H+L), Dylight 549-labeled anti-sheep IgG(H+L), and Dylight 405-labeled anti-rabbit antibodies (Jackson ImmunoResearch) were applied as secondary antibodies. Thereafter, the cells were incubated with Alexa Fluor 647-conjugated mouse anti-β-tubulin (Cell Signaling) for staining of ciliated cells. For in vivo examination of the TMPRSS2 distribution in the human airway epithelium, primary bronchial segments were fixed with 4% PFA (FormaFix) for 30 min at room temperature, followed by rinsing of the samples three times with PBS. Fixed samples were mounted in Tissue-Tek OCT medium and snap-frozen in liquid nitrogen, and 10-μm-thick horizontal sections were cut. Sheep polyclonal anti-CD13 (R&D Systems) and rabbit polyclonal anti-TMPRSS2 (Abcam) were applied as primary antibodies. Donkey-derived Dylight 488-labeled anti-rabbit IgG(H+L) and Dylight 549-labeled anti-sheep IgG(H+L) (Jackson ImmunoResearch) were applied as secondary antibodies, using the procedure described above. Thereafter, the cells were incubated with Alexa Fluor 647-conjugated mouse anti-β-tubulin (Cell Signaling) for staining of ciliated cells and then counterstained with DAPI (4′,6-diamidino-2-phenylindole; Invitrogen). Fluorescence images were acquired using an EC Plan-Neofluor 40×/1.30 oil differential interference contrast (DIC) M27 or Plan-Apochromat 63×/1.40 oil DIC M27 objective on a Zeiss LSM 710 confocal microscope. Image capture, analysis, and processing were performed using ZEN 2010 software (Zeiss).
Statistical analysis.
Statistical significance was calculated by employing two-tailed Student's t test for correlated samples.
RESULTS
TMPRSS2 and HAT cleave the S-protein of HCoV-229E.
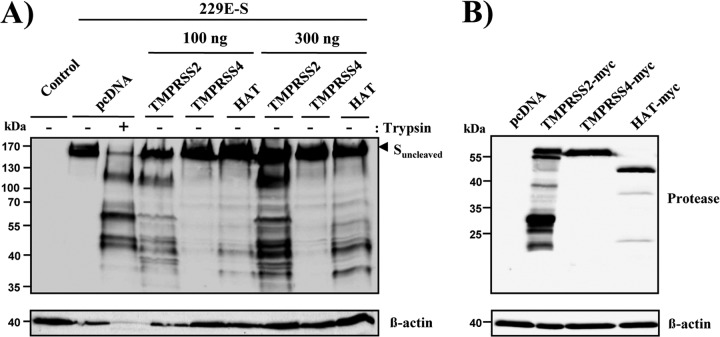
The type II transmembrane serine proteases TMPRSS2 and HAT activate SARS-CoV (30–33). To assess whether these proteases can also activate HCoV-229E, we first asked whether 229E-S is cleaved by TMPRSS2 and HAT. Cleavage by the related protease TMPRSS4 was also tested, since TMPRSS4 can activate SARS-S for cell-cell but not virus-cell fusion (32). To analyze S-protein cleavage, the 229E-S-protein was coexpressed with TMPRSS2, TMPRSS4, or HAT in 293T cells, and cleavage was analyzed by Western blotting. Trypsin treatment of 229E-S-expressing cells served as a positive control, since trypsin is known to cleave 229E-S (29). As a negative control, 229E-S was expressed in the absence of cotransfected protease (pcDNA).
A single band of approximately 160 kDa was detected upon expression of 229E-S in the absence of protease. Trypsin treatment of 229E-S-expressing cells generated prominent S-protein fragments of approximately 120, 65, 50, and 47 kDa (Fig. 1A). A similar cleavage pattern was detected upon coexpression of 229E-S with TMPRSS2 and HAT, indicating that these proteases and trypsin cleave the S-protein at similar or identical sites. Finally, coexpression of TMPRSS4 did not result in cleavage of 229E-S. Western blot analysis of myc-tagged proteins revealed that TMPRSS2, TMPRSS4, and HAT were robustly expressed in transfected 293T cells (Fig. 1B), indicating that the lack of S-protein cleavage by TMPRSS4 was not due to inefficient TMPRSS4 expression. Collectively, these results demonstrate that TMPRSS2 and HAT cleave 229E-S, likely at the same site(s) as that recognized by trypsin.
Fig 1.
TMPRSS2 and HAT cleave the S-protein of HCoV-229E. (A) An expression plasmid for 229E-S was transiently cotransfected with expression plasmids for the indicated proteases or with empty plasmid (pcDNA) into 293T cells. The cells were harvested and treated with PBS or TPCK-trypsin, as indicated, and 229E-S expression was analyzed by Western blotting employing a goat anti-HCoV-229E serum. The detection of β-actin served as a loading control. (B) Plasmids encoding the indicated TTSPs containing an N-terminal myc tag were transiently transfected into 293T cells. Cells transfected with empty plasmid served as a negative control. Protein expression was detected with anti-myc antibody, and detection of β-actin served as a loading control.
TMPRSS2 and HAT activate the S-protein of HCoV-229E for cell-cell fusion.
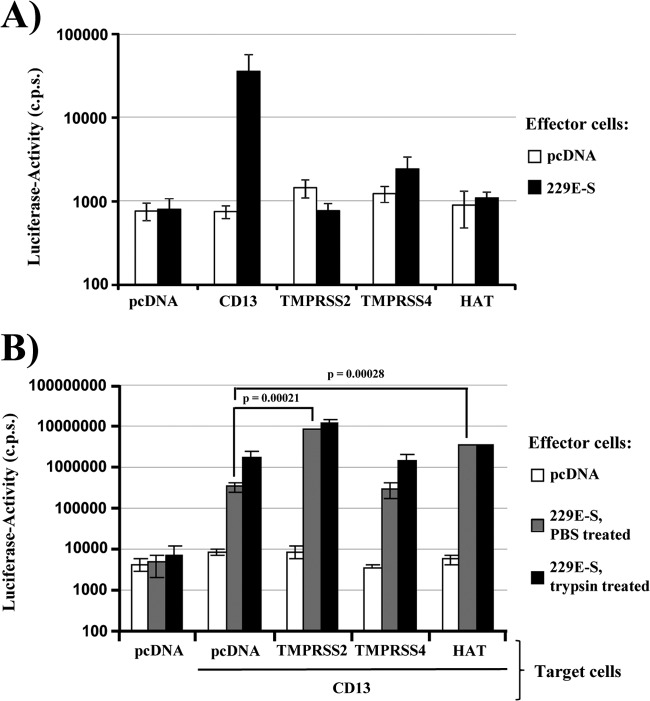
We next asked if cleavage of 229E-S by TMPRSS2 and HAT results in the activation of the S-protein for cell-cell fusion. Employing a previously described cell-cell fusion assay (31, 43), we first investigated whether transfection of 293T target cells with a plasmid encoding either a protease or CD13 was sufficient to allow fusion with 293T effector cells transfected to express 229E-S. Target and effector cells transfected with empty plasmid (pcDNA) served as negative controls. Effector cells expressing 229E-S fused efficiently with target cells expressing CD13, while fusion with target cells producing TMPRSS2, TMPRSS4, or HAT was within the background range measured upon transfection of empty plasmid (Fig. 2A). Similarly, background signals were detected when effector cells were transfected with empty plasmid instead of the plasmid encoding 229E-S (Fig. 2A). Thus, engineered expression of CD13 is essential for 229E-S-mediated fusion with 293T cells, which lack endogenous CD13 (43), and fusion cannot be rescued by expression of TMPRSS2, TMPRSS4, or HAT. We next determined whether 229E-S-mediated fusion with CD13-positive target cells can be enhanced by coexpression of protease. Indeed, coexpression of CD13 with TMPRSS2 or HAT, but not TMPRSS4, increased cell-cell fusion relative to that with cells expressing CD13 alone, and a similar augmentation was detected upon trypsin treatment (Fig. 2B). These observations are consistent with cleavage and activation of 229E-S by TMPRSS2 and HAT.
Fig 2.
TMPRSS2 and HAT increase cell-cell fusion driven by the S-protein of HCoV-229E. (A) Effector 293T cells were cotransfected with the 229E-S plasmid or empty plasmid (pcDNA), jointly with a plasmid encoding VP16 fused to GAL4. In parallel, 293T target cells were transiently cotransfected with plasmids encoding either human CD13 or the indicated proteases or with empty plasmid, jointly with a plasmid containing a GAL4-VP16-activated luciferase expression cassette. Effector and target cells were mixed, and luciferase activities in cell lysates were determined at 48 h postinfection. The results of a representative experiment performed in triplicate are shown; error bars indicate standard deviations (SD). Similar results were obtained in three separate experiments. (B) Experiments were conducted as described for panel A, but target cells expressing CD13 alone or in conjunction with the indicated proteases were examined. Eight hours after mixing of cells, the effector cell-target cell cocultures were treated with PBS or trypsin for 8 h. The results of a representative experiment performed in triplicate are shown; error bars indicate SD. The results were confirmed in two independent experiments.
TMPRSS2 and HAT activate authentic HCoV-229E for cathepsin-independent virus-cell fusion.
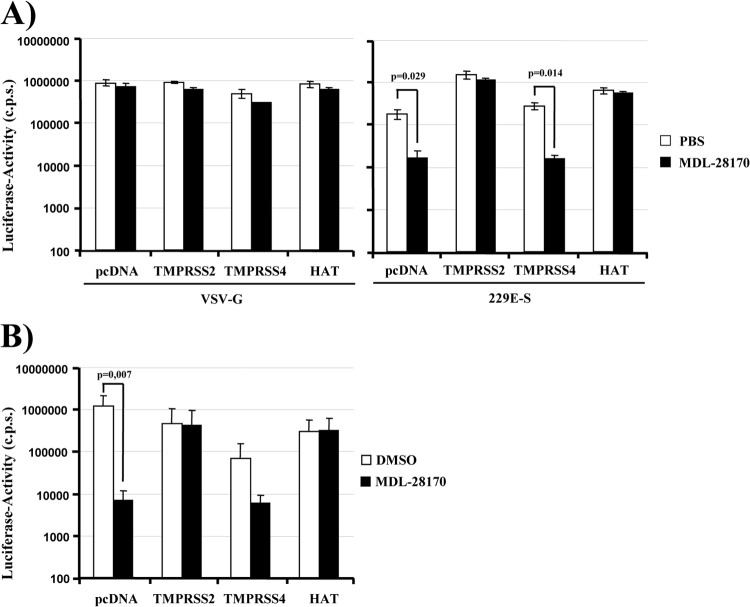
The endosomal/lysosomal cysteine protease cathepsin L can activate HCoV-229E for virus-cell fusion (29). We asked whether expression of TMPRSS2 and HAT on target cells allows for 229E-S-driven entry in the absence of cathepsin L activity. We first addressed this question by analyzing whether expression of TMPRSS2 and HAT facilitates 229E-S-dependent transduction of cells treated with the cathepsin L inhibitor MDL-28170. Transduction of cells with a lentiviral vector pseudotyped with the G-protein of VSV was not modulated by MDL-28170 treatment or expression of TMPRSS2 and HAT, as expected (Fig. 3A, left panel) (31). 229E-S-mediated transduction of 293T cells transiently expressing CD13 and transfected with empty plasmid (pcDNA) was efficiently inhibited by MDL-28170 (Fig. 3A, right panel), again in accord with published data (29). In stark contrast, expression of TMPRSS2 and HAT in target cells fully rescued 229E-S-dependent transduction from inhibition by MDL-28170 (Fig. 3A, right panel), while no rescue was observed upon expression of TMPRSS4. Notably, identical results were obtained when the experiment was repeated with replication-competent HCoV-229E expressing Renilla luciferase (46) (Fig. 3B), indicating that proteolytic activation of the virion-associated 229E-S-protein by TMPRSS2 and HAT can bypass the requirement for activation by cathepsin L.
Fig 3.
TMPRSS2 and HAT allow for cathepsin L-independent host cell entry of HCoV-229E. (A) 293T cells were transiently transfected with plasmids encoding human CD13 and the indicated proteases or with empty plasmid (pcDNA). The cells were seeded into 96-well plates, preincubated with PBS or the cathepsin L inhibitor MDL-28170 (10 μM), and transduced with firefly luciferase-encoding lentiviral vectors bearing VSV-G or 229E-S. Luciferase activities in cell lysates were measured at 72 h postransduction. The results of a representative experiment performed in triplicate are shown; error bars indicate SD. Comparable results were obtained in three independent experiments. (B) Experiments were carried out as described for panel A, but target cells were seeded into 24-well plates, preincubated with 20 μM MDL-28170 or DMSO, and then infected with HCoV-229E encoding Renilla luciferase. Luciferase activities in cell lysates were quantified at 8 h postinfection. The averages for three independent experiments performed in triplicate are shown; error bars indicate SD.
An inhibitor of TMPRSS2 blocks 229E-S-driven entry into Caco-2 cells.
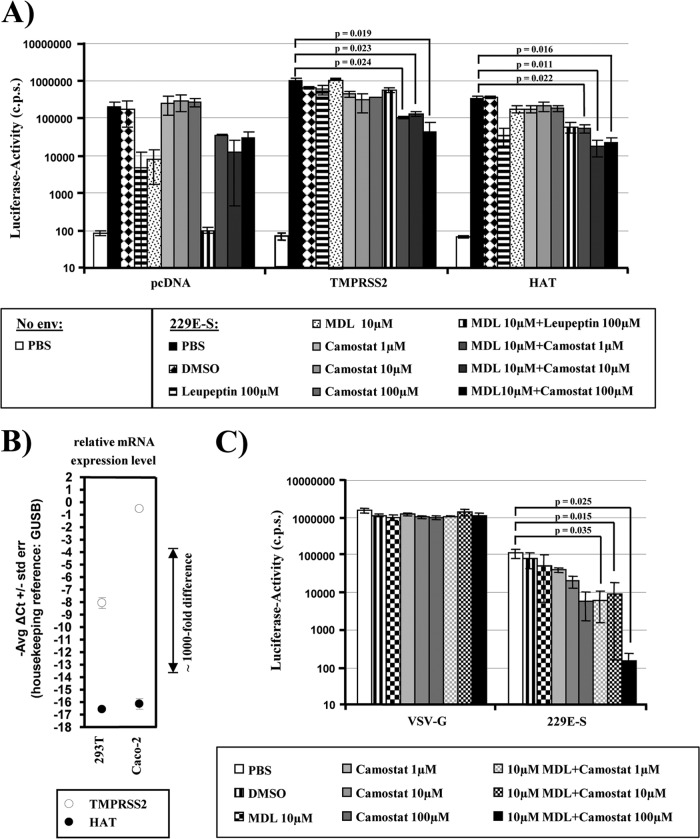
The observation that engineered expression of TMPRSS2 and HAT activates 229E-S for cathepsin L-independent host cell entry triggered the question of which proteolytic pathway is dominant in cells naturally susceptible to HCoV-229E infection. Addressing this question requires the availability of specific inhibitors. A previous study reported that the serine protease inhibitor camostat mesylate (camostat) inhibits the enzymatic activity of TMPRSS2 (50). We therefore investigated whether camostat blocks 229E-S activation by TMPRSS2 and HAT. For this purpose, we transiently transfected 293T-CD13 cells with plasmids encoding TMPRSS2, HAT, or no protease (pcDNA). The transfected cells were then incubated with MDL-28170, increasing amounts of camostat, or the indicated inhibitor combinations (Fig. 4A). In addition, we tested the inhibitory activity of leupeptin, which blocks the activity of several serine, threonine, and cysteine proteases, including cathepsin L (51). The 229E-S-mediated transduction of cells transfected with empty plasmid was not inhibited by camostat, while leupeptin and MDL-28170 reduced the transduction efficiency, in particular when applied in combination (Fig. 4A), in agreement with 229E-S activation by cathepsin L. In sharp contrast, MDL-28170 and leupeptin did not inhibit transduction of TMPRSS2-expressing target cells, in accordance with our previous observation that TMPRSS2 expression rescues 229E-S entry from blockade by cathepsin inhibitors (Fig. 3). The transduction of TMPRSS2-expressing cells was modestly reduced by camostat alone, while a combination of MDL-28170 and camostat significantly inhibited transduction. Similar results were obtained for HAT-positive cells, with the exception that leupeptin treatment alone reduced transduction, suggesting that leupeptin is active against HAT but not TMPRSS2, at least at the concentration used. In sum, these results show that 229E-S can be activated by TMPRSS2/HAT or cathepsin L and that these proteolytic pathways can be specifically inhibited by camostat or MDL-28170, respectively.
Fig 4.
Redundant proteolytic pathways activate the S-protein of HCoV-229E for virus-cell fusion. (A) 293T cells stably expressing CD13 were transiently transfected with plasmids encoding the indicated proteases or with empty plasmid (pcDNA) and were used as target cells for transduction by lentiviral vectors bearing 229E-S. Prior to infection, target cells were incubated with PBS, DMSO, or the indicated protease inhibitors. Luciferase activities in cell lysates were measured at 72 h postinfection. The results of a representative experiment performed in triplicate are shown; error bars indicate SD. Comparable results were observed in three independent experiments. (B) Analysis of TMPRSS2 and HAT expression by quantitative RT-PCR. Total RNAs were prepared from the indicated cell lines, treated with DNase, and reverse transcribed. TMPRSS2, HAT, and GUSB (housekeeping control) were amplified from cDNA by TaqMan gene expression assays. Similar results were obtained in an independent experiment. (C) Caco-2 cells, which express endogenous TMPRSS2, were incubated with PBS, DMSO, or the indicated protease inhibitors, followed by transduction with lentiviral pseudoparticles bearing 229E-S or VSV-G, as a control. At 72 h postinfection, luciferase activities in cell lysates were measured. The results of a representative experiment performed in triplicate are shown; error bars indicate SD. Comparable results were observed in five independent experiments.
The human intestinal epithelial cell line Caco-2 robustly expresses endogenous TMPRSS2 (40) but not HAT, as determined by quantitative reverse transcription-PCR (RT-PCR), while both proteases are not appreciably expressed in 293T cells (Fig. 4B) (40). In addition, Caco-2 cells are susceptible to HCoV-229E infection (52). We therefore asked which proteolytic pathway is used by 229E-S for activation in Caco-2 cells. Treatment of Caco-2 cells with camostat or MDL-28170 had no impact on VSV-G-driven transduction. Similarly, MDL-28170 had little effect on 229E-S-driven transduction (Fig. 4C) at a concentration which robustly inhibited transduction of 293T-CD13 cells (Fig. 3 and 4A). In contrast, 229E-S-dependent transduction was dose-dependently blocked by camostat, indicating that 229E-S is preferentially activated by TMPRSS2 or related serine proteases in this cell line. However, a combination of camostat and MDL-28170 was required to reduce transduction to background levels (Fig. 4C), demonstrating that the TMPRSS2/serine protease and cathepsin L pathways are both operational in Caco-2 cells.
TMPRSS2 expression partially rescues entry driven by the S-protein of HCoV-229E from inhibition by IFITM proteins.
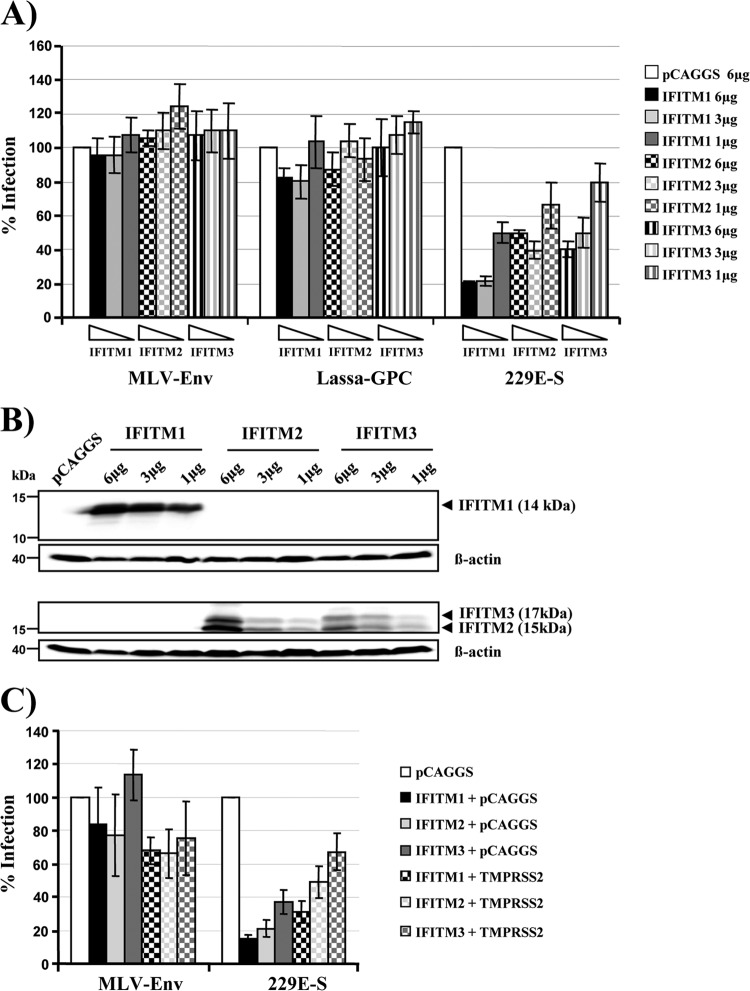
IFITM proteins are interferon-inducible host cell factors which inhibit diverse enveloped viruses, including influenza A viruses, HIV-1, filoviruses, flaviviruses, and SARS-CoV (34, 35, 53, 54). IFITM proteins inhibit viral entry (34, 35, 54, 55), potentially by interfering with clathrin-mediated uptake or vacuolar ATPase activity (56). In order to investigate whether IFITM proteins inhibit infection driven by 229E-S, we transiently expressed IFITM-1, -2, and -3 in 293T-CD13 cells and transduced these cells with pseudotypes bearing 229E-S. Pseudotypes bearing the glycoproteins of MLV and Lassa virus were also tested, since entry driven by these glycoproteins is not inhibited by IFITM proteins (35). Indeed, expression of IFITM proteins in target cells had no appreciable impact on transduction by pseudotypes bearing the MLV or Lassa virus glycoprotein (Fig. 5A). In contrast, transduction mediated by 229E-S was inhibited by expression of all IFITM proteins tested, with IFITM-1 displaying the highest antiviral activity (Fig. 5A). Western blot analysis of transfected target cells showed a dose-dependent expression of IFITM proteins (Fig. 5B), in agreement with the results obtained in the infection experiment (Fig. 5A). Considering that IFITMs are believed to exert their antiviral activity at a late stage (34, 35, 55), likely after uptake of virions into host cell endosomes, we reasoned that expression of TMPRSS2 might protect 229E-S-mediated entry from inhibition by IFITM proteins. Indeed, coexpression of TMPRSS2 increased the transduction efficiency of IFITM-expressing cells approximately 2-fold (Fig. 5C), suggesting that TMPRSS2 activates 229E-S for membrane fusion before IFITM proteins can exert their full antiviral activity.
Fig 5.
TMPRSS2 partially rescues entry driven by the S-protein of HCoV-229E from inhibition by IFITM proteins. (A) 293T-CD13 target cells were transfected with the indicated amounts of IFITM expression plasmids or empty plasmid (pCAGGS) and subsequently transduced with infectivity-normalized pseudotypes carrying the glycoproteins of MLV, Lassa fever virus, or HCoV-229E. Luciferase activities in cell lysates were determined at 72 h postinfection. The results are presented as % infection and represent the averages for three independent experiments; error bars indicate standard errors of the means (SEM). (B) 293T-CD13 cells were transfected with the indicated amounts of expression plasmids encoding IFITM proteins. Cell lysates were prepared after 48 h and analyzed by Western blotting using IFITM-1- and IFITM-2/3-reactive antibodies. (C) 293T-CD13 target cells expressing the indicated IFITM proteins either alone or in combination with TMPRSS2 were transduced with pseudoparticles bearing the glycoprotein of MLV or HCoV-229E, respectively. Cells were processed as described for panel A. The infection efficiency was calculated as % infection based on the infectivity for pCAGGS-transfected target cells, which was set as 100%. Comparable results were obtained in two independent experiments.
TMPRSS2 is coexpressed with CD13 in HCoV-229E target cells in human respiratory epithelium.
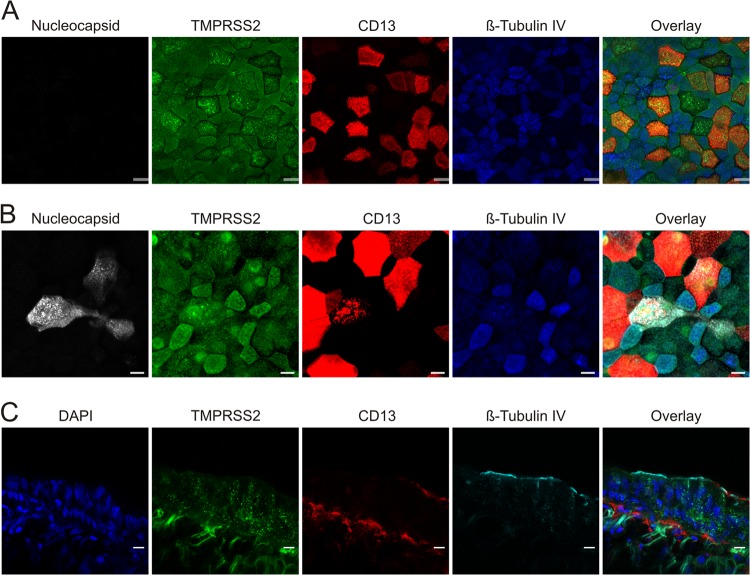
HCoV-229E targets the respiratory tract, and in order to promote viral spread in patients, TMPRSS2 must be expressed in viral target cells in the respiratory epithelium. Analysis of uninfected HAE cell cultures revealed that TMPRSS2 is coexpressed with CD13 and that both proteins are largely absent from ciliated cells, which were identified by staining for tubulin IV (Fig. 6A). Upon infection of HAE cultures with authentic HCoV-229E, nucleocapsid antigen was detected mainly in cells coexpressing TMPRSS2 and CD13 but not tubulin IV (Fig. 6B), in agreement with a recent study showing that HCoV-229E targets nonciliated CD13+ cells (47). In some of the nucleocapsid-positive cells, CD13 expression was reduced and largely confined to intracellular vesicles, which might hint toward downregulation of CD13 expression by HCoV-229E. Examination of bronchial tissue confirmed that CD13+ cells coexpress TMPRSS2 and largely lack tubulin IV, although TMPRSS2 expression did not seem to be limited to tubulin IV− cells. Expression of tubulin IV was confined mainly to the luminal side, while TMPRSS2 and CD13 were detected at the luminal and basolateral membranes (Fig. 6C). Thus, TMPRSS2 is expressed in HCoV-229E target cells in the human respiratory epithelium and promotes viral spread in this tissue.
Fig 6.
TMPRSS2 expression on differentiated HAE cells colocalizes with CD13-expressing cells. The in vitro distribution of TMPRSS2 expression (green) was analyzed in mock (A)- and HCoV-229E (B)-infected HAE cultures that were fixed and immunostained for colocalization with nucleocapsid protein (white) and for CD13 expression (red). To discriminate between ciliated and nonciliated populations, cells were stained with antibodies directed against β-tubulin IV (cilia; blue). Confocal images representative of 4 randomly selected fields from 3 independent experiments are shown. (C) For the in vivo distribution in primary bronchial tissue, TMPRSS2 (green) expression was analyzed in conjunction with CD13 (red), β-tubulin IV/cilia (cyan), and nuclei acids (blue). Confocal images representative of 4 randomly selected fields from 2 independent experiments are shown. Bars, 20 μm.
DISCUSSION
The activation of viral membrane fusion proteins by host cell proteases is essential for the infectivity of many human viruses, and the responsible proteases are potential targets for antiviral intervention. However, in several cases, their nature is unclear. Recent studies suggest a role for cathepsins B and L and/or TMPRSS2/HAT in the activation of human influenza viruses, human metapneumovirus, and SARS-CoV (30, 31, 33, 41, 57–59). Here we show that TMPRSS2 and HAT cleave and activate the HCoV-229E S-protein for cathepsin L-independent virus-cell fusion. In addition, TMPRSS2 expression was found to rescue 229E-S-driven entry from blockade by cathepsin inhibitors and antiviral IFITM proteins. Finally, immunohistochemistry revealed that HCoV-229E preferentially infects CD13+ TMPRSS2+ HAE cells, indicating that TMPRSS2 could activate HCoV-229E for spread in the infected host.
A prerequisite to the activation of 229E-S by TMPRSS2 and HAT is the cleavage of the S-protein by these proteases. Coexpression of TMPRSS2 and trypsin treatment induced 229E-S proteolysis, resulting in the production of prominent S-protein fragments of 120, 65, 50, and 47 kDa. Cleavage of 229E-S by HAT yielded an identical cleavage pattern, except that the 120-kDa fragment was not detected upon 229E-S cleavage by HAT, indicating subtle differences in the substrate specificities of HAT and TMPRSS2, which were already noted during the analysis of SARS-S activation by these proteases (32). The sensitivity of 229E-S to cleavage by trypsin is in agreement with published results (29). Differences in the size and number of cleavage products—the previous study by Kawase and coworkers detected fragments of 150 and 80 to 85 kDa (29)—likely reflect differences in experimental conditions and, more importantly, in the reagents used for S-protein detection: Kawase and coworkers employed a serum directed against the C terminus of the S-protein (29), while we used a serum raised against intact HCoV-229E. Finally, no cleavage was observed upon coexpression of TMPRSS4, which belongs to the same TTSP subfamily as TMPRSS2. Whether the failure of TMPRSS4 but not TMPRSS2 to cleave 229E-S reflects differences in substrate specificities or spatial orientations of the respective protease domains remains to be elucidated.
The coexpression of CD13 with TMPRSS2 or HAT in target cells increased cell-cell fusion driven by the HCoV-229E S-protein, suggesting that 229E-S cleavage by TMPRSS2 and HAT results in S-protein activation. However, transient expression of CD13 on target cells was sufficient for robust 229E-S-mediated membrane fusion, suggesting that cell-cell fusion can proceed in the absence of proteolytic activation of 229E-S or, more likely, that 293T cells express a protease which can activate 229E-S for cell-cell fusion, at least under conditions of high expression of the S-protein and its receptor. Notably, fusion of effector cells expressing the SARS-CoV S-protein with 293T target cells transfected to express only ACE2, the viral receptor, depends on the activity of a so far unidentified leupeptin-sensitive protease that is different from cathepsin L (60). It is thus tempting to speculate that the same protease might also activate 229E-S and that this activation is increased by directed expression of TMPRSS2 and HAT.
Expression of TMPRSS2 and HAT in target cells robustly activated 229E-S for infectious viral entry into target cells pretreated with a cathepsin L inhibitor. This observation, which was made with a 229E-S-bearing vector as well as authentic HCoV-229E, unambiguously demonstrates that 229E-S cleavage by TMPRSS2 and HAT results in S-protein activation and renders viral entry independent from S-protein processing by cathepsin L. The failure of HAT to cleave 229E-S into a 120-kDa fragment is thus compatible with 229E-S activation, indicating that production of the 65-, 50-, or 47-kDa fragment, but not the 120-kDa fragment, might be critical for S-protein activation. Activation of virus-cell fusion by TMPRSS2 has also been reported for SARS-S (30, 32). In contrast, HAT failed to activate SARS-S for cathepsin L-independent entry (31), suggesting that differential S-protein processing by TMPRSS2 and HAT affects activation of SARS-S but not 229E-S.
Activation of 229E-S by transfected TMPRSS2 raised the questions of whether endogenous TMPRSS2 can exert the same function and whether activation by TMPRSS2 predominates over activation by cathepsin L in cells naturally permissive to HCoV-229E infection. Only a few cell lines express robust amounts of endogenous TMPRSS2, limiting the choice of target cells for infection experiments. We opted for the intestinal cell line Caco-2, since these cells express endogenous TMPRSS2 to levels sufficient for activation of influenza virus (40) and are also susceptible to infection by authentic HCoV-229E (52). Activity of cathepsin L was largely dispensable for 229E-S-driven entry into Caco-2 cells. In contrast, treatment of Caco-2 cells with camostat, which was previously shown to inhibit SARS-S activation by TMPRSS2 (50), markedly reduced 229E-S-driven transduction of 293T cells. Thus, activation of 229E-S by TMPRSS2 (or a closely related TTSP) might predominate over activation by cathepsin L in the Caco-2 cell model of HCoV-229E infection. Nevertheless, simultaneous inhibition of cathepsin L and TMPRSS2 was required to fully suppress 229E-S-dependent transduction of Caco-2 cells, demonstrating that both proteolytic systems need to be targeted to obtain a profound antiviral effect.
IFITM proteins are interferon-inducible host cell factors which inhibit the cellular entry of diverse enveloped viruses, including SARS-CoV, likely after viral uptake into host cell endosomes (34, 35, 53, 55). Expression of IFITM-1, -2, and -3 in target cells markedly reduced 229E-S-dependent transduction, suggesting that the antiviral activity of IFITM proteins extends to HCoV-229E infection. Whether subtle differences in HCoV-229E inhibition by the different IFITM proteins tested are biologically meaningful or simply reflect differences in IFITM expression upon transient transfection is, at present, unclear. The coexpression of TMPRSS2 partially rescued 229E-S-driven transduction from inhibition by IFITM-1 to -3, as previously reported for SARS-S (34), indicating that the protease activates 229E-S for membrane fusion before virions reach the late endosome, where IFITMs may exert their antiviral activity. Thus, TMPRSS2 not only allows HCoV-229E to infect cells lacking cathepsin L activity but also might protect the virus from inhibition by IFITM proteins, thereby allowing infectious viral entry despite the onset of the interferon response.
A prerequisite for a role of TMPRSS2 in HCoV-229E infection is the expression of the protease in viral target cells in the human lung. Immunofluorescence analysis of HAE cultures revealed coexpression of CD13 and TMPRSS2 in nonciliated cells, which were targeted by authentic HCoV-229E, in agreement with a recent study (47). In addition, evidence for coexpression of TMPRSS2 and CD13 in bronchial epithelium was obtained, indicating that TMPRSS2 could activate HCoV-229E for pulmonary spread in human patients. In sum, our results demonstrate that redundant proteolytic systems, i.e., TMPRSS2 and cathepsin L, can activate the S-protein of HCoV-229E in cell culture and, most likely, in infected humans. In addition, our findings and previous studies show that different respiratory viruses hijack TMPRSS2 to ensure their activation. Considering that knockout of the Tmprss2 gene in mice shows no phenotype in the absence of infection (61), the TMPRSS2 protease might constitute an attractive target for therapeutic intervention.
ACKNOWLEDGMENTS
We thank Kathryn Holmes for providing the polyclonal 229E antiserum, Andrea Marzi for cloning of the CD13 expression plasmid, Abraham Brass for IFITM expression plasmids, and Graham Simmons and Michael Farzan for advice. We also thank Ulrike Goedecke and Nadine Meyer for expert technical assistance.
This work was funded by BMBF (SARS-Verbund grant 01KI1005C) and the Leibniz Foundation.
Footnotes
Published ahead of print 27 March 2013
REFERENCES
- 1. Bermingham A, Chand M, Brown C, Aarons E, Tong C, Langrish C, Hoschler K, Brown K, Galiano M, Myers R, Pebody R, Green H, Boddington N, Gopal R, Price N, Newsholme W, Drosten C, Fouchier R, Zambon M. 2012. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 17:20290. [PubMed] [Google Scholar]
- 2. Corman V, Eckerle I, Bleicker T, Zaki A, Landt O, Eschbach-Bludau M, BSvan Gopal R, Ballhause M, Bestebroer T, Muth D, Muller M, Drexler J, Zambon M, Osterhaus A, Fouchier R, Drosten C. 2012. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 17:20285. [DOI] [PubMed] [Google Scholar]
- 3. Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguiere AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967–1976 [DOI] [PubMed] [Google Scholar]
- 4. Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953–1966 [DOI] [PubMed] [Google Scholar]
- 5. Osterhaus AD, Fouchier RA, Kuiken T. 2004. The aetiology of SARS: Koch's postulates fulfilled. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359:1081–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367:1814–1820 [DOI] [PubMed] [Google Scholar]
- 7. Al-Ahdal MN, Al-Qahtani AA, Rubino S. 2012. Coronavirus respiratory illness in Saudi Arabia. J. Infect. Dev. Ctries. 6:692–694 [DOI] [PubMed] [Google Scholar]
- 8. World Health Organization 2012. Coronaviruses. Global alert and response (GAR). WHO, Geneva, Switzerland: http://www.who.int/csr/don/2012_11_30/en/index.html [Google Scholar]
- 9. World Health Organization 2013. Novel coronavirus infection—update. http://www.who.int/csr/don/2013_02_21/en/index.html
- 10. Bradburne AF, Bynoe ML, Tyrrell DA. 1967. Effects of a “new” human respiratory virus in volunteers. Br. Med. J. 3:767–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Holmes KV. 2001. Coronaviruses, p 1187–1203 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 12. van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, Wertheim-van Dillen PM, Kaandorp J, Spaargaren J, Berkhout B. 2004. Identification of a new human coronavirus. Nat. Med. 10:368–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, Huang Y, Wong BH, Poon RW, Cai JJ, Luk WK, Poon LL, Wong SS, Guan Y, Peiris JS, Yuen KY. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chiu SS, Chan KH, Chu KW, Kwan SW, Guan Y, Poon LL, Peiris JS. 2005. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin. Infect. Dis. 40:1721–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gorse GJ, O'Connor TZ, Hall SL, Vitale JN, Nichol KL. 2009. Human coronavirus and acute respiratory illness in older adults with chronic obstructive pulmonary disease. J. Infect. Dis. 199:847–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jean A, Quach C, Yung A, Semret M. 2013. Severity and outcome associated with human coronavirus OC43 infections among children. Pediatr. Infect. Dis. J. 32:325–329 [DOI] [PubMed] [Google Scholar]
- 17. Jevsnik M, Ursic T, Zigon N, Lusa L, Krivec U, Petrovec M. 2012. Coronavirus infections in hospitalized pediatric patients with acute respiratory tract disease. BMC Infect. Dis. 12:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van der Hoek L, Sure K, Ihorst G, Stang A, Pyrc K, Jebbink MF, Petersen G, Forster J, Berkhout B, Uberla K. 2005. Croup is associated with the novel coronavirus NL63. PLoS Med. 2:e240 doi:10.1371/journal.pmed.0020240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gaunt ER, Hardie A, Claas EC, Simmonds P, Templeton KE. 2010. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J. Clin. Microbiol. 48:2940–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hayden FG. 2006. Respiratory viral threats. Curr. Opin. Infect. Dis. 19:169–178 [DOI] [PubMed] [Google Scholar]
- 21. Prill MM, Iwane MK, Edwards KM, Williams JV, Weinberg GA, Staat MA, Willby MJ, Talbot HK, Hall CB, Szilagyi PG, Griffin MR, Curns AT, Erdman DD. 2012. Human coronavirus in young children hospitalized for acute respiratory illness and asymptomatic controls. Pediatr. Infect. Dis. J. 31:235–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hamre D, Procknow JJ. 1966. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 121:190–193 [DOI] [PubMed] [Google Scholar]
- 23. Yeager CL, Ashmun RA, Williams RK, Cardellichio CB, Shapiro LH, Look AT, Holmes KV. 1992. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357:420–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bonavia A, Zelus BD, Wentworth DE, Talbot PJ, Holmes KV. 2003. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J. Virol. 77:2530–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lendeckel U, Kahne T, Riemann D, Neubert K, Arndt M, Reinhold D. 2000. Review: the role of membrane peptidases in immune functions. Adv. Exp. Med. Biol. 477:1–24 [DOI] [PubMed] [Google Scholar]
- 26. Holmes KV, Compton SR. 1995. Coronavirus receptors, p 55–71 In Siddell SG. (ed), Coronaviridae. Plenum Press, New York, NY [Google Scholar]
- 27. Hofmann H, Pöhlmann S. 2004. Cellular entry of the SARS coronavirus. Trends Microbiol. 12:466–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kielian M, Rey FA. 2006. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat. Rev. Microbiol. 4:67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kawase M, Shirato K, Matsuyama S, Taguchi F. 2009. Protease-mediated entry via the endosome of human coronavirus 229E. J. Virol. 83:712–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F. 2010. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 84:12658–12664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bertram S, Glowacka I, Muller MA, Lavender H, Gnirss K, Nehlmeier I, Niemeyer D, He Y, Simmons G, Drosten C, Soilleux EJ, Jahn O, Steffen I, Pöhlmann S. 2011. Cleavage and activation of the severe acute respiratory syndrome coronavirus spike protein by human airway trypsin-like protease. J. Virol. 85:13363–13372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Glowacka I, Bertram S, Muller MA, Allen P, Soilleux E, Pfefferle S, Steffen I, Tsegaye TS, He Y, Gnirss K, Niemeyer D, Schneider H, Drosten C, Pöhlmann S. 2011. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 85:4122–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T. 2011. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 85:873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang IC, Bailey CC, Weyer JL, Radoshitzky SR, Becker MM, Chiang JJ, Brass AL, Ahmed AA, Chi X, Dong L, Longobardi LE, Boltz D, Kuhn JH, Elledge SJ, Bavari S, Denison MR, Choe H, Farzan M. 2011. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 7:e1001258 doi:10.1371/journal.ppat.1001258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, Adams DJ, Xavier RJ, Farzan M, Elledge SJ. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dijkman R, Koekkoek SM, Molenkamp R, Schildgen O, van der Hoek L. 2009. Human bocavirus can be cultured in differentiated human airway epithelial cells. J. Virol. 83:7739–7748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hofmann H, Pyrc K, van der Hoek L, Geier M, Berkhout B, Pöhlmann S. 2005. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U. S. A. 102:7988–7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Simmons G, Reeves JD, Grogan CC, Vandenberghe LH, Baribaud F, Whitbeck JC, Burke E, Buchmeier MJ, Soilleux EJ, Riley JL, Doms RW, Bates P, Pöhlmann S. 2003. DC-SIGN and DC-SIGNR bind Ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 305:115–123 [DOI] [PubMed] [Google Scholar]
- 39. Hofmann H, Geier M, Marzi A, Krumbiegel M, Peipp M, Fey GH, Gramberg T, Pöhlmann S. 2004. Susceptibility to SARS coronavirus S-protein-driven infection correlates with expression of angiotensin converting enzyme 2 and infection can be blocked by soluble receptor. Biochem. Biophys. Res. Commun. 319:1216–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bertram S, Glowacka I, Blazejewska P, Soilleux E, Allen P, Danisch S, Steffen I, Choi SY, Park Y, Schneider H, Schughart K, Pöhlmann S. 2010. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J. Virol. 84:10016–10025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chaipan C, Kobasa D, Bertram S, Glowacka I, Steffen I, Tsegaye TS, Takeda M, Bugge TH, Kim S, Park Y, Marzi A, Pöhlmann S. 2009. Proteolytic activation of the 1918 influenza virus hemagglutinin. J. Virol. 83:3200–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wentworth DE, Holmes KV. 2001. Molecular determinants of species specificity in the coronavirus receptor aminopeptidase N (CD13): influence of N-linked glycosylation. J. Virol. 75:9741–9752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hofmann H, Simmons G, Rennekamp AJ, Chaipan C, Gramberg T, Heck E, Geier M, Wegele A, Marzi A, Bates P, Pöhlmann S. 2006. Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors. J. Virol. 80:8639–8652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stamminger T, Gstaiger M, Weinzierl K, Lorz K, Winkler M, Schaffner W. 2002. Open reading frame UL26 of human cytomegalovirus encodes a novel tegument protein that contains a strong transcriptional activation domain. J. Virol. 76:4836–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Connor RI, Chen BK, Choe S, Landau NR. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935–944 [DOI] [PubMed] [Google Scholar]
- 46. van den Worm SH, Eriksson KK, Zevenhoven JC, Weber F, Zust R, Kuri T, Dijkman R, Chang G, Siddell SG, Snijder EJ, Thiel V, Davidson AD. 2012. Reverse genetics of SARS-related coronavirus using vaccinia virus-based recombination. PLoS One 7:e32857 doi:10.1371/journal.pone.0032857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dijkman R, Jebbink MF, Koekkoek SM, Deijs M, Jónsdóttir HR, Molenkamp R, Ieven M, Goossens H, Thiel V, van der Hoek L. 2013. Isolation and characterization of current human coronavirus strains in primary human epithelial cell cultures reveals differences in target cell tropism. J. Virol. 87:6081–6090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dijkman R, Mulder HL, Rumping L, Kraaijvanger I, Deijs M, Jebbink MF, Verschoor EJ, van der Hoek L. 2009. Seroconversion to HCoV-NL63 in rhesus macaques. Viruses 1:647–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sastre P, Dijkman R, Camunas A, Ruiz T, Jebbink MF, van der Hoek L, Vela C, Rueda P. 2011. Differentiation between human coronaviruses NL63 and 229E using a novel double-antibody sandwich enzyme-linked immunosorbent assay based on specific monoclonal antibodies. Clin. Vaccine Immunol. 18:113–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kawase M, Shirato K, van der Hoek L, Taguchi F, Matsuyama S. 2012. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 86:6537–6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Katunuma N, Kominami E. 1985. Lysosomal sequestration of cytosolic enzymes and lysosomal thiol cathepsins. Adv. Enzyme Regul. 23:159–168 [DOI] [PubMed] [Google Scholar]
- 52. Wang G, Deering C, Macke M, Shao J, Burns R, Blau DM, Holmes KV, Davidson BL, Perlman S, McCray PB., Jr 2000. Human coronavirus 229E infects polarized airway epithelia from the apical surface. J. Virol. 74:9234–9239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ, Wise HM, Kane L, Goulding D, Digard P, Anttila V, Baillie JK, Walsh TS, Hume DA, Palotie A, Xue Y, Colonna V, Tyler-Smith C, Dunning J, Gordon SB, Smyth RL, Openshaw PJ, Dougan G, Brass AL, Kellam P. 2012. IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484:519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lu J, Pan Q, Rong L, He W, Liu SL, Liang C. 2011. The IFITM proteins inhibit HIV-1 infection. J. Virol. 85:2126–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li K, Markosyan RM, Zheng YM, Golfetto O, Bungart B, Li M, Ding S, He Y, Liang C, Lee JC, Gratton E, Cohen FS, Liu SL. 2013. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 9:e1003124 doi:10.1371/journal.ppat.1003124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wee YS, Roundy KM, Weis JJ, Weis JH. 2012. Interferon-inducible transmembrane proteins of the innate immune response act as membrane organizers by influencing clathrin and v-ATPase localization and function. Innate Immun. 18:834–845 [DOI] [PubMed] [Google Scholar]
- 57. Böttcher E, Matrosovich T, Beyerle M, Klenk HD, Garten W, Matrosovich M. 2006. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 80:9896–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shirogane Y, Takeda M, Iwasaki M, Ishiguro N, Takeuchi H, Nakatsu Y, Tahara M, Kikuta H, Yanagi Y. 2008. Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J. Virol. 82:8942–8946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Watanabe R, Matsuyama S, Shirato K, Maejima M, Fukushi S, Morikawa S, Taguchi F. 2008. Entry from the cell surface of severe acute respiratory syndrome coronavirus with cleaved S-protein as revealed by pseudotype virus bearing cleaved S-protein. J. Virol. 82:11985–11991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Simmons G, Bertram S, Glowacka I, Steffen I, Chaipan C, Agudelo J, Lu K, Rennekamp AJ, Hofmann H, Bates P, Pöhlmann S. 2011. Different host cell proteases activate the SARS-coronavirus spike-protein for cell-cell and virus-cell fusion. Virology 413:265–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kim TS, Heinlein C, Hackman RC, Nelson PS. 2006. Phenotypic analysis of mice lacking the Tmprss2-encoded protease. Mol. Cell. Biol. 26:965–975 [DOI] [PMC free article] [PubMed] [Google Scholar]